Abstract
The yields of egg-grown influenza vaccines are maximized by the production of a seed strain using a reassortment of the seasonal influenza virus isolate with a highly egg-adapted strain. The seed virus is selected based on high yields of viral hemagglutinin (HA) and expression of the surface antigens from the seasonal isolate. The remaining proteins are usually derived from the high-growth parent. However, a retrospective analysis of vaccine seeds revealed that the seasonal PB1 gene was selected in more than 50% of reassortment events. Using the model seasonal H3N2 virus A/Udorn/307/72 (Udorn) virus and the high-growth A/Puerto Rico/8/34 (PR8) virus, we assessed the influence of the source of the PB1 gene on virus growth and vaccine yield. Classical reassortment of these two strains led to the selection of viruses that predominantly had the Udorn PB1 gene. The presence of Udorn PB1 in the seed virus, however, did not result in higher yields of virus or HA compared to the yields in the corresponding seed virus with PR8 PB1. The 8-fold-fewer virions produced with the seed virus containing the Udorn PB1 were somewhat compensated for by a 4-fold increase in HA per virion. A higher HA/nucleoprotein (NP) ratio was found in past vaccine preparations when the seasonal PB1 was present, also indicative of a higher HA density in these vaccine viruses. As the HA viral RNA (vRNA) and mRNA levels in infected cells were similar, we propose that PB1 selectively alters the translation of viral mRNA. This study helps to explain the variability of vaccine seeds with respect to HA yield.
INTRODUCTION
Seasonal influenza is a highly contagious acute respiratory disease, with the severity of symptoms varying greatly from year to year. Globally, an estimated 250,000 to 500,000 people die from seasonal influenza epidemics annually, with 90% of these deaths and more than half of hospitalizations occurring in the elderly (1). Vaccination has been shown to significantly decrease both deaths and hospitalization caused by seasonal influenza, thus markedly reducing the impact of the disease in the elderly (2). Current forms of influenza vaccines, including split-inactivated virus preparations, induce strain-specific neutralizing antibodies against the viral surface glycoproteins HA and neuraminidase (NA). These antibodies efficiently mediate high levels of protection against homologous infection (3). However, constant drift occurs in the antigenic regions of the immunodominant HA (4–6) and, also, the NA (6, 7) of influenza viruses and can render the protection induced by previous vaccination incomplete. For this reason, vaccines need to be continually updated to contain virus strains that are predicted to antigenically resemble those viruses that will be circulating in the human population during the oncoming influenza season.
Seasonal influenza vaccines are trivalent formulations containing two influenza A viruses of subtypes H3N2 and H1N1 and an influenza B virus. Although cell culture-grown influenza vaccine is now available, the majority of manufacturers still produce the annual influenza vaccine in eggs because of the high yields from this source. As clinical isolates chosen for potential inclusion in the vaccine often grow to only low titers in eggs, these viruses are first manipulated by specialist laboratories to improve egg growth and, thus, antigen yield. This process, using gene reassortment, was first described in the 1960s (8) and occurs when an egg is infected with both the seasonal isolate and an egg-adapted high-growth parent virus. Coinfected cells contain copies of both of the viral genomes, each comprising eight independent segments of viral RNA (vRNA), from which a range of viral progeny can potentially be packaged and released (9, 10). Reassortant viruses containing the antigenic HA and NA of the seasonal strain are selected in the presence of antisera to the HA and NA of the egg-adapted strain. The viruses predominating after egg passage of the antibody-selected population will have the high-growth properties of the egg-adapted virus and, thus, some gene segments from this parent. From this population, a virus is cloned by limiting dilution and stored as a seed for vaccine production.
While this “classical reassortment” process has been used for over 40 years, the factors driving the selection of genes other than HA/NA are still not well understood (11–13). Nor have detailed investigations into the impact of different gene constellations of reassortants on viral growth or antigen yields been reported (13, 14). A strong bias for the selection of viruses with the HA and NA genes alone or the HA and NA genes and one other gene from the seasonal virus parent has been noted, with the most frequently selected additional gene of the seasonal virus being the PB1 gene (11, 12). Here, we examine the gene constellations of previously selected candidate vaccine seed strains and confirm the dominance of particular genotypes. We then investigate whether these different genotypes affect antigen yield. We used a model seasonal H3N2 strain reassorted with a high-growth H1N1 parent and also recreated the potential progeny by reverse genetics to investigate specific genotypes with respect to relative growth and antigen yields.
MATERIALS AND METHODS
Influenza A viruses.
In this study, we used the highly egg-adapted A/Puerto Rico/8/34 (PR8; H1N1) virus that is used in classical reassortment to produce high-yield H3N2 viruses for vaccine production. The A/Udorn/307/72 (Udorn; H3N2) virus was used as a model seasonal isolate. These viruses were subjected to classical reassortment using a modification of the method devised by Kilbourne (9) as described below. A clone of the virus containing Udorn HA, NA, and PB1 with the remaining genes from PR8, referred to as PR8(Ud-HA,NA,PB1), was isolated using limiting dilution in eggs as a result of this process. Similar nomenclature is used below for all wild-type and reverse genetics-derived viruses. Other viral progeny present in the allantoic fluid prior to limiting dilution were cloned by plaque formation, and the origins of the HA, NA, and PB1 genes were determined using quantitative reverse transcriptase PCR (RT-PCR).
Eight-plasmid reverse genetics (15) was used to generate viruses corresponding to the parent PR8 and Udorn viruses and the PR8(Ud-HA,NA), PR8(Ud-HA,NA,PB1), PR8(Ud-HA,NA,PB2), and PR8(Ud-HA,NA,PA) viruses. Rescued viruses were passaged from transfection supernatants in 10- to 12-day-old embryonated hen's eggs. All viruses were then inoculated into multiple eggs at a constant dose of infectious virus (100 PFU/egg), and the allantoic fluid was stored separately for analysis.
Madin-Darby canine kidney (MDCK) cell-derived viruses were prepared by infecting cells at a multiplicity of infection (MOI) of 3 PFU/cell for 1 h at 37°C. Following absorption, the inoculum was removed and cells were washed with serum-free RPMI 1640 (Gibco, Gaithersburg, MD, USA) supplemented with antibiotics. Cells were incubated at 37°C and 5% CO2, and cell culture supernatants were harvested at various times postinfection as specified in the figures.
Classical reassortment.
Ten-day-old embryonated hen's eggs were coinfected with PR8 and Udorn and incubated for 3 days. Allantoic fluid was harvested and passaged twice in eggs in the presence of polyclonal antiserum to purified PR8 virus, raised in goats, in order to remove viruses containing the HA and NA from the high-growth parent. On the first passage, eggs were exposed to the virus for 1 h prior to the addition of the antiserum to the eggs, whereas for the second passage, the virus was preincubated with the antiserum for 1 h and the mixture subsequently inoculated into eggs. The eggs were incubated for 48 h, after which the allantoic fluid was harvested and used as a source of reassorted viruses.
Virus quantitation.
Hemagglutination assays were carried out with 1% chicken erythrocytes using the original method (16) adapted to microtiter plates. The infectious viral titer was determined by the quantitation of plaques on confluent MDCK cell monolayers as previously described (17). Viral particles were assumed to contain one copy of each vRNA segment for purposes of enumeration (18). vRNA was detected by quantitative RT-PCR of RNA released from virions in clarified allantoic fluid after 0.05% Triton X-100 disruption (adapted from reference 19) or extracted from cell culture supernatant using the QIAamp viral RNA minikit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions.
Quantitative RT-PCR.
Detection of vRNA and mRNA was undertaken by polarity-specific quantitative RT-PCR using a TaqMan one-step RT-PCR master mix kit (Applied Biosystems, Carlsbad, CA, USA). Each 25-μl reaction mixture contained 5 μl of RNA, 12.5 μl of 2× AmpliTaq gold DNA polymerase mix, 0.625 μl of 40× RT enzyme mix, 1.25 μl of each 10 μM gene-specific forward or reverse primer, and 0.25 μl of 25 μM gene-specific probe. The RT reaction mixture was incubated for 30 min at 50°C. Following the reverse transcription step, 1.25 μl of the primer of opposite polarity at 10 μM was added. Amplification and detection were performed using an Applied Biosystems 7500 fast RT-PCR system. In all quantitative RT-PCR assays, serially diluted plasmids containing the corresponding influenza virus genes at known copy numbers were used as standards. The reaction conditions, primers (Geneworks, Adelaide, South Australia, Australia), and TaqMan (Applied Biosystems) probe sequences are available on request.
Minigenome assay for polymerase activity.
A β-lactamase reporter assay (20) was used to compare the activities of viral polymerase complexes. Briefly, pCAGGS-BLA (10 ng/well) was transfected into a subconfluent monolayer of 293T cells grown in 96-well plates, together with 10 ng of four pHW2000 plasmids, each expressing one of the three influenza virus polymerase genes (PB1, PB2, or PA) or the nucleoprotein (NP) gene. Transfected cells were incubated at 35°C and 5% CO2. At 24 h posttransfection, the β-lactamase produced was detected, after lysis of cells, by the addition of the LyticBLazer-FRET (Förster resonance energy transfer) B/G substrate (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. β-Lactamase cleaves the substrate, turning it from green to blue as measured by optical density with excitation at 405 nm and emission at 445 nm and 520 nm. Specific β-lactamase activity and, thus, relative polymerase activity, was calculated as follows: (520 nm/445 nm ratio of the sample)/(520 nm/445 nm ratio of BLA plasmid alone).
Electrophoresis of viral proteins and Western blot analysis.
The relative viral protein contents of infected MDCK cells, semipurified allantoic fluid (clarified by ultracentrifugation, pelleted at 31,000 × g, and resuspended in phosphate-buffered saline [PBS]), and monovalent split vaccine preparations were analyzed. Samples were disrupted in SDS sample buffer (Invitrogen) for 10 min at 95°C in the presence or absence of reducing agent. Split virus preparations were then deglycosylated with PNGaseF (peptide N-glycosidase F) according to the manufacturer's instructions (Sigma-Aldrich, St. Louis, MO, USA). Samples were separated on either precast 4 to 20% Tris-Glycine gels or 10% Bis-Tris gels in the presence or absence of the NuPAGE antioxidant.
Following electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) membranes using the iBlot dry blotting system (Invitrogen). The membranes were blocked using 1% casein in PBS for 1 h at room temperature and probed for 1 h at room temperature with monoclonal antibodies recognizing influenza A virus NP (clone F8; Abd Serotech), influenza A virus M1 (clone GA2B; Abd Serotech), Udorn HA (clone 36; previously characterized in reference 21), or the C-terminal region of the HA2 protein (clone FLUB25.10C9.3C10; CSL Ltd.). Blots were washed and incubated for 1 h with a horseradish peroxidase (HRP)-conjugated secondary antibody and were developed using an Opti-4CN substrate kit (Bio-Rad) according to the manufacturer's instructions. Densitometry of Western blots was performed using ImageQuant TL software.
Statistical analysis.
Data were analyzed for statistical significance using either a one-way analysis of variance (ANOVA) with Tukey's multiple comparison posttest or a two-way ANOVA with the Bonferroni posttest, calculated using Prism 5 software (GraphPad Software, Inc., San Diego, CA, USA).
RESULTS
The seasonal PB1 gene is selected through classical reassortment at relatively high frequency in vaccine seed viruses.
The gene composition of potential influenza virus candidate vaccine seed strains derived by classical reassortment from 2003 to 2009 was determined (Table 1). The data reveal that the majority of the genes from the highly egg-adapted parent are required for the high-yield phenotype. During reassortment of H3N2 seasonal viruses with the egg-adapted parent, the PB2, PA, and NS genes of the seasonal parent were selected on only 7%, 13%, and 13% of occasions, respectively, and the seasonal NP and M genes were never selected (Table 2). In all five of the H1N1 events analyzed, the seasonal PB2, PA, NP, M, and NS genes were never selected. For both subtypes, the PB1 gene of the seasonal strain was present at much higher frequency than other seasonal genes: 50% in H3N2 and 60% in H1N1 seed viruses.
Table 1.
Genotypes of viruses selected through classical reassortment in a retrospective study of potential vaccine seed strains from 2003 to 2009
| Parental seasonal isolate | Reassortant | Sub type | Sourcea of indicated gene |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | |||
| A/Wyoming/3/03 | X147 | H3N2 | P | P | P | S | P | S | P | P |
| A/Wyoming/3/03 | IVR-134 | H3N2 | P | S | P | S | P | S | P | P |
| A/California/7/04 | IVR-141 | H3N2 | P | S | P | S | P | S | P | P |
| A/New York/55/04 | X157 | H3N2 | P | P | P | S | P | S | P | P |
| A/New York/55/04 | X157b | H3N2 | P | P | P | S | P | S | P | P |
| A/Wellington/1/04 | IVR-139 | H3N2 | P | P | P | S | P | S | P | P |
| A/Hiroshima/52/05 | IVR-142 | H3N2 | P | S | P | S | P | S | P | P |
| A/Wisconsin/67/05 | IVR-143 | H3N2 | P | S | P | S | P | S | P | P |
| A/Wisconsin/67/05 | X161 | H3N2 | P | S | S | S | P | S | P | S |
| A/Nepal/921/06b | H3N2 | P | S | S | S | P | S | P | P | |
| A/Victoria/500/06 | IVR-144 | H3N2 | P | S | P | S | P | S | P | S |
| A/Brisbane/10/07 | IVR-147 | H3N2 | S | P | P | S | P | S | P | P |
| A/Uruguay/716/07 | IVR-149 | H3N2 | P | P | P | S | P | S | P | P |
| A/Brisbane/24/08 | IVR-151 | H3N2 | ND | P | P | S | P | S | P | P |
| A/Victoria/210/09 | IVR-155 | H3N2 | P | S | P | S | P | S | P | P |
| A/Singapore/37/09b | H3N2 | P | P | P | S | P | S | P | P | |
| A/Fukishima/141/06 | IVR-146 | H1N1 | P | S | P | S | P | S | P | P |
| A/Brisbane/59/07 | IVR-148 | H1N1 | P | S | P | S | P | S | P | P |
| A/Guam/1/07 | IVR-150 | H1N1 | P | P | P | S | P | S | P | P |
| A/Pennsylvania/8/08 | IVR-152 | H1N1 | P | P | P | S | P | S | P | P |
| A/California/7/09 | X181 | H1N1 | P | S | P | S | P | S | P | P |
P, gene originates from high-growth virus; S, gene originates from seasonal virus; ND, not determined.
Virus was not made as a seedlot virus and, hence, was not allocated a reassortant number.
Table 2.
Frequencies of genes in viruses selected through classical reassortment in our retrospective study of potential vaccine seed strains from 2003 to 2009 and in reassortant viruses from 1968 to 2009 reported in the literature
| Source of reassortment events | Viral subtype | No. of times gene originated from seasonal parent/total no. of reassortants |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | ||
| Analyzed in this study | H3N2 | 1/15 | 8/16 | 2/16 | 16/16 | 0/16 | 16/16 | 0/16 | 2/16 |
| H1N1 | 0/5 | 3/5 | 0/5 | 5/5 | 0/5 | 5/5 | 0/5 | 0/5 | |
| Reported in the literaturea | H3N2 | 13/92 | 41/92 | 11/92 | 92/92 | 19/92 | 91/92 | 4/92 | 9/92 |
| H1N1 | 1/24 | 15/24 | 1/24 | 24/24 | 3/24 | 24/24 | 0/24 | 1/24 | |
| Combined totalb | H3N2 | 14/107 | 49/108 | 13/108 | 108/108 | 19/108 | 107/108 | 4/108 | 11/108 |
| H1N1 | 1/29 | 18/29 | 1/29 | 29/29 | 3/29 | 29/29 | 0/29 | 1/29 | |
The reassortant viruses analyzed from the literature were collated from references 11–14, 22, and 23.
Combined data from this and previously published studies.
The genotypes of a number of reassortant viruses selected as potential vaccine strains from 1968 to the present were also collated from the literature (11–14, 22, 23). Of the additional 92 H3N2 and 24 H1N1 reassortant viruses analyzed, 45% and 63%, respectively (in total, 48%) contained the seasonal PB1 gene (Table 2). As with our own observations, selection for other seasonal genes was less frequent than for PB1 (NP, 19%; PB2, 12%; PA, 10%; NS, 9%; and M, 3%). This broader analysis, incorporating other reassortment and selection events, confirmed that, while the incorporation of other genes of the seasonal virus occurred only with low frequencies, the seasonal PB1 gene was present in about half (67/137) of the potential vaccine seeds. This might indicate that the PB1 of the egg-adapted parent does not contribute to the high-growth phenotype and so this gene is selected randomly. However, for each individual reassortment event, viruses with either the seasonal or the egg-adapted PB1 dominated and were selected.
Udorn PB1 was selected at a higher frequency than was PR8 PB1 in a model system of classical reassortment.
To investigate the selection of PB1 for a particular combination of seasonal virus and high-growth parent in more detail, classical reassortment was undertaken between the model seasonal strain Udorn and the high-growth parent strain PR8. Following the second antiserum passage to select for the HA and NA of Udorn virus, progeny viruses were isolated by plaque assay and the origins of the HA, NA, and PB1 genes were determined for each plaque by virus-specific RT-PCR. Of the 72 cloned viruses, each of which contained Udorn HA and NA, 54 (75%) contained Udorn PB1, whereas only 18 (25%) contained PR8 PB1, mimicking a reassortment event where viruses containing the seasonal PB1 dominated the viral progeny.
The presence of the Udorn PB1 gene resulted in a low-HA-yield virus.
The wild-type parental Udorn and PR8 viruses and the virus PR8(Ud-HA,NA,PB1), which was selected by the classical reassortment procedure following limiting dilution in eggs, were inoculated into replicate eggs, and the virus yield from each egg determined by HA titration (Fig. 1A). The seasonal parent virus had a significantly lower mean HA titer than did the high-growth parent (P < 0.001, one-way ANOVA), while the reassortant showed an intermediate mean HA titer, with this virus not being significantly different from the Udorn parent, indicating that the reassortment process did not substantially improve the HA yield.
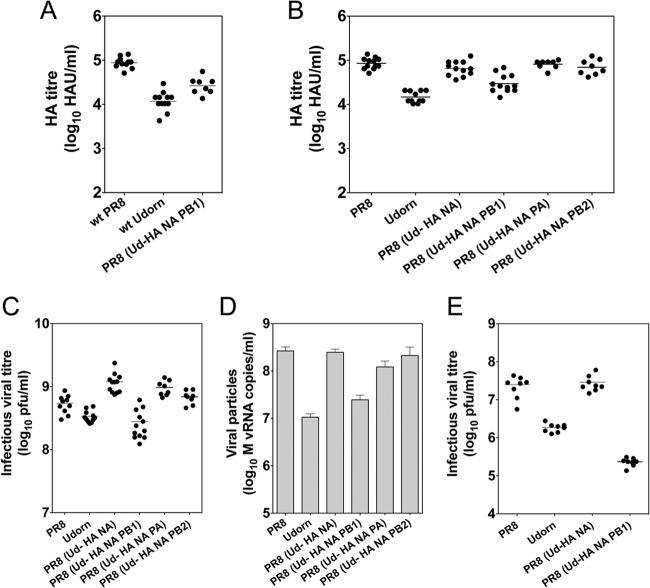
Fig 1.
The genetic makeup of reassortant viruses affects HA titer and viral growth. The hemagglutination titers of individual eggs infected with a constant dose (100 PFU) of the wild-type (wt) virus parents PR8 and Udorn were compared to those of the PR8(Ud-HA,NA,PB1) virus selected by classical reassortment of the two parental viruses (A). The hemagglutination titers (B) and infectious virus titers, determined by plaque formation in MDCK cells (C), were measured in eggs infected with a constant dose (100 PFU) of the reverse genetics-derived viruses corresponding to the PR8 and Udorn parent viruses and additional viruses containing Udorn HA and NA with or without the Udorn PB1, PB2, or PA (with the remaining genes originating from PR8). The titers of viral particles (D), measured using copy numbers of M vRNA in the allantoic fluid samples used in the assay whose results are shown in panel C, were determined by quantitative RT-PCR as a surrogate. The yields of infectious virus from individual MDCK cell cultures infected with 100 PFU of virus were determined by plaque assay of cell culture supernatants (E). In panels A, B, C, and E, each symbol represents the titer of an individual egg or culture supernatant and the line is the geometric mean titer. In panel D, each bar represents the mean particle number of all the eggs in that group and the error bar shows the standard error of the mean.
Reverse genetics was used to produce the equivalent viruses and, also, other viruses with gene constellations that were not selected during the classical reassortment procedure. The viruses were also grown in replicate eggs and the individual HA titers of the allantoic fluids determined (Fig. 1B). Reverse genetics-derived parents and PR8(Ud-HA,NA,PB1) virus exhibited HA titers similar to those seen in the experiments whose results are shown in Figure 1A. However, the PR8(Ud-HA,NA) virus containing PB1 from the high-growth parent had HA titers that were significantly increased compared to those in Udorn virus (P < 0.001, one-way ANOVA) and more comparable to those of PR8 virus.
As the PB1 protein is the core component of the heterotrimeric influenza virus polymerase complex, which is responsible for the transcription and replication of the viral genome, we examined whether the inclusion of the other components of the polymerase complex, PB2 and PA, would have similar effects on HA yield (Fig. 1B). PR8(Ud-HA,NA,PB2) and PR8(Ud-HA,NA,PA) showed HA titers comparable to those of PR8(Ud-HA,NA) (P > 0.05, one-way ANOVA) rather than to the HA titer of PR8(Ud-HA,NA,PB1) (P < 0.01, one-way ANOVA), indicating that inclusion of the seasonal PB1 but not other polymerase subunits negates the high-yield phenotype.
The presence of the Udorn PB1 gene resulted in a low-growth virus.
To establish whether differences observed in HA titer were due to differences in viral replication, we measured the infectious viral titers (Fig. 1C) and particle numbers (Fig. 1D) in the allantoic fluid samples by plaque formation and quantitative RT-PCR of the M gene, respectively. The PR8(Ud-HA,NA,PB1) virus showed 4-fold less infectious virus (P < 0.0001, one-way ANOVA) and 8-fold fewer viral particles (P < 0.05, one-way ANOVA) than the PR8(Ud-HA,NA) virus. The PR8(Ud-HA,NA,PB2) and PR8(Ud-HA,NA,PA) viruses showed growth characteristics similar to those of PR8(Ud-HA,NA) virus.
Thus, consistently across all three measurements (HA titer, infectious virus titer, and virus particle number), the inclusion of Udorn PB1 in the model vaccine seed virus significantly reduced viral yield, despite the selection of this virus by the classical reassortment process. The reduced-growth phenotype was unique to the inclusion of Udorn PB1 and was not due to the presence of a hybrid polymerase complex per se. The phenotypes observed were not specific to egg-grown viruses but were also observed for viruses grown in MDCK cells (Fig. 1E).
The presence of the Udorn PB1 gene did not alter vRNA and viral mRNA production in infected MDCK cells.
To examine whether the poor growth of the PR8(Ud-HA,NA,PB1) virus was due to a poorly functioning polymerase, a minigenome assay was used. This involved the transfection of a β-lactamase reporter gene along with the viral polymerase and NP genes, which together with the vRNA make up the ribonucleoprotein (RNP) complex. The intrinsic activities of the Udorn and PR8 RNPs, as well as RNPs with either Udorn NP or PB1 and the remaining proteins of PR8, were examined (Fig. 2A). The Udorn RNP was approximately two-thirds as active as the PR8 RNP (mean of three experiments, 66%; range, 62.5 to 70%). This appeared to be irrespective of the source of the NP and, therefore, independent of polymerase complex-NP interactions. Furthermore, the presence of Udorn PB1 in an otherwise PR8 RNP reduced the polymerase activity to levels not statistically different from that of the Udorn RNP (P > 0.05 in each of 3 experiments). These data imply that reduced intrinsic activity of this hybrid polymerase complex relative to the activity of the PR8 polymerase complex may be responsible for the poorer replication of PR8(Ud-HA,NA,PB1) virus. Nevertheless, comparison of the levels of production of vRNA or viral mRNA in MDCK cells after infection with PR8(Ud-HA,NA) virus or PR8(Ud-HA,NA,PB1) virus showed this to be unlikely (Fig. 2B to D). Whole RNA was extracted from infected MDCK cells, and the amount of M vRNA, as an indicator of overall vRNA production, was determined by quantitative RT-PCR (Fig. 2B). No statistically significant difference was observed between the two viruses in the copy number of the M vRNA produced (P > 0.05, two-way ANOVA). This observation also held true for the production of NP, PA, and NS vRNA (data not shown). The ability of the polymerase complexes from the two different viruses to produce mRNA following infection of MDCK cells was also measured. No statistically significant difference was observed in the ability of the polymerase complexes to produce matrix M1/M2 mRNA from the M gene (Fig. 2C), HA mRNA (Fig. 2D), or NS1 mRNA (data not shown). These findings indicate that, despite the polymerase complex of PR8(Ud-HA,NA,PB1) virus having a lower intrinsic activity than the polymerase complex of PR8(Ud-HA,NA) virus, this did not result in a difference in the production of vRNA or viral mRNA in infected cells.
Fig 2.
The composition of the RNP complex affects the intrinsic polymerase activity in a minigenome assay but not the production of vRNA or viral mRNA in infected cells. (A) Minigenome assays were performed in 293T cells transfected with the pCAGGS-BLA reporter gene and pHW2000 plasmids that each expressed a component of the influenza virus RNP complex. The polymerase activities of the PR8 and Udorn RNP complexes were measured, in addition to those of the complexes expressing the Udorn NP or PB1 with the remaining RNP proteins from PR8, referred to as PR8(Ud-NP) RNP and PR8(Ud-PB1) RNP, respectively. Each bar represents the mean of three independent experiments, and the error bars show the standard errors of the means, each normalized to the activity of the PR8 RNP complex within each experiment. (B to D) MDCK cells were infected with PR8(Ud-HA,NA) virus (white) or PR8(Ud-HA,NA,PB1) virus (gray) at an MOI of 3 PFU per cell. Total RNA was extracted from 1 × 106 infected cells at 0, 4, 8, and 24 h after a 1-h virus absorption period, and the copy numbers of the M vRNA (B) and the viral mRNA resulting from the transcription of the M gene (M1 and M2 mRNA) (C) and HA mRNA (D) were assessed by quantitative RT-PCR. Each bar represents the geometric mean of three independent experiments, and the error bars represent the standard errors of the means.
The inclusion of Udorn PB1 results in altered ratios of viral proteins.
The readout from the minigenome assay and the output of progeny virus from infected cells is dependent not only on nucleic acid production but also on viral protein production, suggesting that the levels of translation of viral proteins should also be investigated. It is well known that preferential translation of viral mRNA over cellular mRNA occurs in infected cells (reviewed in reference 24), and recently, the viral polymerase complex has been shown to play a role in this process (25). The effect of the inclusion of Udorn PB1 in the model seed virus on protein synthesis in infected cells was therefore examined. Western blot analysis of infected MDCK cells (Fig. 3) revealed that PR8(Ud-HA,NA,PB1)-infected cells contained approximately 2-fold more HA protein than PR8(Ud-HA,NA)-infected cells at 4, 8, and 24 h postinfection (Fig. 3A and B). In contrast, similar levels of M1 protein were observed in cells infected with the two viruses (Fig. 3A and C). In the absence of differences in transcription levels, these data suggest that the inclusion of Udorn PB1 resulted in differential translation of viral mRNA to yield higher levels of HA but not M1, resulting in an increased HA/M1 protein ratio within infected cells.
Fig 3.
Viral protein expression in cells infected with viruses differing in the origin of the PB1 protein. Aliquots of 1 × 105 MDCK cells infected with PR8(Ud-HA,NA) virus or PR8(Ud-HA,NA,PB1) virus from the preparations used in the experiments whose results are shown in Figure 2B, C, and D were disrupted and separated on 4 to 20% Tris-glycine gels under nonreducing conditions. Proteins were transferred to PVDF membranes and probed with anti-Udorn HA and anti-M1 monoclonal antibodies. pi, postinfection. (A). The relative contents of HA (B) and M1 (C) proteins were determined by densitometry of stained bands and analyzed using the ImageQuant TL software. White bars, PR8(Ud-HA,NA) virus; gray bars, PR8(Ud-HA,NA,PB1) virus. Each bar represents the geometric mean of two independent experiments, and the error bars represent the standard errors of the means.
The inclusion of Udorn PB1 results in a higher HA density in the virion.
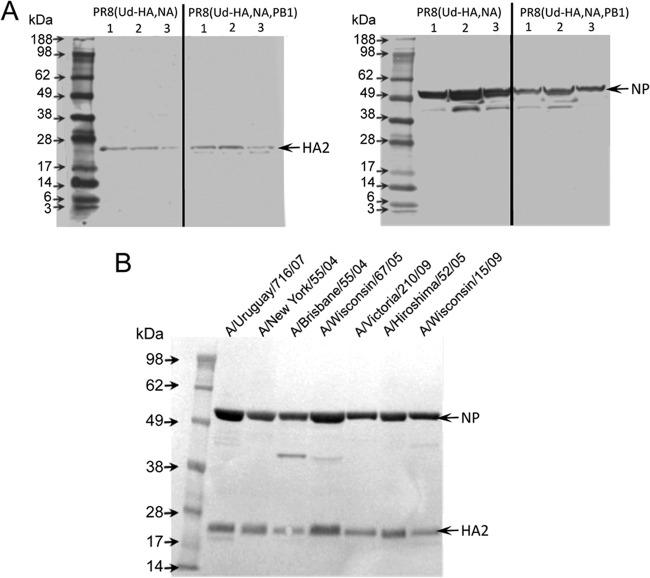
Further analysis of the yields of hemagglutination units (HAU) (Fig. 1B), PFU (Fig. 1C), and particle numbers (Fig. 1D) of PR8(Ud-HA,NA) and PR8(Ud-HA,NA,PB1) viruses showed that the inclusion of Udorn PB1 increased the ratio of HAU/PFU 2-fold (from 5.82 × 10−5 to 1.18 × 10−4) and the ratio of HAU/particle number 4-fold (from 2.81 × 10−4 to 1.34 × 10−3). This implied that the PR8(Ud-HA,NA,PB1) virus, in addition to yielding more HA in infected cells, also has a higher density of HA in the virion. To confirm this, the ratios of HA/NP in semipurified allantoic fluid samples of PR8(Ud-HA,NA) and PR8(Ud-HA,NA,PB1) viruses were analyzed by Western blotting (Fig. 4A). Densitometry measurements (Table 3) revealed that the PR8(Ud-HA,NA) virus sample had 2-fold more NP than the PR8(Ud-HA,NA,PB1) virus sample but 2-fold less HA protein. PR8(Ud-HA,NA,PB1) virions therefore had a 4-fold greater HA/NP protein ratio than PR8(Ud-HA,NA) virions. This is consistent with the 4-fold higher HAU/particle ratio described above. These data together indicated that the inclusion of Udorn PB1 resulted in a higher HA density on the surface of the virion.
Fig 4.
Western blots used for the determination of relative viral protein contents of samples reported in Tables 3 and 4. (A) Eggs were inoculated with 100 PFU of either PR8(Ud-HA,NA) or PR8(Ud-HA,NA,PB1). Allantoic fluid was clarified by ultracentrifugation, and the virus was pelleted at 31,000 × g and then resuspended in PBS. Tris-glycine gels (4 to 20%) were loaded with 20,000 HAU of virus sample and separated by SDS-PAGE under reducing conditions. Proteins were transferred to a PVDF membrane and probed with either anti-NP or anti-HA2 monoclonal antibodies. The relative protein contents were determined by densitometry of stained bands analyzed using the ImageQuant TL software and are presented in Table 3. (B) Monovalent split virus vaccine preparations were deglycosylated, and samples containing 21 μg of viral protein, as determined by Lowry assay, were separated on 10% bis-Tris gels under reducing conditions in the presence of NuPAGE antioxidant. Proteins were transferred to a PVDF membrane and probed with either anti-NP or anti-HA2 monoclonal antibodies. The relative protein contents were determined by densitometry of stained bands analyzed using the ImageQuant TL software and are presented in Table 4.
Table 3.
Relative protein content of reassortant viruses differing in the origin of PB1
| Virus | Mean amt (±SD) ofa: |
HA/NP ratio (10−2) | |
|---|---|---|---|
| HA (×105) | NP (×106) | ||
| PR8(Ud-HA, NA) | 2.7 (± 1.7) | 18.6 (± 4.6) | 1.5 (± 1.2) |
| PR8(Ud-HA, NA, PB1) | 5.4 (± 0.4) | 8.3 (± 1.9) | 6.52 (± 4.3) |
The relative protein content of Western blots (shown in Figure 4A) was determined by densitometry of stained bands and analyzed using ImageQuant TL software. Relative units of band intensity (arbitrary values) are expressed as the mean of three different egg preparations.
The inclusion of the seasonal PB1 gene in vaccine seed viruses selected through classical reassortment results in higher HA density in virions.
To investigate whether phenotypic differences observed in the HA density of the model vaccine seed viruses were representative of vaccine candidate seed viruses selected through classical reassortment, Western blot analysis of purified monovalent split vaccine virus preparations was undertaken (Fig. 4B). Again, HA/NP ratios were calculated. In the vaccine preparations analyzed, those containing PR8 PB1 had HA/NP ratios between 1.63 and 3.86, whereas the inclusion of the seasonal PB1 resulted in higher HA/NP ratios of between 4.10 and 5.31 (Table 4). Therefore, each candidate vaccine virus containing the seasonal PB1 had a higher HA density per virion than those containing PR8 PB1, indicating that the finding obtained with the model vaccine seed strain Udorn was consistent for recent vaccine seed viruses.
Table 4.
Relative viral protein content of vaccine seed viruses
| Split virus vaccine | Seasonal virus genes | Amt ofa: |
HA/NP ratio (×10−1) | |
|---|---|---|---|---|
| HA protein (×106)a | NP protein (×107) | |||
| A/New York/55/2004 | HA, NA | 5.3 | 1.3 | 3.86 |
| A/Uruguay/716/2007 | HA, NA | 6.2 | 2.1 | 2.96 |
| A/Brisbane/10/2007 | HA, NA, PB2 | 1.9 | 1.2 | 1.63 |
| A/Wisconsin/15/2009 | HA, NA | 3.7 | 1.3 | 2.90 |
| A/Wisconsin/67/2005 | HA, NA, PB1 | 9.2 | 1.7 | 5.31 |
| A/Hiroshima/52/2005 | HA, NA, PB1 | 6.5 | 1.4 | 4.72 |
| A/Victoria/210/2009 | HA, NA, PB1 | 4.7 | 1.1 | 4.10 |
The relative protein content of the Western blot shown in Figure 4B was determined by densitometry of stained bands and analyzed using the ImageQuant TL software. Relative units of band intensity (arbitrary values) are shown.
DISCUSSION
In this study, we have investigated the prevalence of the seasonal PB1 gene in vaccine seed strains and found that, compared to other non-HA and NA genes, it was present at much higher frequencies (approximately 50%), confirming the observations of others (11–14, 22, 23). We also established that Udorn undergoing classical reassortment with the highly egg-adapted PR8 virus modeled a situation where the seasonal PB1 dominated the high-growth progeny viruses. This model revealed that the inclusion of the H3N2 seasonal PB1 gene in the model seed virus increased the HA protein per virion 4-fold compared to its levels when the PR8 PB1 gene was included but also reduced the virus particle production 8-fold. A 2-fold difference in the yield of HA protein was thus observed in egg-grown virus. In the context of vaccine production, this is highly significant and translates to a requirement for double the number of eggs per manufacturing cycle to achieve the same number of vaccine doses, for the H3N2 component at least.
As vaccine seed strains are selected for high-growth properties (11), as well as for the presence of the seasonal HA and NA, one might assume that when viruses containing the seasonal PB1 gene were selected, they would exhibit higher-growth properties than other possible alternative progeny. However, in the model system, the comparison of potential progeny derived by reverse genetics revealed that the inclusion of Udorn PB1 had a significantly negative effect on the viral growth compared to the inclusion of PR8 PB1, resulting in lower yields of viral particles, infectious virions, and HAU. PB1, as part of the trimeric polymerase complex, is known to drive the transcription and replication of the viral genome (26), and previous studies have shown that viral growth can be altered by the presence of a hybrid polymerase containing components from viruses of different subtypes (27, 28). Poor growth as a result of hybrid polymerase complexes has been attributed to reductions in vRNA replication or mRNA transcription that limit the ability of viruses to package (29). As in other studies (27, 28, 30), we used a minigenome reporter assay to measure the activity of hybrid polymerase complexes, and we showed a reduction in polymerase activity when the Udorn PB1 gene was included in PR8 RNP complexes. However, we showed that this reduced intrinsic polymerase activity did not result in a decrease in the level of vRNA or mRNA in cells infected with the corresponding viruses. Therefore, differences in the growth characteristics could not be attributed to limiting amounts of viral genome available for packaging or of mRNA to produce viral protein.
Despite the poorer growth, PR8(Ud-HA,NA,PB1) exhibited 4-fold higher HAU/particle and HA/NP protein ratios than PR8(Ud-HA,NA). Retrospective analysis of seven H3N2 viruses selected as candidate vaccine strains also exhibited similar patterns of protein expression, with those containing the seasonal PB1 gene having higher HA/NP ratios, suggesting that these viruses also had higher HA protein content in virions. In addition, a similar trend of poor growth but increased HA/NP ratio was described when the PB1 of an H5N1 virus was included with its HA and NA in reverse genetics-derived H5N1 vaccine candidates containing all other genes from PR8 virus (31).
Supporting our findings, a recent study (32) used cryoelectron microscopy to determine the effect of gene constellation on virion morphology. The study showed that the inclusion of the PB1 protein with the HA and NA proteins from an H3N2 parent in a PR8 backbone altered the density of the surface glycoproteins. While the researchers were unable to distinguish between HA and NA on the surface of the virion, they showed more glycoprotein spikes and reduced glycoprotein spacing on the virion surface when the PB1 from the H3N2 parent was included.
The molecular mechanisms driving the increased density of HA on the virion surface upon inclusion of the seasonal PB1 gene are of interest to vaccine manufacturers, as a greater understanding could lead to further increases in antigen yield if the growth differences could be overcome. It has been reported previously (29) that the higher intrinsic viral polymerase activity of an H3N2 virus compared to that of an H1N1 virus led to enhancement of the replication and transcription of viral RNA and greater expression of the viral HA protein and its accumulation on the cell surface late during virus replication. This triggered stronger extracellular signal-regulated kinase (ERK) activation to provide more-efficient nuclear RNP export, which in turn led to an increase in the formation of infectious progeny virions (29). This contrasts with our observations with the model vaccine seed viruses, where the increased polymerase activity of the RNP of PR8(Ud-HA,NA) compared to that of PR8(Ud-HA,NA, PB1) did result in greater growth of the PR8(Ud-HA,NA) virus, but this was not due to greater viral RNA or HA production in cells. Instead, the increased HA/NP ratio in vaccine seed viruses incorporating seasonal PB1 and the greater amount of HA protein in cells infected with these viruses imply a direct link between the PB1 gene and/or protein and the amount of HA protein produced. In the absence of any difference in cellular vRNA and mRNA, the mechanism for altered amounts of HA protein is likely to act posttranscriptionally, altering viral protein translation.
Upon influenza virus infection, the cellular translation initiation factor eIF4E, which recognizes the cap of mRNA, may become dephosphorylated and so host-cellular cap-dependent translation is inhibited (33–36). Nevertheless, translation of the capped viral mRNA still occurs independently of eIF4E (37), and recently, it has been shown that the viral polymerase complex can act as a substitute for eIF4E (25). These observations enable us to understand a potential mechanism by which PB1 plays a role in modulating the translation of viral mRNA. It is known that sequences present within the 5′ untranslated region (UTR) of influenza viral genes are responsible for directing the selective translation of viral mRNA over the host cell mRNA (38) and, also, that the first 12 nucleotides, common to all viral 5′ UTRs, are known to bind the PB1 component of the influenza virus polymerase complex (39). Hence, the ability of the PB1 to bind this sequence and sequences downstream has the potential to affect translation. We propose here that it is the PB1 subunit that is critical for the ability of the viral polymerase to function as a substitute for eIF4E, because differences in HAU per particle were not observed in viruses with hybrid polymerases containing a heterologous PB2 or PA subunit. We further propose that binding of the hybrid polymerase to viral gene segment-specific nucleotide sequences downstream from the conserved 5′ UTR accounts for the selective modulation of HA translation relative to the translation of M1 and NP that we observed.
In summary, these results demonstrate that the inclusion of the seasonal PB1 gene in candidate vaccine seed strains, shown here for the H3N2 subtype, can alter the phenotypic characteristics of viral growth and HA content. Our findings indicate that the inclusion of the seasonal PB1 gene results in a decrease in viral replication but an increase in the density of HA in the virion. Our data suggest that the PB1 gene can directly modulate the relative translation of viral mRNAs, and further studies are aimed at understanding the mechanism controlling this. Our study also raises the question of why more-poorly growing viruses can, in some circumstances, dominate the viral progeny after classical reassortment.
ACKNOWLEDGMENTS
This work was supported by project grant 509281 from the National Health and Medical Research Council of Australia and by CSL Ltd.
We thank Jesse Bodle for help with the Western blotting technique and St. Jude Children's Hospital, Memphis, TN, for providing the pHW2000 plasmid for reverse genetics.
Footnotes
Published ahead of print 6 March 2013
REFERENCES
- 1. World Health Organization 2009. Influenza (seasonal). Fact sheet no. 211. http://www.who.int/mediacentre/factsheets/fs211/en/index.html
- 2. Nordin J, Mullooly J, Poblete S, Strikas R, Petrucci R, Wei F, Rush B, Safirstein B, Wheeler D, Nichol KL. 2001. Influenza vaccine effectiveness in preventing hospitalizations and deaths in persons 65 years or older in Minnesota, New York, and Oregon: data from 3 health plans. J. Infect. Dis. 184:665–670 [DOI] [PubMed] [Google Scholar]
- 3. Couch RB, Kasel JA. 1983. Immunity to influenza in man. Annu. Rev. Microbiol. 37:529–549 [DOI] [PubMed] [Google Scholar]
- 4. Wiley D, Wilson I, Skehel J. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373. [DOI] [PubMed] [Google Scholar]
- 5. Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza-virus A/PR/8/34 hemagglutinin (H-1 subtype). Cell 31:417–427 [DOI] [PubMed] [Google Scholar]
- 6. Webster RG, Laver WG, Air GM, Schild GC. 1982. Molecular mechanisms of variation in influenza viruses. Nature 296:115–121 [DOI] [PubMed] [Google Scholar]
- 7. Laver WG, Air GM, Webster RG, Markoff LJ. 1982. Amino acid sequence changes in antigenic variants of type A influenza virus N2 neuraminidase. Virology 122:450–460 [DOI] [PubMed] [Google Scholar]
- 8. Kilbourne ED, Murphy JS. 1960. Genetic studies of influenza viruses. I. Viral morphology and growth capacity as exchangeable genetic traits. Rapid in ovo adaptation of early passage Asian strain isolates by combination with PR8. J. Exp. Med. 111:387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kilbourne ED. 1969. Future influenza vaccines and the use of genetic recombinants. Bull. World Health Org. 41:643–645 [PMC free article] [PubMed] [Google Scholar]
- 10. Kilbourne ED, Schulman JL, Schild GC, Schloer G, Swanson J, Bucher D. 1971. Related studies of a recombinant influenza-virus vaccine. I. Derivation and characterization of virus and vaccine. J. Infect. Dis. 124:449–462 [DOI] [PubMed] [Google Scholar]
- 11. Bergeron C, Valette M, Lina B, Ottmann M. 2010. Genetic content of influenza H3N2 vaccine seeds. PLoS Curr. 2:RRN1165 doi:10.1371/currents.RRN1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fulvini AA, Ramanunninair M, Le J, Pokorny BA, Arroyo JM, Silverman J, Devis R, Bucher D. 2011. Gene constellation of influenza A virus reassortants with high growth phenotype prepared as seed candidates for vaccine production. PLoS One 6:e20823 doi:10.1371/journal.pone.0020823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oxford JS, Corcoran T, Newman R, Major D, Schild GC. 1986. Biochemical and antigenic analysis using monoclonal antibodies of a series of influenza A (H3N2) and (H1N1) virus reassortants. Vaccine 4:9–14 [DOI] [PubMed] [Google Scholar]
- 14. Baez M, Palese P, Kilbourne ED. 1980. Gene composition of high-yielding influenza vaccine strains obtained by recombination. J. Infect. Dis. 141:362–365 [DOI] [PubMed] [Google Scholar]
- 15. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Groth FdS, Webster RG. 1966. Disquisitions of original antigenic sin. I. Evidence in man. J. Exp. Med. 124:331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tannock GA, Paul JA, Barry RD. 1984. Relative immunogenicity of the cold-adapted influenza virus A/Ann Arbor/6/60 (A/AA/6/60-ca), recombinants of A/AA/6/60-ca, and parental strains with similar surface antigens. Infect. Immun. 43:457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei Z, McEvoy M, Razinkov V, Polozova A, Li E, Casas-Finet J, Tous GI, Balu P, Pan AA, Mehta H, Schenerman MA. 2007. Biophysical characterization of influenza virus subpopulations using field flow fractionation and multiangle light scattering: correlation of particle counts, size distribution and infectivity. J. Virol. Methods 144:122–132 [DOI] [PubMed] [Google Scholar]
- 19. van Elden LJ, Nijhuis M, Schipper P, Schuurman R, van Loon AM. 2001. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 39:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cavrois M, De Noronha C, Greene WC. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151–1154 [DOI] [PubMed] [Google Scholar]
- 21. Brown LE, Murray JM, White DO, Jackson DC. 1990. An analysis of the properties of monoclonal antibodies directed to epitopes on influenza virus hemagglutinin. Arch. Virol. 114:1–26 [DOI] [PubMed] [Google Scholar]
- 22. Florent G. 1980. Gene constellation of live influenza A vaccines. Arch. Virol. 64:171–173 [DOI] [PubMed] [Google Scholar]
- 23. Xu X, Kilbourne ED, Hall HE, Cox NJ. 1994. Nonimmunoselected intrastrain genetic variation detected in pairs of high-yielding influenza A (H3N2) vaccine and parental viruses. J. Infect. Dis. 170:1432–1438 [DOI] [PubMed] [Google Scholar]
- 24. Yángüez E, Nieto A. 2011. So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus Res. 156:1–12 [DOI] [PubMed] [Google Scholar]
- 25. Yángüez E, Rodriguez P, Goodfellow I, Nieto A. 2012. Influenza virus polymerase confers independence of the cellular cap-binding factor eIF4E for viral mRNA translation. Virology 422:297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. González S, Ortín J. 1999. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18:3767–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li C, Hatta M, Nidom CA, Muramoto Y, Watanabe S, Neumann G, Kawaoka Y. 2010. Reassortment between avian H5N1 and human H3N2 influenza viruses creates hybrid viruses with substantial virulence. Proc. Natl. Acad. Sci. U. S. A. 107:4687–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murakami S, Horimoto T, Mai LQ, Nidom CA, Chen H, Muramoto Y, Yamada S, Iwasa A, Iwatsuki-Horimoto K, Shimojima M, Iwata A, Kawaoka Y. 2008. Growth determinants for H5N1 influenza vaccine seed viruses in MDCK cells. J. Virol. 82:10502–10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marjuki H, Yen Franks H-LJ, Webster RG, Pleschka S, Hoffmann E. 2007. Higher polymerase activity of a human influenza virus enhances activation of the hemagglutinin-induced Raf/MEK/ERK signal cascade. Virol. J. 4:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li C, Hatta M, Watanabe S, Neumann G, Kawaoka Y. 2008. Compatibility among polymerase subunit proteins is a restricting factor in reassortment between equine H7N7 and human H3N2 influenza viruses. J. Virol. 82:11880–11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abt M, de Jonge J, Laue M, Wolff T. 2011. Improvement of H5N1 influenza vaccine viruses: influence of internal gene segments of avian and human origin on production and hemagglutinin content. Vaccine 29:5153–5162 [DOI] [PubMed] [Google Scholar]
- 32. Moulès V, Terrier O, Yver M, Riteau B, Moriscot C, Ferraris O, Julien T, Giudice E, Rolland Erny J-PA, Bouscambert-Duchamp M, Frobert E, Rosa-Calatrava M, Pu Lin Y, Hay A, Thomas D, Schoehn G, Lina B. 2011. Importance of viral genomic composition in modulating glycoprotein content on the surface of influenza virus particles. Virology 414:51–62 [DOI] [PubMed] [Google Scholar]
- 33. Scheper GC, Proud CG. 2002. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 269:5350–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuberek J, Jemielity J, Jablonowska A, Stepinski J, Dadlez M, Stolarski R, Darzynkiewicz E. 2004. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5′ terminus. Biochemistry 43:5370–5379 [DOI] [PubMed] [Google Scholar]
- 35. Ling J, Morley SJ, Traugh JA. 2005. Inhibition of cap-dependent translation via phosphorylation of eIF4G by protein kinase Pak2. EMBO J. 24:4094–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ross G, Dyer JR, Castellucci VF, Sossin WS. 2006. Mnk is a negative regulator of cap-dependent translation in Aplysia neurons. J. Neurochem. 97:79–91 [DOI] [PubMed] [Google Scholar]
- 37. Burgui I, Yanguez E, Sonenberg N, Nieto A. 2007. Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 81:12427–12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garfinkel MS, Katze MG. 1993. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5′-untranslated region. J. Biol. Chem. 268:22223–22226 [PubMed] [Google Scholar]
- 39. González S, Ortín J. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]