Abstract
Human cytomegalovirus (HCMV) encodes the UL37 exon 1 protein (pUL37x1), which is the potent viral mitochondrion-localized inhibitor of apoptosis (vMIA), to increase survival of infected cells. HCMV vMIA traffics from the endoplasmic reticulum (ER) to ER subdomains, which are physically linked to mitochondria known as mitochondrion-associated membranes (MAM), and to mitochondria. The antiapoptotic function of vMIA is thought to primarily result from its ability to inhibit Bax-mediated permeabilization of the outer mitochondrial membrane (OMM). Here, we establish that vMIA retargets Bax to the MAM as well as to the OMM from immediate early through late times of infection. However, MAM localization of Bax results in its increased ubiquitination and proteasome-mediated degradation. Surprisingly, HCMV infection does not increase OMM-associated degradation (OMMAD) of Bax, even though the ER and mitochondria are physically connected at the MAM. It was recently found that lipid rafts at the plasma membrane can connect extrinsic and intrinsic apoptotic pathways and can serve as sites of apoptosome assembly. In transfected permissive human fibroblasts, vMIA mediates, through its cholesterol affinity, association of Bax and apoptosome components with MAM lipid rafts. While Bax association with MAM lipid rafts was detected in HCMV-infected cells, association of apoptosome components was not. These results establish that Bax recruitment to the MAM and its MAM-associated degradation (MAMAD) are a newly described antiapoptotic mechanism used by HCMV infection to increase cell survival for its growth.
INTRODUCTION
Human cytomegalovirus (HCMV) encodes multiple antiapoptotic proteins in the UL36 to UL38 loci, including the viral mitochondrion-localized inhibitor of apoptosis (vMIA) or UL37 exon 1 (pUL37x1), UL36, and UL38 proteins and in the immediate early (IE) 1-2 locus (1–4). Both vMIA and another antiapoptotic product, beta 2.7 RNA, inhibit mitochondrion-mediated apoptosis in HCMV-infected cells (5–11).
vMIA is the predominant UL37 isoform from IE times and throughout HCMV infection (12–16). vMIA is synthesized at the endoplasmic reticulum (ER) membrane, remains membrane anchored by its hydrophobic N-terminal leader, and traffics from the ER to mitochondria (17, 18). Its antiapoptotic activity is currently ascribed to its inhibition of Bax-induced permeabilization of the outer mitochondrial membrane (OMM) (5, 7, 8, 19, 20). However, unlike various BH3 domain-containing antiapoptotic proteins, regulation of Bax function by vMIA remains poorly understood (21).
HCMV vMIA traffics to ER-mitochondrion contacts known as mitochondrion-associated membranes (MAM) (16, 17, 22, 23). The MAM provides high-calcium (Ca2+) microdomains for ER to mitochondrial Ca2+ signaling needed for basal metabolism and sites for Ca2+ homeostasis (24–27). Nonetheless, mitochondrial Ca2+ overload can initiate mitochondrion-mediated apoptosis (28–30). HCMV vMIA causes Ca2+ efflux from ER stores during infection (31), consistent with its ability to stimulate mitochondrial metabolism and to blunt stress responses (32–37).
The MAM is notable for its striking enrichment of lipid synthetic proteins and of internal lipid rafts (38–40). Cholesterol binding by HCMV vMIA enables its association with MAM lipid rafts (41). Furthermore, the MAM has been associated with protein degradation (42, 43). Erlin-1 and -2 (SPFH1 and -2), which localize in ER lipid rafts, associate with inositol 1,4,5-trisphosphate receptors (IP3Rs) prior to their ubiquitination, p97 association, and degradation by the ER-associated degradation (ERAD) pathway. gp78 (also known as AMFR), an E3 ubiquitin ligase, partially localizes in the MAM, initiates ubiquitination of targeted substrates in the peripheral ER, and transfers the ubiquitinated substrates to the central ER for their degradation by the ERAD pathway (44–48).
It was recently found that lipid rafts at the plasma membrane can connect extrinsic and intrinsic apoptotic pathways and can serve as sites of apoptosome assembly, including Apaf1, Bid, cytochrome c (Cyt c), procaspase 9, and caspase 3 (49, 50). Apoptosis induction is regulated by ubiquitination and proteasomal degradation of activated Bax at the OMM, known as OMM-associated degradation (OMMAD) (51–54).
The ubiquitin-proteasome system facilitates various stages of HCMV infection (55–58). HCMV evades the immune response by US2- and US11-mediated targeting of the major histocompatibility complex class I (MHC-I) heavy chain to the ERAD pathway for degradation (59–61). Furthermore, HCMV infection results in relocalization of specific 19S proteasome subunits to the periphery of nuclear viral replication centers (58).
Using quantitative proteomic analyses, we recently found that HCMV infection dramatically alters the MAM proteome at late times, notably increasing the abundance of Ca2+ homeostasis proteins, chaperones, and metabolic enzymes (37). Given that vMIA binds Bax (5, 7, 8, 19, 20, 62) and is partially localized in the MAM (18, 22, 23, 37, 41), we hypothesized that HCMV vMIA recruits Bax to the MAM and thereby partially neutralizes its activity in the infected cell. Indeed, our studies show that Bax is partially retargeted to the MAM and its lipid rafts throughout infection in HCMV-infected human foreskin fibroblasts (HFFs) in a vMIA-dependent manner. Proteasome-mediated degradation of MAM-localized but not of mitochondrion-localized Bax is increased in HCMV-infected HFFs. These findings establish a novel mechanism for Bax regulation by MAM-associated degradation (MAMAD) and that HCMV uses this mechanistically distinct pathway to regulate Bax levels and consequently function in the infected cell.
MATERIALS AND METHODS
Cells and viruses.
HFFs were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS) (HyClone), 100 U/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen, Gibco) as previously described (13).
The HCMV ΔUL37x1 mutant (ΔUL37x1 Mut or BADsubstitutionUL37x1 [BADsubUL37x1]) and its wild-type (wt) parent control HCMV (BADwt) were graciously provided by Tom Shenk (31, 63) for these studies. HCMV wt and HCMV ΔUL37x1 Mut stocks were grown in HFFs, and titers were determined as previously described (64, 65). HFFs were uninfected or infected with HCMV wt and HCMV ΔUL37x1 Mut at a multiplicity of infection (MOI) of 1 or 3 PFU/cell.
Transient transfections.
For fractionations, HFFs were transfected using Lipofectamine 2000 (Invitrogen) suspended in Optimem (Gibco) according to the manufacturer's protocols as previously described (17, 18, 22). Twenty-four hours after transfection, adherent cells were harvested and either stored on ice before fractionation or stored at −80°C for subsequent fractionation as described previously (41).
Subcellular fractionation.
HCMV-infected and uninfected HFFs were fractionated to isolate heavy MAM, mitochondrial, microsomal, and cytosolic fractions as described previously (22, 23, 37, 66). Briefly, cells were pelleted, resuspended in sucrose homogenization medium (SHM) (0.25 M sucrose, 10 mM Tris [pH 7.4], and/or 10 mM HEPES [pH 7.4], Roche complete protease inhibitor cocktail) and lysed using a motor-driven Potter-Elvehjem homogenizer. Homogenates were pelleted by centrifugation at 600 × g for 5 min at 4°C, resuspended in SHM, and homogenized again. The homogenates were pooled and pelleted by centrifugation as described above. The cycle was repeated 2 to 4 times until a pellet no longer formed, and aliquots of this homogenate were retained as total protein. Crude mitochondria and MAM (pellet) were separated from microsomes and cytosol (supernatant) by centrifugation at 10,300 × g for 10 min at 4°C. Ultracentrifugation (95,000 × g, 60 min, 4°C) further separated microsomes (pellet) from cytosol (supernatant). Cytosol was concentrated using 4-ml Amicon filter units (low-binding regenerated cellulose Ultracel-4 membrane, 3 kDa) by centrifugation (7,500 × g, 30 min, 4°C). Pelleted MAM and mitochondria were resuspended in 300 μl of ice-cold mannitol buffer A (0.25 M d-mannitol, 5 mM Tris [pH 7.4], and/or 5 mM HEPES, 0.5 mM EGTA, 1 mM phenylmethylsulfonyl fluoride [PMSF]), subjected to homogenization, and layered onto a 10-ml 30% Percoll suspension in mannitol buffer B (0.225 M d-mannitol, 25 mM Tris [pH 7.4], and/or 25 mM HEPES, 1 mM EGTA, 1 mM PMSF). Ultracentrifugation (95,000 × g, 65 min, 4°C) was used to band mitochondria and MAM. Collected mitochondria and MAM bands were diluted 5-fold in either 1× phosphate-buffered saline (PBS) plus 1 mM PMSF or SHM and subjected to centrifugation to pellet mitochondria (6,150 × g, 10 min, 4°C) or heavy MAM (10,300 × g, 10 min, 4°C). All of the fractions were resuspended in a minimal volume (50 to 200 μl) of SHM and assayed for protein concentration using a bicinchoninic acid (BCA) reagent kit (Pierce) according to the manufacturer's recommended protocol.
For ER/MAM and mitochondrial fractionations, pelleted cells were resuspended in 2 ml of MTE buffer (270 mM mannitol, 10 mM Tris-HCl, 0.1 mM EDTA [pH 7.4]) supplemented with Complete protease inhibitor cocktail (Roche) and 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma), and lysed by sonication (at an output power of 3.5). Cellular debris and intact cells were removed by centrifugation at 700 × g for 10 min at 4°C. Crude mitochondria were pelleted by centrifugation at 15,000 × g for 10 min at 4°C, and the postmitochondrial supernatant was used for purification of ER/MAM fractions. Purified mitochondria were obtained by banding in discontinuous sucrose gradients consisting of 1.0 M and 1.7 M sucrose steps in 10 mM Tris-HCl (pH 7.6) at 40,000 × g for 22 min at 4°C. Postmitochondrial supernatants were layered onto discontinuous sucrose gradients (1.3, 1.5, and 2.0 M sucrose in 10 mM Tris-HCl [pH 7]) and banded by centrifugation at 100,000 × g for 45 min at 4°C. The purified ER/MAM and mitochondrial pellets were resuspended in 1× PBS and stored at −20°C until use. Protein concentrations of isolated subcellular fractions were determined using a BCA assay kit (Pierce), following the manufacturer's recommendations.
Lipid raft isolation.
Detergent-resistant membranes (DRMs) were isolated on flotation sucrose gradients as previously described (41). Briefly, transiently transfected HFFs (6 × 107 cells) were harvested by scrapping or by trypsinization, resuspended in MBS (25 mM 4-morpholineethanesulfonic acid, 150 mM sodium chloride [pH 6.5] containing protein inhibitor cocktail [Roche at 1 tablet/50 ml or 1 mM PMSF), lysed by Dounce homogenization and sonication (Polytron; 3× 30-s bursts), and treated with 1% Triton X-100 on ice for 30 min. Following detergent treatment, permeabilized cell lysates were mixed 1:1 with a 90% sucrose solution and overlaid with discontinuous layers of 35% and 5% sucrose. Gradients were resolved by centrifugation in a Beckman SW41 rotor at 160,030 × g for 18 h or at 187,813 × g for 16 h at 4°C. Twelve 1-ml fractions were sequentially collected starting from the top to the bottom of the tube after centrifugation. Aliquots (20 to 30 μl) of each gradient fraction were resolved by SDS-PAGE and Western analyses.
Western analyses.
Total or fractionated proteins were resolved by SDS-PAGE in 4 to 12% Bis-Tris NuPage gels (Invitrogen) and analyzed by Western blotting as previously described (17, 22). Blots were probed with rabbit anti-UL37x1 (amino acids 27 to 40) antiserum (DC35; 1:2,500) (17, 22), rabbit anti-Bax (Millipore; 1:500), goat anti-Bid (Santa Cruz; 1:100); rabbit anti-Apaf1 (Santa Cruz; 1:100); rabbit anti-procaspase 9 (GeneTex; 1:500), rabbit anti-caspase 3 (Millipore; 1:500), mouse anti-Cyt c (Abcam; 1:200), rabbit anti-fatty acid coenzyme A ligase 4 (anti-FACL4) (Abgent; 1:200), mouse anti-mitofusin 2 (anti-Mfn2) (AbCam; 1:200), rabbit anti-PACS-2 (GeneTex; 1:500), rabbit antiprohibitin (GeneTex; 1:500), mouse anti-caveolin 1 (anti-Cav1) (BD Biosciences; 1:500), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Sigma; 1:5,000), or mouse anti-α-tubulin (Sigma; 1:2,000) and with the corresponding horseradish peroxidase-conjugated secondary antibody (1:2,500 or 1:5,000). Reactive proteins were detected using an ECL enhanced chemiluminescence detection kit (Pierce). Blots were exposed to film (Denville Scientific), and the exposures were scanned using a Bio-Rad GS-800 calibrated densitometer and analyzed using Image J Software. Digital images were generated by using Scan Wizard Pro version 1.21 and processed in Adobe Photoshop version CS5, 12.0.3.
Immunoprecipitation of ubiquitinated proteins.
HCMV wt-infected HFFs (MOI of 3) and uninfected HFFs were untreated or treated with 2.5 μM MG132 (Calbiochem) from 48 to 72 h postinfection (hpi). Subcellular fractions including MAM and mitochondria were isolated at 72 hpi. MAM (200 μg) and mitochondrial (200 μg) proteins were immunoprecipitated using ubiquitin affinity resin as recommended by manufacturer (Pierce ubiquitin enrichment kit from ThermoScientific). Flotation gradient fractions 4 (DRMs, 200 μg) and 12 (soluble proteins, 200 μg) from uninfected, staurosporine (STS)-treated HFFs and from HCMV-infected HFFs were also immunoprecipitated using the ubiquitin affinity resin (ThermoScientific).
Flow cytometry analysis.
Flow cytometry was performed as described previously (67). Briefly, HFFs were washed twice in 1 × PBS supplemented with 2.5% FCS, pelleted (290 × g for 5 min) after each wash, and resuspended to a final concentration of 1 × 106 cells/ml in 1× annexin V (AV) binding buffer (BD Biosciences). HFFs (3 × 105 to 5 × 105 cells) were stained with 5 μl allophycocyanin-conjugated AV (BD Biosciences) and propidium iodide (PI; 50 μg/ml) (BD Biosciences) for 15 min at room temperature. Cells were examined by flow cytometry within 1 h of staining on a FACSCaliber (BD Biosciences) fluorescence-activated cell sorter (FACS), and data acquisition files were analyzed using FlowJo version 7.6 (TreeStar).
RESULTS
HCMV vMIA-mediated Bax recruitment to the MAM of infected HFFs.
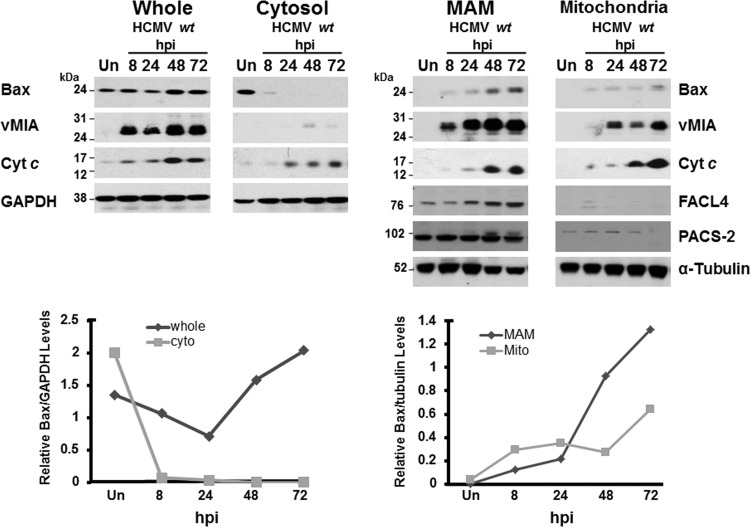
HCMV vMIA, a potent antiapoptotic protein in HeLa cells and HFFs, recruits Bax to the OMM and blocks Bax-mediated OMM permeabilization (1, 5, 8, 9, 19, 68). vMIA, the predominant UL37 product, traffics through the MAM (17, 22, 23), a site which affects mitochondrion-mediated apoptosis (28, 29, 69). We therefore tested whether Bax is also recruited to the MAM during HCMV infection. To that end, we isolated subcellular fractions, including the MAM, mitochondria, and cytosol, from uninfected and HCMV-infected HFFs throughout all temporal phases of infection, from IE through late times of infection (Fig. 1). The abundance of Bax increased in the MAM and mitochondrial fractions from HCMV-infected HFFs at IE (8 hpi), early (24 hpi), early/late (48 hpi), and late (72 hpi) times of infection compared to that in uninfected HFFs. Furthermore, the relative Bax levels, normalized to tubulin, showed increasing levels of Bax in the MAM as well as mitochondria (bottom right). Bax recruitment to the MAM and mitochondria paralleled the presence of vMIA in those fractions. Conversely, Bax levels in the cytosol of HCMV-infected cells decreased from IE to late times of infection compared to those in uninfected HFFs. The relative Bax levels in the cytosol from HCMV-infected HFFs, normalized to GAPDH in the corresponding fractions, progressively decreased throughout infection (bottom left). MAM markers, including FACL4 and PACS-2, verified the identity of the MAM fraction. Importantly, Cyt c levels increased progressively in the MAM and mitochondrial fractions of HCMV-infected HFFs compared to uninfected cells. Together, these results establish that Bax is increasingly recruited to the MAM and mitochondria of permissively HCMV-infected HFFs, paralleling the presence of vMIA in those subcellular compartments.
Fig 1.
Bax recruitment to the MAM of HCMV wt-infected HFFs. HFFs (four roller bottles each) were uninfected (Un) or infected with HCMV wt (BADwt; MOI of 3 PFU/cell). HCMV wt-infected HFFs were harvested at 8, 24, 48, or 72 hpi. Uninfected HFFs were harvested at 72 hpi. Cells were fractionated as described previously (22, 37, 66). Whole-cell (30 μg) and fractionated (10 μg) proteins were resolved by gel electrophoresis, blotted, and probed using antibodies to Bax (1:500; Millipore), vMIA (DC35; 1:2,500), FACL4 (1:200; Abgent), PACS-2 (1:500; GeneTex), Cyt c (1:200; Abcam), α-tubulin (1:2,000; Sigma), and GAPDH (1:5,000; Sigma). The relative levels of Bax were quantified by Image J and normalized to GAPDH (left) or to α-tubulin (right). The normalized Bax levels are shown below the corresponding Western blots. Cyto, cytosol; Mito, mitochondria.
HCMV UL37 proteins are its only proteins encoded by the virus, which are currently known to traffic to the MAM (22, 23). To determine if vMIA plays a role in Bax recruitment to the MAM and to mitochondria, we examined Bax levels in the MAM and mitochondria of HCMV ΔUL37x1 Mut-infected HFFs compared to uninfected HFFs and parent HCMV wt-infected HFFs (31) (Fig. 2A). Normalized Bax levels in the MAM (3.3×) and in mitochondria (not detected [ND]) of HCMV ΔUL37x1 Mut-infected HFFs were similar to the Bax levels in MAM (4.4×) and mitochondria (ND) of uninfected HFFs. Moreover, the Bax levels in HCMV ΔUL37x1 Mut-infected MAM and mitochondria were markedly lower than those in the MAM (16.9×) and mitochondria (4.1×) of HCMV wt-infected HFFs. The absence of vMIA in all fractions from HCMV ΔUL37x1 Mut-infected HFFs verified the mutant's identity. Mfn2 and prohibitin served as controls for ER-mitochondrion contacts (37). The inability of the HCMV ΔUL37x1 Mut to recruit Bax to the MAM and mitochondria established the requirement for vMIA in Bax recruitment to the MAM and to mitochondria of HCMV-infected HFFs.
Fig 2.
(A) vMIA recruits Bax to the MAM and mitochondria during HCMV infection. HFFs were uninfected (left) or infected with HCMV wt (BADwt; middle) or the HCMV ΔUL37x1 Mut (BADsubUL37x1; right) at an MOI of 1 PFU/cell. Subcellular fractions were isolated at 72 hpi as previously described (22, 37, 66). Total protein (TP; 30 μg) and fractionated proteins (10 μg) from purified microsomes (Mi), cytosol (Cy), MAM (M), and mitochondria (Mt) were resolved by SDS-PAGE and examined by Western analyses for the presence of Bax (1:500; Millipore), Mfn2 (1:200; Cell Signal), vMIA (1:2,500; DC35), and prohibitin (1:500; GeneTEX). α-Tubulin (1:2,000; Invitrogen) served as a loading control. The relative levels of Bax in each fraction were normalized to the corresponding levels of α-tubulin. ND, not detected. (B) HCMV ΔUL37x1 Mut is defective in antiapoptotic protection of infected cells. HFFs were infected with HCMV wt (BADwt) or HCMV ΔUL37x1 Mut (BADsubUL37x1) at an MOI of 1 or were mock infected and analyzed by flow cytometry as previously described (67). Viable cells (lower left quadrant, AV−/PI−), nonapoptotic cell death (upper left, AV−/PI+), and apoptotic cells (lower right, AV+/PI−, and upper right, AV+/PI+) were examined in mock-infected HFFs (left column), HCMV wt-infected HFFs (middle column), and HCMV ΔUL37x1 Mut-infected HFFs (right column) at 3 dpi (top row) and 5 dpi (bottom row).
Apoptosis of HCMV ΔUL37x1 Mut-infected HFFs.
As the HCMV ΔUL37x1 Mut is defective in Bax recruitment to the MAM and to mitochondria, we tested the induction of apoptosis in infected HFFs using AV binding, PI staining, and flow cytometry (Fig. 2B). A modest increase in AV binding (AV+) cells (26.4%) was detected in HCMV ΔUL37x1 Mut-infected HFFs at 3 days postinfection (dpi) compared to mock-infected HFFs (9.2%) and HCMV wt-infected HFFs (8.6%). With increasing time of infection (5 dpi), the antiapoptotic activity of vMIA during HCMV infection of HFFs was more clearly evidenced by the increase in AV+ cells in HCMV ΔUL37x1 Mut-infected (53.6%) compared to HCMV wt-infected (9.1%) and mock-infected (8.8%) HFFs. Thus, HCMV ΔUL37x1 Mut, which does not detectably recruit Bax to the MAM or mitochondria, is functionally defective for antiapoptotic activity in HCMV-infected HFFs.
MAM-localized Bax is targeted for MAMAD.
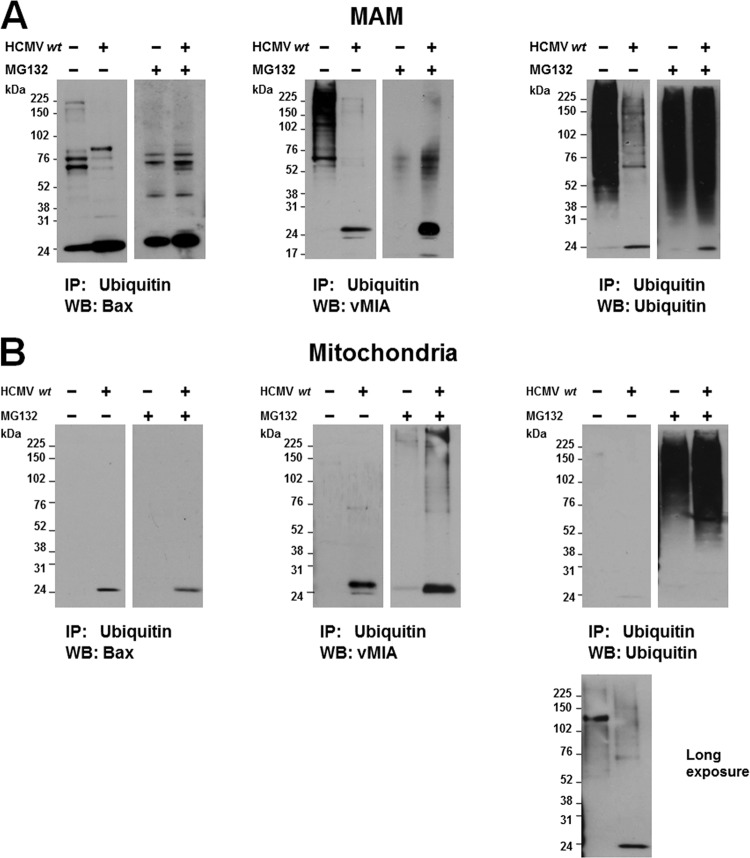
The MAM localization of ubiquitin-proteasome system components and the regulation of activated Bax by OMMAD (37, 43, 47, 51, 52) led us to ask whether Bax in the MAM and mitochondrial fractions is targeted for ubiquitination and proteasomal degradation (MAM-associated degradation [MAMAD]) (Fig. 3). Ubiquitinated proteins were enriched from MAM and mitochondrial fractions from uninfected and HCMV wt-infected HFFs at 72 hpi. To measure proteasome-mediated degradation of proteins, uninfected and HCMV-infected HFFs were untreated or treated with MG132, a proteasome inhibitor, from 48 to 72 hpi.
Fig 3.
Proteasome inhibition increases the abundance of ubiquitinated Bax in MAM (A) but not in mitochondria (B) of HCMV-infected cells. HFFs were uninfected or infected with HCMV wt (MOI of 3). Cells were treated with the proteasome inhibitor MG132 (2.5 μM) from 48 to 72 hpi. Uninfected (4 roller bottles) and HCMV-infected (4 roller bottles) HFFs were harvested and fractionated as described previously (22, 66). MAM proteins (200 μg) (A) or mitochondrial proteins (200 μg) (B) were immunoprecipitated (IP) with ubiquitin affinity resin (Pierce ubiquitin enrichment kit; Thermo Scientific). Immunoprecipitated proteins were resolved by SDS-PAGE and examined by Western blot (B) analyses for the presence of Bax (left), vMIA (middle), or total ubiquitinated proteins (right). In panel B, a longer exposure (1 min) of the mitochondrial proteins from untreated, uninfected, and HCMV-infected cells is shown below the original exposure (5 s), corresponding to the exposure time of mitochondrial proteins from MG132-treated cells (5 s).
Polyubiquitinated Bax species in the MAM were more abundant (∼2.9-fold) in MG132-treated, HCMV-infected HFFs than in untreated, HCMV-infected HFFs (Fig. 3A, left). These results show that ubiquitinated Bax is degraded more rapidly in HCMV-infected HFFs in a proteasome-dependent manner. Consistent with this finding, polyubiquitinated species of vMIA in the MAM from HCMV-infected cells (middle) increased (∼13-fold) following MG132 treatment, suggesting that ubiquitinated vMIA species were partly targeted for MAMAD.
Importantly, proteasomal degradation of numerous ubiquitinated MAM proteins was markedly increased during HCMV infection (Fig. 3A, right). Ubiquitinated MAM proteins were more abundant (∼3.3×) in untreated, uninfected HFFs than in untreated, HCMV-infected HFFs. Furthermore, MG132 treatment increased the abundance (∼4-fold) of ubiquitinated proteins in the MAM fraction of HCMV-infected HFFs compared to untreated, HCMV-infected HFFs, whereas MG132 treatment did not increase the abundance (∼1.1×) of ubiquitinated MAM proteins in the uninfected HFFs. Thus, HCMV infection of HFFs increases the proteasomal degradation of numerous MAM substrate proteins, including the recruited Bax, at late times of infection. Together, these results suggest that ubiquitinated species of Bax and vMIA are targeted for MAMAD. Moreover, the MAMAD of numerous MAM proteins is markedly increased at late times of HCMV infection.
Bax is recruited to the OMM by HCMV vMIA, where it blocks Bax-dependent OMM permeabilization (5, 8, 9, 19). Bax proapoptotic activity in mitochondria is regulated by its parkin-dependent ubiquitination and proteasomal degradation (52). Therefore, we also examined the ubiquitination and proteasome-mediated degradation of mitochondrion-localized Bax and vMIA during HCMV infection (Fig. 3B). In contrast to the MAM fraction, HCMV infection did not specifically alter OMMAD of ubiquitinated Bax in mitochondria. MG132 treatment did not markedly alter the abundance of ubiquitinated Bax levels in mitochondria of uninfected cells or of HCMV-infected HFFs (Fig. 3B, left). Nonetheless, MG132 treatment modestly increased the abundance of ubiquitinated vMIA species in the mitochondrial fraction of HCMV-infected HFFs (Fig. 3B, middle), suggesting its proteasome-mediated degradation at the OMM. Total ubiquitinated mitochondrial proteins were similar in untreated, uninfected and untreated, HCMV-infected HFFs (Fig. 3B, right) (∼0.97×). Furthermore, MG132 treatment similarly increased the abundances of ubiquitinated proteins in mitochondria of uninfected HFFs (∼27.1×) and of HCMV-infected HFFs (∼32.6×). The latter result is consistent with the previously detected ubiquitin-proteasome pruning of the OMM or OMMAD in uninfected cells (53, 70, 71). Nonetheless, HCMV does not markedly alter the OMMAD of Bax or of total ubiquitinated mitochondrial proteins during permissive infection of HFFs.
MAM-localized Bax is partially associated with internal lipid rafts.
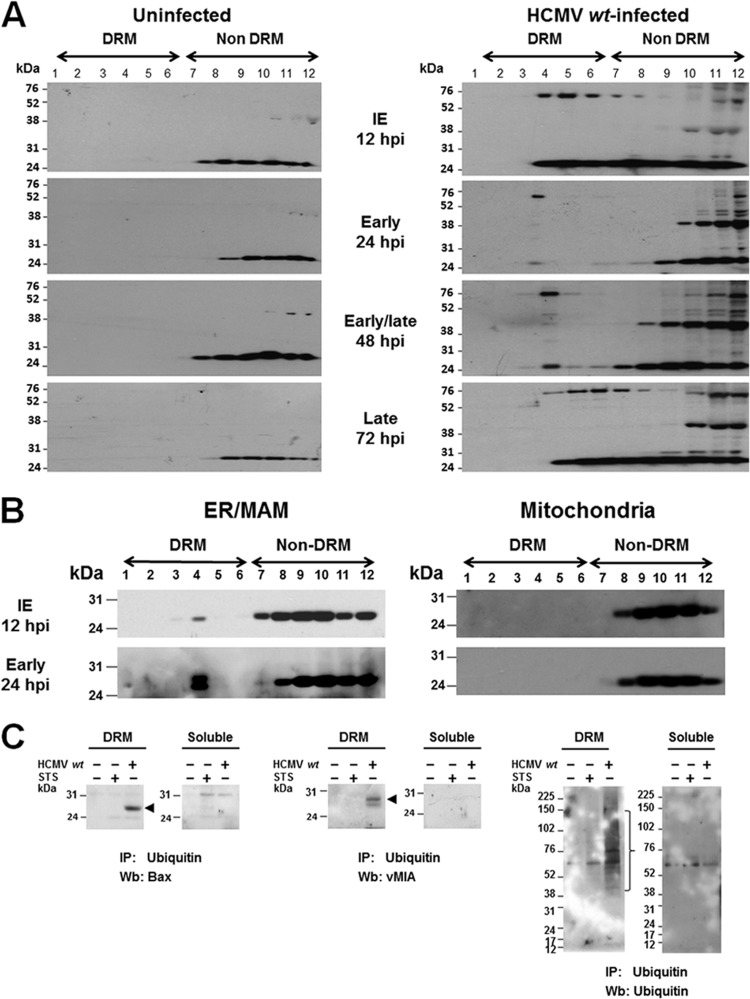
vMIA partially associates with MAM lipid rafts by its ability to bind cholesterol (41). Lipid rafts can serve as sites of apoptosome assembly and connections between extrinsic and intrinsic apoptotic pathways (49, 50). To determine if Bax is recruited by vMIA to MAM lipid rafts, we examined DRM fractions from HCMV wt-infected HFFs and uninfected HFFs previously characterized for the presence of vMIA and control Lyn (a lipid raft marker) (41) for the presence of Bax (Fig. 4A). Bax was not detectably associated with DRMs in uninfected HFFs (Fig. 4A, left). Conversely, Bax association with DRMs was detected in HCMV wt-infected HFFs (right) from IE (12 hpi) until late times of HCMV infection (72 hpi).
Fig 4.
(A) Bax is lipid raft associated during HCMV infection. DRMs isolated from uninfected HFFs (left) or HCMV wt-infected HFFs (right) (HCMV wt, MOI of 3), harvested at IE (12 hpi), early (24 hpi), early/late (48 hpi), and late (72 hpi) times of infection, and characterized for the presence of vMIA and Lyn (41) were examined for the association of Bax with DRMs. Fifteen microliters of each gradient fraction was resolved by SDS-PAGE and examined by Western analysis using anti-Bax antibody (1:500; Millipore). (B) Bax is associated with ER/MAM lipid rafts in HCMV-infected HFFs. Flotation gradient fractions from ER/MAM and mitochondria isolated at 12 hpi and 24 hpi from HCMV wt-infected HFFs (MOI of 3) and previously characterized for the presence of Sig-1R, erlin 2, and prohibitin (41) were examined for the presence of Bax. Each gradient fraction (20 μl) was resolved by SDS-PAGE and analyzed by Western blotting using anti-Bax antibody. (C) Ubiquitinated Bax, ubiquitinated vMIA, and total ubiquitinated proteins were detected in the DRMs of HCMV-infected HFFs. DRMs were isolated from uninfected, HCMV-infected (72 hpi, MOI = 3), or STS-treated (50 nM, 20 h) HFFs (four roller bottles each) by cold detergent extraction and banding on flotation gradients as previously described (41). Fractions 4 (DRM) and 12 (soluble proteins) were immunoprecipitated (IP) as described in Fig. 3. The presence of ubiquitinated Bax (left), ubiquitinated vMIA (middle), and total ubiquitinated proteins (right) was verified by Western blot (Wb) analyses.
Bax associates with ER/MAM lipid rafts.
vMIA localizes to MAM lipid rafts (41). To determine if Bax is recruited by vMIA to MAM lipid rafts during infection, DRM fractions isolated from the ER/MAM and mitochondrial fractions from HCMV wt-infected HFFs at IE (12 hpi) and early (24 hpi) times of infection previously characterized for the presence of vMIA, and control Sigma-1 receptor (Sig-1R), erlin-2, and prohibitin (lipid raft markers) (41) were examined for the presence of Bax (Fig. 4B). Bax was associated with lipid rafts in the ER/MAM fractions but not with lipid rafts in mitochondrial fractions isolated from HCMV-infected HFFs. Thus, in a pattern similar to that of vMIA (41), Bax associates with ER/MAM lipid rafts but not in mitochondrial lipid rafts from HCMV-infected HFFs.
Ubiquitinated Bax is associated with DRMs.
To determine if ubiquitinated Bax and ubiquitinated vMIA associate with DRMs, we immunoprecipitated ubiquitinated proteins in DRM (fraction 4) and soluble (fraction 12) proteins and examined the presence of Bax, vMIA, and total ubiquitinated proteins (Fig. 4C). Ubiquitinated Bax was detected in DRMs from HCMV wt-infected HFFs but not in those from uninfected HFFs or from STS-treated HFFs (Fig. 4C, left). Similarly, ubiquitinated vMIA was detected in the DRMs from HCMV wt-infected HFFs. As expected, ubiquitinated vMIA was not found in DRMs from uninfected HFFs or STS-treated HFFs (Fig. 4C, middle). Total ubiquitinated proteins were readily detected in the DRM fraction of HCMV-infected cells but not in DRMs from uninfected or STS-treated HFFs (Fig. 4C, right). Furthermore, total ubiquitinated proteins were less abundant in the soluble fractions from uninfected, STS-treated HFFs and HCMV wt-infected HFFs. Taken together, the presence of ubiquitinated Bax, ubiquitinated vMIA, and total ubiquitinated proteins in DRMs suggest that their proteasome-mediated degradation is associated with their MAM lipid raft localization.
Bax association with DRMs in infected cells is vMIA dependent.
To determine whether vMIA is responsible for Bax recruitment to MAM lipid rafts during HCMV infection, we examined the association of Bax with DRMs from HCMV ΔUL37x1 Mut-infected HFFs from early (24 hpi) to very late (120 hpi) times of infection since the HCMV ΔUL37x1 Mut is delayed in growth in HFFs (31). In control HFFs, Bax association with DRM was detected from IE through late times of HCMV wt infection (Fig. 4A). In contrast, Bax association with lipid rafts was not detected in HCMV ΔUL37x1 Mut-infected HFFs at any of the times examined (Fig. 5). Cav1 served as a control for lipid raft fractions. These results show that Bax recruitment to MAM lipid rafts during HCMV infection is vMIA dependent.
Fig 5.
HCMV ΔUL37x1 Mut does not recruit Bax to MAM lipid rafts. HFFs were infected with the HCMV ΔUL37x1 Mut (BADsubUL37x1) at an MOI of 1.5. DRMs were harvested from the cells at early (24 hpi), late (72 and 96 hpi), and very late (120 hpi) times of infection and purified as in Fig. 4 and as described previously (41). Each flotation gradient fraction (30 μl) was resolved by SDS-PAGE and analyzed by Western blotting using anti-Bax (1:500; Millipore) or mouse anti-Cav1 (1:500; BD Biosciences) antibodies.
vMIA is sufficient for Bax recruitment to lipid rafts and is dependent on its cholesterol binding.
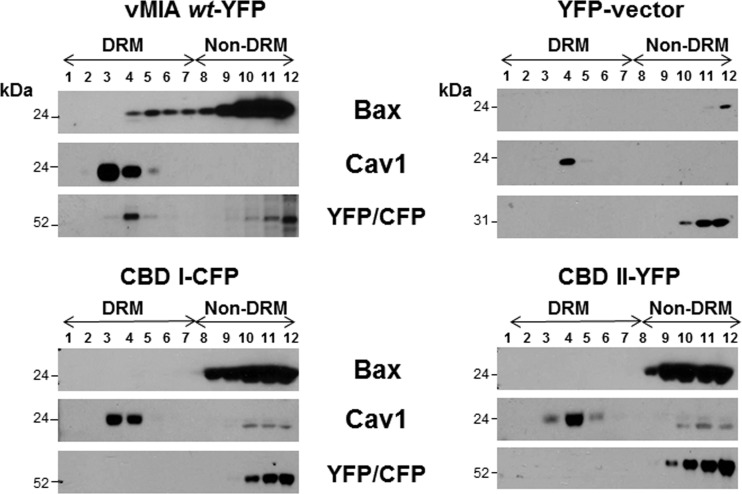
As vMIA binds Bax and its association with lipid rafts is dependent upon its cholesterol affinity (41), we tested whether vMIA is sufficient for Bax recruitment to lipid rafts and whether this recruitment is dependent upon vMIA's cholesterol binding (Fig. 6). To that end, we transfected HFFs with expression vectors of the vMIA wt (vMIA-yellow fluorescent protein [YFP]), the cholesterol binding I (CBD I-cyan fluorescent protein [CFP]) and II (CBD II-YFP) mutants, or YFP (vector control) (41). We then tested Bax recruitment to lipid rafts. Bax was recruited to DRMs of HFFs transfected with expression vector of vMIA-YFP wt. However, the CBD I and CBD II mutants, which are severely defective in MAM lipid raft association (41), were unable to recruit Bax to DRMs. The YFP vector control did not recruit Bax to DRMs of transfected HFFs. Caveolin 1 was used as a lipid raft fraction marker and the fluorophores were used to detect vMIA wt-YFP, CBDI-CFP, and CBDII-YFP. These results establish that vMIA is sufficient for Bax recruitment to DRMs and that this recruitment is dependent upon the cholesterol binding of vMIA.
Fig 6.
vMIA is sufficient for Bax recruitment to MAM lipid rafts. HFFs (2 × 107 cells) were transfected with vectors expressing vMIA1–163-YFP, CBD I1–163-CFP, or CBDII1–163-YFP (41). Controls were transfected with YFP vector alone. Twenty-four hours after transfection, DRMs were isolated and banded on flotation gradients as described previously (41). Each gradient fraction (20 μl) was examined by Western blotting using rabbit anti-Bax (1:500; Millipore), mouse anti-Cav1 (1:500; BD Biosciences), and mouse anti-GFP/YFP/CFP (1:200; Santa Cruz).
vMIA-dependent recruitment of apoptosome components and downstream effectors to lipid rafts.
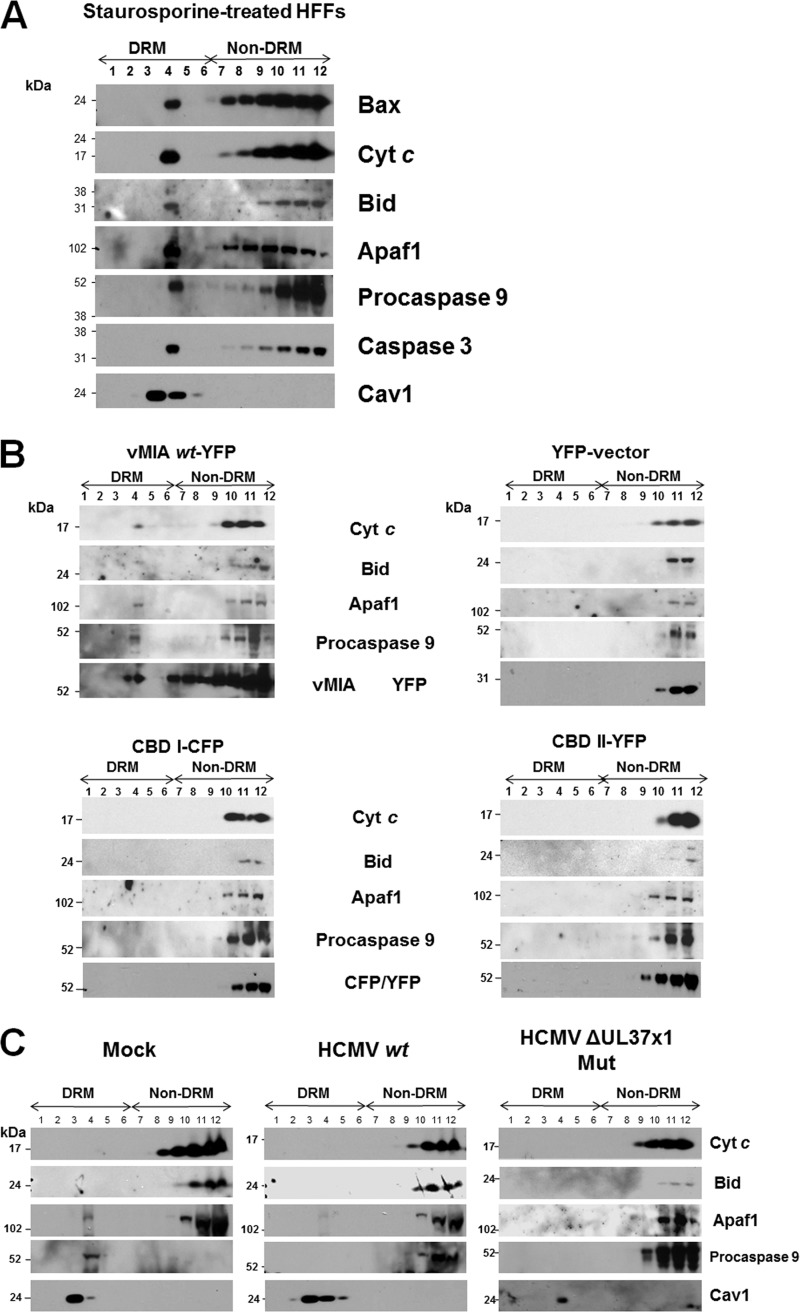
It has recently been established that lipid rafts can serve as apoptosis-prone sites for apoptosome assembly in cancer cells (49, 50). However, HFFs are primary, untransformed cells. We therefore tested whether recruitment of Bax and apoptosome components and effectors to DRMs could be detected in STS-treated HFFs (Fig. 7A). Recruitment of Bax, Cyt c, Bid, Apaf1, procaspase 9, and caspase 3 was detected in DRMs from STS-treated HFFs. Cav1 served as a lipid raft marker. These results show that we could detect lipid raft association of these apoptosome components and effectors in apoptotic HFFs.
Fig 7.
(A) Association of Bax and apoptosome components and effectors with DRMs following STS treatment of HFFs. HFFs were treated with STS (50 nM) for 20 h. DRMs were isolated from the STS-treated cells and banded on flotation gradients as described previously (41). Aliquots of each fraction (30 μl) were separated by SDS-PAGE and examined for the presence of Bax (1:500; Millipore), Cyt c (1:200; Abcam), Bid (1:100; Santa Cruz), Apaf1 (1:100; Santa Cruz), procaspase 9 (1:500; GeneTex), caspase 3 (1:500; Millipore), and Cav1 (1:500; BD Biosciences) antibodies. (B) Lipid raft association of downstream apoptosome components requires vMIA lipid raft association. HFFs (1 × 107 cells) were transfected with vectors expressing pUL37x11–163-YFP, CBD I1–163-CFP, or CBDII1–163-YFP (41) and control YFP alone as in Fig. 6. Each gradient fraction (30 μl) was resolved by SDS-PAGE and examined using antibodies to Cyt c (1:200; Abcam), Bid (1:100; Santa Cruz), Apaf1 (1:100; Santa Cruz), procaspase 9 (1:500; GeneTex), and vMIA (DC35, 1:2,500) or mouse anti-GFP/YFP/CFP antibodies (1:200; Santa Cruz). Control anti-Cav1 is shown in Fig. 6. (C) HCMV infection inhibits recruitment of apoptosome components to lipid rafts. HFFs were mock infected or infected with HCMV wt (BADwt) or the HCMV ΔUL37x1 Mut (BADsubUL37x1) at an MOI of 1.5. Lipid rafts were isolated at 72 hpi, and gradient fractions (30 μl) were resolved by SDS-PAGE. Western blots performed using antibodies to Cyt c (1:200; Abcam), Bid (1:100; Santa Cruz), Apaf1 (1:100; Santa Cruz), procaspase 9 (1:500; GeneTex), and Cav1 (1:500; BD Biosciences).
We then examined whether vMIA is sufficient to recruit apoptosome components, in addition to Bax, to DRMs and whether its ability to bind cholesterol is required for recruitment of apoptosome components. DRMs were isolated from HFFs transfected with vMIA wt or the cholesterol binding mutants, CBD I and II (Fig. 7B). Cyt c, Bid, Apaf1, and procaspase 9 were weakly detected in lipid rafts from transfected HFFs expressing vMIA (Fig. 7B, upper left). Similar to Bax (Fig. 6), recruitment of Cyt c, Bid, Apaf1, or procaspase 9 was not detected in HFFs expressing CBD I or CBD II mutants or in vector-transfected HFFs. Cav1 (Fig. 6) and vMIA (Fig. 7B) served as lipid raft markers. Together, these results show that vMIA can recruit Bax and induce the association of apoptosome components to lipid rafts by its affinity for cholesterol.
Lipid raft association of apoptosome components is not detected in HCMV-infected HFFs.
As vMIA recruited Bax to DRMs of transfected cells and of HCMV-infected HFFs (Fig. 5 and 6) and apoptosome components and effectors in transfected cells (Fig. 7B), we tested whether apoptosomes were also recruited to DRMs of HCMV-infected HFFs (Fig. 7C). Apaf1 and procaspase 9 were weakly detected in DRMs from mock-infected HFFs. In contrast, Cyt c, Bid, Apaf1, and procaspase 9 were not detected in the DRMs of either HCMV wt- or HCMV ΔUL37x1 Mut-infected HFFs at late times of infection (72 hpi), despite the detection of Bax recruitment to DRMs of HCMV wt-infected cells (Fig. 4A). Cav1 served as a lipid raft marker (Fig. 7C). Taken together, these results suggest that HCMV infection recruits Bax to MAM DRMs using vMIA but inhibits the recruitment of downstream apoptosome assembly on MAM DRMs.
DISCUSSION
While the ability of vMIA to inhibit Bax proapoptotic activity is partly responsible for its antiapoptotic activity, the mechanisms underlying this inhibition are poorly understood and just being deciphered (5, 7–9, 19). Herein, we demonstrate that HCMV vMIA can inhibit apoptosis by reducing Bax levels at ER-mitochondrion contacts of the infected cell. We detected the expected increased levels of Bax recruited to mitochondria during HCMV infection, which correlate well with the previous detection of Bax recruitment by vMIA to the OMM (5, 8, 9). In addition, we also found an even more marked increase in Bax associated with the MAM from IE through late times of HCMV infection.
Bax transcription does not appear to be markedly altered (≤2-fold) during IE or early to early/late times of HCMV infection (72, 73). Consistently, we observe minor increases of total Bax (maximally ∼1.4-fold over uninfected cells) during HCMV infection, but most of this is recruited to the MAM or mitochondria, while free Bax decreases in the cytosol.
We show that Bax is retargeted to the MAM in a vMIA-dependent manner and that vMIA is sufficient to recruit Bax to ER/MAM lipid rafts. Significantly, Bax retargeting to the MAM resulted in its increased ubiquitination and proteasome-mediated degradation. To our knowledge, this is the first report of Bax recruitment to the MAM and its targeting for proteasomal degradation in association with the MAM. Our results establish MAM-associated, ubiquitin-proteasome degradation as a mechanism for the regulation of Bax and suggest that HCMV infection globally increases proteasome-mediated degradation of targeted MAM proteins, MAMAD, by late times of infection. Previously, ubiquitin-proteasome-mediated degradation of Bax has been demonstrated to occur by the human papillomavirus type 16 E5 protein (74). While it is not clear if Bax degradation by HPV E5 is dependent of MAM localization, together these studies suggest that ubiquitin-proteasome-mediated Bax degradation may be a common mechanism employed by viruses to prevent host cell apoptosis.
HCMV infection is known to modulate the ubiquitin-proteasome system to enhance its replication (55–58). HCMV infection causes relocalization of some 19S proteasome subunits to the periphery of nuclear viral replication compartments (58). We detected a global increase in proteasome activity in the MAM of HCMV-infected cells at late times of infection. Consistent with this finding, we detected using quantitative proteomics an increased abundance of proteasome non-ATPase regulatory 19S subunits (subunits 1, 2, 3, 11, and 14 with 2- to 3.5-fold increases) in the MAM fraction of HCMV-infected HFFs at late times of infection (37). We also found increases in ubiquitin ligase E2 UBE2N, UBE2D2, UBE2D3 (1.3- to 2.4-fold increases) but not UBE2J1 (required for MHC class I degradation) in the MAM at late times. Importantly, we detected an ∼2-fold increase in the abundance of an E3 ubiquitin ligase (HRD1, synoviolin) required for ERAD (75) in the MAM of HCMV-infected HFFs, but we did not detect gp78, a known E3 ubiquitin ligase, which partially localizes to the MAM (47) and the other ortholog of yeast Hrd1p (37). HCMV US11 targets MHC I for ERAD degradation using derlin-1, which is in a complex with HRD1 and gp78 (75). Together, our findings suggest that Bax ubiquitination is occurring in the MAM and does not require gp78.
Some ubiquitin ligases (CHIP) link chaperones and the 26S proteasome machinery by ubiquitinating chaperone substrates and channeling them toward the proteasome (76–78). Chaperones are among the proteins increased to the highest levels (∼16.7- to 27-fold) in the MAM of HCMV-infected cells (37). Furthermore, we detected increased levels of hsc71 (5-fold increase) in the MAM of infected cells. Thus, increased MAM-associated chaperones may play roles in the induction of proteasomal activity of the MAM during HCMV infection.
In the MAM, Bax becomes associated with lipid rafts and depends on the ability of vMIA to associate with cholesterol. vMIA associates with MAM lipid rafts, as do Sig-1R and erlins (39, 41, 42). MAM lipid raft proteins erlin-1 and -2 (SPFH1 and -2) have been found to target IP3Rs for ubiquitination and ERAD (43), suggesting the possibility that Bax degradation in the MAM is associated with its lipid raft location. Our findings of increased abundance of ubiquitinated Bax, ubiquitinated vMIA, and total ubiquitinated proteins in the MAM but not mitochondrial fractions following MG132 treatment are consistent with this possibility. Furthermore, we found that ubiquitinated Bax and ubiquitinated vMIA are associated with DRMs, consistent with the possibility that Bax degradation in the MAM is associated with its lipid raft location.
Despite the finding of recruitment of Bax and of apoptosome components to MAM lipid rafts following vMIA expression in transfected permissive HFFs, only Bax recruitment was detected in HCMV-infected HFFs. This finding suggests that although vMIA is sufficient to recruit Bax and induce apoptosome association with MAM lipid rafts, other HCMV products or HCMV-induced cell proteins can inhibit the association of apoptosome components with MAM lipid rafts. Reduction of apoptosome component recruitment to DRMs will predictably reduce the propensity of these proapoptotic platforms to initiate mitochondrion-mediated apoptosis of HCMV-infected cells, consistent with vMIA antiapoptotic activities in the infected cells (9, 10).
Apoptosome assembly is already known to be inhibited by a number of proteins. For example, an alternatively spliced caspase 3 short isoform (caspase 3s) binds full-length procaspase 3 and blocks its interaction with caspase 9 (79). Thus, caspase 3s inhibits apoptosome assembly. Similarly, cdc6, an AAA+ ATPase with multiple roles in the cell cycle, forms stable complexes with activated Apaf1 and thereby inhibits apoptosome assembly (80). While cdc6 was not detected in our quantitative analyses of the MAM proteome of HCMV-infected cells, we found that HCMV infection greatly increases the abundance (∼12.7-fold) of two members of the ATPase family AAA domain-containing proteins 3A and 3B in the MAM (37) and may thereby inhibit apoptosome assembly. Alternatively, MAMAD of the apoptosome components studied may be induced in HCMV-infected HFFs, which would result in their reduced level in MAM DRMs, thereby protecting infected HFFs from apoptosis. Consistent with this possibility is our finding that increased MAMAD in HCMV-infected cells targets many proteins that localize to the MAM during infection. We do not yet know if the half-life of apoptosome components is decreased during HCMV infection.
Our current model is that HCMV vMIA recruits Bax to the MAM and then targets it for proteasome-mediated degradation. However, steadily increasing Bax levels, which occur during infection, overwhelm the MAM system and are recruited to the OMM, where vMIA blocks its permeabilization of the OMM. Even though Bax is increasingly associated with the MAM of HCMV-infected cells and degraded, as infection progresses, Bax is also detected in increasing levels in mitochondria. It appears that the increasing levels of activated Bax cannot be neutralized solely by recruitment to the MAM and MAMAD. Thus, HCMV has evolved complementary yet redundant mechanisms to block Bax proapoptotic activity in the infected cell.
Although Bax is increasingly recruited to the OMM during infection, we did not detect HCMV induction of OMMAD degradation of Bax, which is known to regulate activated Bax activity in uninfected cells (51–54). In the absence of HCMV infection, ubiquitin proteasome degradation of Bax is known to be induced by mitochondrial ubiquitin E3 ligase parkin (52). This mechanism is known to regulate Bax proapoptotic function and to underlie the antiapoptotic function of parkin. In mitochondria, the activated form of Bax is selectively targeted for degradation (54). Thus, HCMV infection seems to selectively increase the ER/MAM-associated ubiquitin-proteasome machinery but not OMMAD.
Together, our results show that HCMV utilizes vMIA to block Bax proapoptotic activities in the infected cell by two distinct mechanisms, the first being its previously recognized inhibition of Bax-mediated permeabilization of the OMM (5, 7–9, 19) and the second, as demonstrated herein, the recruitment and increased degradation of Bax in the MAM subcompartment of the ER. Furthermore, the HCMV antiapoptotic program also involves the reduction of apoptosis-prone platforms in the MAM DRMs by a mechanism reducing the association of apoptosome components. Thus, HCMV infection independently blocks mitochondrion-mediated apoptosis by a previously unrecognized mechanism using MAM-associated proteasome-mediated degradation.
ACKNOWLEDGMENTS
We thank Jyoti Jaiswal and Chad Williamson for thoughtful comments on the manuscript and Tom Shenk for the gift of the recombinant HCMV viruses. We thank Eric P. Hoffman and Kanneboyina Nagaraju for unwavering support.
These studies were supported by Public Health Service grants R01 AI057906 and R21 AI081957 from the National Institute for Allergy and Infectious Diseases and Children's Research Institute funds to A.C.P.
Footnotes
Published ahead of print 13 March 2013
REFERENCES
- 1. Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han JW, Lutz RJ, Watanabe S, Cahir McFarland ED, Kieff ED, Mocarski ES, Chittenden T. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. U. S. A. 96:12536–12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U. S. A. 98:7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu H, Shen Y, Shenk T. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arnoult D, Bartle LM, Skaletskaya A, Poncet D, Zamzami N, Park PU, Sharpe J, Youle RJ, Goldmacher VS. 2004. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. U. S. A. 101:7988–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brune W. 2011. Inhibition of programmed cell death by cytomegaloviruses. Virus Res. 157:144–150 [DOI] [PubMed] [Google Scholar]
- 7. Norris KL, Youle RJ. 2008. Cytomegalovirus proteins vMIA and m38.5 link mitochondrial morphogenesis to Bcl-2 family proteins. J. Virol. 82:6232–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poncet D, Larochette N, Pauleau AL, Boya P, Jalil AA, Cartron PF, Vallette F, Schnebelen C, Bartle LM, Skaletskaya A, Boutolleau D, Martinou JC, Goldmacher VS, Kroemer G, Zamzami N. 2004. An anti-apoptotic viral protein that recruits Bax to mitochondria. J. Biol. Chem. 279:22605–22614 [DOI] [PubMed] [Google Scholar]
- 9. Poncet D, Pauleau AL, Szabadkai G, Vozza A, Scholz SR, Le Bras M, Briere JJ, Jalil A, Le Moigne R, Brenner C, Hahn G, Wittig I, Schagger H, Lemaire C, Bianchi K, Souquere S, Pierron G, Rustin P, Goldmacher VS, Rizzuto R, Palmieri F, Kroemer G. 2006. Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J. Cell Biol. 174:985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reboredo M, Greaves RF, Hahn G. 2004. Human cytomegalovirus proteins encoded by UL37 exon 1 protect infected fibroblasts against virus-induced apoptosis and are required for efficient virus replication. J. Gen. Virol. 85:3555–3567 [DOI] [PubMed] [Google Scholar]
- 11. Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. 2007. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316:1345–1348 [DOI] [PubMed] [Google Scholar]
- 12. Adair R, Liebisch GW, Colberg-Poley AM. 2003. Complex alternative processing of human cytomegalovirus UL37 pre-mRNA. J. Gen. Virol. 84:3353–3358 [DOI] [PubMed] [Google Scholar]
- 13. Adair R, Liebisch GW, Su Y, Colberg-Poley AM. 2004. Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection. J. Gen. Virol. 85:3541–3553 [DOI] [PubMed] [Google Scholar]
- 14. Al-Barazi HO, Colberg-Poley AM. 1996. The human cytomegalovirus UL37 immediate-early regulatory protein is an integral membrane N-glycoprotein which traffics through the endoplasmic reticulum and Golgi apparatus. J. Virol. 70:7198–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kouzarides T, Bankier AT, Satchwell SC, Preddy E, Barrell BG. 1988. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology 165:151–164 [DOI] [PubMed] [Google Scholar]
- 16. Mavinakere MS, Colberg-Poley AM. 2004. Dual targeting of the human cytomegalovirus UL37 exon 1 protein during permissive infection. J. Gen. Virol. 85:323–329 [DOI] [PubMed] [Google Scholar]
- 17. Mavinakere MS, Williamson CD, Goldmacher VS, Colberg-Poley AM. 2006. Processing of human cytomegalovirus UL37 mutant glycoproteins in the endoplasmic reticulum lumen prior to mitochondrial importation. J. Virol. 80:6771–6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williamson CD, Colberg-Poley AM. 2010. Intracellular sorting signals for sequential trafficking of human cytomegalovirus UL37 proteins to the endoplasmic reticulum and mitochondria. J. Virol. 84:6400–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma J, Edlich F, Bermejo GA, Norris KL, Youle RJ, Tjandra N. 2012. Structural mechanism of Bax inhibition by cytomegalovirus protein vMIA. Proc. Natl. Acad. Sci. U. S. A. 109:20901–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pauleau A-L, Larochette N, Giordanetto F, Scholz SR, Poncet D, Zamzami N, Golmacher VS, Koemer G. 2007. Structure-function analysis of the interaction between Bax and the cytomegalovirus-encoded protein vMIA. Oncogene 26:7067–7080 [DOI] [PubMed] [Google Scholar]
- 21. Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. 2011. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell 44:517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bozidis P, Williamson CD, Colberg-Poley AM. 2008. Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells. J. Virol. 82:2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bozidis P, Williamson CD, Wong DS, Colberg-Poley AM. 2010. Trafficking of UL37 proteins into mitochondrion-associated membranes during permissive human cytomegalovirus infection. J. Virol. 84:7898–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, Vais H, Cheung KH, Yang J, Parker I, Thompson CB, Birnbaum MJ, Hallows KR, Foskett JK. 2010. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142:270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayashi T, Su TP. 2007. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131:596–610 [DOI] [PubMed] [Google Scholar]
- 26. Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763–1766 [DOI] [PubMed] [Google Scholar]
- 27. Simmen T, Lynes EM, Gesson K, Thomas G. 2010. Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM). Biochim. Biophys. Acta 1798:1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mendes CC, Gomes DA, Thompson M, Souto NC, Goes TS, Goes AM, Rodrigues MA, Gomez MV, Nathanson MH, Leite MF. 2005. The type III inositol 1,4,5-trisphosphate receptor preferentially transmits apoptotic Ca2+ signals into mitochondria. J. Biol. Chem. 280:40892–40900 [DOI] [PubMed] [Google Scholar]
- 29. Rizzuto R, Marchi S, Bonora M, Aguiari P, Bononi A, De Stefani D, Giorgi C, Leo S, Rimessi A, Siviero R, Zecchini E, Pinton P. 2009. Ca(2+) transfer from the ER to mitochondria: when, how and why. Biochim. Biophys. Acta 1787:1342–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ. 2003. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300:135–139 [DOI] [PubMed] [Google Scholar]
- 31. Sharon-Friling R, Goodhouse J, Colberg-Poley AM, Shenk T. 2006. Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc. Natl. Acad. Sci. U. S. A. 103:19117–19122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alwine JC. 2008. Modulation of host cell stress responses by human cytomegalovirus. Curr. Top. Microbiol. Immunol. 325:263–279 [DOI] [PubMed] [Google Scholar]
- 33. Chambers JW, Maguire TG, Alwine JC. 2010. Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 84:1867–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Isler JA, Skalet AH, Alwine JC. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79:6890–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. 2006. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2:e132 doi:10.1371/journal.ppat.0020132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26:1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang A, Williamson CD, Wong DS, Bullough MD, Brown KJ, Hathout Y, Colberg-Poley AM. 2011. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol. Cell. Proteomics 10:M111.009936. doi:10.1074/mcp.M111.009936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bionda C, Portoukalian J, Schmitt D, Rodriguez-Lafrasse C, Ardail D. 2004. Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 382:527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hayashi T, Fujimoto M. 2010. Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol. Pharmacol. 77:517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stone SJ, Vance JE. 2000. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 275:34534–34540 [DOI] [PubMed] [Google Scholar]
- 41. Williamson CD, Zhang A, Colberg-Poley AM. 2011. The human cytomegalovirus protein UL37 exon 1 associates with internal lipid rafts. J. Virol. 85:2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Browman DT, Resek ME, Zajchowski LD, Robbins SM. 2006. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119:3149–3160 [DOI] [PubMed] [Google Scholar]
- 43. Wang Y, Pearce MM, Sliter DA, Olzmann JA, Christianson JC, Kopito RR, Boeckmann S, Gagen C, Leichner GS, Roitelman J, Wojcikiewicz RJ. 2009. SPFH1 and SPFH2 mediate the ubiquitination and degradation of inositol 1,4,5-trisphosphate receptors in muscarinic receptor-expressing HeLa cells. Biochim. Biophys. Acta 1793:1710–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benlimame N, Simard D, Nabi IR. 1995. Autocrine motility factor receptor is a marker for a distinct membranous tubular organelle. J. Cell Biol. 129:459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 98:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goetz JG, Genty H, St-Pierre P, Dang T, Joshi B, Sauve R, Vogl W, Nabi IR. 2007. Reversible interactions between smooth domains of the endoplasmic reticulum and mitochondria are regulated by physiological cytosolic Ca2+ levels. J. Cell Sci. 120:3553–3564 [DOI] [PubMed] [Google Scholar]
- 47. St Pierre P, Nabi IR. 2011. The Gp78 ubiquitin ligase: probing endoplasmic reticulum complexity. Protoplasma 249(Suppl 1):S11–S1822045301 [Google Scholar]
- 48. Wang HJ, Benlimame N, Nabi I. 1997. The AMF-R tubule is a smooth ilimaquinone-sensitive subdomain of the endoplasmic reticulum. J. Cell Sci. 110:3043–3053 [DOI] [PubMed] [Google Scholar]
- 49. Gajate C, Gonzalez-Camacho F, Mollinedo F. 2009. Lipid raft connection between extrinsic and intrinsic apoptotic pathways. Biochem. Biophys. Res. Commun. 380:780–784 [DOI] [PubMed] [Google Scholar]
- 50. Gajate C, Mollinedo F. 2007. Edelfosine and perifosine induce selective apoptosis in multiple myeloma by recruitment of death receptors and downstream signaling molecules into lipid rafts. Blood 109:711–719 [DOI] [PubMed] [Google Scholar]
- 51. Benard G, Neutzner A, Peng G, Wang C, Livak F, Youle RJ, Karbowski M. 2010. IBRDC2, an IBR-type E3 ubiquitin ligase, is a regulatory factor for Bax and apoptosis activation. EMBO J. 29:1458–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Johnson BN, Berger AK, Cortese GP, Lavoie MJ. 2012. The ubiquitin E3 ligase parkin regulates the proapoptotic function of Bax. Proc. Natl. Acad. Sci. U. S. A. 109:6283–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karbowski M, Youle RJ. 2011. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr. Opin. Cell Biol. 23:476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu FT, Agrawal SG, Gribben JG, Ye H, Du MQ, Newland AC, Jia L. 2008. Bortezomib blocks Bax degradation in malignant B cells during treatment with TRAIL. Blood 111:2797–2805 [DOI] [PubMed] [Google Scholar]
- 55. Kaspari M, Tavalai N, Stamminger T, Zimmermann A, Schilf R, Bogner E. 2008. Proteasome inhibitor MG132 blocks viral DNA replication and assembly of human cytomegalovirus. FEBS Lett. 582:666–672 [DOI] [PubMed] [Google Scholar]
- 56. Prosch S, Priemer C, Hoflich C, Liebenthaf C, Babel N, Kruger DH, Volk HD. 2003. Proteasome inhibitors: a novel tool to suppress human cytomegalovirus replication and virus-induced immune modulation. Antivir. Ther. 8:555–567 [PubMed] [Google Scholar]
- 57. Sadanari H, Tanaka J, Li Z, Yamada R, Matsubara K, Murayama T. 2009. Proteasome inhibitor differentially regulates expression of the major immediate early genes of human cytomegalovirus in human central nervous system-derived cell lines. Virus Res. 142:68–77 [DOI] [PubMed] [Google Scholar]
- 58. Tran K, Mahr JA, Spector DH. 2010. Proteasome subunits relocalize during human cytomegalovirus infection, and proteasome activity is necessary for efficient viral gene transcription. J. Virol. 84:3079–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tortorella D, Gewurz B, Schust D, Furman M, Ploegh H. 2000. Down-regulation of MHC class I antigen presentation by HCMV: lessons for tumor immunology. Immunol. Invest. 29:97–100 [DOI] [PubMed] [Google Scholar]
- 60. Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769–779 [DOI] [PubMed] [Google Scholar]
- 61. Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432–438 [DOI] [PubMed] [Google Scholar]
- 62. Cam M, Handke W, Picard-Maureau M, Brune W. 2010. Cytomegaloviruses inhibit Bak- and Bax-mediated apoptosis with two separate viral proteins. Cell Death Differ. 17:655–665 [DOI] [PubMed] [Google Scholar]
- 63. Yu D, Smith GA, Enquist LW, Shenk T. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Colberg-Poley AM, Santomenna LD. 1988. Selective induction of chromosomal gene expression by human cytomegalovirus. Virology 166:217–228 [DOI] [PubMed] [Google Scholar]
- 65. Santomenna LD, Colberg-Poley AM. 1990. Induction of cellular hsp70 expression by human cytomegalovirus. J. Virol. 64:2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bozidis P, Williamson CD, Colberg-Poley AM. 2007. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.27. doi:10.1002/0471143030.cb0327s37 [DOI] [PubMed] [Google Scholar]
- 67. Hildreth RL, Bullough MD, Zhang A, Chen HL, Schwartz PH, Panchision DM, Colberg-Poley AM. 2012. Viral mitochondria-localized inhibitor of apoptosis (UL37 exon 1 protein) does not protect human neural precursor cells from human cytomegalovirus-induced cell death. J. Gen. Virol. 93:2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hayajneh WA, Colberg-Poley AM, Skaletskaya A, Bartle LM, Lesperance MM, Contopoulos-Ioannidis DG, Kedersha NL, Goldmacher VS. 2001. The sequence and antiapoptotic functional domains of the human cytomegalovirus UL37 exon 1 immediate early protein are conserved in multiple primary strains. Virology 279:233–240 [DOI] [PubMed] [Google Scholar]
- 69. Hayashi T, Rizzuto R, Hajnoczky G, Su TP. 2009. MAM: more than just a housekeeper. Trends Cell Biol. 19:81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cuconati A, Mukherjee C, Perez D, White E. 2003. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 17:2922–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, Wang X. 2003. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 17:1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Browne EP, Wing B, Coleman D, Shenk T. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Challacombe JF, Rechtsteiner A, Gottardo R, Rocha LM, Browne EP, Shenk T, Altherr MR, Brettin TS. 2004. Evaluation of the host transcriptional response to human cytomegalovirus infection. Physiol. Genomics 18:51–62 [DOI] [PubMed] [Google Scholar]
- 74. Oh JM, Kim SH, Cho EA, Song YS, Kim WH, Juhnn YS. 2010. Human papillomavirus type 16 E5 protein inhibits hydrogen-peroxide-induced apoptosis by stimulating ubiquitin-proteasome-mediated degradation of Bax in human cervical cancer cells. Carcinogenesis 31:402–410 [DOI] [PubMed] [Google Scholar]
- 75. Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ. 2011. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. U. S. A. 108:2034–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ahmed SF, Deb S, Paul I, Chatterjee A, Mandal T, Chatterjee U, Ghosh MK. 2012. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 287:15996–16006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19:4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Paul I, Ahmed SF, Bhowmik A, Deb S, Ghosh MK. 2012. The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity. Oncogene 32:1284–1295 [DOI] [PubMed] [Google Scholar]
- 79. Vegran F, Boidot R, Solary E, Lizard-Nacol S. 2011. A short caspase-3 isoform inhibits chemotherapy-induced apoptosis by blocking apoptosome assembly. PLoS One 6:e29058 doi:10.1371/journal.pone.0029058 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80. Niimi S, Arakawa-Takeuchi S, Uranbileg B, Park JH, Jinno S, Okayama H. 2012. Cdc6 protein obstructs apoptosome assembly and consequent cell death by forming stable complexes with activated Apaf-1 molecules. J. Biol. Chem. 287:18573–18583 [DOI] [PMC free article] [PubMed] [Google Scholar]