Abstract
Human papillomaviruses (HPVs) modulate expression of host microRNAs. Our deep-sequencing analysis of organotypic raft cultures identified microRNA 145 (miR-145) as a differentiation-dependent microRNA that has functionally active target sequences in the HPV-31 E1 and E2 open reading frames. Overexpression of miR-145 in HPV-positive cells resulted in reduced genome amplification and late gene expression, along with decreased levels of cellular transcription factor KLF-4. Our studies show that HPV modulates miR-145 expression to control its own life cycle.
TEXT
High-risk human papillomaviruses (HPVs) are the causative agents of cervical and most other anogenital malignancies. Infection by HPVs occurs in the basal layers of stratified epithelia that become exposed due to microabrasions. Following entry, viral genomes are established as low-copy nuclear episomes at approximately 10 to 100 copies per cell (1, 2). In differentiated suprabasal layers, high levels of late gene expression are induced along with genome amplification and virion assembly (3). Two major viral promoters, designated early and late, control transcription during differentiation, but additional regulation also occurs through alternative splicing and other posttranscriptional mechanisms (1, 2, 4). One important mechanism of posttranscriptional control is mediated by small RNAs that are 22 nucleotides in size, referred to as microRNAs (miRNAs).
miRNAs bind to partially complementary target sites in mRNA that are often located in the 3′UTR (untranslated regions) but can also be found in coding sequences and the 5′UTR (5). Binding by miRNAs to target sequences results in either the degradation of the target mRNAs or translational repression of the encoded protein (6). A single miRNA can repress the expression of over 100 genes, and a single gene can be repressed by multiple miRNAs. miRNAs often target multiple genes in a pathway and can act cooperatively with other miRNAs (7). Many miRNAs target developmental (8–10) and differentiation pathways (11–14), as well as cell proliferation and transformation pathways (15–17). Herpesviruses and other small DNA viruses encode their own miRNAs (18); however, this is not the case for HPVs (19). Instead, HPVs modulate expression of host miRNAs to control various aspects of their viral life cycle (20, 21).
Several miRNAs have been shown to be modulated by HPV proteins and to play key roles in the viral life cycle. miRNA 203 (miR-203) is a key regulator of epithelial differentiation and is downregulated in HPV-positive cells upon differentiation. This repression allows for the maintenance of high levels of p63, which is normally repressed by miR-203 in differentiating, infected suprabasal cells and is important for maintaining cells in an active state in the cell cycle (21). Additional studies demonstrated miR-218 to be downregulated by HPV E6, which correlates with increased levels of LAMB3, a translational target of this miRNA (22). A number of reports have documented changes in levels of cellular microRNAs in HPV-positive cells using microarray analyses, but only a limited number of studies has examined the significance of these changes in the viral life cycle (23–25).
Since HPVs modulate the expression of cellular microRNAs to regulate aspects of their differentiation-dependent life cycle, we sought to identify cellular miRNAs that were altered by HPV proteins in a more physiological context than that reflected in monolayer cultures alone. For this analysis, we performed deep sequencing using organotypic raft cultures of a matched set of normal (viv) and HPV-31-transfected human foreskin keratinocytes isolated from the same donor (4). The HPV31 viv line (viv-31gen) contains episomal copies of HPV-31 at approximately 10 to 20 copies per cell and activates late viral functions, such as genome amplification, as well as late gene expression upon differentiation. Small RNAs of less than 50 nucleotides in size were isolated from viv and viv-31gen raft cultures, and libraries were made using the Illumina small RNA library kit. Solexa deep sequencing of the libraries was performed, and the reads were matched to human and HPV genomes. No HPV-related sequences were present in the viv-31gen raft library, consistent with previous reports showing that the HPV-31 genome does not encode miRNAs (19). Our studies further demonstrated that HPVs do not express noncoding small RNAs that are larger than microRNAs (i.e., 22 nucleotides) but less than 50 nucleotides in size. A total of 862 cellular miRNAs were found to be differentially expressed between the two sets of libraries. We further refined the list by using a threshold of at least 100 reads and a fold difference of at least 2. Setting a read threshold eliminated any miRNAs that were expressed at low levels and unlikely to be significant regulators of HPV activities. This analysis resulted in the identification of 93 miRNAs that had the potential to be regulators of the HPV differentiation-dependent life cycle. Of this group, 55 miRNAs were repressed and 38 were expressed at higher levels in viv-31gen rafts than in viv keratinocytes. The 10 top-ranked miRNAs activated and repressed by HPVs are shown in Table 1. The complete list of 93 miRNAs we found to be significantly altered in HPV-positive organotypic rafts is shown in Fig. S1 in the supplemental material.
Table 1.
Ten cellular miRNAs most activated/repressed by HPV-31 in organotypic raft culturesa
| Rank | High in viv | Fold difference | High in viv-31gen | Fold difference |
|---|---|---|---|---|
| 1 | hsa-miR-582-3p | 9.185 | hsa-miR-1246 | 31.693 |
| 2 | hsa-miR-199a-5p | 8.668 | hsa-miR-335* | 10.612 |
| 3 | hsa-miR-214 | 8.432 | hsa-miR-1260b | 8.691 |
| 4 | hsa-miR-143 | 7.278 | hsa-miR-3613-5p | 5.855 |
| 5 | hsa-miR-145 | 7.096 | hsa-miR-1260 | 4.6 |
| 6 | hsa-miR-145* | 6.894 | hsa-miR-1285 | 4.111 |
| 7 | hsa-miR-369-3p | 6.522 | hsa-miR-576-5p | 4.015 |
| 8 | hsa-miR-655 | 6.377 | hsa-miR-615-5p | 3.921 |
| 9 | hsa-miR-493 | 6.211 | hsa-miR-92b | 3.692 |
| 10 | hsa-miR-493* | 5.318 | hsa-miR-25* | 3.316 |
A small RNA population was isolated using a Roche small RNA isolation kit, and small RNA libraries were made using the Illumina small RNA library kit. Solexa deep sequencing of the libraries was performed. The resulting miRNAs were refined by using a threshold of at least 100 reads and a fold difference of at least 2. High in viv, having increased levels in viv normal keratinocytes compared to HPV-31-positive cells; high in viv-31gen, having increased levels in HPV-31-positive cells compared to viv normal keratinocytes.
We screened the seed sequences of the differentially expressed miRNAs for ones that could potentially directly target HPV mRNAs. The 8-mer seed sequences for miRNA 145 (miR-145) are present in the coding regions of HPV-31 E1 (nucleotides 2623 to 2630) and E2 (nucleotides 3089 to 3096). Similar miR-145 seed sequences are present in most HPV types, as shown in the Table 2, suggesting that they are functionally significant. Our deep-sequencing analysis showed that miR-145 was suppressed 7-fold in HPV-31 rafts compared to the normal rafts. For further validation, we performed quantitative PCR (qPCR) analysis of three independently grown sets of raft cultures and demonstrated that miR-145 was suppressed 10-fold, on average, in viv-31gen rafts (Fig. 1A), which is comparable to deep-sequencing data. We also demonstrated the suppression of miR-145 in raft cultures from three sets of HPV-31-positive and normal keratinocytes isolated from a different donor background and found similar results (data not shown).
Table 2.
Target sites of miR-145 across HPV subtypes
| Target HPV subtype for coding regiona: | ||||
|---|---|---|---|---|
| E1 | E2 | E4 | L1 | L2 |
| 2 | 7 | 87 | 97 | 119 |
| 3 | 31 | 108 | 123 | 126 |
| 6 | 52 | 128 | 148 | |
| 11 | 80 | 134 | ||
| 13 | 86 | |||
| 16 | 87 | |||
| 27 | 108 | |||
| 30 | 112 | |||
| 31 | ||||
| 34 (2) | ||||
| 35 | ||||
| 39 | ||||
| 53 | ||||
| 56 | ||||
| 57 | ||||
| 59 | ||||
| 60 | ||||
| 62 | ||||
| 68 (2) | ||||
| 70 (2) | ||||
| 73 | ||||
| 77 | ||||
| 81 | ||||
| 82 | ||||
| 83 | ||||
| 88 | ||||
| 90 | ||||
| 91 | ||||
| 92 | ||||
| 121 (2) | ||||
| 128 (2) | ||||
| 131 | ||||
| 149 | ||||
Numbers in parentheses indicate the number of target sites within the coding region.
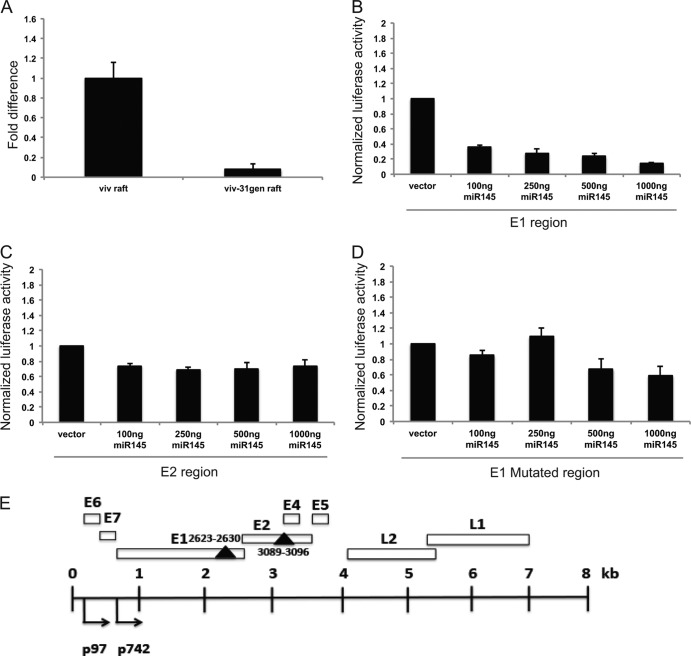
Fig 1.
Levels of miR-145 are repressed in HPV-31-positive organotypic rafts. (A) qPCR analysis of miR-145 levels in 13-day-old organotypic raft cultures of matched normal keratinocytes (viv) and HPV-positive cells that stably maintain episomes (viv-31gen). The data are normalized to a commonly used control, U6 small RNA levels, and are represented as fold changes with respect to miR-145 levels in viv raft cultures, which were set to 1.0. U6 RNA is commonly used as a normalization control for miRNA studies. (B and C) Luciferase reporter assays measuring responsiveness to miR-145 of sequences in HPV-31 E1 and E2. The two miR-145 target regions were individually cloned into a 3′UTR-luciferase reporter (psiCheck-2; Promega), and the relative luciferase activity (Renilla/firefly luciferase) of each region in the presence of increasing concentrations of miR-145 expression plasmid was assayed in transient assays. The miR-145 sequence in E1 showed a significant reduction (80%) in luciferase activity with increasing levels of miR-145 (B), whereas the E2 region showed a slight reduction in luciferase activity (C). The data are from three independent experiments, and standard errors are shown. (D) Mutation of miR-145 seed sequence in the E1 region (AAC TGG AAA converted to AAT TGG AAA) resulted in loss of responsiveness to miR-145. The data shown are averages with standard errors from three independent experiments. (E) Schematic representation of HPV-31 genome with miR-145 target sequences indicated. The early and late promoters, designated p97 and p742, are indicated along with various open reading frames, which are shown as open boxes. The two solid triangles represent the miR-145 seed sequences, and the nucleotide positions are indicated.
It was important to determine if the miR-145 seed sequences in HPV-31 actually functioned as target sequences for this miRNA. A commonly used method to screen for functional, reactive sequences makes use of a luciferase reporter in which potential miRNA target sequences are cloned into the 3′UTR. The luciferase reporter is then cotransfected along with expression vectors for the miRNA and assayed for luciferase expression. The miR-145 seed sequence containing E1 and E2 regions of HPV-31 were individually cloned into psiCheck-2 luciferase vector. These clones were transfected into HEK293T cells along with increasing concentrations of an miR-145 expression plasmid. The results of three independent experiments, expressed as Renilla luciferase activity relative to that of control firefly luceriferase, are shown in Fig. 1. The miR-145 site in the E1 region showed a pronounced decrease in luciferase activity in a dose-dependent manner with cotransfection of miR-145 (Fig. 1B). In contrast, the E2 region displayed a statistically significant but only moderate decrease in relative luciferase activity (Fig. 1C). Single-nucleotide mutations made in the E1 seed region abolished responsiveness to miR-145 (Fig. 1D). We conclude that the seed sequence in the E1 open reading frame is a bona fide miR-145 reactive element, while the sequence in E2 is only modestly responsive. Since our studies indicated that HPV-31 suppresses the expression of miR-145, we investigated if either of the two major oncoproteins, E6 and E7, contributes to this effect. We used retroviruses expressing either E6 or E7 alone to infect primary keratinocytes and then selected for stable cell lines. Expression of E6 alone did not alter miR-145 levels significantly, whereas E7 was able to suppress miR-145 to lower levels, comparable to those seen in viv-31gen rafts (Fig. 2A).
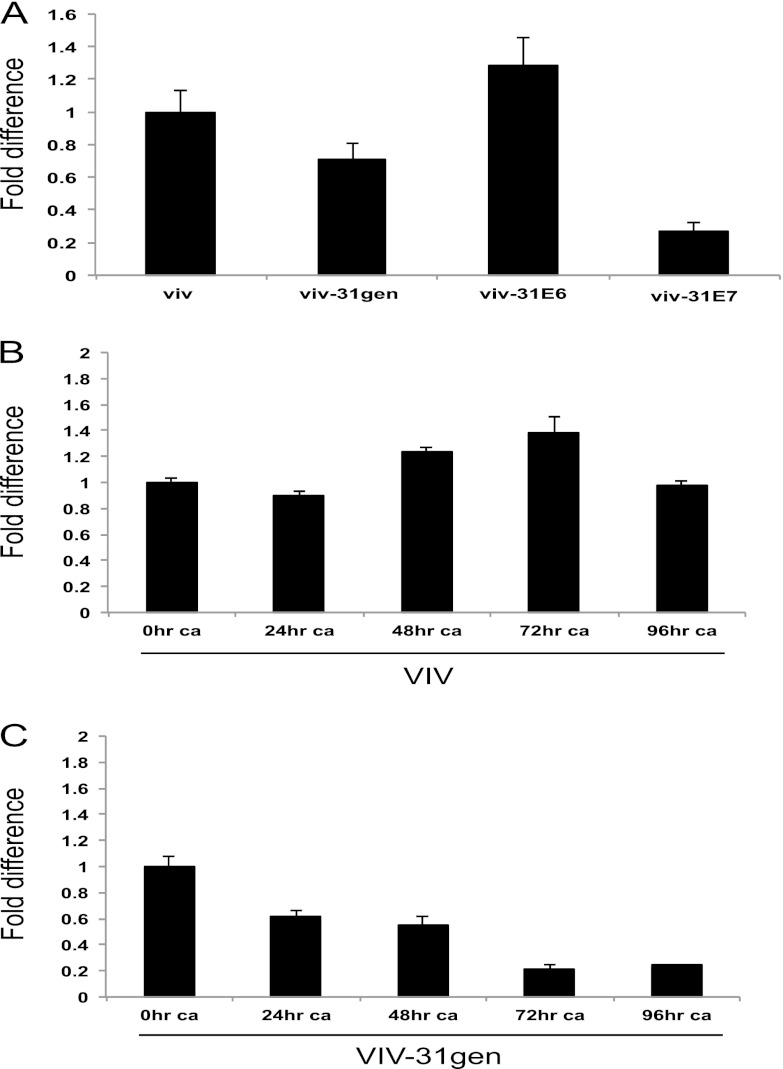
Fig 2.
Levels of miR-145 decrease upon differentiation of HPV-31-positive cells. (A) E7 protein mediates repression of miR-145. Shown is the qPCR analysis of miR-145 levels in monolayer cultures of viv, viv-31gen, viv-31E6, and viv-31E7 cells. The data are normalized to U6 levels and are represented as fold difference from miR-145 levels in normal keratinocytes (viv), which were set to 1.0. (B) qPCR analysis of miR-145 levels in normal keratinocytes induced to differentiate in high calcium media. The data are normalized to U6 levels and are represented as fold change relative to miR-145 levels in undifferentiated cultures (set to 1.0). (C) qPCR analysis of miR-145 levels in cells stably maintaining HPV-31 episome (viv-31gen) monolayer cultures upon differentiation in high calcium. The data are normalized to U6 levels and are represented as fold change from levels seen in undifferentiated cells. Results are averages from three independent experiments, and standard errors are shown. ca, calcium-induced differentiation.
To determine if miR-145 played any role in the viral life cycle, we first investigated if expression of miR-145 in HPV-positive cells varied with differentiation. For this analysis, we grew viv-31gen cells and viv normal keratinocytes in high calcium medium, which induces differentiation and late viral functions. We observed that the levels of miR-145 were maintained at similar levels in normal keratinocytes throughout the time course of high calcium treatment (Fig. 2B). We do not believe that the small variations in miR-145 levels in viv cells seen at 48 and 72 h are significant. In contrast, the levels of miR-145 were found to be high in undifferentiated monolayer cultures of viv-31gen cells and decreased by approximately 80% upon differentiation in high calcium. Undifferentiated viv-31gen cells exhibited a 1.5-fold, statistically significant decrease in miR-145 levels compared to levels in viv cells, and the most prominent differences were seen upon differentiation. The reduction was observed as early as 24 h of calcium treatment, and the levels were further reduced at later time points (Fig. 2C). This differentiation-dependent reduction in miR-145, seen specifically in HPV-positive cells, suggests that modulation of the levels of this microRNA could contribute to regulating viral activities during the various stages of differentiation.
We next sought to determine the effect of forced expression of miR-145 on early or late viral events. For this analysis, we used lentiviruses expressing miR-145 to infect HPV-31-positive CIN-612 cells and used drug selection to isolate cells stably expressing this miRNA (CIN-612 miR-145). CIN-612 cells are HPV-31-positive cells derived from a patient biopsy specimen and do not express any drug resistance markers, in contrast to cell lines such as viv-31gen. CIN-612 miR-145 cells were first grown in monolayers, and the overexpression of miR-145 was confirmed by qPCR. The cells were then grown either in normal growth medium or high calcium medium to induce differentiation. HPV-positive cells amplify episomal DNA upon differentiation in high calcium media within 48 h (26, 27). Expression of miR-145 from a heterologous promoter resulted in reduced episomal viral DNA in the undifferentiated cells. In addition, we observed reduced amplification in CIN-612 miR-145 cells upon differentiation (Fig. 3A). Similar results were seen in three independent experiments, and the results are shown in the bar diagram (Fig. 3A, inset). The reduced levels of episomes in undifferentiated cells may contribute to the impaired amplification seen upon differentiation.
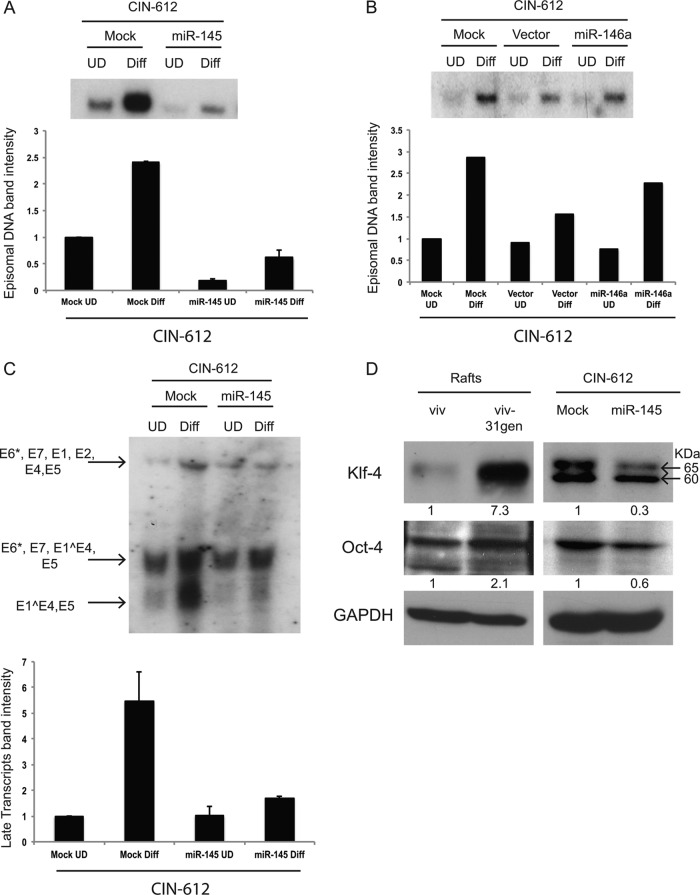
Fig 3.
High-level expression of miR-145 from heterologous expression vectors blocks HPV genome amplification, late gene expression, and induction of KLF-4. (A) Southern blot analysis of supercoiled episomal viral DNA levels upon differentiation (Diff) of CIN-612 cells with forced expression of miR-145. CIN-612 miR-145 cells and control mock-transfected cells were induced to differentiate in high calcium, and DNA was harvested after 48 h. Quantification of band intensities was done using Image J, and averages from three independent experiments with standard errors are shown in the bar graph. UD, undifferentiated. (B) Southern blot analysis of supercoiled episomal viral DNA levels following differentiation of CIN-612 cells expressing high levels of miR-146a (CIN-612 miR-146a cells), vector control, and mock-transfected cells. Quantification of the band intensities is shown in the bar graph. No effect of miR-146a on amplification is seen. (C) Northern blot analysis of early and late viral transcripts during differentiation of CIN-612 miR-145 cells and mock-infected control cells. Arrows indicate early transcripts encoding E6*, E7, E1, E2, E5, E6*, E7, and E1Ê4, as well as late transcripts encoding E1Ê4 and E5. The late transcript bands were quantified using Image J, and results from three independent experiments with standard errors are shown. (D) Western blot analysis showing levels of KLF-4 and Oct-4 in cell extracts from raft cultures of viv, viv-31gen, and CIN-612 control cells, as well as CIN-612 miR-145 cells. A nonspecific band (at 60 kDa) was detected in CIN-612 cultures which was not seen in the viv or viv-31gen rafts. The individual KLF-4 and Oct-4 bands were quantified by Image J and normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels, and they are represented as fold differences under each band.
MicroRNA-146a is another miRNA that we found to be suppressed in viv-31gen rafts, but in contrast to miR-145, it lacked any identifiable seed sequences in the HPV-31 genome. We sought to determine if overexpression of this microRNA had similar effects on HPV maintenance and amplification. CIN-612 cells were transfected with a plasmid expressing miR-146a, and stable drug-resistant cell lines were expanded. In contrast to our observations with CIN 612 miR-145 cells, there was no significant change in the levels of viral DNA in the CIN-612 miR-146a cells compared to the mock and vector control cells (Fig. 3B). The miR-146a studies provide a control for nonspecific effects that could be the result of high-level miRNA expression. We conclude that miR-145 has a specific effect on HPV genome maintenance and amplification.
Late gene expression is a second late viral event that is associated with differentiation status (4, 28). Activation of the HPV-31 late promoter induces expression of transcripts encoding E1̂E4, E1, E2, and E5 and can occur independently of amplification, which acts primarily to increase total transcript levels (28). To determine if miR-145 had an effect on the levels of viral transcripts, total RNA was isolated from CIN-612 and CIN-612 miR-145 cells and examined by Northern blotting for viral transcripts. Heterologous expression of miR-145 significantly decreased expression of late transcripts (Fig. 3C). Since miR-145 was shown here to target E1, it likely affects either the stability of messages encoding full-length E1 or the translation of E1, E6, or E7. Since no antibodies are available for these HPV-31 proteins, we were unable to determine which ones are the primary targets of this microRNA.
One previously identified cellular target of miR-145 is the p63-regulated transcription factor KLF-4 (29), and we investigated if its levels are altered in HPV-positive cells. KLF-4 is one of four transcription factors, along with Oct-4, SOX-2, and c-Myc, that act together to induce stem cell pluripotency (30). We first investigated if KLF-4 protein levels were altered in HPV-positive rafts. When total proteins were isolated from viv-31gen and viv rafts, we found KLF-4 levels to be significantly increased in viv-31gen rafts. Examination of CIN-612 miR-145 cells grown in a monolayer culture showed decreased levels of KLF-4 compared to those of control CIN-612 cells. In CIN-612 and CIN-612 miR-145 cells grown in monolayer cultures, we observed a nonspecific cross-reacting band of slightly lower mobility whose levels were not altered by miR-145 expression (Fig. 3D). Our identification of the KLF-4 band has been confirmed with knockdowns (unpublished data). These results suggest that KLF-4 is a cellular target of miR-145 and contributes to the control of viral life cycle during differentiation. Oct-4 has also been reported to be regulated by miR-145 (30), and, consistent with previous reports, we observed a modest reduction of Oct-4 levels in CIN-612 miR-145 cells.
In this study, we identified miR-145 as a regulator of the HPV life cycle. Our analyses identified two sites in HPV-31 that match to miR-145 seed sequences, and these sites are conserved in many other HPV subtypes, suggesting miR-145 is an important regulator. The miR-145 site in E1 was highly responsive to repression by miR-145, while the site in E2 was only modestly reactive. Importantly, heterologous expression of miR-145 significantly reduced stable viral copy numbers and blocked differentiation-dependent viral genome amplification. This indicates that HPVs downregulate miR-145 expression to allow for persistent viral infections.
The question arises as to why HPVs retain microRNA-responsive sequences in their genomes if viral proteins act to downregulate expression of certain microRNAs. We observed that in normal keratinocytes, miR-145 levels remain high and relatively constant throughout differentiation. In contrast, in HPV-positive cells, miR-145 levels are high in undifferentiated cells and decrease upon differentiation. This suggests a mechanism whereby higher levels of miR-145 contribute to controlling low-copy maintenance in undifferentiated cells. Upon differentiation, the levels of miR-145 decrease, resulting in a loss of suppression of replication proteins and facilitating viral genome amplification. Our studies indicate that differential expression of miR-145 is important for the virus life cycle.
As discussed above, miR-145 targets sequences in HPV-31 E1 and E2 to control either the stability of messages encoding E6, E7, E1, and E2 or the translatability of the corresponding viral proteins. Since HPV messages are all polycistronic, it is possible that either E6, E7, or E2 is the target of miR-145 action, and all of them may play roles in viral replication. The most likely candidate is E1, but there are no effective antibodies to detect HPV-31 E1 proteins in cells that stably maintain episomes. miRNA target sequences do not necessarily have to be located in 3′UTR, as they can also be located in 5′UTR or in the middle of coding regions (5). While it is unclear which specific viral proteins are targeted by miR-145, high levels of miR-145 can inhibit viral amplification upon differentiation and reduce viral copy numbers in undifferentiated cells. This is consistent with our model that high levels of miR-145 in undifferentiated cells help control the stable replication of episomes to keep the viral copy number low, while upon differentiation, levels of miR-145 decrease to permit robust viral replication. The miR-145 and miR-143 loci are located on chromosome 5q32, and these two miRNAs are expressed as bicistronic transcripts, often acting synergistically (31–33). Our studies show reductions in miR-143 levels in viv-31gen raft cultures similar to those seen with miR-145. In contrast to miR-145, no sequences reactive to miR-143 are present in HPV genomes. Future studies will investigate the effects of mutating each of the miR-145 reactive sequences in HPV-31 genomes on the differentiation-dependent life cycle.
The reduction in miR-145 levels by HPV-31 appears to be mediated by the oncoprotein E7. E7 can suppress the activity of several key cellular tumor suppressors, such as pRb (34), NF-κB (35, 36), and p300 (37). An examination of the promoter of miR-145 revealed that p65 (Rel A), part of the NF-κB transcriptional machinery, has two binding regions. E7 reduces NF-κB activity in HPV-positive cells (35, 36), and this may contribute to repression of miR-145 expression.
miR-145 has been reported to act as a tumor suppressor, since its expression is repressed in a number of tumors and overexpression can inhibit tumor growth (38–40). However, in our studies, we did not see any effect of miR-145 overexpression on the growth properties of HPV-positive cells. miR-145 also targets the pluripotency factors KLF-4, SOX-2, and Oct-4, and it functions to regulate the proliferative capacity of embryonic stem cells (30). The effect of miR-145 on Oct-4 has been reported to be modest, and we observed similar minimal reductions in our studies. In contrast, KLF-4 is highly responsive to miR-145, as confirmed in our analyses. KLF-4 is a downstream mediator of p63 action (29), and our previous studies indicate that p63 is necessary for activation of late viral events upon differentiation (28). Future studies will investigate the effects of knocking down KLF-4 and other pluripotency factors on the viral life cycle. We show here that repression of miR-145 regulates viral replication as well as KLF-4 protein levels, and we believe that both activities are important to allow for genome amplification upon differentiation. Overall, our studies identify miR-145 as an important regulator of the HPV life cycle both by directly targeting viral sequences and by acting on cellular factors, such as KLF-4.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kavi Mehta and Archana Raja for helping with the bioinformatics analysis of deep-sequencing data. We also sincerely appreciate Eva Gottwein and Jovanka Koo for sharing their expertise in different stages of the project.
This work was supported by a grant from the NCI to L.A.L.
Footnotes
Published ahead of print 6 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00153-13.
REFERENCES
- 1. Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10:550–560 [DOI] [PubMed] [Google Scholar]
- 2. Munger K, Howley PM. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213–228 [DOI] [PubMed] [Google Scholar]
- 3. Wang HK, Duffy AA, Broker TR, Chow LT. 2009. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 23:181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gunasekharan V, Hache G, Laimins L. 2012. Differentiation-dependent changes in levels of C/EBPbeta repressors and activators regulate human papillomavirus type 31 late gene expression. J. Virol. 86:5393–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin HR, Ganem D. 2011. Viral microRNA target allows insight into the role of translation in governing microRNA target accessibility. Proc. Natl. Acad. Sci. U. S. A. 108:5148–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 7. Uhlmann S, Mannsperger H, Zhang JD, Horvat EA, Schmidt C, Kublbeck M, Henjes F, Ward A, Tschulena U, Zweig K, Korf U, Wiemann S, Sahin O. 2012. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 8:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gokey NG, Srinivasan R, Lopez-Anido C, Krueger C, Svaren J. 2012. Developmental regulation of microRNA expression in Schwann cells. Mol. Cell. Biol. 32:558–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854 [DOI] [PubMed] [Google Scholar]
- 10. Zhao Y, Srivastava D. 2007. A developmental view of microRNA function. Trends Biochem. Sci. 32:189–197 [DOI] [PubMed] [Google Scholar]
- 11. Bentwich I. 2005. A postulated role for microRNA in cellular differentiation. FASEB J. 19:875–879 [DOI] [PubMed] [Google Scholar]
- 12. Guo L, Zhao RC, Wu Y. 2011. The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. Exp. Hematol. 39:608–616 [DOI] [PubMed] [Google Scholar]
- 13. Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. 2009. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol. Cell. Biol. 29:5290–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schoolmeesters A, Eklund T, Leake D, Vermeulen A, Smith Q, Force Aldred S, Fedorov Y. 2009. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One 4:e5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. 2012. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One 7:e34150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838 [DOI] [PubMed] [Google Scholar]
- 17. Stahlhut Espinosa CE, Slack FJ. 2006. The role of microRNAs in cancer. Yale J. Biol. Med. 79:131–140 [PMC free article] [PubMed] [Google Scholar]
- 18. Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 19. Cai X, Li G, Laimins LA, Cullen BR. 2006. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J. Virol. 80:10890–10893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McKenna DJ, McDade SS, Patel D, McCance DJ. 2010. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J. Virol. 84:10644–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melar-New M, Laimins LA. 2010. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 84:5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. 2008. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 27:2575–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greco D, Kivi N, Qian K, Leivonen SK, Auvinen P, Auvinen E. 2011. Human papillomavirus 16 E5 modulates the expression of host microRNAs. PLoS One 6:e21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lajer CB, Garnaes E, Friis-Hansen L, Norrild B, Therkildsen MH, Glud M, Rossing M, Lajer H, Svane D, Skotte L, Specht L, Buchwald C, Nielsen FC. 2012. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br. J. Cancer 106:1526–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng ZM, Wang X. 2011. Regulation of cellular miRNA expression by human papillomaviruses. Biochim. Biophys. Acta 1809:668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong S, Mehta KP, Laimins LA. 2011. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J. Virol. 85:9486–9494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moody CA, Laimins LA. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5:e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mighty KK, Laimins LA. 2011. p63 is necessary for the activation of human papillomavirus late viral functions upon epithelial differentiation. J. Virol. 85:8863–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA. 2012. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell 22:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. 2009. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137:647–658 [DOI] [PubMed] [Google Scholar]
- 31. Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, Lagna G, Hata A. 2011. Down-regulation of Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-beta and bone morphogenetic protein 4. J. Biol. Chem. 286:28097–28110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takaoka Y, Shimizu Y, Hasegawa H, Ouchi Y, Qiao S, Nagahara M, Ichihara M, Lee JD, Adachi K, Hamaguchi M, Iwamoto T. 2012. Forced expression of miR-143 represses ERK5/c-Myc and p68/p72 signaling in concert with miR-145 in gut tumors of Apc(Min) mice. PLoS One 7:e42137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. 2009. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 23:2166–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giarre M, Caldeira S, Malanchi I, Ciccolini F, Leao MJ, Tommasino M. 2001. Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a-imposed G1 cell cycle arrest. J. Virol. 75:4705–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spitkovsky D, Hehner SP, Hofmann TG, Moller A, Schmitz ML. 2002. The human papillomavirus oncoprotein E7 attenuates NF-kappa B activation by targeting the Ikappa B kinase complex. J. Biol. Chem. 277:25576–25582 [DOI] [PubMed] [Google Scholar]
- 36. Vandermark ER, Deluca KA, Gardner CR, Marker DF, Schreiner CN, Strickland DA, Wilton KM, Mondal S, Woodworth CD. 2012. Human papillomavirus type 16 E6 and E7 proteins alter NF-kB in cultured cervical epithelial cells and inhibition of NF-kB promotes cell growth and immortalization. Virology 425:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bernat A, Avvakumov N, Mymryk JS, Banks L. 2003. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene 22:7871–7881 [DOI] [PubMed] [Google Scholar]
- 38. Cho WC, Chow AS, Au JS. 2009. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur. J. Cancer 45:2197–2206 [DOI] [PubMed] [Google Scholar]
- 39. Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. 2009. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. U. S. A. 106:3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J, Guo H, Zhang H, Wang H, Qian G, Fan X, Hoffman AR, Hu JF, Ge S. 2011. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1 gene. Cancer 117:86–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.