Abstract
Hepatitis C virus (HCV) causes chronic hepatitis, cirrhosis, and liver cancer. cis-acting RNA elements of the HCV genome are critical for translation initiation and replication of the viral genome. We hypothesized that the coding regions of nonstructural proteins harbor enhancer and essential cis-acting replication elements (CRE). In order to experimentally identify new cis RNA elements, we utilized an unbiased approach to introduce synonymous substitutions. The HCV genome coding for nonstructural proteins (nucleotide positions 3872 to 9097) was divided into 17 contiguous segments. The wobble nucleotide positions of each codon were replaced, resulting in 33% to 41% nucleotide changes. The HCV genome containing one of each of 17 mutant segments (S1 to S17) was tested for genome replication and infectivity. We observed that silent mutations in segment 13 (S13) (nucleotides [nt] 7457 to 7786), S14 (nt 7787 to 8113), S15 (nt 8114 to 8440), S16 (nt 8441 to 8767), and S17 (nt 8768 to 9097) resulted in impaired genome replication, suggesting CRE structures are enriched in the NS5B region. Subsequent high-resolution mutational analysis of NS5B (nt 7787 to 9289) using approximately 51-nucleotide contiguous subsegment mutant viruses having synonymous mutations revealed that subsegments SS8195-8245, SS8654-8704, and SS9011-9061 were required for efficient viral growth, suggesting that these regions act as enhancer elements. Covariant nucleotide substitution analysis of a stem-loop, JFH-SL9098, revealed the formation of an extended stem structure, which we designated JFH-SL9074. We have identified new enhancer RNA elements and an extended stem-loop in the NS5B coding region. Genetic modification of enhancer RNA elements can be utilized for designing attenuated HCV vaccine candidates.
INTRODUCTION
Hepatitis C virus (HCV) affects up to 170 million people worldwide, and about 350,000 people die every year from HCV-mediated liver ailments (1–4). Although some HCV-infected patients clear their infections spontaneously, most become chronic carriers, with a risk of serious complications, including cirrhosis, liver failure, and hepatocellular carcinoma (HCC). Hepatitis C is the leading indication for liver transplants in developed countries and is the major etiologic factor responsible for the recent doubling of HCC (5). The current standard of care for the treatment of HCV infection, HCV NS3/4A protease inhibitors in combination with pegylated alpha interferon (PEG-IFN) and ribavirin, has led to an increased sustained virological response (SVR) rate of up to 75% for genotype 1 (GT1) HCV but has also caused increased adverse events, including gastrointestinal symptoms and anemia (6, 7). There is no vaccine available against HCV. Upon exposure, HCV replicates mainly in hepatocytes and a small proportion in mononuclear cells, biliary epithelial cells, and sinusoidal-lining cells (8). HCV, an enveloped virus, belongs to the family Flaviviridae. HCV strains are clustered into six major genotypes. The hepatitis C virion contains a 9.6-kb single-stranded, positive-sense RNA genome (9). The genome codes for a single polyprotein, which is mainly proteolytically processed into 10 proteins having structural and nonstructural (NS) functions (10–12). The polyprotein-coding region is flanked by 5′ and 3′ nontranslated regions (NTR). HCV subgenomic replicons have been predominantly used for studying viral genome replication (13, 14). Genotype 2a JFH-1 strain-based viruses have been useful for studying various steps of viral replication (15–17).
The 5′ NTR contains RNA elements that are critical for viral genomic RNA replication and cap-independent translation initiation (18–21). The internal ribosomal entry site (IRES) in the 5′ NTR acts as the landing site for ribosomal subunits and other cellular proteins involved in translation. MicroRNA-122 binds to the 5′ NTR and enhances viral genome translation and genome replication by possibly preventing viral genome degradation (22, 23). Conserved stem-loop structures SL47 and SL87 in the core protein-coding region have been shown to be required for HCV genome translation and viral replication (24). The 3′ NTR is comprised of three segments, a proximal variable region, a polypyrimidine tract (polyU/UC), and a 3′ X tail (25–28). Earlier studies proposed the presence of stem-loop structures in the 3′ terminus of the HCV genome (29–31). This region is critical for the initiation of negative-strand genome replication and stimulating translation through long-range RNA-RNA interaction (32–34), as well as binding to cellular and viral proteins, including PTBP1, ribosomal proteins, NS3, and NS5A (35–41).
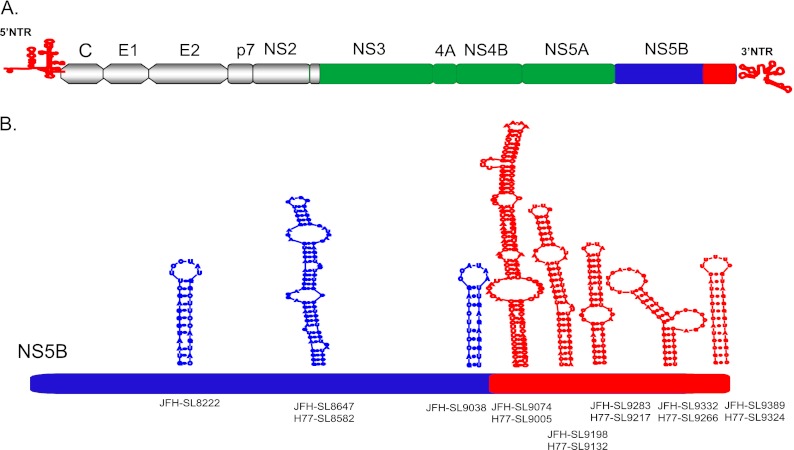
NS5B, which codes for RNA-dependent RNA polymerase, is critical for viral genome replication (42–49). Detailed thermodynamic predictions and phylogenetic analyses, as well as experimental analyses, revealed the presence of conserved stem-loop structures in the NS5B-coding region across all the genotypes (50–54). Smith and Simmonds described potential RNA structures at the 3′ end of the NS5B region (53), and this was later experimentally tested by You and colleagues using a genotype 1b Con1 subgenomic replicon system to designate the stem-loop structures 5BSL3.1, 5BSL3.2, and 5BSL3.3 (55). Earlier, Hofacker et al. predicted 5BSL1 and SL8828 using computational methods (52). Tuplin and colleagues presented a total of six potential RNA structures in the NS5B region, i.e., SL7730 (5BSL1), SL8376 (5BSL2), SL8828, SL8926, SL9011 (5BSL3.1), and SL9118 (5BSL3.3) (50). In an independent experimental analysis using a genotype 1b replicon, Lee et al., described SL-V (5BSL3.3), SL-VI (5BSL3.2), and SL-VII (5BSL3.1) in the NS5B region (56). Limited mutagenesis analysis using a Con1 replicon showed that 5BSL1, 5BSL2, 5BSL3.1, and 5BSL3.3 structures were not required for genome replication (55). However, another report indicated that SL-V, or 5BSL3.3, was critical for genome replication of the genotype 1b replicon (56). Several studies employed different methods to designate the stem-loop structures in the HCV genome, resulting in multiple nomenclatures for the same structure. In this report, we followed the recently adopted standardized system for naming the stem-loop based on the genomic position of the first 5′ paired base of the RNA structure (57–60). We summarize all the homologous stem-loop structures described in the NS5B coding region below. cis-acting replication element (CRE) SL9266 (5BSL3.2, or SL-VI) has been extensively characterized by mutagenesis studies and is critical for viral genome replication. Reverse genetic analysis in the genotype 1b Con1 replicon system revealed that SL9266 (5BSL3.2) forms a long-range interaction with an unpaired sequence downstream of SL9033 and that the interaction is critical for viral genome replication (57); however, a new study reported that this interaction was not critical for viral production of an intragenotype 2a chimeric virus, J6/JFH-1, in Huh-7.5 cells (60). Furthermore, SL9266 forms a kissing-loop interaction with 3′ NTR SL2 (SL9571) (32, 34). These long-range interactions formed by stem-loop structures have been shown to be critical for HCV genome replication. It has been recently reported that the 5BSL3.2 CRE is involved in a long-range interaction with subdomain IIId of the 5′ NTR IRES region, and the resulting interaction negatively influences the IRES-mediated translation (61, 62). It is postulated that the 5BSL3.2 CRE could function as a switch modulating the translation and genome replication of the sense-stranded HCV genome (60). The RNA structure of 5BSL3.2 was determined by both enzymatic primer extension analysis and selective 2′-hydroxyl acylation analyzed through primer extension (SHAPE) methods (55, 60).
The structural proteins core, E1, and E2, as well as p7 and NS2, are not required for HCV genome replication, whereas the NS3, NS4A, NS4B, NS5A, and NS5B proteins are (13, 14). The C-terminal domains of the NS4B and NS5A proteins can be trans-complemented (63, 64). Since the genome replication functions of most of these nonstructural proteins cannot be trans-complemented by exogenously expressing a functional protein, these nonstructural proteins must act in a cis manner during genome replication. This cis nature of the proteins limits the experimental options for identifying cis-acting RNA structural elements present in the coding region. HCV CRE structures have been identified through computational phylogenetic analysis and thermodynamic predictions. However, phylogenetically nonconserved sequences forming conserved RNA structures may not be predicted by computational methods. We hypothesized that the coding regions of nonstructural proteins harbor enhancer and essential cis-acting replication elements. To test this hypothesis, we utilized an unbiased approach to introduce synonymous mutations in the NS3, NS4A, NS4B, NS5A, and NS5B coding regions. This mutational approach preserved the amino acid sequence of the HCV polyprotein but could alter the RNA structures. We studied the effects of these synonymous mutations on HCV genome replication and de novo infection to identify new cis-acting RNA elements in the coding region of HCV.
MATERIALS AND METHODS
Cells.
The Huh-7.5.1 cell line (kindly provided by F. Chisari) was cultured in complete Dulbecco's modified Eagle's medium (DMEM) containing 10% to 15% fetal bovine serum (FBS), 10 mM nonessential amino acids (Invitrogen, Carlsbad, CA), 10 mM HEPES, penicillin (100 units/ml), streptomycin (100 mg/ml), and 2 mM l-glutamine at 37°C with 5% CO2 (27).
Virus and plasmid constructs.
An intragenotype 2a chimeric virus, pJ6/JFH-C, and a monocistronic chimeric reporter virus, pNRLFC, based on the pJ6/JFH-C parental virus, were described previously (27). For the current study, we used a chemically synthesized plasmid, pFNX-HCV, and a pFNX-Rluc reporter (which have sequences similar to those of pJ6/JFH-C and pNRLFC, respectively). In brief, the intragenotype 2a chimeric virus pFNX-HCV, comprising the 5′ NTR, structural regions, and part of the nonstructural regions (p7 and partial NS2) of the J6CF strain (the genomic region, nucleotides [nt] 1 to 2878; NCBI accession no. AF177036) and the nonstructural regions of strain JFH-1 (NCBI accession no. AB047639), was generated. The monocistronic chimeric reporter virus pFNX-Rluc, based on the pFNX-HCV parental virus, was generated by inserting a Renilla luciferase gene between the 5′ NTR and core gene. The luciferase gene and core gene are joined by the foot and mouth disease virus 2A (F2A) peptide gene sequence to serve as a cleavage signal. To generate an envelope-null mutant virus, an in-frame deletion of nt 1040 to 2215 was engineered in the FNX-HCV genome. This deletion removed most of the E1 and E2 coding regions. An RNA polymerase-null (Pol−) virus with a pFNX-HCV or pFNX-Rluc background was generated by replacing the catalytic residues GDD with AAG amino acid residues. pFNX-HCV and pFNX-Rluc plasmids were used for construction of recombinant viruses. The sequence information for the primers used for the construction of mutant viruses is available upon request. The engineered mutations in the plasmids were verified by sequencing.
In vitro transcription and RNA transfection.
The viral plasmids were first linearized with the XbaI restriction enzyme and treated with mung bean nuclease (New England BioLabs, Beverly, MA) before being subjected to in vitro transcription using the T7 Ribomax Express Large Scale RNA Production System according to the manufacturer's instructions (Promega Corporation, Madison, WI). The DNase-treated RNA was purified, quantified, and stored at −80°C in aliquots. To minimize the variation in RNA quality, for each experiment, RNA production (transcription, purification, quantification, and storage) for all the mutant and wild-type viruses was done at the same time. To increase the survivability of Huh-7.5.1 cells after electroporation, the cells were cultured in 15% FBS. Before electroporation, the Huh-7.5.1 cells were briefly trypsinized and washed twice with ice-cold Opti-MEM transfection medium (Invitrogen) and resuspended in Opti-MEM at 1 × 107 cells per ml. Ten micrograms of in vitro-transcribed RNA was mixed with 400 μl of cells in 0.4-cm electroporation cuvettes. Electroporation was conducted by using a Bio-Rad elecroporator at 270 V, 100 Ω, and 950 μF. Subsequently, the cells were resuspended in 40 ml of complete DMEM with 15% FBS and plated in T-25 or T-75 flasks and 48-well plates. At 8 h posttransfection, media containing dead cell debris in the culture flasks and plates were replaced with fresh complete DMEM with 10% FBS. The cells were lysed for RNA (reverse transcription-quantitative PCR [RT-qPCR]) and protein (Western blotting and luciferase assay) at the indicated time points.
RT-qPCR.
A two-step RT-qPCR was carried out to determine the HCV RNA copy number. Briefly, 1 μg of total cellular RNA was reverse transcribed by using Superscript III Reverse Transcriptase (Invitrogen) and random hexamers. For qPCR, 100 ng of the resulting cDNA was used as a template with HCV 5′ UTR-specific primers (JFH RTQ F, 5′-CTGGGTCCTTTCTTGGATAA-3′, and JFH RTQ R, 5′-CCTATCAGGCAGTACCACA) and a SYBR green qPCR SuperMix-UDG with Rox kit (Invitrogen). In vitro-transcribed JFH-1 genomic RNA (101 to 107 copies) was reverse transcribed along with the samples and included as a standard for copy number determination during qPCR. The reaction was run at 95°C for 15 s and 60°C for 30 s (40 cycles) using a ViiA 7 real-time PCR system (Applied Biosystems).
Multiple-cycle growth curve analysis.
To assess virus growth, naïve Huh-7.5.1 cells were plated in a 48-well plate at a density of 1 × 104 per well. At 4 h postplating, the cells were infected with each virus in duplicate at a multiplicity of infection (MOI) of 0.1. At 6 h postinfection, the cells were washed twice with PBS, and 500 μl of fresh medium was added to each well. The cell-free supernatants collected at 2 days postinfection (p.i.), 4 days p.i., and 6 days p.i. were assessed for virus titer.
Measuring virus titer.
The virus titer was measured by calculating the number of focus-forming units (FFU) of infectious viral particles per ml of cell-free culture supernatant. The infected culture supernatant was 10-fold serially diluted in complete DMEM and inoculated in triplicate onto naïve Huh-7.5.1 cells (3 × 103 cells/well) in 96-well plates. At 72 h p.i., the cells were fixed and immunostained for HCV NS5A antigen. The NS5A antigen-positive foci were counted at the highest dilution, and the average number of focus-forming units per ml was calculated.
Renilla luciferase reporter assay for viral genome replication and infectivity.
For viral genome replication assays, the HCV RNA-electroporated cells were plated in triplicate in 48-well plates. The cells were lysed with passive lysis buffer (Promega) at the indicated time points. The culture plates were gently rocked at room temperature for 15 min and then stored at −80°C. To determine the supernatant infectivity, 500 μl of cell-free supernatant obtained from HCV RNA-transfected cells at the indicated time points was inoculated in triplicate onto naïve Huh-7.5.1 cells in 48-well plates. At 6 h p.i., the viral inoculum was replaced with 500 μl of fresh medium per well. At 48 h p.i., the cells were lysed and stored at −80°C. Ten microliters of lysate was used to measure the Renilla luciferase activity using a Renilla Luciferase Assay System kit (Promega).
Western blotting.
For Western blotting, the cell lysates were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked (5% skim milk, 0.2% Tween 20 in PBS) and probed with mouse monoclonal antibody NS3 (clones 8G-2 and H23 [Abcam]) and beta-actin (Sigma). Goat anti-mouse IgG secondary antibody conjugated with horseradish peroxidase (HRP) (Amersham Pharmacia Biotech) was detected by chemiluminescence (ECL Plus; Amersham Pharmacia Biotech).
Immunofluorescence assay.
The HCV-infected and -transfected cells were fixed with methanol. Following three PBS washes, the cells were blocked (10% fetal bovine serum, 3% bovine serum albumin [BSA], 0.1% Triton X-100 in PBS) and incubated with rabbit polyclonal anti-NS5A primary antibody (a kind gift from Asim Dasgupta, UCLA, Los Angeles, CA) at a dilution of 1:200 for 5 h to overnight at 4°C. The goat anti-rabbit IgG polyclonal antibody conjugated to Alexa Fluor 594 was added as a secondary antibody (Invitrogen) at 1:1,000 dilution and incubated for 1 h at room temperature. Between antibody changes, the cells were washed thrice with PBS. The nucleus was stained with Hoechst dye (Invitrogen).
Sequence analysis.
For sequence alignment, we used the sequences of genotype 1a H77 (GenBank accession no. AF009606), genotype 1b Con1 (AJ238799), genotype 2a JFH-1 (AB047639), genotype 3 (NC_009824), genotype 4 (NC_009825), genotype 5 (NC_009826), and genotype 6 (NC_009827). In this study, unless otherwise stated, the numbering of nucleotide positions is based on the JFH-1 (AB047639) reference sequence. For stem-loop nomenclature, according to current standard practice, we numbered the stem-loops based on the first 5′ paired base position of genomic sequence (58).
RNA structure analysis.
We utilized the mfold Web server (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) for RNA structure predictions using the default settings (65).
Statistical analysis.
The error bars indicate standard deviations. P values were determined by the unpaired t test.
RESULTS
Strategy for engineering silent mutations in the HCV genome.
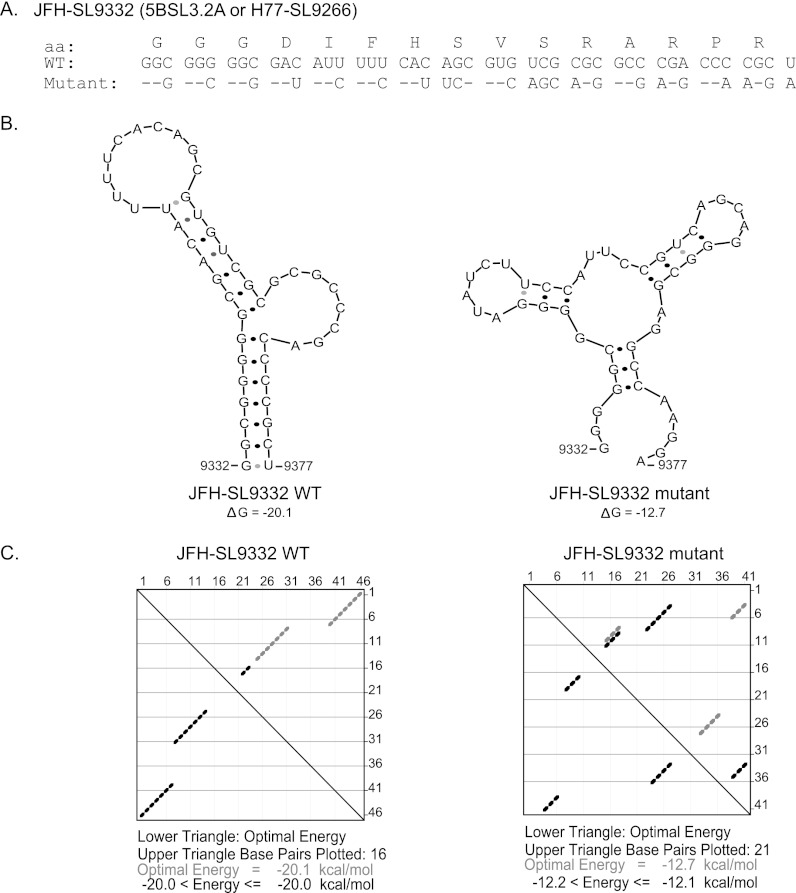
Identification of cis elements in the nonstructural-protein-coding region (about 6 kb) is limited by the inability to trans-complement the functions of most nonstructural proteins. Existing computational programs have limitations in predicting nonconserved nucleotide sequences forming conserved structures. To identify HCV RNA elements involved in genome replication and viral de novo infectivity, we utilized an unbiased approach to introduce HCV genome scale silent mutations. Substitutions were made at the wobble nucleotide positions of each tricodon. The mutations change Watson-Crick base pairing of the single-stranded RNA genome in the formation of higher-order structures. Thus, the silent mutations affect the structure and function of RNA elements while preserving the protein sequence and function. To minimize the introduction of suboptimal codons, the percentage of codon use by HCV genotypes was taken into account. The frequency of codon use by strain JFH-1 was calculated using an online program (http://www.kazusa.or.jp/codon/countcodon.html). Amino acids with more than four codons, such as leucine, serine, and arginine, allowed more than one nucleotide substitution per codon. Single-codon-containing residues, methionine and tryptophan, cannot be mutated. Both of the codons of amino acids with two codons (asparagine, aspartic acid, cysteine, glutamine, glutamic acid, histidine, lysine, phenylalanine, and tyrosine) are represented in the JFH-1 genome at a frequency of 5.3 to 29.7 per 1,000 codons. Once the parameters were set for introducing the silent mutations into the viral genome, we computationally modeled the generated silent mutations in the context of previously well-characterized cis elements, and Fig. 1 shows an example using CRE 5BSL3.2 (H77-SL9266 or JFH-SL9332). The mutant JFH-SL9332 sequence had 22 nucleotide changes (Fig. 1A). mfold analysis of predicted structures showed that the introduced nucleotide substitutions clearly destabilized RNA structures by forming mispaired stems and changing nucleotides involved in kissing-loop interactions (Fig. 1B). The energy dot blot analysis indicated that the synonymous mutations altered the optimal energy from −20.1 kcal/mol to −12.7 kcal/mol and made changes in the base pairing (Fig. 1C). Subsequently, we experimentally tested this mutational approach in the HCV coding region. A summary list of previously described stem-loop structures in the NS5B coding region, as well as homologous structures present in the genotype 1a H77 strain and genotype 2a JFH-1 strain, is provided in Table 1.
Fig 1.
Computationally modeling the effects of synonymous mutations in a well-characterized CRE, 5BSL3.2a (H77-SL9266 or JFH-SL9332). (A) JFH-1 SL9332 wild-type (WT) and mutant sequences are shown. aa, amino acids. (B) Predicted stem-loop structures of wild-type and mutated sequences using RNA mfold analysis. Note the altered Watson-Crick base pair interactions in the mutant structure and lowered folding energy, resulting in a destabilized RNA structure. The folding Gibbs free energy (ΔG, in kcal/mol) for the predicted stem-loops is shown under each structure. (C) Energy dot plots for optimal and suboptimal folding of stem-loop wild-type and mutant structures. The upper triangle displays possible base pair combinations at various energy levels. The nucleotide positions of each base are displayed on the top axis and right axis of the upper triangle. The paired bases are shown as diamond dots in the plot. For example, wild-type base 46 (top axis) pairs with base 1 (right axis). The lower triangle shows the paired bases with optimal folding energy at 37°C to form a stem-loop structure.
Table 1.
Summary of stem-loop structures described in the NS5B coding region
| Stem-loop | ||||
|---|---|---|---|---|
| GT2-JFH-1a | Diviney et al., GT1-H77 (57) | You et al., GT1-Con1 (55) | Tuplin et al. (50) | Lee et al. (56) |
| JFH-SL8001 | 5BSL1 | SL7730 | ||
| JFH-SL8647 | 5BSL2 | SL8376 | ||
| JFH-SL9098 | H77-SL9033 | SL8828 | ||
| JFH-SL9198 | H77-SL9132 | SL8926 | ||
| JFH-SL9283 | H77-SL9217 | 5BSL3.1 | SL9011 | SL-VII |
| JFH-SL9332 | H77-SL9266 | 5BSL3.2 | SL-VI | |
| JFH-SL9389 | H77-SL9324 | 5BSL3.3 | SL9118 | SL-V |
Present study.
Systematic mutational analysis of the HCV NS3-NS5B coding region to identify RNA elements.
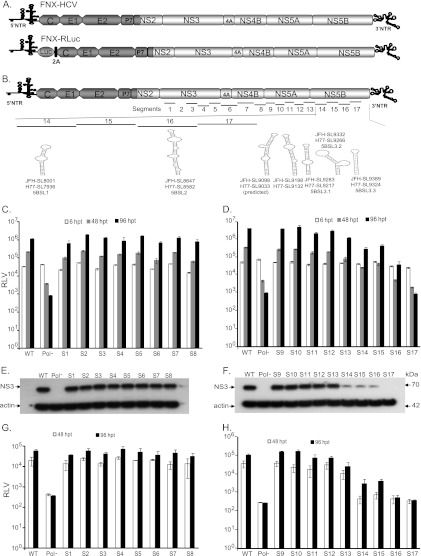
To identify RNA elements present in the HCV nonstructural-protein-coding region (NS3 to NS5B) covering nucleotides 3872 to 9097, we initially performed a low-resolution screening experiment using reporter HCV. For this study, we used an intra-genotype 2a chimeric virus, FNX-HCV, and a monocistronic Renilla luciferase reporter virus, FNX-Rluc (Fig. 2A). The reporter virus is useful for screening large numbers of individual mutant viruses. The FNX-Rluc virus has a Renilla luciferase gene fused in frame to the core gene through an F2A cleavage signal sequence (Fig. 2A). We observed that the luciferease reporter HCV had a level of genome replication similar to that of the wild-type parental virus but was 10- to 100-fold attenuated in infectious-particle production (27). The FNX-Rluc virus is also very sensitive in detecting changes in viral genome replication (27, 66, 67). The NS3-to-NS5B region was divided into 17 segments (S1 to S17) 285 to 360 nucleotides in size (Table 2 and Fig. 2). A total of 34.4% to 41.7% of the nucleotides were substituted in each segment. On average, every third nucleotide was mutated, except for methionine and tryptophan codons. The nature of nucleotide substitutions engineered for each segment is provided in Table S1 in the supplemental material. HCV NS3 (S1 to S5), NS4A/B (S6), NS4B (S7), NS4B/5A (S8), NS5A (S9 to S12), NS5A/5B (S13), and NS5B (S14 to S17) gene regions were individually evaluated. No previously predicted RNA secondary structure was present in the segments tested, except in S14 and S16. The S13 segment comprised the 3′ end of NS5A (nt 7457 to 7666) and 120 nucleotides from the 5′ end of NS5B (nt 7667 to 7786). Segment S14 contains a predicted CRE, JFH-SL8001 (a homologue of H77-SL7936), and segment S16 encompasses another predicted CRE, JFH-SL8647 a (homologue of H77-SL8582).
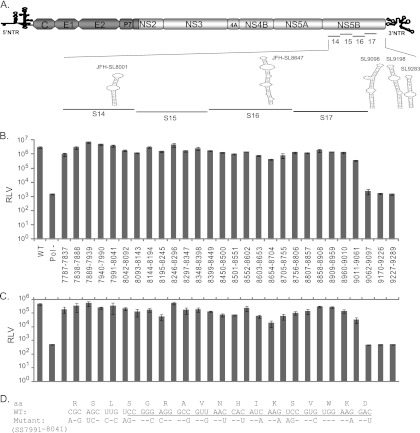
Fig 2.
RNA cis element analysis of the HCV nonstructural-protein-coding region. (A) Schematic diagram of hepatitis C viruses used in this study. FNX-HCV is a synthetic version of a genotype 2a J6CF and JFH-1 chimera (the J6CF strain region is shown in dark gray, including the 5′ NTR, and the JFH-1 strain region is shown in light gray). A monocistronic Renilla luciferase (luc) reporter virus (FNX-Rluc) based on the J6/JFH chimeric virus is shown. The luciferase gene is fused in frame with the core gene through foot and mouth disease virus 2A sequence. (B) Schematic representation of the HCV genome showing the locations of 17 mutated segments in the nonstructural-protein-coding region. The stem-loop structures described in the NS5B region corresponding to the genome positions of JFH-1 and H77 are noted. (C and D) Analysis of viral genome replication of mutant reporter viruses. Huh-7.5.1 cells were electroporated with 10 μg of in vitro-transcribed genomic RNA of wild-type FNX-Rluc reporter virus and individual S1 to S17 mutant reporter viruses. A control reporter virus with nonfunctional polymerase activity (Pol−) is included. The transfected cells were lysed at 6, 48, and 96 h p.t. using Promega Renilla luciferase assay lysis buffer, and the levels of Renilla luciferase were quantified. The experiment was performed in triplicate. The mean Renilla luciferase values (RLV) with standard deviations are shown in log10 scale as a bar graph. The replication-deficient mutant S17 showed luciferase activity similar to that of the Pol− control virus. At 96 h p.t., mutant viruses S14, S15, S16, and S17 had significant reductions in genome replication compared to that of wild-type virus (P value < 0.0001 by unpaired t test). (E and F) Western blot of the expression of HCV protein. The 70-kDa NS3 antigen was detected from the protein lysates obtained at 96 h posttransfection by primary mouse monoclonal antibody and secondary goat-anti mouse IgG conjugated with HRP. As a loading control, β-actin was included. (G and H) Infectivities of mutant viruses. Naïve Huh-7.5.1 cells were infected with the cell-free supernatants harvested at 48 and 96 h posttransfection. Renilla luciferase activities were measured from the lysates harvested at 48 h postinfection. The mean RLV with standard deviations are depicted in the bar graph. Mutant reporter viruses S13, S14, S15, S16, and S17 exhibited significant reductions in infectivity at 48 h p.t. (P value ≤ 0.005) and 96 h p.t. (P value < 0.005) compared to that of wild-type virus. The experiment was repeated three times, and data from a representative experiment are shown.
Table 2.
Summary of mutant segments and introduced nucleotide changes
| Gene | Segment | JFH-1 genome location | Size (nt) | No. of nt with: |
Total no. of changes (%) | ||
|---|---|---|---|---|---|---|---|
| No change | Conserved change (%) | Nonconserved change (%) | |||||
| NS3 | S1 | 3872–4156 | 285 | 170 | 85 (29.9) | 30 (10.5) | 115 (40.4) |
| NS3 | S2 | 4157–4441 | 285 | 178 | 88 (30.9) | 19 (6.6) | 107 (37.5) |
| NS3 | S3 | 4442–4726 | 305 | 200 | 85 (27.9) | 20 (6.5) | 105 (34.4) |
| NS3 | S4 | 4727–5011 | 285 | 171 | 86 (30.2) | 28 (9.8) | 114 (40.0) |
| NS3 | S5 | 5012–5299 | 288 | 186 | 87 (30.2) | 15 (5.2) | 102 (35.4) |
| NS4A/B | S6 | 5300–5650 | 351 | 225 | 97 (27.7) | 29 (8.2) | 126 (35.9) |
| NS4B | S7 | 5651–6010 | 360 | 218 | 108 (30.0) | 34 (9.4) | 142 (39.4) |
| NS4B/5A | S8 | 6011–6301 | 291 | 173 | 92 (31.6) | 26 (8.9) | 118 (40.5) |
| NS5A | S9 | 6302–6592 | 291 | 187 | 83 (28.5) | 21 (7.2) | 104 (35.7) |
| NS5A | S10 | 6593–6880 | 288 | 178 | 78 (27.1) | 32 (11.1) | 110 (38.2) |
| NS5A | S11 | 6881–7168 | 288 | 168 | 73 (25.4) | 47 (16.3) | 120 (41.7) |
| NS5A | S12 | 7169–7456 | 288 | 174 | 93 (32.4) | 21 (7.2) | 114 (39.6) |
| NS5A/5B | S13 | 7457–7786 | 330 | 199 | 73 (22.2) | 58 (17.5) | 131 (39.7) |
| 5B | S14 | 7787–8113 | 327 | 193 | 94 (28.8) | 40 (12.2) | 134 (41.0) |
| 5B | S15 | 8114–8440 | 327 | 202 | 91 (27.9) | 34 (10.3) | 125 (38.2) |
| 5B | S16 | 8441–8767 | 327 | 200 | 91 (27.8) | 36 (11.0) | 127 (38.8) |
| 5B | S17 | 8768–9097 | 330 | 201 | 91 (27.6) | 38 (11.5) | 129 (39.1) |
| Total | 6,179 | 3,791 | 1,782 | 606 | 2,388 | ||
To determine whether the engineered nucleotide substitution in the JFH-1 genome is conserved or nonconserved between HCV genotypes, we analyzed the sequence alignment of a mutated segment with the corresponding regions from six major genotypes (genotype 1a H77 [GenBank accession no. AF009606], genotype 1b Con1 [AJ238799], genotype 2a JFH-1 [AB047639], genotype 3 [NC_009824], genotype 4 [NC_009825], genotype 5 [NC_009826], and genotype 6 [NC_009827]). If the substituted nucleotide in the JFH-1 genome was also represented in any of the genotypes, we considered the mutation a conserved change. The number of conserved (natural) and nonconserved (arbitrary) mutations for each segment is given in Table 2.
Individual mutant reporter viruses in the background of FNX-Rluc were constructed in order to assess viral replication fitness. The location of each mutant segment is shown in Fig. 2B, along with previously described CRE structures. Genomic RNAs generated from the 17 mutant reporter constructs were individually electroporated into Huh-7.5.1 cells in two batches. The wild-type FNX-Rluc and Pol− reporter viruses were included as controls. The genome replication of each virus was measured by assaying luciferase activity at 6 h posttransfection (p.t.), 48 h p.t., and 96 h p.t. (Fig. 2C and D). Western blotting was performed to detect the expression of viral antigen NS3 at 96 h p.t. (Fig. 2E and F). The infectious viral particles produced by mutant viruses at 48 and 96 h p.t. were evaluated by infecting naïve Huh-7.5.1 cells with cell-free supernatant collected from transfected cells. Renilla luciferase activity was measured from infected cells at 48 h p.i., and mean values were plotted as a bar graph (Fig. 2G and H).
Our results indicate that the mutant S17 had a replicatory phenotype similar to that of replication-incompetent Pol− reporter virus, suggesting an essential role of the RNA elements in the S17 segment. Furthermore, we observed that the mutant viruses S13, S14, S15, and S16 had exhibited reduced genome replication kinetics compared to that of wild-type FNX-Rluc virus. At 96 h p.t., S13, S14, S15, and S16 mutants had 3.4-, 13.7-, 9.3-, and 103.8-fold reductions (P value < 0.0001 by unpaired t test), respectively, in genome replication compared to wild-type replication. The mutants S13, S14, S15, and S16 also exhibited 4.1-, 35-, 25-, and 202-fold reductions (P value < 0.005 by unpaired t test), respectively, in virus infectivity compared to the wild type at 96 h p.t. At 6 h p.t., these S13, S14, S15, and S16 mutants had luciferase values similar to that of wild-type virus, indicating that there were similar levels of translation of input-transfected viral genomes. For mutants S1 to S12, which span NS3 to NS5A, we did not consistently observe any significant growth difference from wild-type virus in independent experiments despite the change of over 34% of nucleotides in each segment. Since S13 to S17 are segments from the NS5B coding region, our systematic experimental analysis suggests that the HCV cis-acting replication elements are concentrated in this area. We reproduced this observation by three independent experiments. During the first experiment, we tested two sequence-verified independent plasmid clones per mutant and obtained a consistent phenotype. For subsequent repetition of experiments, we used a single sequence-verified clone for each mutant. This redundant approach provided high-confidence results.
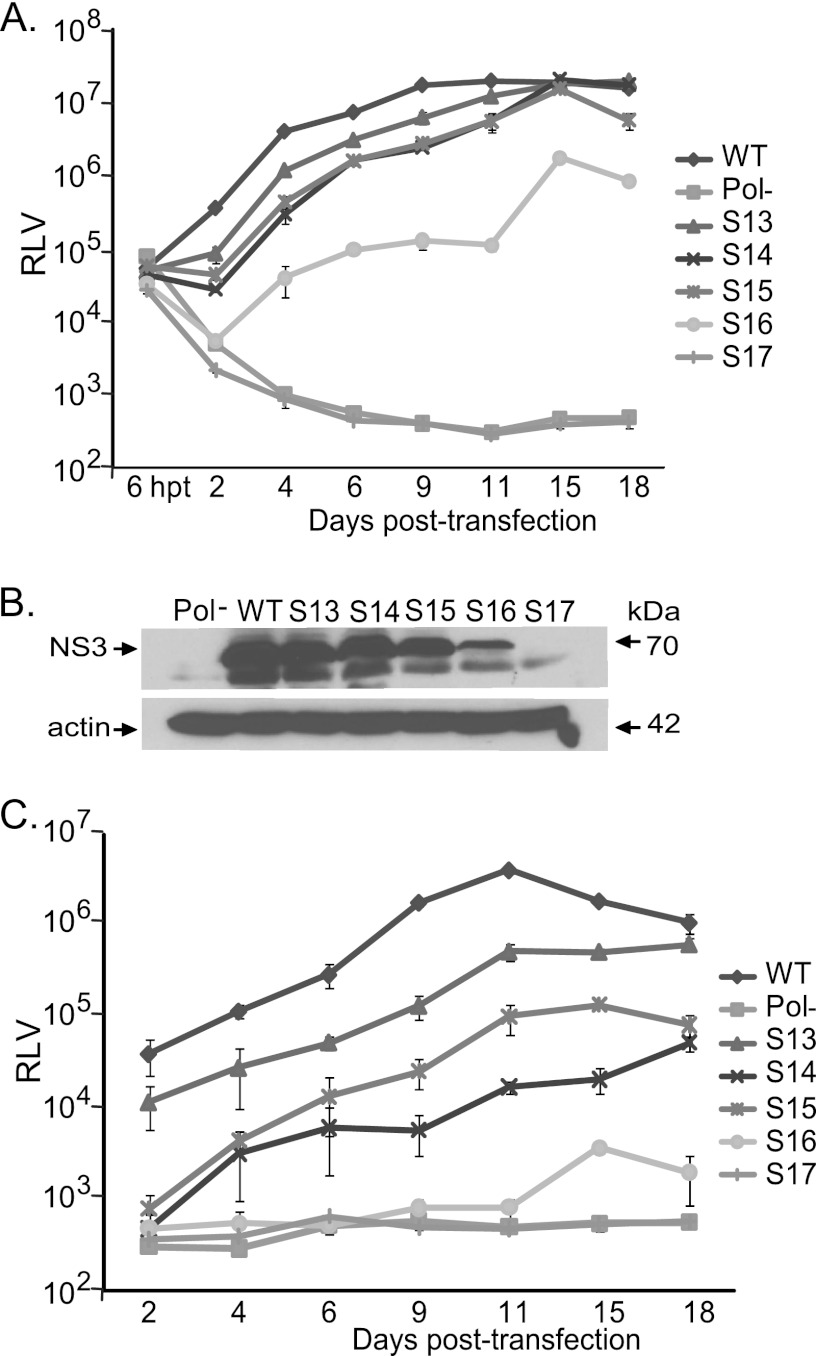
We focused on S13, S14, S15, S16, and S17 mutant reporter viruses for further study involving their long-term replicatory phenotypes (Fig. 3). The genome replication and infectivity of these mutants were studied on days 2, 4, 6, 9, 11, 15, and 18 posttransfection. The mutants S13, S14, S15, and S16 were able to establish persistent infection. S17 virus replication was critically impaired and had similar phenotypes of replication-defective Pol− reporter virus at all the time points tested. The wild-type FNX-Rluc virus reached peak genome replication at 11 days p.t., and at this time point, the S13, S14, S15, and S16 mutants exhibited 1.6-, 3.5-, 3.6-, and 181.3-fold reductions, respectively, in genome replication compared to that of the wild type. At 15 days p.t., the genome replication levels of mutants S13, S14, and S15 had caught up with wild-type replication levels. The mutant virus S16 showed an attenuated phenotype at all the time points studied but maintained persistent infection. In parallel, we analyzed the abilities of viral particles produced by the various mutant viruses to establish de novo infection at the indicated time points. At 11 days p.t., the S13, S14, S15, and S16 mutants had 7.7-, 228.1-, 39.4-, and 4,666.1-fold reductions, respectively, in infectivity compared to that of the wild type, which exhibited peak infectivity (Fig. 3C). Mutant viruses S14 and S15 showed similar infectivity to each other at days 2, 4, and 6 posttransfection but began to exhibit different patterns from day 9 posttransfection on. In conclusion, mutant viruses S13, S14, and S15 had delayed genome replication kinetics and the S16 mutant exhibited attenuated replication kinetics during long-term culture.
Fig 3.
Genome replication and infectious virus production kinetics during long-term culturing of CRE mutant reporter viruses. (A) Genome replication kinetics of wild-type reporter virus, as well as mutant reporter viruses S13, S14, S15, S16, and S17, at the indicated time points posttransfection. Cell lysates were harvested at various time points to measure Renilla luciferase activity. The mean and standard deviation calculated from triplicate RLV for each mutant are shown in the graph. (B) Western blots showing the expression of NS3 antigen at 9 days p.t. and control β-actin. The Pol− and S17 mutants lack NS3 expression. (C) Analysis of virus production kinetics. Huh-7.5.1 cells were inoculated with the supernatant harvested from each virus-infected culture at the indicated time points. The Renilla luciferase activities of infected cells were measured. Mean values with standard deviations are depicted in the graph.
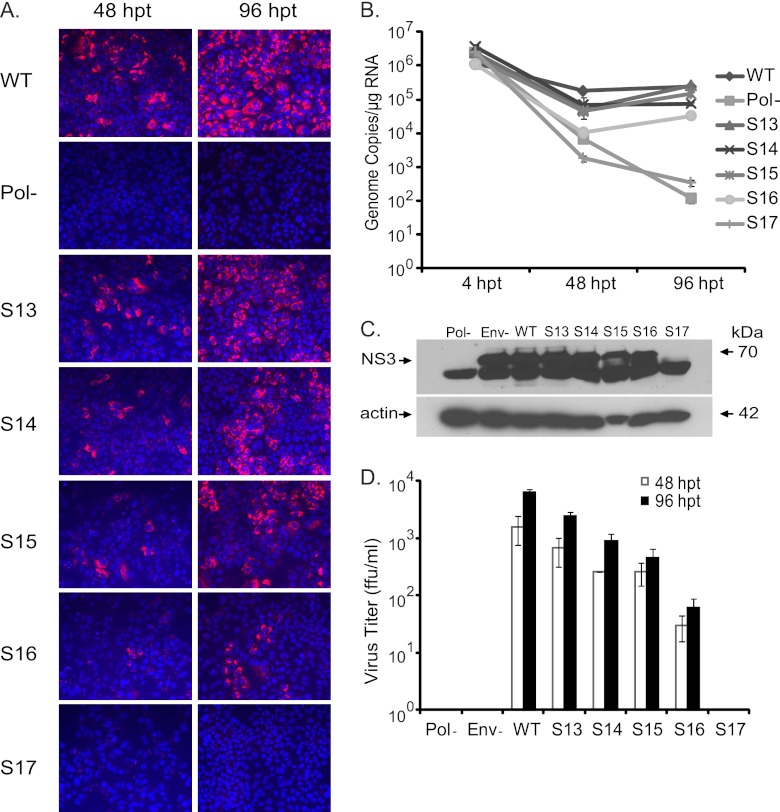
The phenotypes of viruses containing mutant segments S1 to S17 were assessed in a Renilla luciferase reporter virus background. The reporter virus facilitated rapid screening of viral mutants by measuring luciferase enzyme activity and indicated the NS5B coding region for new CREs. Thus, independently, we verified the growth phenotypes of S13, S14, S15, S16, and S17 mutant viruses in an FNX-HCV background (Fig. 4). We confirmed that the S17 mutant was a replication-defective virus using both viral backgrounds. Measuring genome copies by RT-qPCR revealed that all the mutants showed reduced genome replication at 48 h p.t. compared to that of the wild-type FNX-HCV (Fig. 4B). At 96 h p.t., S13 replicated its genome at levels similar to that of the wild-type virus. At the same time point, S13, S14, S15, and S16 had 2.7-, 7.2-, 14.4-, and 108.3-fold reductions of de novo infectivity, respectively, compared to that of the wild type, as determined by using focus-forming units (Fig. 4D). As a further control for infectivity, we included envelope-null (Env−) virus (lacking E1 and E2 genes), which is defective in producing infectious particles (27). Immunofluorescence assays and Western blotting showed that all the tested viruses except the Pol− control and mutant S17 expressed various levels of HCV antigens NS5A and NS3 (Fig. 4A and C). Taken together, we observed that silent mutations in segment S13 (nucleotides 7557 to 7786, the NS5A/5B junction region) resulted in mild attenuation, mutations in the S14 (nt 7787 to 8113) and S15 (nt 8114 to 8440) segments led to moderate attenuation, mutations in segment S16 (nt 8441 to 8767) resulted in severe attenuation, and silent mutations in S17 (nt 8768 to 9097) resulted in a lethal phenotype. S14 and S16 each have a previously predicted CRE structure, while the S13, S15, and S17 regions have no known cis RNA elements.
Fig 4.
Confirming the growth phenotypes of CRE mutant viruses in the NS5B region. (A) Immunofluorescence assay to detect viral antigen. The in vitro-transcribed RNA genomes of FNX wild-type virus and individual mutant viruses S13, S14, S15, S16, and S17 were electroporated into Huh-7.5.1 cells. At 48 and 96 h posttransfection, the cells were fixed and immunostained for HCV NS5A antigen (red). Hoechst staining was used to visualize cell nuclei (blue). (B) Genome replication kinetics of mutant viruses. RNA was harvested from transfected cells at 4, 48, and 96 h posttransfection and subjected to RT-quantitative PCR. The genome copy numbers per microgram of RNA were quantified and are depicted in the graph. The FNX-HCV mutant lacking polymerase activity was included as a control for the genome replication-deficient phenotype. (C) Analysis of HCV antigen expression. Western blotting was performed to detect NS3 antigen and β-actin. (D) Infectivities of various mutant viruses. The virus titer (focus-forming units/ml) was measured by infecting naïve Huh-7.5.1 cells with the cell-free supernatants collected at 48 and 96 h posttransfection from each mutant-virus-transfected cell culture. The mean values and standard deviations of the titers are shown in the graph. All the mutants had significantly reduced infectivity at 96 h posttransfection compared to that of the wild type (P value < 0.05 by unpaired t test). To assess infectivity, we included an FNX mutant virus lacking envelope genes (Env-) as an additional control. The experiment was done three times, and data from a representative experiment are shown.
High-resolution screening of the NS5B region for CRE identification.
Our mapping study for identification of CRE structures in the HCV NS3-NS5B coding region indicated that NS5B likely contains additional RNA elements. Silent mutations introduced in the NS5B segments S14, S15, S16, and S17 resulted in moderate to severe impairment of genome replication and de novo infectivity. Each of these segments spans about 327 nucleotides (Table 2). In order to obtain a fine map of cis elements present in this NS5B coding region (nucleotides 7787 to 9289), we further divided the region into 28 subsegments (SS) approximately 51 nucleotides (17 codons) in size (Fig. 5A and B). Each subsegment was named according to its genomic location in the JFH-1 strain reference sequence. The NS5B segments (S14, S15, S16, and S17) corresponding to each subsegment division are illustrated in Fig. 5A. The details of the nucleotide substitutions for each segment are available in Table S2 in the supplemental material. We also included subsegments SS9170-9226 and SS9227-9289, which are predicted to have critical CREs in the HCV genome. SS9170-9226 (57 nucleotides; 19 codons) contains a conserved sequence, 5′-CGGGC-3′ (nt 9173 to 9177), between HCV genotypes and was shown to base pair with a downstream stem-loop, Con1-SL9266 (5BSL3.2, or JFH-SL9332), in the Con1 replicon system (57). This subsegment is also part of a stem-loop structure, JFH-SL9198 (H77-SL9132) (Fig. 5A). SS9227-9289 (63 nucleotides; 21 codons) includes parts of both stem-loop structures, JFH-SL9198 and JFH-SL9283 (H77-SL9217).
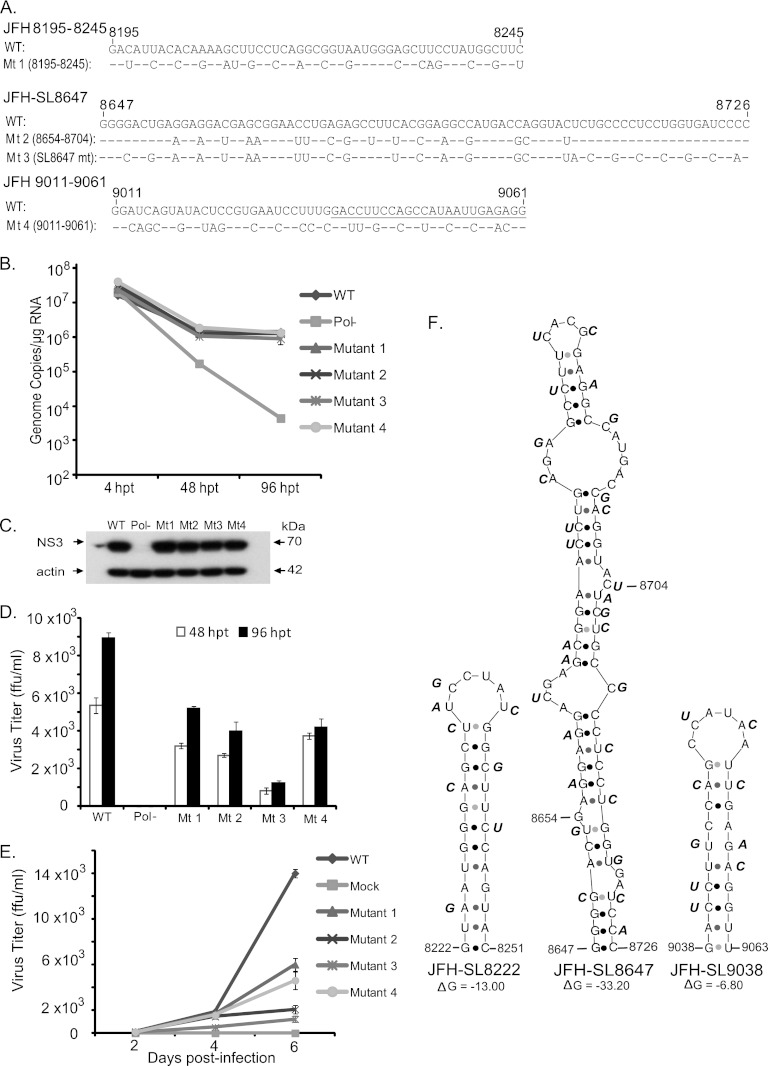
Fig 5.
High-resolution mutational analysis for identification of additional cis-acting replication elements in the NS5B protein-encoding region. (A) Schematic diagram of HCV genome depicting the mutated subsegments in NS5B. JFH-1 CRE stem-loops present in and adjacent to the mutated subsegments are presented. SL9332 and SL9389 are not shown. (B) Genome replication of mutant reporter viruses. A total of 28 contiguous mutant reporter viruses were constructed in NS5B (covering nucleotide positions 7787 to 9289), and a full-length RNA genome was generated for each mutant reporter virus by in vitro transcription. Huh-7.5.1 cells were electroporated with individual mutant RNA genomes, and 72 h posttransfection, cell lysates were harvested for Renilla luciferase assay. Means of triplicate values were calculated and are presented in the bar graph with standard deviations. The nucleotide positions of each mutant are given below the bar graph. FNX-Rluc wild-type and polymerase activity-null mutant reporter viruses were included as controls. (C) The corresponding infectivities of mutant reporter viruses presented in panel B are shown in the bar graph as mean values with standard deviations. The Renilla luciferase activities were measured from the Huh-7.5.1 cell lysates harvested 48 h postinfection. Mutants SS8195-8245, SS8654-8704, SS9011-9061, SS9062-9097, and SS9170-9226, exhibiting over 5-fold reductions (P value < 0.005) in infectivity compared to that of the wild-type virus, were selected for detailed study. (D) Nucleotide sequence of subsegment SS7991-8041. The nucleotide substitutions engineered in the mutant virus are depicted. The sequence forming the predicted stem-loop JFH-SL8001 is underlined. The screening experiment was done twice, and data from a representative experiment are shown.
For initial screening, Huh-7.5.1 cells were electroporated with genomic RNAs of each mutant subsegment reporter virus. At 72 h p.t., the cells were lysed and Renilla luciferase activity was measured. The mean luciferase values thus obtained indicated the extent of genome replication for each mutant (Fig. 5B). The infectivity of each mutant in comparison to the wild-type virus was assayed in Huh-7.5.1 cells as shown in Fig. 5C. As expected, Pol−, SS9170-9226, and SS9227-9289 mutants failed to replicate the genome and produce infectious particles. Interestingly, the subsegment SS7991-8041 mutant containing a predicted stem-loop structure, JFH-SL8001 (H77-SL7936), exhibited a replication phenotype similar to that of wild-type FNX-Rluc virus (Fig. 5). Modeling the engineered silent mutations in JFH-SL8001 revealed that the mutations could lead to lack of or altered base pairing and loss of a stable, high-energy stem-loop structure. Furthermore, at 72 h p.t., we observed that compared to the genome replication level of wild-type virus, the subsegment mutants SS8195-8245, SS8654-8704, and SS9011-9061 exhibited mild reductions of 2.0-, 7.0-, and 8.0-fold, respectively, in genome replication. For the same time point, mutants SS8195-8245, SS8654-8704, and SS9011-9061 had 8.0-, 23.5-, and 13.0-fold reductions, respectively, in infectivity or infectious virus production compared to that of the wild-type virus. Surprisingly, the SS9062-9097 mutant virus showed replication deficiency, implying the presence of a possible critical cis element in the region that might explain the lethal phenotype exhibited by the S17 segment (Fig. 2, 3, and 4). Based on this screening experiment, we followed up with selected mutants that exhibited reductions in both genome replication and infectivity. The mutant subsegments SS8195-8245, SS8654-8704, SS9011-9061, and SS9062-9097 were selected, as they exhibited over 5-fold reduction (P value < 0.005 by unpaired t test) in infectivity compared to that of wild-type virus.
Verification of functional RNA cis elements in NS5B subsegments SS8195-8245, SS8654-8704, and SS9011-9061.
The high-resolution screening of NS5B region RNA elements described above was carried out using mutant Renilla luciferase reporter viruses. Hence, for subsequent detailed verification, we engineered the silent mutations present in each of the mutant subsegments SS8195-8245, SS8654-8704, and SS9011-9061 into the FNX-HCV background (nonreporter wild-type virus) (Fig. 6). The analysis of the viral genome replication, infectivity, and growth curve of each subsegment is described below.
Fig 6.
Detailed analysis of JFH-SL8647 (H77-SL8582) and RNA elements in nucleotide positions 8195 to 8245 and 9011 to 9061. (A) Nucleotide sequence alignments of JFH-SL8647, various subsegments, and mutants. The nucleotide substitutions engineered in the individual FNX-HCV mutant viruses are depicted. The underlined nucleotide sequence in JFH 9011-9061 forms part of JFH-SL9038. Mt, mutant. (B) Replication kinetics of mutant viral genomes. The in vitro-transcribed RNA genomes were electroporated into Huh-7.5.1 cells. Total RNAs harvested at 4, 48, and 96 h posttransfection from cells were subjected to RT-qPCR. The genome copy numbers are depicted in the graph. Mutant 3 (the SL8647 mutant) had significant reduction in genome replication at 96 h p.t. (P value < 0.05 by unpaired t test) compared to that of the wild-type virus. (C) Western blotting of HCV antigen expression. Protein lysates obtained at 96 h posttransfection were used. HCV NS3 antigen and loading control β-actin are shown. (D) Titers of various mutant viruses at 48 and 96 h posttransfection. The cell-free supernatants collected at the indicated time points were inoculated onto naïve Huh-7.5.1 cells, and viral titers (focus-forming units/ml) were quantified as described in Materials and Methods. Mean values and standard deviations of the titers are shown in the graph. All the mutants had significantly reduced infectivities at 48 and 96 h posttransfection compared to that of the wild type (P value < 0.005 by unpaired t test). (E) Growth kinetics of mutant viruses. For multistep growth curve data, the Huh-7.5.1 cells were infected with the indicated viruses at an MOI of 0.1. Cell-free supernatants were harvested at 2, 4, and 6 days postinfection, and the virus titers were measured. Mean values and standard deviations of the titers are shown in the line graph. Compared to wild-type virus growth, all the mutants showed significantly attenuated growth at 4 days postinfection (P value < 0.05 by unpaired t test) and 6 days postinfection (P value < 0.0001 by unpaired t test). (F) Predicted stem-loop structures of JFH-SL8222, JFH-SL8647, and JFH-SL9038. Each nucleotide substitution engineered in the mutant viruses is shown beside the wild-type nucleotide sequence in the stem-loop. The folding Gibbs free energy (ΔG, in kcal/mol) for the predicted stem-loops is depicted under each structure. The experiment was repeated three times, and data from a representative experiment are presented.
(i) Characterization of JFH-1 NS5B region subsegment 8195 to 8245.
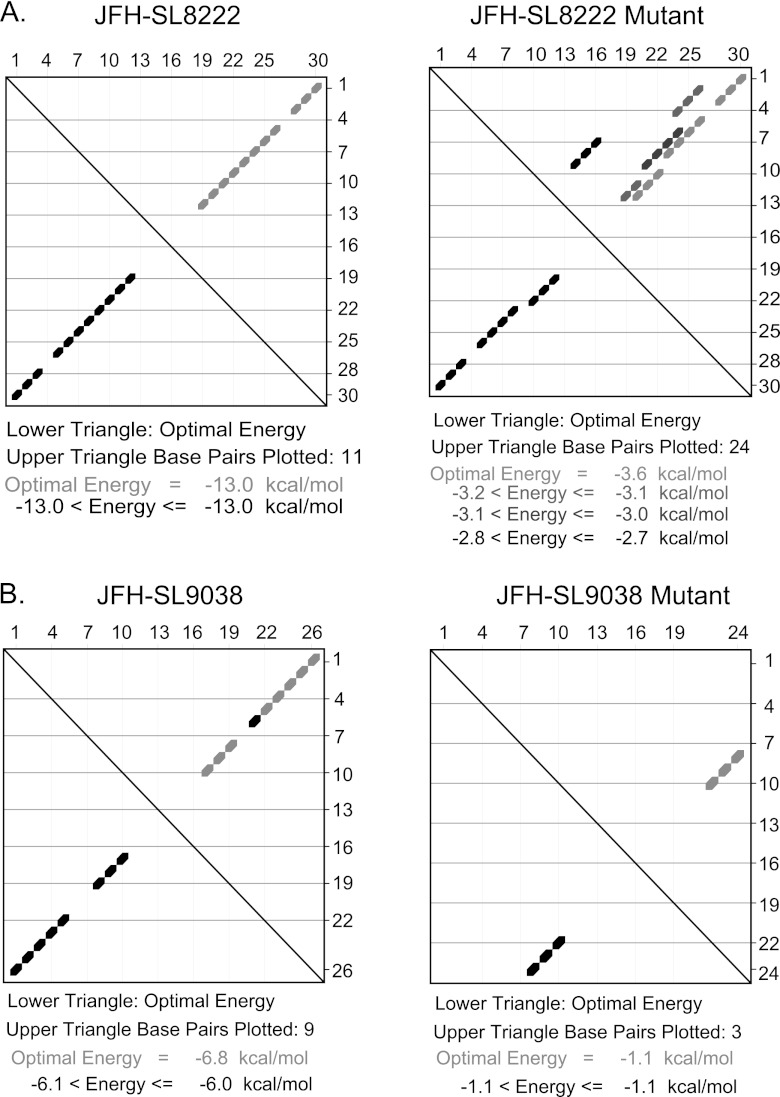
Mutational analysis of the S15 segment (nucleotides 8114 to 8440) revealed a moderately attenuated phenotype with significant reductions in both genome replication and de novo infectivity (Fig. 2, 3, and 4). Subsequent high-resolution screening of the S15 region showed that silent mutations in subsegment SS8195-8245 resulted in reduction in the infectivity level (Fig. 5). We hypothesized that subsegment SS8195-8245 may contain a novel RNA element. SS8195-8245 comprises 51 nucleotides (17 amino acids), and 18 of the 51 nucleotides were replaced to form the mutant FNX-HCV (Fig. 6A). Interestingly, the SS8195-8245 mutant did not show statistically significant changes in the genome replication level at 48 h p.t. and 96 h p.t. in Huh-7.5.1 cells compared to the wild-type virus (Fig. 6B). However, compared to the infectivity titers of wild-type virus at 48 h p.t. and 96 h p.t., the mutant SS8195-8245 showed significant reduction in infectious virus production (P value < 0.05 by unpaired t test) (Fig. 6D). For further confirmation, we performed a multistep growth curve analysis using a multiplicity of infection of 0.1 as a starting point for both the wild type and mutant SS8195-8245 (Fig. 6E). We observed that at 4 days p.i. and 6 days p.i., the mutant SS8195-8245 exhibited significant growth reduction compared to the wild type (P values by unpaired t test for 4 days p.i., <0.05, and 6 days p.i., <0.0001). At 6 days p.i., the mutant SS8195-8245 had 2.3-fold growth attenuation compared to wild-type viral growth (Fig. 6E). We reproduced these results in three independent experiments. Subsequently, we analyzed the SS8195-8245 region for the presence of any RNA secondary structures using the mfold computational prediction Web server (65). The JFH-1 genome sequence was predicted to have a stem-loop at nucleotides 8222 to 8251 with the folding Gibbs free energy (ΔG) of −13.0 kcal/mol (Fig. 6F). Further mfold analysis of homologous sequences from other genotypes revealed possible stem-loop structures in all the genotypes in this region (data not shown). In all six genotypes, this predicted RNA secondary structure contains a conserved sequence, 5′-AUGGG-3′ (JFH-1, nt 8225 to 8229; H77, nt 8160 to 8164). This sequence may have a role in cis element function, besides coding for conserved methionine and glycine amino acids. Energy dot plots for possible optimal and suboptimal folding of the predicted stem-loop JFH-SL8222 are shown in Fig. 7A. mfold analysis of engineered nucleotide substitutions in mutant SS8195-8245 sequence revealed that the mutations could destabilize the predicted stem-loop SL8222 (Fig. 7A). Further study is required to experimentally verify the JFH-SL8222 stem-loop structure. Alternate RNA secondary and tertiary structures in this region (nucleotides 8195 to 8245) of the HCV genome are also possible. In summary, the mutant SS8195-8245 reporter virus exhibited reductions in both genome replication and infectivity (Fig. 5). The SS8195-8245 mutant in the nonreporter FNX-HCV viral background showed significant reduction in infectivity, and thus, the cis element present in the SS8195-8245 region is required for optimum virus growth and can act as an enhancer element. It is possible that the effects of nucleotide changes in SS8195-8245 on viral genome replication in the FNX-HCV background may be very subtle, but the effects may be amplified during de novo infection. Moreover, the SS8195-8245 region cis element may have a role in efficient packaging of the viral genome. Taken together, we have identified a novel RNA cis element in the JFH-1 genome that is required for optimum de novo infectivity.
Fig 7.
Energy dot plot analysis of predicted stem-loops JFH-SL8222 and JFH-SL9038. mfold analysis was performed to generate energy dot plots for SL8222 and SL9038 using the respective wild-type and mutant nucleotide sequences shown in Fig. 6F. (A) Energy dot blots for SL8222 wild-type and mutant sequences. The wild-type SL8222 has an optimal energy of −13.0 kcal/mol, and the mutant SL8222 has a significantly lower free energy of −3.6 kcal/mol. Also note that mutant SL8222 could form various base-pairing combinations, as shown in the upper triangle. (B) Energy dot plots for wild-type and mutant SL9038 sequences. Mutations destabilize the stem-loop by altering base pairing, resulting in a lower optimal free-energy level.
(ii) Genetic analysis of the predicted stem-loop structure JFH-SL8647 (H77-SL8582).
Silent mutations in an engineered S16 (nt 8441 to 8767) mutant virus exhibited stronger attenuation of genome replication and de novo infectivity than in the S14 and S15 mutants (Fig. 2, 3, and 4). Fine mapping of the S16 segment with additional subsegment mutants showed that the SS8654-8704 virus had reduced genome replication and infectivity (Fig. 5). A previous study based on thermodynamic and phylogenetic prediction of RNA stem-loop structures in the HCV protein-coding region revealed a conserved stem-loop structure, JFH-SL8647 (H77-SL8582), in this region. The 17 nucleotide substitutions we introduced in subsegment SS8654-8704 could potentially destabilize this JFH-SL8647 structure (Fig. 6). For further verification, we constructed a mutant HCV (SL8647 mutant) that had 26 silent mutations spanning the entire JFH-SL8647 sequence (Fig. 6A). The SL8647 mutant virus exhibited a significant reduction in genome replication in Huh-7.5.1 cells at 96 h p.t. compared to that of wild-type virus (P value < 0.05 by unpaired t test) (Fig. 6B). The mutant SS8654-8704 showed a trend of reduction in genome replication, but it was not statistically significant. The expression of HCV antigen NS3 at 96 h p.t. was detected by Western blotting (Fig. 6C). Both mutants showed significant reduction in infectivity at 48 h p.t. and 96 h p.t. compared to that of the wild-type virus (P value < 0.0005 by unpaired t test) (Fig. 6D). At 96 h p.t., the SS8654-8704 and SL8647 mutants had 2.2- and 7.2-fold reductions in infectivity, respectively. The attenuated phenotype of the mutants was also corroborated by multistep growth curve results (Fig. 6E). Compared to wild-type viral growth at 6 days p.i., the SS8654-8704 and SL8647 mutants had 6.7- and 11.5-fold reductions, respectively (P value < 0.0001 by unpaired t test). The SL8647 mutant exhibited stronger growth attenuation than SS8654-8704, which corresponds to the fact that the entire stem-loop sequence was mutated in the former virus. The predicted JFH-SL8647 secondary RNA structure, along with the introduced nucleotide changes, is depicted in Fig. 6F. These results suggest that JFH-SL8647 is required for optimum levels of viral genome replication, but its alteration still allows the virus to grow, albeit at attenuated levels. Our study provides experimental verification of the role of SL8647 during genome replication and de novo infection.
(iii) Analysis of the RNA element in nucleotides 9011 to 9061.
High-resolution screening for potential CRE sequence in the S17 segment (nt 8768 to 9097) revealed that the RNA element in the subsegment SS9011-9061 was required for genome replication and efficient viral infectivity (Fig. 5). The SS9011-9061 mutant virus contained 22 nucleotide substitutions (Fig. 6A). The mutant SS9011-9061 viral genome replication level did not show a statistically significant difference from that of the wild type (Fig. 6B). Detailed analysis of infectivity at 48 h p.t. and 96 h p.t. revealed that the SS9011-9061 mutant had significant reduction in infectious virus production compared to the wild type (Fig. 6D) (P value < 0.005 by unpaired t test). The multistep growth curve analysis also showed the important role of the cis element in SS9011-9061 for optimum viral growth (Fig. 6E). mfold analysis predicted a possible stem-loop structure at JFH-1 genome positions 9038 to 9063 with a ΔG of −6.8 kcal/mol (Fig. 6F). Extending the mfold computational analysis to the homologous regions in other genotypes revealed a semiconserved RNA stem-loop secondary structure (data not shown). Energy dot blots of the predicted wild-type JFH-SL9038 and the mutant SL9038 revealed that the synonymous mutations could destabilize the RNA structure by loss of base pairing (Fig. 7). Further structure-function analysis is required for verification of stem-loop JFH-SL9038.
Our validation study thus revealed that the mutant virus (the SL8647 mutant) with 26 silent mutations spanning the entire JFH-SL8647 sequence exhibited reductions in both genome replication and virus growth, while the SS8654-8704 mutant with 17 nucleotide changes showed only attenuated virus growth. It has been reported that genetic analysis of NS5B-SL2 (a JFH-SL8647 homologue in the genotype 1b Con1 strain) by introducing 6 silent mutations in the tip of the stem-loop structure did not affect the genome replication of the Con1 replicon (55). These results imply that in order to observe the effects of cis elements in SS8195-8245, SS8654-8704, and SS9011-9061 on genome replication in the context of the FNX-HCV background, the nucleotide substitutions need to be extended to flanking sequences. The FNX-Rluc reporter virus can be very sensitive for amplifying subtle alteration in genome replication due to increased length of the genome caused by luciferase gene insertion. Taken together, the minor RNA elements in NS5B regions SS8195-8245, SS8654-8704, and SS9011-9061 are required for efficient de novo HCV infectivity and may affect virus growth by subtle alteration of viral genome replication. The predicted JFH-SL8647 plays an important role in efficient viral genome replication and infectivity.
Identification of an extended stem structure in JFH-SL9098 (H77-SL9033).
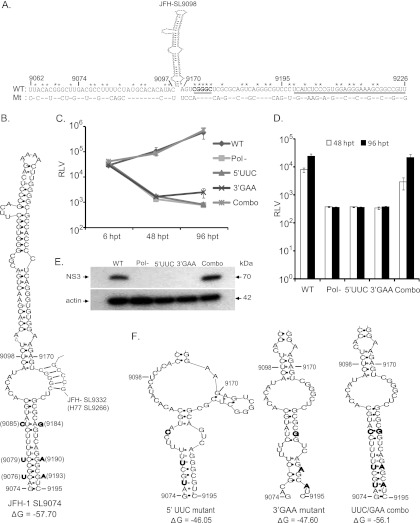
Our mutational analysis of NS5B locations 8768 to 9097 showed a replication-incompetent phenotype for S17 mutant virus (Fig. 2, 3, and 4), indicating the presence of a critical CRE structure. Our screening using the subsegment SS9062-9097 mutant reporter virus (Fig. 5) further reinforced the notion that a functional CRE is more than likely present in this locus; however, this subsegment does not contain any previous experimentally verified or predicted RNA secondary structure. We also observed that the SS9170-9226 subsegment mutant exhibited a lethal phenotype, as the wild-type subsegment contained a conserved 5′-CGGGC-3′ (nt 9173 to 9177) sequence and a stem-loop structure, JFH-SL9198 (H77-SL9132) (Fig. 5 and 8). A predicted stem-loop structure, JFH-SL9098, homologous to genotype 1 H77-9033, is located between the subsegments SS9062-9097 and SS9170-9226 (Fig. 8A). The search for additional RNA secondary structures in the critical subsegment SS9062-9097 and flanking sequences using mfold computational analysis revealed a possible extended stem structure for the predicted stem-loop JFH-SL9098 (H77-9033) (Fig. 8B). In order to experimentally characterize the extended stem structure, we generated mutant HCV with covariant nucleotide substitutions. In the 5′ end of the stem, nucleotide positions 9076 (C→U), 9079 (C→U), and 9085 (U→C) were changed, and the resulting reporter virus was named the 5′ UUC mutant. In the 3′ end of the stem, we mutated positions 9184 (A→G), 9190 (G→A), and 9193 (G→A) and named the virus the 3′ GAA mutant. mfold analysis showed that the silent covariant substituted nucleotides in positions 9076 to 9193, 9079 to 9190, and 9085 to 9184 could form Watson-Crick base pair interactions and should maintain the stem secondary structure. The UUC/GAA combination mutant virus was engineered with the complementing nucleotide changes at both the 5′ and 3′ ends of the stem. Modeling the 5′ UUC and 3′ GAA nucleotide substitutions using the mfold Web server showed that these mutations could destabilize the extended stem structure (Fig. 8F).
Fig 8.
Genetic characterization of extended stem structure formed by JFH-SL9098 (H77-SL9033). (A) JFH-SL9098 stem-loop flanking nucleotide sequences of WT and mutant subsegments (9062 to 9097 and 9170 to 9226). The underlined nucleotide sequence forms part of JFH-SL9198. Sequence forming the long-range interaction with JFH-SL9332 is in boldface and underlined. Asterisks indicate conserved nucleotides. (B) Predicted extended stem structure formed by JFH-SL9098. The previously reported stem-loop structure is formed by nucleotide positions 9098 to 9170. The silent complementing nucleotide substitutions engineered on the left and right sides of the extended stem are shown. Sequences predicted to form long-range interactions between JFH-SL9074 and JFH-SL9332 are shown in the stem-loop. (C) Analysis of genome replication of mutant reporter viruses at the indicated times posttransfection. Mutants 5′ UCC and 3′ GAA exhibited significant reductions in genome replication at 48 h p.t. (P value = 0.0093 by unpaired t test) and 96 h p.t. (P value < 0.05) compared to that of wild-type virus. (D) Infectivity of each virus. The infectivities of mutants 5′ UCC and 3′ GAA were significantly reduced at 48 h p.t. (P value = 0.0018) and 96 h p.t. (P value = 0.0021) compared to that of wild-type virus. (E) Western blotting of HCV antigen expression at 96 h p.t. Western blotting was performed to detect NS3 antigen and β-actin. The detailed methodology for panels C, D, and E is given in the legend to Fig. 1. (F) Predicted extended stem structures formed by mutant sequences. Due to space constraints, only the lower halves of the stems are shown. Note that the 5′ UCC and 3′ GAA mutant sequences have altered base pairing in the nucleotide-substituted stem region. In the UUC/GAA combination mutant, the stem structure is restored. The folding Gibbs free energy (ΔG, in kcal/mol) for the stem-loops is given under each structure. The data presented are from one of three independent experiments.
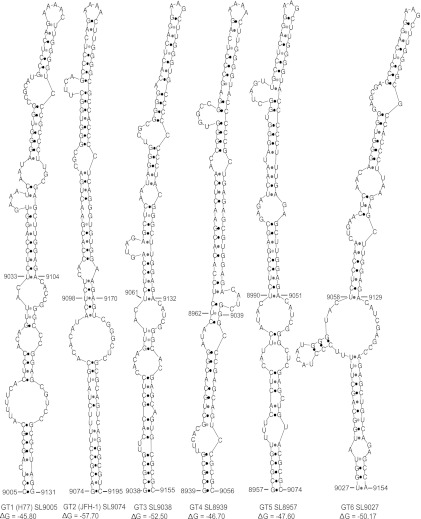
The genomic RNAs generated from wild-type, Pol−, 5′ UUC, 3′ GAA, and UUC/GAA combination reporter viral constructs were individually electroporated into Huh-7.5.1 cells. At 6 h, 48 h, and 96 h posttransfection, cells were harvested in order to measure Renilla luciferase activity (Fig. 8C). Our results indicated that the viruses having mutations in either the 5′ end (5′ UCC mutant) or the 3′ end (3′ GAA mutant) of the stem had defects in genome replication, whereas the complementary mutation containing the virus UUC/GAA combination had genome replication restored to levels similar to that of the wild type. At 96 h p.t., the 3′ GAA mutant showed a low level of genome replication, which was about 3.2-fold over that of the replication-incompetent Pol− mutant. Both the 5′ UUC and 3′ GAA mutant viruses had no detectable infectivity (Fig. 8D). The UUC/GAA combination virus had a level of infectivity similar to that of the wild type. The production of NS3 antigen by replication-competent UUC/GAA combination virus was verified with Western blotting (Fig. 8E). In conclusion, our genetic analysis confirmed the presence of an extended stem structure in the JFH-1 CRE SL-9098. Based on the position of the first paired nucleotide at the 5′ end, the stem-loop structure is named JFH-SL9074 (homologous structure in genotype 1, H77-SL9005). mfold analysis of homologous sequences in all six genotypes revealed conservation of extended stem structure across the genotypes (Fig. 9).
Fig 9.
Predicted stem-loop structures homologous to extended JFH-SL9074. The extended stem-loop structures formed by all six genotypes are depicted. The genome positions of the respective genotypes are displayed for each stem-loop. Previously predicted genotype 1 (GT1) stem-loop H77 SL9033 nucleotide positions, 9033 to 9104, are indicated. The folding Gibbs free energy (ΔG, in kcal/mol) for each stem-loop structure is shown.
DISCUSSION
This investigation describes an unbiased genome scale approach to identifying RNA elements in the hepatitis C virus nonstructural protein-coding region, which modulates genome replication. In order to identify CRE structures, we engineered 2,388 (38.6%) silent point mutations in the genomic region encoding the NS3 to NS5B proteins (nt 3872 to 9097). This region is spanned by 17 contiguous mutant-segment-containing hepatitis C viruses. By this approach, every third nucleotide in the NS3-to-NS5B region was mutated except in the codons for methionine and tryptophan. This is a powerful approach to destabilize the RNA secondary and tertiary structures that form short- or long-range interactions during the dynamic phases of viral growth: viral genome translation, replication, and packaging.
Based on our experimental conditions using Huh-7.5.1 cells and infectious intra-genotype 2a chimeric hepatitis C virus, we did not observe significant replication defects in the viruses that had mutant segments from NS3, NS4A, NS4B, and NS5A, consistent with previous computational studies (50, 52, 53). On the contrary, introduction of silent mutations in the NS5B region resulted in moderate to severe impairment of genome replication and infectivity in the tested mutant viruses (Fig. 2, 3, and 4). Each of the 17 mutant viruses tested in the initial round had 34.4% to 41.7% nucleotide substitutions (Table 2), but only the NS5B region mutants exhibited a reduced genome replication phenotype. Of the total nucleotide changes, 27.6% to 32.4% are conserved substitutions, and the substitutions can be found in naturally occurring HCV strains. The conserved changes may have less influence on modifying RNA structures. However, the nonconserved (arbitrary) synonymous mutations could have major effects on RNA structure and function. Most of the nonconserved changes were introduced in the NS5A and NS5B region (Table 2), and under our experimental conditions, the phenotypic effects of silent mutations were observed only in the NS5B region. The growth-attenuated phenotype of NS5B mutants was independently verified by using mutant viruses generated in the background of the FNX-Rluc reporter virus, as well as the FNX-HCV wild-type virus (Fig. 2 and 4).
Long-term culturing of mutant viruses (S13, S14, S15, and S16) for up to 18 days revealed that all the mutants were able to establish persistent infection under cell culture conditions, albeit at reduced levels compared to wild-type viruses. Genome replication of S13, S14, and S15 mutants gradually reached that of the wild-type virus on day 15 p.t., suggesting delayed genome replication kinetics (Fig. 3). The S16 mutant exhibited genome replication at levels 1 to 2 log units lower than the wild type at all the time points tested. This observation suggests that the cis elements present in the S16 segment positively influence genome replication during both short- and long-term infection. The RNA structures present in S16 could modulate HCV persistent infection (54). During long-term growth, the infectivities of S13 and S14 mutant viruses were similar but began to differentiate between 9 days p.t. and 15 days p.t. It is possible that the rates of accumulation of compensatory mutations influencing infectivity may be different in the two mutants. Further investigation of the nature of compensatory mutations accumulated by these RNA element mutants during long-term growth could yield new mechanistic information.
Segment S14 (NS5B nt 7787 to 8113) mutant HCV had moderate growth attenuation, suggesting the presence of a cis-acting replication element. Interestingly, while screening for CRE structures in this segment using 51-nucleotide contiguous subsegment mutant viruses (7 mutant viruses) (Fig. 5), we did not observe a significant growth defect in genome replication and infectivity even though previous studies predicted the presence of a CRE. A previous study using thermodynamic predictions and phylogenetic analysis of major HCV genotypes reported a possible stem-loop secondary structure, 5BSL1, H77-SL7936, or JFH-SL8001, in this region (50, 52). You et al., reported that introduction of 7 synonymous mutations in 5BSL1 did not affect genome replication of a genotype 1b Con1 replicon (55). Consistent with this report, in our screen, subsegment mutant SS7991-8041 encompassing JFH-SL8001 (5BSL1) had levels of genome replication and infectivity similar to those of the wild-type virus. Modeling the silent mutations engineered in SS7991-8041 mutant virus showed destabilization of the predicted stem-loop structure. These observations suggest that the segment S14 may have minor cis elements embedded across the segment with a collective function during genome replication. It is possible that the phenotype can be observed only when most of the cis elements are mutated. Moreover, since only the wobble nucleotide of the codon was mutated in our approach, it is possible that RNA elements made up of nonwobble nucleotides may be present in the coding region forming a functional CRE structure.
Segment S15 in the NS5B region does not have any predicted or experimentally verified CRE structures, but we observed that the virus containing the S15 mutant segment (nt 8114 to 8440) showed reduced replication fitness. Further fine-mapping study of this segment revealed that silent mutations in subsegment SS8195-8245 resulted in lower genome replication and infectivity of mutant reporter virus. However, the SS8195-8245 mutant in the background of FNX-HCV (nonreporter virus) exhibited reduction only in de novo infectivity. Our results suggest that the RNA cis element present in SS8195-8245 may have a very subtle effect on viral genome replication, but during a de novo infectious cycle, the impairment phenotype may be amplified. We consistently observed that the SS8195-8245 mutant had impaired infectious viral particle production and growth kinetics. A role of this RNA cis element in viral genome packaging cannot be ruled out. Since, the RNA elements located in this region are required for efficient viral growth, they may function as enhancer elements. Furthermore, computational mfold analysis of the SS8195-8245 region predicted a semiconserved stem-loop structure in all six genotypes. The functional significance of this stem-loop structure requires further genetic and biochemical investigation.
In our study, HCV with silent mutations in the segment S16 exhibited the strongest growth attenuation compared to all the replication-competent segment mutant viruses (S1 to S15) tested. The S16 mutant virus had an attenuated level of genome replication during both short-term infection and long-term persistent infection. The S16 segment contains a previously predicted stem-loop structure, JFH-SL8647, homologous to H77-SL8582 or 5BSL2 (50, 55). Subsequent high-resolution mapping of the S16 segment using contiguous short-segment mutant viruses showed that the subsegment SS8654-8704 mutant containing substitutions in JFH-SL8647 had replication impairment. The mutant SL8647 virus with 26 synonymous mutations spanning the entire stem-loop had significant reductions in both genome replication and infectivity (Fig. 6). Interestingly, the SS8654-8704 mutant with 17 nucleotide changes in this stem-loop structure exhibited reduction only in de novo infectivity and did not show statistically significant reduction in genome replication. A study reported by You et al. revealed that 6 silent mutations introduced in the tip of 5BSL2 did not affect viral genome replication in a Con1 subgenomic replicon system (55). These observations suggest that more nucleotide substitutions create larger changes in RNA secondary and tertiary structures and ultimately result in greater growth inhibition. These observations imply that the JFH-SL8647 CRE is required for efficient genome replication and de novo infectivity. This JFH-SL8647 CRE may have a possible enhancer role by finely altering genome replication.
The SS9062-9097 mutant showed a replication-defective phenotype. This observation surprised us, as there was no predicted stem-loop structure in this subsegment. Upon further investigation, a stem-loop structure, JFH-SL9098 (nt 9098 to 9170), homologous to the previously predicted stem-loop structure H77-SL9033, was found downstream of subsegment SS9062-9097. Our experimental approach using covariant mutational analysis confirmed that JFH-SL9098 has an extended stem structure, and we designated it JFH-SL9074. mfold analysis showed a conserved nature of the extended stem formed by homologous stem-loop structures in the other six genotypes. The extended JFH-SL9074 (nt 9074 to 9195) is flanked by the upstream enhancer CRE subsegment SS9011-9061 (the predicted JFH-SL9038) and the downstream critical stem-loop structure JFH-SL9198. A previous study using a genotype 1b Con1 replicon demonstrated that Con1-SL9033 downstream conserved nucleotides form a long-range interaction with Con1-SL9266 (JFH-SL9332) (57). A follow-up structure-function study by the same group showed that this long-range interaction could be genotype specific, as they did not observe any defect in the growth of the intragenotype chimeric virus J6/JFH-1 after genetically abolishing the long-range interaction (60). The extended stem-loop structure JFH-SL9074 is critical for optimum genome replication.
The significance of our study is underscored by the identification of new cis-acting RNA replication elements with enhancer roles and critical roles for genome replication. The enhancer cis element identification required the introduction of multiple nucleotide substitutions that facilitated the detection of a growth-attenuated phenotype. Further study is needed to characterize the minimum number of nucleotide changes required to identify the phenotype. We found that the enhancer elements are enriched in the NS5B coding sequence upstream of JFH-SL9074. In the hammerhead ribozyme, the tertiary interactions by loop-bulge or loop-loop structures in nonconserved regions located far from the active site can stabilize the active conformation of catalytic nucleotides (68). Thus, the HCV conserved and nonconserved enhancer elements may cooperatively function to form proper folding or maintain the alignment of major structural elements in three dimensions for efficient viral genome replication. The additive or synergistic functional nature of the identified enhancer CREs can be elucidated by using various combinations of enhancer mutant CREs. Further comparative structure-function study is warranted to verify whether the identified enhancer elements are genotype specific or conserved across all the major genotypes using available replicon systems.
The RNA viral genome forms higher-order structures during genome translation and replication and in the encapsidated state. The high-resolution structure of the identified enhancer and critical CRE can be elucidated by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) technology and by deep sequencing RNA fragments using the parallel analysis of RNA structure (PARS) method (69, 70). Furthermore, long- and short-range RNA-RNA interactions formed by CRE structures, as well as RNA-protein interactions, are crucial for viral genome replication. The mutant CRE viruses can be used for identification of proteins interacting with CREs through protein-RNA pulldown and mass spectrometry identifications. These experiments will provide mechanistic insights into viral genome replication.
The current study characterizing the growth phenotypes of various cis element mutant viruses was carried out in the human hepatoma cell line Huh-7.5.1, which lacks the innate immune sensor RIG-I (71). The RNA secondary and tertiary structures at the genome level of HCV evolved to evade detection by PAMP (pathogen-associated molecular patterns) recognizing cellular innate immune sensors. HCV poly(dU) sequence at the 3′ NTR has been shown to act as an HCV PAMP (72). We engineered hundreds of nucleotide substitutions in the NS3, NS4A/4B, NS5A, and NS5B coding regions. Thus, this collection of hepatitis C viruses with silent mutations can be tested in primary hepatocytes and humanized mice with xenografted human primary liver cells and immune cells to elucidate the role of the HCV genome structure in immune evasion, persistent infection, and hepatitis C pathogenesis. These studies can provide vital information for developing attenuated vaccine candidates by modifying HCV RNA structure.
In summary, we utilized an unbiased approach to identify cis-acting RNA elements in the nonstructural-protein-coding region of the HCV genome. We observed that the NS5B coding region is enriched with RNA elements and that the coding region for NS3, NS4A/4B, and NS5A does not have any critical RNA functional elements (Fig. 10A). Though dispensable, the NS5B 5′ coding region contains RNA elements that are necessary for efficient genome replication and infectious progeny production and thus have an enhancer function. We have identified new enhancer cis RNA elements predicted to have conserved and nonconserved stem-loop structures, JFH-SL8222 and JFH-SL9038 (Fig. 10B). These RNA elements may play a role in genome packaging and the assembly of viral particles. Furthermore, we have experimentally verified the presence of a previously predicted stem-loop structure, JFH-SL8647, and demonstrated that this stem-loop is required for efficient HCV genome replication. We have identified an extended stem structure, JFH-SL9074, that plays a critical role during HCV genome replication and infectivity. We have elucidated the roles of several cis-acting RNA elements in the protein-coding region of the HCV genome during viral growth, and this knowledge will further facilitate the understanding of the mechanism of HCV replication and the design of attenuated vaccine candidates.
Fig 10.
Schematic diagram of enhancer and critical RNA cis elements in the HCV genome. (A) HCV genome regions are color coded based on the nature of the CREs. Both 5′ and 3′ NTRs and the 3′ end of NS5B (the red region) harbor essential CREs. The 5′ end of NS5B (blue) contains cis elements with enhancer functions. NS3, NS4A, NS4B, and NS5A coding regions (green) may not harbor RNA structures involved in viral genome replication. The C, E1, E2, p7, and NS2 coding regions (gray) are dispensable for genome replication. (B) The NS5B coding region harbors both enhancer (blue) and critical (red) RNA cis elements. The enhancer elements are clustered in the 5′ two-thirds of the NS5B coding region and may be involved in stabilizing RNA-RNA and RNA-protein interactions, as well as efficient packaging of the viral genome. Predicted stem-loop structures in the NS5B enhancer subsegments SS8195-8245 (JFH-SL8222) and SS9011-9061 (JFH-SL9038) are depicted. We observed that stem-loop JFH-SL8647 is required for efficient genome replication. The location of the extended stem-loop JFH-SL9074 (homologous to H77-SL9005) is shown in the context of upstream enhancer structures and the downstream critical CREs. Not drawn to scale.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Chisari for the Huh-7.5.1 cell line. We also thank Yu-Chen Cheng for editing the manuscript.
This work was supported by a Cedars-Sinai Medical Center Programmatic Research Award to V.A.
Footnotes
Published ahead of print 13 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00840-12.
REFERENCES
- 1. Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705–714 [DOI] [PubMed] [Google Scholar]
- 2. Alter MJ. 1997. Epidemiology of hepatitis C. Hepatology 26:62S–65S [DOI] [PubMed] [Google Scholar]
- 3. Wasley A, Grytdal S, Gallagher K. 2008. Surveillance for acute viral hepatitis—United States, 2006. MMWR Surveill Summ. 57:1–24 [PubMed] [Google Scholar]
- 4. Perz JF, Alter MJ. 2006. The coming wave of HCV-related liver disease: dilemmas and challenges. J. Hepatol. 44:441–443 [DOI] [PubMed] [Google Scholar]
- 5. El-Serag HB. 2002. Hepatocellular carcinoma: an epidemiologic view. J. Clin. Gastroenterol. 35:S72–S78 [DOI] [PubMed] [Google Scholar]
- 6. Ciesek S, Manns MP. 2011. Hepatitis in 2010: the dawn of a new era in HCV therapy. Nat. Rev. Gastroenterol. Hepatol. 8:69–71 [DOI] [PubMed] [Google Scholar]
- 7. Ghany MG, Nelson DR, Strader DB, Thomas DL, Seeff LB, American Association for the Study of Liver Diseases 2011. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology 54:1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nouri-Aria KT, Sallie R, Mizokami M, Portmann BC, Williams R. 1995. Intrahepatic expression of hepatitis C virus antigens in chronic liver disease. J. Pathol. 175:77–83 [DOI] [PubMed] [Google Scholar]
- 9. Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: viruses and their replication, p 1101–1152 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 10. Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. U. S. A. 88:5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartenschlager R, Ahlborn-Laake L, Yasargil K, Mous J, Jacobsen H. 1995. Substrate determinants for cleavage in cis and in trans by the hepatitis C virus NS3 proteinase. J. Virol. 69:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 14. Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 15. Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 16. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959–1962 [DOI] [PubMed] [Google Scholar]
- 19. Brown EA, Zhang H, Ping LH, Lemon SM. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20:5041–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friebe P, Lohmann V, Krieger N, Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047–12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 23. Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. 2012. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U. S. A. 109:941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccalà G, Leroux-Roels G, Mavromara P, Bartenschlager R. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J. Virol. 82:11503–11515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blight KJ, Rice CM. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. U. S. A. 96:2291–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arumugaswami V, Remenyi R, Kanagavel V, Sue EY, Ngoc Ho T, Liu C, Fontanes V, Dasgupta A, Sun R. 2008. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog. 4:e1000182 doi:10.1371/journal.ppat.1000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yi M, Lemon SM. 2003. 3′ Nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han JH, Houghton M. 1992. Group specific sequences and conserved secondary structures at the 3′ end of HCV genome and its implication for viral replication. Nucleic Acids Res. 20:3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kolykhalov AA, Feinstone SM, Rice CM. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tanaka T, Kato N, Cho MJ, Sugiyama K, Shimotohno K. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. You S, Rice CM. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J. Virol. 82:184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 80:11579–11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gontarek RR, Gutshall LL, Herold KM, Tsai J, Sathe GM, Mao J, Prescott C, Del Vecchio AM. 1999. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 27:1457–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chung RT, Kaplan LM. 1999. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem. Biophys. Res. Commun. 254:351–362 [DOI] [PubMed] [Google Scholar]
- 38. Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417–36428 [DOI] [PubMed] [Google Scholar]
- 39. Kanai A, Tanabe K, Kohara M. 1995. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 376:221–224 [DOI] [PubMed] [Google Scholar]
- 40. Wood J, Frederickson RM, Fields S, Patel AH. 2001. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 75:1348–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Banerjee R, Dasgupta A. 2001. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 75:1708–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 96:13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Behrens SE, Tomei L, De Francesco R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12–22 [PMC free article] [PubMed] [Google Scholar]
- 44. Lohmann V, Korner F, Herian U, Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937–943 [DOI] [PubMed] [Google Scholar]
- 46. Lee H, Liu Y, Mejia E, Paul AV, Wimmer E. 2006. The C-terminal hydrophobic domain of hepatitis C virus RNA polymerase NS5B can be replaced with a heterologous domain of poliovirus protein 3A. J. Virol. 80:11343–11354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ivashkina N, Wolk B, Lohmann V, Bartenschlager R, Blum HE, Penin F, Moradpour D. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76:13088–13093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adachi T, Ago H, Habuka N, Okuda K, Komatsu M, Ikeda S, Yatsunami K. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38–48 [DOI] [PubMed] [Google Scholar]
- 49. Oh JW, Ito T, Lai MM. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694–7702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8:824–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tuplin A, Evans DJ, Simmonds P. 2004. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 85:3037–3047 [DOI] [PubMed] [Google Scholar]
- 52. Hofacker IL, Fekete M, Flamm C, Huynen MA, Rauscher S, Stolorz PE, Stadler PF. 1998. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 26:3825–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith DB, Simmonds P. 1997. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J. Mol. Evol. 45:238–246 [DOI] [PubMed] [Google Scholar]
- 54. Simmonds P, Tuplin A, Evans DJ. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: implications for virus evolution and host persistence. RNA 10:1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. You S, Stump DD, Branch AD, Rice CM. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee H, Shin H, Wimmer E, Paul AV. 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 78:10865–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ. 2008. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 82:9008–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kuiken C, Combet C, Bukh J, Shin IT, Deleage G, Mizokami M, Richardson R, Sablon E, Yusim K, Pawlotsky JM, Simmonds P, Los Alamos HIV Database Group 2006. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology 44:1355–1361 [DOI] [PubMed] [Google Scholar]
- 59. Kuiken C, Mizokami M, Deleage G, Yusim K, Penin F, Shin IT, Charavay C, Tao N, Crisan D, Grando D, Dalwani A, Geourjon C, Agrawal A, Combet C. 2006. Hepatitis C databases, principles and utility to researchers. Hepatology 43:1157–1165 [DOI] [PubMed] [Google Scholar]
- 60. Tuplin A, Struthers M, Simmonds P, Evans DJ. 2012. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 40:6908–6921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Romero-Lopez C, Berzal-Herranz A. 2009. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 15:1740–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Romero-Lopez C, Berzal-Herranz A. 2012. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 69:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jones DM, Patel AH, Targett-Adams P, McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Appel N, Herian U, Bartenschlager R. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zuker M. 2003. mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gonzalez O, Fontanes V, Raychaudhuri S, Loo R, Loo J, Arumugaswami V, Sun R, Dasgupta A, French SW. 2009. The heat shock protein inhibitor Quercetin attenuates hepatitis C virus production. Hepatology 50:1756–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Khachatoorian R, Arumugaswami V, Ruchala P, Raychaudhuri S, Maloney EM, Miao E, Dasgupta A, French SW. 2012. A cell-permeable hairpin peptide inhibits hepatitis C viral nonstructural protein 5A-mediated translation and virus production. Hepatology 55:1662–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Martick M, Scott WG. 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126:309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wilkinson KA, Merino EJ, Weeks KM. 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J. Am. Chem. Soc. 127:4659–4667 [DOI] [PubMed] [Google Scholar]
- 71. Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.