Abstract
The novel human coronavirus EMC (hCoV-EMC), which recently emerged in Saudi Arabia, is highly pathogenic and could pose a significant threat to public health. The elucidation of hCoV-EMC interactions with host cells is critical to our understanding of the pathogenesis of this virus and to the identification of targets for antiviral intervention. Here we investigated the viral and cellular determinants governing hCoV-EMC entry into host cells. We found that the spike protein of hCoV-EMC (EMC-S) is incorporated into lentiviral particles and mediates transduction of human cell lines derived from different organs, including the lungs, kidneys, and colon, as well as primary human macrophages. Expression of the known coronavirus receptors ACE2, CD13, and CEACAM1 did not facilitate EMC-S-driven transduction, suggesting that hCoV-EMC uses a novel receptor for entry. Directed protease expression and inhibition analyses revealed that TMPRSS2 and endosomal cathepsins activate EMC-S for virus-cell fusion and constitute potential targets for antiviral intervention. Finally, EMC-S-driven transduction was abrogated by serum from an hCoV-EMC-infected patient, indicating that EMC-S-specific neutralizing antibodies can be generated in patients. Collectively, our results indicate that hCoV-EMC uses a novel receptor for protease-activated entry into human cells and might be capable of extrapulmonary spread. In addition, they define TMPRSS2 and cathepsins B and L as potential targets for intervention and suggest that neutralizing antibodies contribute to the control of hCoV-EMC infection.
INTRODUCTION
Human coronaviruses were long considered agents of mild respiratory disease, with the prototype viruses 229E and OC43 being responsible for up to 30% of common cold cases requiring medical attention (1, 2). However, the outbreak of the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002–2003 abruptly changed this view. The spread of the novel virus claimed more than 700 lives, predominantly elderly and immunocompromised individuals, and caused massive economic damage (3). SARS-CoV-related viruses were detected in bats, and it is believed that these animals served as a natural reservoir (4, 5) from which the virus was transmitted via intermediate hosts, such as palm civets (6), to humans. Thus, although most human coronaviruses known today (OC43, 229E, NL63, and HKU1) circulate worldwide and cause mild respiratory disease (7), the zoonotic transmission of novel coronaviruses to humans can pose a significant threat to public health.
A novel coronavirus, termed hCoV-EMC (8), recently emerged in the Middle East, and so far 13 laboratory-confirmed cases have been reported to the WHO, including 6 from Saudi Arabia, 2 from Qatar, 2 from Jordan, and 3 from the United Kingdom (9, 10). The cases from the United Kingdom cluster within one family, with the initial individual but not the subsequent ones having a history of travel to Pakistan and Saudi Arabia (10), suggesting that human-to-human transmission occurred. Disquietingly, the new virus shares several similarities with SARS-CoV. First, hCoV-EMC appears to be highly pathogenic, with 7 of the 13 identified cases having a fatal outcome, and infection induces a severe acute respiratory disease (8, 9). Second, the virus, like SARS-CoV, belongs to the betacoronavirus genus and might have been transmitted from bats to humans (8), as suggested by its close relatedness to the bat coronaviruses HKU4 and HKU5 and the isolation of hCoV-EMC-related viruses in bats from Ghana and Europe (11). At present, there is no evidence for efficient interindividual transmission of hCoV-EMC (9). However, a few adaptive amino acid changes might be sufficient to allow hCoV-EMC to spread rapidly within the human population, with potentially severe consequences. Therefore, it is imperative to elucidate hCoV-EMC interactions with host cells and to transform this knowledge into effective antiviral strategies.
The interaction of the coronavirus spike (S) protein with host cell receptors and proteases is essential for the first step in coronavirus infection, i.e., viral invasion of host cells (12, 13). The binding of the S protein to host cell receptors attaches viruses to target cells and is a major determinant of the viral cell and organ tropism (14). Two receptors for human coronaviruses have been identified so far, namely, CD13 (used by hCoV-229E) (15) and ACE2 (used by SARS-CoV and hCoV-NL63) (16, 17). Moreover, sialic acid has been described as a receptor determinant of hCoV-OC43 (18), and the coronavirus murine hepatitis virus (MHV) was shown to engage murine but not human CEACAM1 for cellular entry (19), although it is worth noting that MHV host range mutants which employ human CEACAM1 for cellular entry have been reported (20, 21). The S proteins are synthesized as inactive precursors and transform into an active state upon proteolytic cleavage (12, 13). The activity of the pH-dependent endosomal cysteine proteases cathepsin B and, particularly, cathepsin L was found to be required for entry of SARS-CoV (22) and hCoV-229E (23) into certain host cells, and evidence for S-protein proteolysis by cathepsins was provided (22). However, recent work indicates that the type II transmembrane serine proteases (TTSPs) TMPRSS2 and HAT can cleave the SARS-CoV S protein (SARS-S) and that SARS-S processing by TMPRSS2 allows for cathepsin B/L-independent virus-cell fusion (24–28). Which receptors and proteases are used by hCoV-EMC for attachment and activation is unknown at present.
Here we investigated the viral and cellular determinants of hCoV-EMC entry into host cells. We show that the hCoV-EMC S protein (EMC-S) mediated transduction of a broad range of human target cells when the protein was incorporated into lentiviral particles. EMC-S did not engage known coronavirus receptors for cellular entry and was activated by TMPRSS2 and endosomal cathepsins. Finally, transduction was inhibited by serum from an hCoV-EMC-infected patient, indicating that S-protein-specific neutralizing antibodies are generated in the context of hCoV-EMC infection.
MATERIALS AND METHODS
Cell culture.
Most of the adherent cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, Invitrogen) supplemented with 5 to 10% fetal calf serum (FCS; Biochrom) and 1% penicillin-streptomycin sulfate (Cytogen). 293T cells genetically engineered to stably express human ACE2 (hACE2; 293T-ACE2 cells) (29) or hCD13 (293T-CD13 cells) (S. Bertram et al., submitted for publication) were maintained in DMEM supplemented with 100 μg/ml Zeocin (Invitrogen). Caco-2 cells were cultured in DMEM-GlutaMAX medium (Invitrogen) supplemented with 10% FCS. The promonocytic cell line THP-1 was grown in RMPI 1640 medium (Gibco) supplemented with 10% FCS and antibiotics and differentiated into a macrophage-like culture by induction with phorbol myristate acetate (PMA) (20 ng/ml; Sigma) for 48 h. For generation of primary human monocytes, peripheral blood mononuclear cells were purified by Ficoll gradient centrifugation and cultured in monocyte adherence medium (RPMI 1640 medium with 7.5% human fibrin-depleted plasma) for 24 h. Thereafter, adherent cells were resuspended in monocyte differentiation medium (X-Vivo 10 medium [Lonza] supplemented with 1% human fibrin-depleted plasma and 1% penicillin-streptomycin sulfate), seeded into 96-well plates at a density of 7.5 × 104 cells/well, and allowed to differentiate into monocyte-derived macrophages for at least 7 days before usage for transduction experiments. Microscopic inspection of cells used for transduction revealed a clear macrophage-like morphology. All cells were grown in a humidified atmosphere at 37°C and 5% CO2.
Plasmids and antibodies.
Expression plasmids for SARS-S, 229E-S, Zaire ebolavirus glycoprotein (EBOV-GP), vesicular stomatitis virus glycoprotein (VSV-G), and murine leukemia virus envelope protein (MLV-Env) have been described previously (30, 31). Plasmids encoding the transmembrane proteases TMPRSS2, TMPRSS4, TMPRSS3, TMPRSS6, and HAT were published earlier (24, 32–34). An expression plasmid encoding EMC-S (EMC-S FLAG) with a C-terminal FLAG tag was generated by reverse transcription, using RNAs from CoV-EMC-infected cell cultures and the oligonucleotides 2c-nhCoV-S-BamHI-F (TACGGATCCGCCACCATGATACACTCAGTGTTTCTACTGATGT) and 2c-nhCoV-SflagC-SalI-R (AGCGTCGACTTACTTGTCATCGTCATCCTTGTAATCGCCTCCGTGAACATGAACCTTATGCGGC), followed by ligation of the PCR fragment into plasmid pCG1. The resulting plasmid was used as a template for amplification of the EMC-S open reading frame without a tag, using oligonucleotides p5-BetaCoV_Kpn (GATCGGTACCACCATGATACACTCAGTGTTTCTACTG) and p3-BetaCoV_Xho (GATCCTCGAGTTAGTGAACATGAACCTTATGCGGCTCTAGG). The PCR amplicon was then inserted into the plasmid pCAGGS by using the KpnI and XhoI restriction sites. The resulting plasmid expresses EMC-S with a sequence identical to that deposited under GenBank accession number AFS88936. To generate EMC-S with a V5 tag, the C-terminal sequence of EMC-S was amplified using oligonucleotides BetaCoV_seq 2567 (CTCAATCATCTCCTATCATACCAGG) and p3-BetaCoV_Xho_V5 (GATCCTCGAGTTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCGTGAACATGAACCTTATGCGGCTCTAGG), followed by replacement of the corresponding sequence within the pCAGGS-EMC-S plasmid by use of SphI and XhoI. The integrity of all PCR-amplified nucleotide sequences was verified by automated sequence analysis. For generation of lentiviral virus-like particles (VLPs), plasmid p96ZM651gag-opt, encoding human immunodeficiency virus type 1 Gag (p55), was employed (35). A V5-reactive monoclonal antibody was obtained from Invitrogen; HIV Gag proteins were detected using the hybridoma 183-H12-5C cell culture supernatant (NIH AIDS Reagent Program). Secondary antibodies were purchased from Dianova. An hCoV-EMC-reactive serum was isolated from a blood sample obtained from an hCoV-EMC-infected patient treated in a hospital in Essen, Germany.
Production and analysis of VLPs.
VLPs were generated as described previously (25). Briefly, plasmid p96ZM651gag-opt was coexpressed with either an empty plasmid as a control or plasmids encoding the S proteins of SARS-CoV and hCoV-EMC, followed by concentration of supernatants by ultrafiltration using Vivaspin centrifugal concentrators (Sartorius) and purification of VLPs by centrifugation of supernatants through a 20% sucrose cushion at 13,000 × g for 2 to 3 h at 4°C. Concentrated VLPs were treated with trypsin (100 μg/ml) for 10 min at room temperature, followed by the addition of the same concentration of soybean trypsin inhibitor (Sigma). The samples were then analyzed by Western blotting, employing a monoclonal antibody directed against the V5 tag.
Production and analysis of lentiviral pseudotypes.
Lentiviral pseudotypes were generated and harvested from the culture supernatants of transfected 293T cells as described previously (31, 36). The supernatants were harvested, passed through 0.45-μm-pore-size filters, and stored at −80°C. Before usage in experiments, pseudotypes were normalized for equal infectivity by transduction of Caco-2 cells with serially diluted supernatants followed by determination of luciferase activity in cell lysates according to the manufacturer's instructions (Promega). Normalization of the viral p24 capsid protein content was performed using a commercially available HIV-1 p24 enzyme-linked immunosorbent assay (ELISA) (HIV-1 p24 antigen capture assay; Advanced BioScience Laboratories).
Neutralization assay.
The neutralizing activity of serum obtained from an hCoV-EMC-infected patient treated in Essen, Germany, was measured by preincubation of lentiviral pseudotypes with defined serum dilutions in triplicate for 30 min at 37°C in a total volume of 50 μl. Thereafter, the mixture was added to 104 Caco-2 cells in 96 wells for 8 h, followed by complete exchange of the culture medium. After 72 h, the reduction in transduction efficiency was measured by quantifying luciferase activities in cell lysates.
Inhibition studies.
The inhibitors MDL28170 (Calbiochem), camostat mesylate (Tocris), leupeptin, and ammonium chloride (Sigma) were diluted in solvent as recommended by the manufacturers and used at the indicated concentrations. Within the experiments, the volume of solvent was kept constant, and solvent without inhibitor was used as a control. Target cells were pretreated with the inhibitors for 60 min at 37°C and transduced with infectivity-normalized pseudotypes in the presence of inhibitor. The inhibitor-containing medium was replaced by fresh culture medium without inhibitor at 8 h postransduction, and transduction efficiency was measured after 72 h.
RESULTS
The human coronavirus EMC spike protein is expressed in transfected 293T cells and incorporated into retroviral particles.
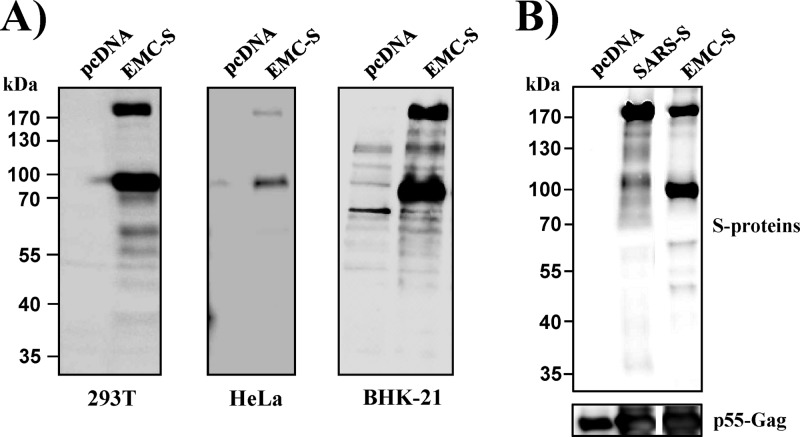
Retroviral particles pseudotyped with S proteins (pseudotypes) have been used successfully to characterize the host cell entry of coronaviruses, including SARS-CoV (37–39). Therefore, we asked if pseudotyping can be employed to study the cellular entry driven by EMC-S. Efficient expression of viral envelope proteins in transfected cells is a prerequisite for pseudotyping. Western blot analysis of 293T, HeLa, and BHK-21 cells transfected to express EMC-S with a C-terminal antigenic tag revealed two prominent bands, of 170 and 100 kDa, and a minor band, of 60 kDa, which were not detected in control cells transfected with empty plasmid, indicating that the S protein was expressed efficiently (Fig. 1A). The detection of two bands is consistent with cleavage of the S protein (170 kDa) into an N-terminal (not detected) and a C-terminal (100 kDa) subunit by host cell proteases. In order to determine whether efficient EMC-S expression allows for virion incorporation, we coexpressed HIV-1 Gag and EMC-S in 293T cells and analyzed the S-protein incorporation into particles released from these cells. SARS-S was included as a positive control, since we and others previously showed that SARS-S is incorporated into retroviral particles (37–39). Western blot analysis of pseudoparticles released from transfected 293T cells indeed revealed efficient particle incorporation of SARS-S, and the S protein was largely uncleaved, in agreement with published data (37–39) (Fig. 1B). A signal of similar intensity was measured for EMC-S, and again, two prominent bands, of 170 and 100 kDa, were detected (Fig. 1B), indicating that EMC-S and SARS-S are incorporated into retroviral particles with similar efficiencies and that the former but not the latter S protein is efficiently processed by host cell proteases.
Fig 1.
EMC-S is expressed in transfected cells and incorporated into lentiviral particles. (A) Plasmids encoding EMC-S with a C-terminal tag were transfected into 293T, HeLa, and BHK-21 cells, as indicated. The transfected cells were lysed, and the lysates were analyzed by Western blotting using tag-specific monoclonal antibodies. Cells transfected with empty plasmid (pcDNA) served as a negative control. (B) SARS-S or EMC-S with a C-terminal V5 tag was coexpressed with HIV-1 Gag protein in 293T cells. Cells expressing Gag-Pol alone were used as a control (pcDNA). At 48 h posttransfection, virus-like particles were harvested, concentrated via ultrafiltration and high-speed centrifugation through a sucrose cushion, and analyzed by Western blotting using a V5-specific antibody and a p55-Gag-reactive antibody as a loading control. The results are representative of at least three independent experiments.
The human coronavirus EMC spike protein mediates entry into cell lines derived from different human tissues.
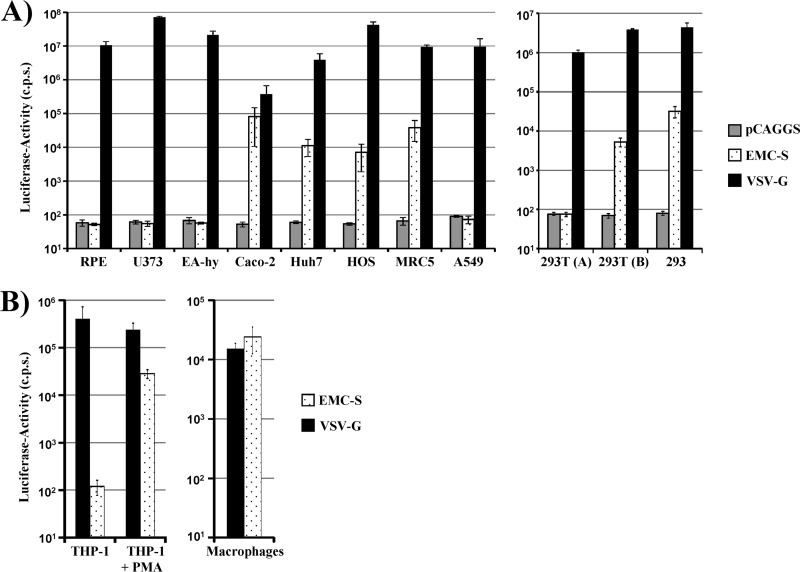
We next investigated if retroviral particles pseudotyped with EMC-S were able to transduce human cells. Pseudotypes bearing the G protein of VSV were used as a positive control, since VSV-G facilitates transduction of an extremely broad range of cell types. Bald particles bearing no glycoprotein were used as negative controls (pCAGGS). A vector encoding luciferase was used for pseudotyping, and transduction efficiency was determined by quantifying luciferase activities in the lysates of transduced cells. Pseudotypes bearing VSV-G were able to robustly transduce all cell types tested, while signals measured upon transduction with particles bearing no glycoprotein were within the background range (Fig. 2A), as expected. EMC-S facilitated transduction of Caco-2 (colon), HOS (osteosarcoma), Huh-7 (liver), and MRC5 (lung) cells, while RPE (retina), U373 (glioblastoma), EA-hy (endothelium), and A549 (lung) cells were not susceptible to EMC-S-mediated transduction (Fig. 2A, left panel). Transduction of 293 and 293T (kidney) cells was also observed (Fig. 2A, right panel), although some batches of these cells were nonsusceptible, potentially due to batch- or culture-dependent differences in the expression of the so far unknown hCoV-EMC receptor. Finally, THP-1 monocytic cells were nonsusceptible to EMC-S-driven transduction but acquired susceptibility upon PMA-induced differentiation into macrophages (Fig. 2B, left panel), suggesting that expression of the EMC-S receptor might be induced upon differentiation of monocytes into macrophages. Indeed, primary human monocyte-derived macrophages could be transduced efficiently by EMC-S-bearing pseudotypes (Fig. 2B, right panel), in agreement with robust receptor expression. In sum, EMC-S mediates transduction of a broad range of target cells, indicating that the viral receptor is expressed in different cell types and organs.
Fig 2.
Lentiviral pseudotypes bearing EMC-S transduce a broad spectrum of human cells. Lentiviral pseudotypes carrying the glycoprotein of VSV or EMC as well as pseudotypes bearing no glycoprotein (pCAGGS) were normalized for equal p24 capsid protein content and used for transduction of the indicated human cell lines (A) or unstimulated and PMA-stimulated THP-1 cells and primary human monocyte-derived macrophages (B). Cells were incubated with pseudotypes for 8 h, followed by replacement of the infection medium with fresh culture medium. After 72 h, cell lysates were prepared and analyzed for luciferase activity. The results shown are representative of three (A) or two (B) experiments performed in triplicate, using two independent pseudotype preparations. Error bars indicate standard deviations (SD).
The spike protein of the human coronavirus EMC does not use known coronavirus receptors for host cell entry.
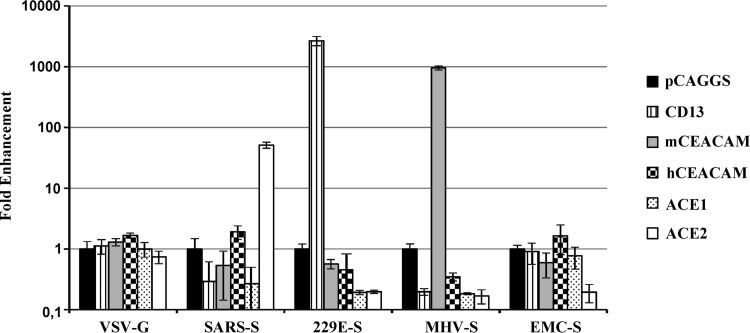
The following proteinaceous receptors for coronavirus have been identified so far: CD13 (used by hCoV-229E and others [15]), murine CEACAM-1 (used by MHV [19]), and ACE2 (used by SARS-CoV and hCoV-NL63 [16, 17]). We investigated if EMC-S was able to employ one of these receptors for host cell entry. For this purpose, the respective receptors were transiently expressed in 293T cells and the cells subsequently transduced with pseudotypes bearing the S proteins of hCoV-229E (229E-S), MHV (MHV-S), and SARS-CoV. ACE1 was also included in this experiment due to its sequence similarity with ACE2. Expression of the respective coronavirus receptors did not modulate host cell entry driven by VSV-G (Fig. 3). In contrast, ACE2 but not ACE1 expression augmented transduction driven by SARS-S, while expression of CD13 and murine but not human CEACAM-1 facilitated entry driven by 229E-S and MHV-S, respectively (Fig. 3), in keeping with published results (15, 19). Notably, expression of none of the receptors tested augmented transduction driven by EMC-S (Fig. 3), indicating that hCoV-EMC uses a different host cell factor for cellular entry.
Fig 3.
EMC-S does not employ previously described coronavirus receptors for entry into target cells. 293T cells were transiently transfected with empty plasmid (pCAGGS) or plasmids expressing the indicated coronavirus receptors. The cells were then transduced with infectivity-normalized pseudotypes bearing VSV-G, SARS-S, 229E-S, MHV-S, or EMC-S. Luciferase activities in cell lysates were determined at 72 h postinfection. Similar results were obtained in an independent experiment. Transduction of cells transfected with receptor-encoding plasmids is shown as fold enhancement relative to transduction of cells transfected with empty plasmid, which was set as 1.
Low pH and cathepsin activity are required for transduction mediated by the human coronavirus EMC spike protein.
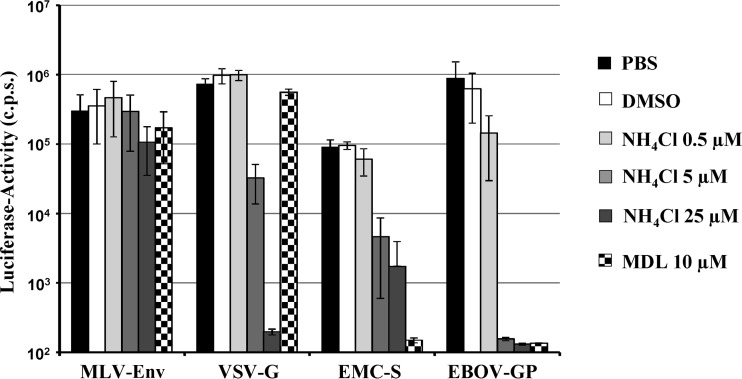
The cellular entry of enveloped viruses requires envelope protein-driven fusion of the viral membrane with a host cell membrane. The membrane fusion reaction is energetically unfavorable and can proceed only once the envelope protein receives a trigger, either receptor engagement, low pH, or both (40, 41). In addition, many viral glycoproteins require activation by host cell proteases, including the pH-dependent endosomal cysteine proteases cathepsins B and L, which activate the Ebola virus glycoprotein (EBOV-GP) (42) and SARS-S (22). Entry of viruses which depend on low pH can be inhibited by lysosomotropic agents such as ammonium chloride, and these agents also block entry of viruses which require cathepsin B/L activity for entry (22). In contrast, protease inhibitors such as MDL28170 specifically block activation of viruses which require cathepsin B/L activity for entry but do not inhibit viruses which are activated by low pH alone (22). We employed ammonium chloride and MDL28170 to determine which host cell factors trigger EMC-S for virus-cell fusion. The murine leukemia virus envelope protein (MLV-Env) is triggered by receptor engagement for membrane fusion (43, 44), and transduction mediated by MLV-Env was not blocked by any of the agents tested (Fig. 4), as expected. In contrast, VSV-G requires low pH as a trigger for membrane fusion (45), and VSV-G-driven transduction was inhibited by ammonium chloride but not by MDL28170 (Fig. 4), in keeping with published results (22). Transduction by EBOV-GP depends on cathepsin B/L activity (42) and was thus inhibited by both ammonium chloride and MDL28170 (Fig. 4), again in accordance with published data (22, 42). Finally, transduction mediated by EMC-S was also blocked by ammonium chloride and MDL28170 (Fig. 4), indicating that EMC-S, like EBOV-GP and SARS-S, depends on cathepsin B/L activity for host cell entry.
Fig 4.
EMC-S-driven cellular entry depends on the activity of cathepsin B and/or L. MRC5 target cells were preincubated with the indicated concentrations of ammonium chloride, MDL28170, dimethyl sulfoxide (DMSO), or phosphate-buffered saline (PBS) for 60 min, followed by inoculation with infectivity-normalized pseudotypes bearing the glycoprotein of VSV, MLV, hCoV-EMC, or EBOV. Luciferase activity in cell lysates was measured at 72 h postinfection. The results of a representative experiment performed in triplicate are shown. Transduction efficiency is shown relative to transduction of PBS-treated cells, which was set as 100%. Error bars indicate SD. Similar results were obtained in another experiment using different pseudotype preparations.
TMPRSS2 activates the spike protein of human coronavirus EMC for cathepsin B/L-independent host cell entry.
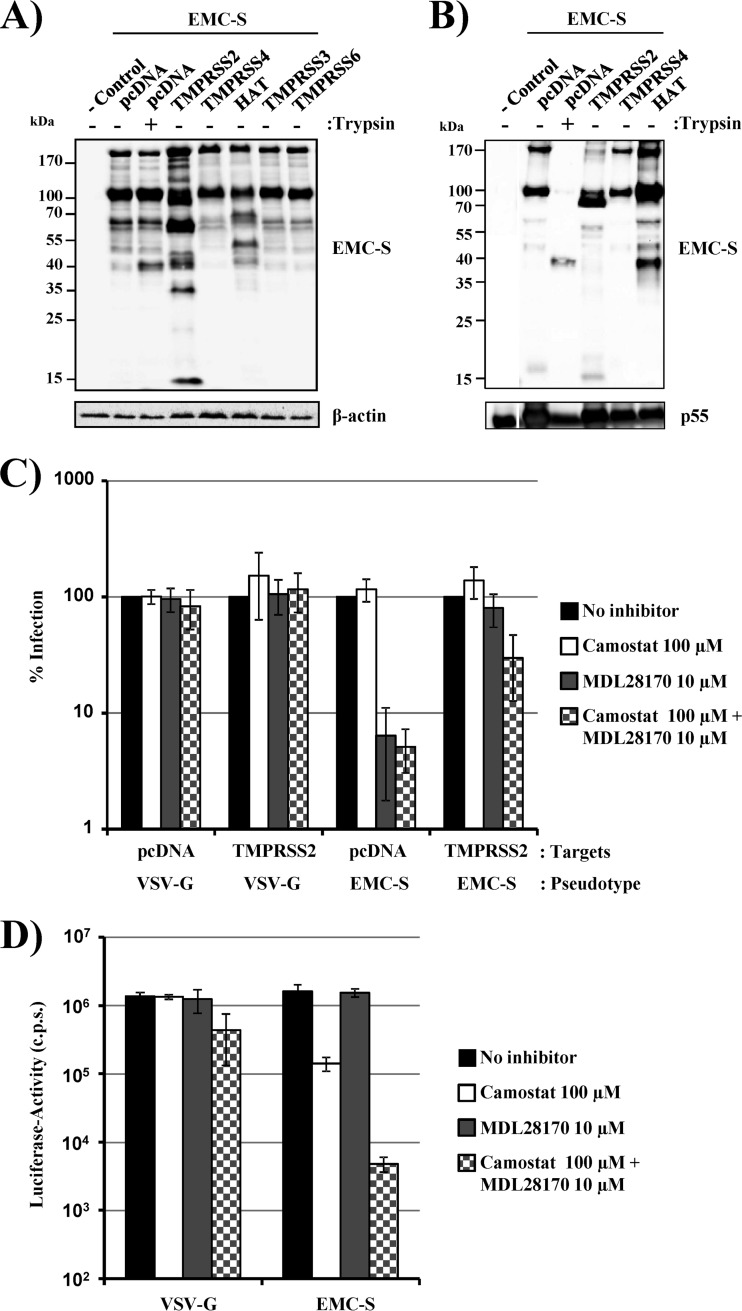
We and others have previously shown that the TTSP TMPRSS2 cleaves SARS-S and that cleavage activates SARS-S for cathepsin B/L-independent host cell entry (25–27). We therefore analyzed if TMPRSS2 can also activate EMC-S for virus-cell fusion. For this purpose, we first asked if TMPRSS2 and other members of the TTSP family cleave EMC-S with a C-terminal antigenic tag when coexpressed in transfected 293T cells. The effect of trypsin on EMC-S was analyzed in parallel. Western blot analysis of cells transfected to express EMC-S in the absence of a protease revealed prominent bands of 170 and 100 kDa and a minor band of approximately 60 kDa (Fig. 5A), in agreement with our previous findings (Fig. 1A). Trypsin treatment produced an additional cleavage fragment of 40 kDa, indicating that a trypsin-sensitive site is located within the S2 portion of EMC-S. Coexpression of TMPRSS2 produced prominent EMC-S cleavage fragments of 90 and 58 kDa, with additional fragments of 15, 35, and 40 kDa (Fig. 1A and 5A). Expression of HAT also resulted in EMC-S proteolysis, but the cleavage fragments differed from those produced by TMPRSS2. Finally, coexpression of TMPRSS3, TMPRSS4, and TMPRSS6 did not result in EMC-S cleavage, despite robust expression of these proteases in the transfected cells, as determined by Western blotting of epitope-tagged proteins (data not shown), indicating that these proteases do not recognize EMC-S as a substrate. Thus, EMC-S is processed by a so far unknown protease expressed in host cells and can additionally be cleaved by trypsin, TMPRSS2, or HAT. Western blot analysis of EMC-S bearing VLPs generated in the presence of trypsin, TMPRSS2, and HAT revealed that EMC-S cleavage products were incorporated into the viral membrane (Fig. 5B). Thus, the EMC-S fragments of 170 and 100 kDa detected in control VLPs were partially replaced by prominent fragments of 40 and 90 kDa in VLPs generated in cells coexpressing HAT and TMPRSS2, respectively (Fig. 5B). Similarly, a predominant fragment of 40 kDa was detected in trypsin-treated VLPs.
Fig 5.
TMPRSS2 activates EMC-S for cathepsin B/L-independent entry into target cells. (A) 293T cells were transfected with empty plasmid (−control) or cotransfected with plasmids encoding EMC-S and the indicated proteases. Forty-eight hours after transfection, cells were either left untreated or treated with trypsin, and lysates were analyzed by Western blotting using a V5 antibody or a β-actin antibody as a loading control. The results are representative of three independent experiments with different plasmid preparations. (B) Incorporation of cleaved EMC-S forms into lentiviral particles was assessed by coexpression of EMC-S (with a C-terminal V5 tag) with the indicated proteases and HIV-1 Gag. Virus-like particles were concentrated from the culture supernatants and analyzed by Western blotting using a V5 antibody, with a p55-Gag antibody used as a loading control. The experiment shown is representative of three independent experiments. (C) 293T cells were transiently transfected with empty plasmid (pcDNA) or a plasmid encoding TMPRSS2 and used as target cells for transduction by lentiviral vectors bearing EMC-S or VSV-G (as a control). Prior to infection, target cells were incubated with the indicated protease inhibitors. Luciferase activities in cell lysates were measured at 72 h postinfection. The average of three experiments (two for VSV-G-mediated transduction of TMPRSS2-expressing cells) performed in triplicate is shown. Transduction of untreated cells was set as 100%. Error bars indicate standard errors of the means (SEM). (D) Caco-2 cells, which express endogenous TMPRSS2, were incubated with DMSO or the indicated protease inhibitors, followed by transduction with lentiviral pseudotypes bearing EMC-S or VSV-G (as a control). At 72 h postinfection, luciferase activities in cell lysates were measured. The results of a representative experiment performed in triplicate are shown; error bars indicate SD. Comparable results were observed in two independent experiments.
In order to assess if cleavage results in EMC-S activation, we performed inhibition analyses with MDL28170, an inhibitor of cathepsin B/L (22), and camostat mesylate (camostat), a serine protease inhibitor active against TMPRSS2 (28). VSV-G-dependent transduction of 293T cells transfected with empty plasmid or a TMPRSS2 expression plasmid was not inhibited by MDL28170, camostat, or a combination of both inhibitors (Fig. 5C), as expected. Similarly, camostat treatment of target cells transfected with empty plasmid had little effect on EMC-S-mediated transduction, although a slight reduction in transduction efficiency was observed. In contrast, incubation of empty plasmid-transfected target cells with MDL28170 alone markedly reduced transduction efficiency, and inhibition was not increased by simultaneous treatment with camostat (Fig. 5C). Notably, inhibition by MDL28170 was fully rescued by expression of TMPRSS2, but entry remained sensitive to inhibition by camostat (Fig. 5C), indicating that TMPRSS2 can activate EMC-S for cathepsin B/L-independent entry. The activation of EMC-S by TMPRSS2 in transfected cells raised the question of which activation pathway dominates in target cells that are naturally susceptible to EMC-S-mediated transduction. We selected Caco-2 cells to address this question, since these cells express endogenous TMPRSS2 (32) and are susceptible to EMC-S-driven transduction (Fig. 2A). None of the inhibitors tested reduced VSV-G-mediated transduction of Caco-2 cells, although a modest reduction was observed when a combination of MDL28170 and camostat was tested (Fig. 5D). Incubation of Caco-2 cells with camostat reduced EMC-S-mediated transduction about 10-fold, while MDL28170 treatment had no effect, indicating that TMPRSS2-dependent activation prevails over cathepsin B/L-mediated activation in Caco-2 cells (Fig. 5D). However, a combination of camostat and MDL28170 was required to reduce transduction efficiency to background levels (Fig. 5D), indicating that both TMPRSS2 and cathepsin B/L can activate EMC-S for entry into naturally susceptible cells.
The serum from a human coronavirus EMC-infected patient contains neutralizing antibodies.
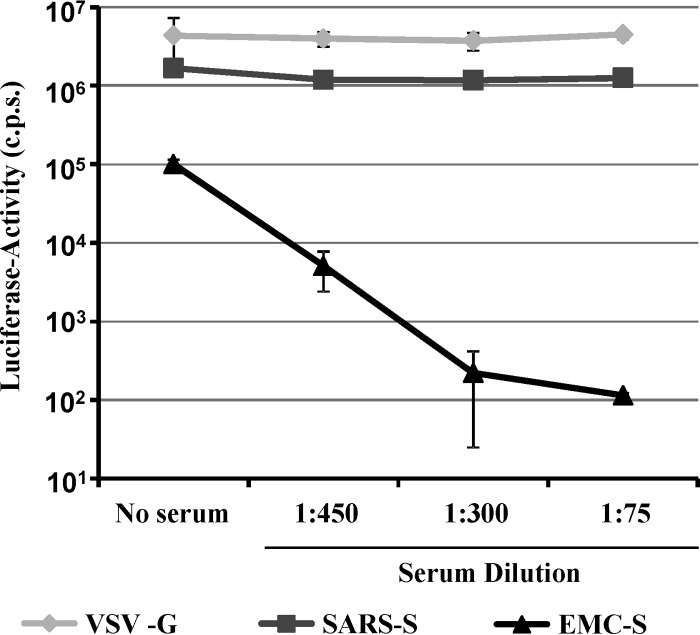
The generation of neutralizing antibodies might be important for the control of hCoV-EMC infection, and an efficient method for the detection of such antibodies could be highly useful for diagnostic efforts. Therefore, we tested if EMC-S-mediated transduction could be blocked by serum obtained from an hCoV-EMC-infected patient who survived the disease. The patient serum did not diminish VSV-G- or SARS-S-mediated transduction of 293T-ACE2 cells but reduced EMC-S-mediated transduction up to 1,000-fold, in a concentration-dependent manner (Fig. 6). Thus, neutralizing antibodies are generated in infected, surviving patients and can be detected in a highly efficient and quantitative fashion in the pseudotype transduction system.
Fig 6.
Host cell entry driven by EMC-S is neutralized by serum from an hCoV-EMC-infected patient. Pseudotypes bearing the glycoproteins of VSV, SARS-CoV, and hCoV-EMC, normalized for equal infectivity, were preincubated with the indicated serum dilutions in triplicate for 30 min at room temperature. Thereafter, the mixtures were added to 293T-ACE2 cells for 8 h, followed by replacement of the infection medium by fresh culture medium. Luciferase activities were determined after 72 h postinfection. The results shown are representative of three independent experiments. Error bars indicate SD.
DISCUSSION
The constantly increasing exploitation of natural resources, personal mobility, and climate change facilitate the zoonotic transmission of novel viruses to humans (46), with potentially dramatic consequences, as exemplified by the SARS pandemic. The outbreak of hCoV-EMC in Saudi Arabia raises concerns that a SARS-like pandemic might be about to unfold (47). Assessment of the pandemic potential and the development of specific antiviral measures require an understanding of hCoV-EMC interactions with host cells. Employing lentiviral pseudoparticles, we investigated the viral and cellular factors contributing to hCoV-EMC entry into host cells. We showed that a broad panel of human cell lines is susceptible to EMC-S-driven transduction, suggesting that the viral receptor is broadly expressed. The nature of the receptor remains to be explored, since none of the known coronavirus receptors augmented EMC-S-dependent transduction. Inhibitors of cathepsins B and L and TMPRSS2 blocked EMC-S-dependent virus-cell fusion, indicating that EMC-S can use redundant proteolytic pathways to ensure its activation. Finally, serum from an hCoV-EMC-infected patient inhibited EMC-S- but not VSV-G- or SARS-S-driven transduction, indicating that S-protein-specific neutralizing antibodies are generated in patients and might contribute to control of hCoV-EMC infection. Our results reveal important insights into the molecular processes underlying hCoV-EMC entry into host cells and define targets for intervention.
The incorporation of foreign glycoproteins into lentiviral particles, termed pseudotyping, offers an efficient means of studying host cell entry of highly pathogenic viruses without the need for a high level of biocontainment. Pseudotyping has been used successfully in the past to study the host cell entry of coronaviruses, including SARS-CoV (37, 39), and proved to be a suitable tool to analyze the entry of hCoV-EMC. Thus, EMC-S was efficiently incorporated into lentiviral particles, and pseudotypes bearing EMC-S were able to readily transduce a panel of human cell lines, including cells derived from the colon, liver, kidney, and lung. Whether these cell lines are also susceptible to infection by authentic hCoV-EMC and adequately mimic infection of primary cells remains to be determined. The susceptibility of lung-derived cell lines to EMC-S-mediated transduction was expected, since the virus causes severe respiratory disease and was isolated from sputum (8). The susceptibility of intestine- and kidney-derived cell lines is notable because these organs are also targeted by SARS-CoV (48, 49). The invasion of the intestines by SARS-CoV might account for the diarrhea frequently observed in SARS cases (50), while viral spread to the kidney might promote renal failure (51), a symptom occasionally associated with SARS-CoV infection. Five of the nine hCoV-EMC-infected patients documented up to December 2012 developed renal failure (9), and it is thus conceivable that hCoV-EMC, like SARS-CoV, is capable of extrapulmonary spread, which might account for certain aspects of the disease presentation.
The observation that SARS-CoV and hCoV-EMC both infect certain kidney- and colon-derived cell lines (8, 17, 52, 53) indicates that these viruses might use the same receptor for host cell entry, i.e., ACE2. Alternatively, hCoV-EMC might have adapted to use one of the other previously identified coronavirus receptor proteins, i.e., CD13 (15) or CEACAM-1 (19). However, directed expression of these receptors did not augment EMC-S-driven transduction of target cells, while transduction mediated by control S proteins was enhanced, demonstrating that EMC-S did not adapt to use any of the known coronavirus receptors. This conclusion is in keeping with results reported by Müller and colleagues, who recently demonstrated that hCoV-EMC does not use ACE2 for entry (53). Which cellular receptor is bound by EMC-S and whether binding is mediated by a receptor binding site proposed on the basis of bioinformatic analysis (54) remain to be determined. As was done for the identification of the SARS-CoV receptor ACE2 (17), coimmunoprecipitation followed by mass spectrometric identification of binding partners might be a suitable way to identify the elusive hCoV-EMC receptor. The observation that THP-1 cells acquire susceptibility to EMC-S-driven transduction upon differentiation into macrophages, most likely because of upregulation of receptor expression, might aid in these endeavors.
The coronavirus S proteins characterized so far are class I membrane fusion proteins. A characteristic of class I membrane fusion proteins is their need for proteolytic activation (40). Thus, the proteins are synthesized as inactive precursors which transit into an active state only upon proteolysis. A seminal study showed that SARS-S, like EBOV-GP, is activated by the pH-dependent endosomal cysteine proteases cathepsins B and L during uptake of virions into target cells (22). Inhibitors of cathepsins B and L, such as MDL28170, are available, and their development as therapeutics for coronavirus and filovirus infection has been proposed (22, 55). However, recent studies demonstrated that the type II transmembrane serine proteases TMPRSS2 and HAT, which were initially shown to activate influenza viruses (56), also activate SARS-S and that activation by TMPRSS2 renders SARS-S-mediated entry cathepsin B/L independent (24–28). TMPRSS2 and HAT are coexpressed with ACE2 in human respiratory epithelium (57), and TMPRSS2 was shown to contribute to SARS-S activation in cultured human airway-derived cells (28), indicating that TMPRSS2-mediated SARS-S activation might be operative in vivo.
In contrast to SARS-S, EMC-S is efficiently cleaved in transfected cells, and likely also in infected cells. The nature of the responsible proteolytic activity and the biological relevance of cleavage are unknown at present. Although EMC-S is cleaved in transfected cells and processed S protein is incorporated into virions, further processing by host cell proteases during viral entry is required for EMC-S activation. A similar observation has been documented for EBOV-GP, which is processed by furin in infected cells (58) but requires cathepsin activity in target cells in order to facilitate infectious entry (42). EMC-S can be activated by cathepsin B/L, as demonstrated by inhibition of EMC-S-dependent transduction by the cathepsin B/L inhibitor MDL28170. In addition, EMC-S can be processed and activated by TMPRSS2 for cathepsin B/L-independent entry. Thus, EMC-S, like SARS-S, is activated by endosomal cathepsins and type II transmembrane serine proteases in cell culture, suggesting that the activities of both proteases must be blocked in patients in order to efficiently suppress viral replication.
Coronavirus S proteins are major targets for neutralizing antibodies (37, 39, 59, 60), and the observation that serum from an hCoV-EMC-infected patient specifically neutralized EMC-S but not VSV-G or SARS-S is in keeping with this notion. Notably, work published during the revision of the present article demonstrated that the patient serum we used also neutralizes authentic hCoV-EMC (61), indicating that the pseudoparticle system adequately mirrors neutralization of authentic hCoV-EMC. Whether the neutralizing antibodies contributed to control of hCoV-EMC infection or were produced largely after the viral spread subsided is at present unknown. Regardless of the role of neutralizing antibodies, the easy detection of neutralizing activity present in human serum by the vector systems used here suggests that this tool might prove useful in determining the frequency of hCoV-EMC infection in patients and the potential cross-reactivity of serum with other coronavirus S proteins. Our initial analysis demonstrates that antibodies generated in hCoV-EMC-infected patients do not cross-neutralize SARS-S. Whether the same is true for the more closely related S proteins of HKU4 and HKU5 remains to be investigated.
In sum, the present study provides important insights into hCoV-EMC cell tropism and should contribute to our understanding of the pathogenesis of this virus. In addition, it demonstrates that hCoV-EMC uses a novel receptor for cellular entry, and it defines host cell enzymes important for activation of EMC-S, which might constitute targets for intervention. Finally, the study identifies pseudotypes as a valuable tool for the detection of neutralizing antibodies directed against EMC-S, a finding that has important implications for diagnostic approaches and efforts to characterize the humoral immune response in hCoV-EMC-infected patients.
ACKNOWLEDGMENTS
We thank A. Krueger and N. Meyer for technical assistance and all members of our group for helpful discussions.
This work was supported by BMBF, SARS Verbund (grant 01KI1005C to S.G., A.H., C.D., and S.P.), the Leibniz Graduate School EIDIS (F.W.), SFB 900 (K.W.), the Leibniz Foundation, and grant R01AI074986 from the National Institute of Allergy and Infectious Diseases (G.S.).
Footnotes
Published ahead of print 6 March 2013
REFERENCES
- 1. Bradburne AF, Bynoe ML, Tyrrell DA. 1967. Effects of a “new” human respiratory virus in volunteers. Br. Med. J. 3:767–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holmes KV. 2001. Coronaviruses, p 1187–1203 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Peiris JS, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 6. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278 [DOI] [PubMed] [Google Scholar]
- 7. Pyrc K, Berkhout B, van der Hoek L. 2007. The novel human coronaviruses NL63 and HKU1. J. Virol. 81:3051–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 [DOI] [PubMed] [Google Scholar]
- 9. World Health Organization 2012. Background and summary of novel coronavirus infection—as of 21 December. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/disease/coronavirus_infections/update_20121221/en/index.html [Google Scholar]
- 10. World Health Organization 21 February2013. Novel coronavirus infection—update. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/don/2013_02_21/en/index.html [Google Scholar]
- 11. Annan A, Baldwin HJ, Corman V, Klose SM, Owusu M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodie YA, Kalko EKV, Lina PHC, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler J. 2013. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 19:456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heald-Sargent T, Gallagher T. 2012. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4:557–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 102:7988–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vlasak R, Luytjes W, Spaan W, Palese P. 1988. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U. S. A. 85:4526–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams RK, Jiang GS, Holmes KV. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 88:5533–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baric RS, Sullivan E, Hensley L, Yount B, Chen W. 1999. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J. Virol. 73:638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McRoy WC, Baric RS. 2008. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion. J. Virol. 82:1414–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 102:11876–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawase M, Shirato K, Matsuyama S, Taguchi F. 2009. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 83:712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bertram S, Glowacka I, Muller MA, Lavender H, Gnirss K, Nehlmeier I, Niemeyer D, He Y, Simmons G, Drosten C, Soilleux EJ, Jahn O, Steffen I, Pöhlmann S. 2011. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 85:13363–13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pöhlmann S. 2011. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85:4122–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84:12658–12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. 2012. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 86:6537–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glowacka I, Bertram S, Herzog P, Pfefferle S, Steffen I, Muench MO, Simmons G, Hofmann H, Kuri T, Weber F, Eichler J, Drosten C, Pöhlmann S. 2010. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 84:1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hofmann H, Simmons G, Rennekamp AJ, Chaipan C, Gramberg T, Heck E, Geier M, Wegele A, Marzi A, Bates P, Pöhlmann S. 2006. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80:8639–8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, Doms RW, Bates P, Pöhlmann S. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115–123 [DOI] [PubMed] [Google Scholar]
- 32. Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi SY, Park Y, Schneider H, Schughart K, Pöhlmann S. 2010. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 84:10016–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung H, Lee KP, Park SJ, Park JH, Jang YS, Choi SY, Jung JG, Jo K, Park DY, Yoon JH, Park JH, Lim DS, Hong GR, Choi C, Park YK, Lee JW, Hong HJ, Kim S, Park YW. 2008. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene 27:2635–2647 [DOI] [PubMed] [Google Scholar]
- 34. Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, Tahara M, Kikuta H, Yanagi Y. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 82:8942–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao F, Li Y, Decker JM, Peyerl FW, Bibollet-Ruche F, Rodenburg CM, Chen Y, Shaw DR, Allen S, Musonda R, Shaw GM, Zajac AJ, Letvin N, Hahn BH. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retroviruses 19:817–823 [DOI] [PubMed] [Google Scholar]
- 36. Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 37. Hofmann H, Hattermann K, Marzi A, Gramberg T, Geier M, Krumbiegel M, Kuate S, Uberla K, Niedrig M, Pöhlmann S. 2004. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 78:6134–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moore MJ, Dorfman T, Li W, Wong SK, Li Y, Kuhn JH, Coderre J, Vasilieva N, Han Z, Greenough TC, Farzan M, Choe H. 2004. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78:10628–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U. S. A. 101:4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McClure MO, Marsh M, Weiss RA. 1988. Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J. 7:513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McClure MO, Sommerfelt MA, Marsh M, Weiss RA. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71:767–773 [DOI] [PubMed] [Google Scholar]
- 45. Superti F, Seganti L, Ruggeri FM, Tinari A, Donelli G, Orsi N. 1987. Entry pathway of vesicular stomatitis virus into different host cells. J. Gen. Virol. 68:387–399 [DOI] [PubMed] [Google Scholar]
- 46. Karesh WB, Dobson A, Lloyd-Smith JO, Lubroth J, Dixon MA, Bennett M, Aldrich S, Harrington T, Formenty P, Loh EH, Machalaba CC, Thomas MJ, Heymann DL. 2012. Ecology of zoonoses: natural and unnatural histories. Lancet 380:1936–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chan JF, Li KS, To KK, Cheng VC, Chen H, Yuen KY. 2012. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 65:477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Farcas GA, Poutanen SM, Mazzulli T, Willey BM, Butany J, Asa SL, Faure P, Akhavan P, Low DE, Kain KC. 2005. Fatal severe acute respiratory syndrome is associated with multiorgan involvement by coronavirus. J. Infect. Dis. 191:193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. 2005. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 202:415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kwan AC, Chau TN, Tong WL, Tsang OT, Tso EY, Chiu MC, Yu WC, Lai TS. 2005. Severe acute respiratory syndrome-related diarrhea. J. Gastroenterol. Hepatol. 20:606–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang JW, Chen KY, Tsai HB, Wu VC, Yang YF, Wu MS, Chu TS, Wu KD. 2005. Acute renal failure in patients with severe acute respiratory syndrome. J. Formos. Med. Assoc. 104:891–896 [PubMed] [Google Scholar]
- 52. Cinatl J, Jr, Hoever G, Morgenstern B, Preiser W, Vogel JU, Hofmann WK, Bauer G, Michaelis M, Rabenau HF, Doerr HW. 2004. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell. Mol. Life Sci. 61:2100–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Müller MA, Raj VS, Muth D, Meyer B, Kallies S, Smits SL, Wollny R, Bestebroer TM, Specht S, Suliman T, Zimmermann K, Binger T, Eckerle I, Tschapka M, Zaki AM, Osterhaus AD, Fouchier RA, Haagmans BL, Drosten C. 2012. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio 3:e00515–12 doi:10.1128/mBio.00515-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jiang S, Lu L, Du L, Debnath AK. 22 December 2012. A predicted receptor-binding and critical neutralizing domain in S protein of the novel human coronavirus HCoV-EMC. J. Infect. doi:10.1016/j.jinf.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou Y, Simmons G. 2012. Development of novel entry inhibitors targeting emerging viruses. Expert Rev. Anti Infect. Ther. 10:1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S, Soilleux EJ. 2012. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One 7:e35876 doi:10.1371/journal.pone.0035876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. 1998. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U. S. A. 95:5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Godet M, Grosclaude J, Delmas B, Laude H. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68:8008–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reguera J, Santiago C, Mudgal G, Ordono D, Enjuanes L, Casasnovas JM. 2012. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 8:e1002859 doi:10.1371/journal.ppat.1002859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Buchholz U, Müller M, Nitsche A, Sanewski A, Wevering N, Bauer-Balci T, Bonin F, Drosten C, Schweiger B, Wolff T, Muth D, Meyer B, Suda S, Krause G, Schade L, Hass W. 2013. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October-November 2012. Euro Surveill. 18:20406 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20406 [PubMed] [Google Scholar]