Abstract
Viruses that replicate in the cytoplasm cannot access the host nuclear capping machinery. These viruses have evolved viral methyltransferase(s) to methylate N-7 and 2′-O cap of their RNA; alternatively, they “snatch” host mRNA cap to form the 5′ end of viral RNA. The function of 2′-O methylation of viral RNA cap is to mimic cellular mRNA and to evade host innate immune restriction. A cytoplasmic virus defective in 2′-O methylation is replicative, but its viral RNA lacks 2′-O methylation and is recognized and eliminated by the host immune response. Such a mutant virus could be rationally designed as a live attenuated vaccine. Here, we use Japanese encephalitis virus (JEV), an important mosquito-borne flavivirus, to prove this novel vaccine concept. We show that JEV methyltransferase is responsible for both N-7 and 2′-O cap methylations as well as evasion of host innate immune response. Recombinant virus completely defective in 2′-O methylation was stable in cell culture after being passaged for >30 days. The mutant virus was attenuated in mice, elicited robust humoral and cellular immune responses, and retained the engineered mutation in vivo. A single dose of immunization induced full protection against lethal challenge with JEV strains in mice. Mechanistically, the attenuation phenotype was attributed to the enhanced sensitivity of the mutant virus to the antiviral effects of interferon and IFIT proteins. Collectively, the results demonstrate the feasibility of using 2′-O methylation-defective virus as a vaccine approach; this vaccine approach should be applicable to other flaviviruses and nonflaviviruses that encode their own viral 2′-O methyltransferases.
INTRODUCTION
Live attenuated vaccine represents the best medical intervention to prevent many viral diseases, such as those caused by vaccinia virus, poliovirus (Sabin), yellow fever virus (YFV; YF-17D), Japanese encephalitis virus (JEV SA14-14-2), and MMR (measles, mumps, and rubella viruses). The attenuated vaccine replicates to a low level but induces immune response and memory that are sufficient to prevent virulent virus infection. The traditional method of developing an attenuated vaccine is by passaging the virus through a foreign host (e.g., tissue culture or live animals). The attenuation of a vaccine strain is empirically achieved through accumulation of random mutations during passaging while maintaining immunogenicity. The function of each accumulated mutation in the vaccine strain needs to be analyzed to understand the mechanism of attenuation. As an alternative approach for vaccine development, viral attenuation could be rationally designed by altering the ability of virus to antagonize innate immunity (1). Such rationally designed virus is replicative and induces protective immunity; however, the virus is quickly eliminated due to its enhanced sensitivity to the antiviral effect of the host innate immune response.
RNA and DNA viruses that replicate in the cytoplasm cannot use the cellular nuclear capping machinery and thus have evolved viral methyltransferase (MTase) to facilitate N-7 and 2′-O capping or a mechanism to “snatch” the cap from cellular mRNA (2). We and others recently demonstrated that the 2′-O methylation of the 5′ cap of viral RNA functions to subvert host innate antiviral responses through escape of IFIT-mediated suppression (3, 4). 2′-O MTase-defective West Nile virus (WNV; flavivirus), vaccinia virus (poxvirus), and mouse hepatitis virus (MHV; coronavirus) are more sensitive to the antiviral effects of murine IFIT-2 (3). Infections by wild-type (WT) and 2′-O MTase mutant WNVs induce equivalent levels of type I interferon (IFN) (3). In contrast, the 2′-O MTase mutant MHV induced a higher level of interferon expression than did the WT MHV (4). These results prompted us to test the hypothesis of using 2′-O methylation-defective virus as a live attenuated vaccine.
Japanese encephalitis virus (JEV) belongs to the genus Flavivirus in the family Flaviviridae. Besides JEV, many flaviviruses are significant human pathogens, including the four serotypes of dengue virus (DENV-1 to -4), West Nile virus (WNV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV). Flavivirus has a single-stranded, positive-sense RNA genome of approximately 11 kb in length. The genomic RNA contains a single long open reading frame (ORF) flanked by the 5′ and 3′ untranslated regions (UTRs). The ORF encodes three structural proteins (capsid [C], premembrane [prM], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The 5′ end of all flavivirus genomic RNAs has a type I cap structure (m7GpppAmp). The MTase domain, located at the N terminus of NS5, catalyzes the N-7 and 2′-O methylations of the 5′ cap (5–7) as well as internal RNA methylation (8). All these methylations use S-adenosyl-l-methionine (SAM) as a methyl donor and generate S-adenosyl-l-homocysteine (SAH) as a by-product (9). The K-D-K-E motif, conserved among all flavivirus MTases, forms the active site of the 2′-O cap and internal adenosine methylation activities (5, 6, 10).
In this study, we used JEV as a model to rationally design an attenuated virus through abolishing the 2′-O methylation of viral RNA. We demonstrate that, as in other flaviviruses, the MTase domain of JEV performs both N-7 and 2′-O cap methylations. A mutant JEV completely defective in 2′-O methylation was more sensitive to IFN inhibition than was the WT virus and thus became attenuated in mice. A single-dose administration of the 2′-O-MTase mutant virus induced robust humoral and cellular immune responses and provided full protection against challenge with lethal JEVs of different genotypes.
MATERIALS AND METHODS
Cells and viruses.
Mammalian cell lines BHK-21, HEK293, A549, and Vero were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). Mosquito C6/36 cells were grown at 28°C in RPMI 1640 medium containing 5% FBS. JEV strains SA14 and SX-06, representative strains of genotypes III and I, respectively (11), were prepared in mouse brain, and their titers were determined in BHK-21 cells by standard plaque formation assay (12).
Expression and purification of JEV MTase domain.
Recombinant MTase representing the N-terminal 300 amino acids of NS5 derived from the JEV strain AJE70 was expressed and purified from Escherichia coli Rosetta 2(DE3)pLysS cells as modified from the work of Ray et al. (6). Briefly, a DNA fragment representing the MTase domain was PCR amplified from the JEV strain genome and cloned into an expression plasmid, pET28(a), at NdeI and XhoI sites; a His tag was added at the N terminus. The E218 was mutated to alanine (E218A) using the QuikChange II XL site-directed mutagenesis kit (Stratagene). The E. coli cells were grown at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8 and then induced by 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C for 18 h. The cells were pelleted at 5,000 rpm for 15 min, resuspended, and sonicated in lysis buffer (25 mM HEPES, pH 7.5, 500 mM NaCl, and 10% glycerol) plus 2 mM β-mercaptoethanol. After centrifugation at 20,000 rpm for 60 min, the lysate supernatant was loaded onto a nickel-nitrilotriacetic acid column for affinity purification. Bound protein was eluted with a lysis buffer containing 200 mM imidazole, dialyzed against the lysis buffer, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The purified protein was concentrated and stored at −80°C before use.
Methylation assay.
The RNA substrate representing the first 190 nucleotides of the 5′ end of the JEV genome was used for the MTase assay. In vitro RNA transcription and the RNA capping were carried out as described previously (10). The 33P-labeled m7G*pppA- and G*pppA-RNA (the asterisk indicates that the following phosphate is labeled) were used for N-7 and 2′-O methylations, respectively. N-7 methylation was performed in a 20-μl volume in the presence of ∼4 pmol of G*pppA-RNA substrate, 50 μM unlabeled SAM (New England BioLabs), and 1 μg of MTase in 50 mM Bis-Tris buffer (pH 6.0) with 2 mM dithiothreitol (DTT) and 300 mM NaCl at 30°C for 1 h. 2′-O methylation was performed in a total volume of 20 μl in buffer containing 50 mM Tris-HCl (pH 9.0), 2 mM DTT, 1 mM MgCl2, 50 μM SAM, 3 to 4 pmol of m7G*pppA-RNA, and 1 μg of MTase protein. The reaction mixture was incubated at 23°C for 15 to 30 min. All the reactions were stopped by phenol-chloroform extraction followed by ethanol precipitation. The methylated RNA products were resuspended in RNase-free H2O, digested with nuclease P1 (US Biological [catalog no. N7000] and Sigma-Aldrich [catalog no. N8630]), and analyzed on polyethyleneimine cellulose thin-layer chromatography (TLC) plates (JT Baker) using a solvent of 0.65 M LiCl. The radioactive cap structure on TLC plates was quantified by a PhosphorImager.
Construction and recovery of JEV MTase mutant (E218A).
A single E218A mutation in NS5 was introduced into the infectious JEV cDNA clone (pAJE70) by PCR-based site-directed mutagenesis to produce the MTase mutant virus as previously described (13). The positive clones were finally sequenced to confirm the designed mutations and designated pAJE70-E218A. The constructed plasmids were linearized by XhoI, treated with mung bean nuclease (NEB), and then subjected to in vitro transcription by using the Ribo Max large-scale RNA production system (Promega) in the presence of the m7G(5′)ppp(5′)G cap analog (Promega). RNA transcripts from pAJE70 and pAJE70-E218A were then transfected into BHK-21 cells with Lipofectamine 2000 (Lipo2000; Invitrogen), and rescued viruses (WT and E218A) were harvested 3 to 4 days after transfection and subjected to the following assays.
Characterization of JEV MTase mutant.
Indirect immunofluorescence assay (IFA) was performed to characterize the expression of JEV E protein in BHK-21 cells transfected with equal amounts of RNA transcripts. Briefly, the transfected BHK-21 cells were fixed with cold acetone and incubated with the mouse monoclonal antibody (MAb) 4D5, specific for JEV E protein (14), followed by goat anti-mouse IgG-fluorescein isothiocyanate (FITC) conjugate as previously described (12). Mutant viruses recovered from viral RNA-transfected cells (passage 0), as well as the viruses after passage on Vero cells for 5 (passage 5) and 10 (passage 10) rounds, were analyzed by plaque assays as previously described (12). One-step growth curves of the WT and E218A viruses in BHK-21, Vero, and C6/36 cells were analyzed as previously described (13). Briefly, cells were preseeded into 48-well plates and then infected with WT and E218A viruses at a multiplicity of infection (MOI) of 1. Culture supernatants were collected at 24, 48, 72, and 96 h postinfection, and the titers were determined by plaque assays.
Mouse neuroinvasiveness and neurovirulence tests.
For neuroinvasiveness tests, groups of 3-week-old female BALB/c mice (n = 6) were intraperitoneally (i.p.) injected with 4 × 107 PFU of WT or E218A virus, respectively. For neurovirulence tests, groups of mice (n = 8) were intracranially (i.c.) injected with WT or mutant viruses with serial 10-fold dilutions from 8 × 107 PFU to 8 PFU. The mortality was then monitored daily for 2 weeks. The 50% lethal dose (LD50) and average survival time (AST) were calculated as previously described (15). All the animal experimental procedures were approved by and carried out in accordance with the guidelines of the Animal Experiment Committee of the State Key Laboratory of Pathogen and Biosecurity, China.
Viremia and tissue distribution.
At least three of the infected mice were bled periorbitally and dissected at 1, 3, 5, and 7 days postinfection. Brains, spinal cords, livers, spleens, and kidneys of mice were collected, homogenized, and diluted with phosphate-buffered saline (PBS) to make the final 10% (wt/vol) suspensions. The viremia and organ distribution of JEV were measured by plaque assay on BHK-21 cells. Student's t test was used to determine whether a significant difference (P < 0.05) existed.
Immunization and challenge experiments in mice.
Groups of 4-week-old female BALB/c mice (n = 10) were subcutaneously (s.c.) immunized with 1 × 104 PFU of E218A virus, and the PBS group was set as a control. Sera were collected at 2 and 4 weeks after immunization for further assay. Four weeks after immunization, the mice in each group were i.p. challenged with lethal doses of JEV strains SX-06 (genotype I) and SA14 (genotype III). The mortality was then monitored for at least 15 days.
A129 mice on the 129/SvEv genetic background were obtained from B&K Universal Ltd. and bred in a specific-pathogen-free facility. Groups of 3-week-old 129 and A129 mice were i.p. injected with 5 × 107 PFU of WT and E218A viruses, respectively, and then monitored daily for 15 days to assess morbidity and mortality. Survival analyses were performed by log-rank tests using GraphPad Prism software 5.0. Average survival times (ASTs) were calculated and analyzed by Student's t test.
IgG and neutralizing antibody assay.
IFA was used to measure the IgG antibody titer against JEV in the sera of immunized mice as previously described (16). Briefly, 2-fold-serially diluted sera were incubated at 37°C for 1 h on the prepared JEV antigenic slide. After that, FITC-conjugated goat anti-mouse IgG in 0.02% Evans blue dilutions was added and incubated at 37°C for another 30 min. Finally, positive cells were detected using a fluorescence microscope.
The neutralizing antibody titer against JEV in the immunized mice was measured by standard 50% plaque reduction neutralization test (PRNT50). Briefly, serial 2-fold dilutions of serum samples were mixed with equal volumes of JEV suspension and inoculated in a 12-well plate containing confluent BHK-21 cells. The plates were then overlaid with 1% agarose-containing medium and incubated at 37°C for 3 days for plaque formation and counting. The PRNT50s were then calculated as previously described (12).
Splenocyte proliferation and cytokine assay.
For the proliferation assay, spleens from immunized and control mice were aseptically harvested, crushed with a sterile glass crusher in RPMI 1640 (Sigma), and filtered with a sterile nylon cell strainer (BD Falcon) to prepare the single-cell suspension. Splenocyte suspensions (2 × 105) were then plated in a 96-well plate in triplicate and were stimulated with 100 μl of heat-inactivated JEV (2.5 μg/ml) for 96 h, and concanavalin A (ConA) (Sigma) was used as a control. The splenocyte proliferation was determined with the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] method using the CellTiter96 nonradioactive cell proliferation assay kit (Promega), the stimulation index (SI) was calculated as mean OD values of stimulated cells/mean OD values of unstimulated cells, and an SI of ≥2 was considered significant. Gamma interferon (IFN-γ) in the culture supernatants was assayed with the mouse IFN-γ enzyme-linked immunosorbent assay (ELISA) kit (Excell).
IFN pretreatment and IFIT overexpression.
BHK-21 cells grown in 96-well plates were infected at an MOI of 5 with WT or E218A virus for 1 h at 37°C. Cells were then treated with various doses of IFN-α A/D (Sigma) (10, 100, or 500 U/ml) and incubated for an additional 48 h. Then, the supernatants were collected and virus titers were determined by plaque assay in BHK-21 cells. Cell viability was determined by the MTS assay (Promega) according to the manufacturer's instructions.
Human IFIT expression plasmids were kindly provided by A. Pichlmair (17). HEK293 cells were transfected with equal amount of plasmids IFIT-1, -2, -3, and -5 using Lipo2000 according to the manufacturers' protocol. After 24 h of incubation, cells were infected with the WT and mutant JEVs at an MOI of 1. The viral titers in the supernatant were then assayed at 24, 48, and 72 h postinfection by plaque assay.
Statistic analysis.
The significance between survival curves from each group was analyzed by Kaplan-Meier survival analysis with log-rank tests with GraphPad Prism software 5.0. Average results were obtained from at least three independent experiments, and Student's t test was used to determine whether a significant difference (P < 0.05) existed.
RESULTS
N-7 and 2′-O methylations of WT and mutant JEV MTase.
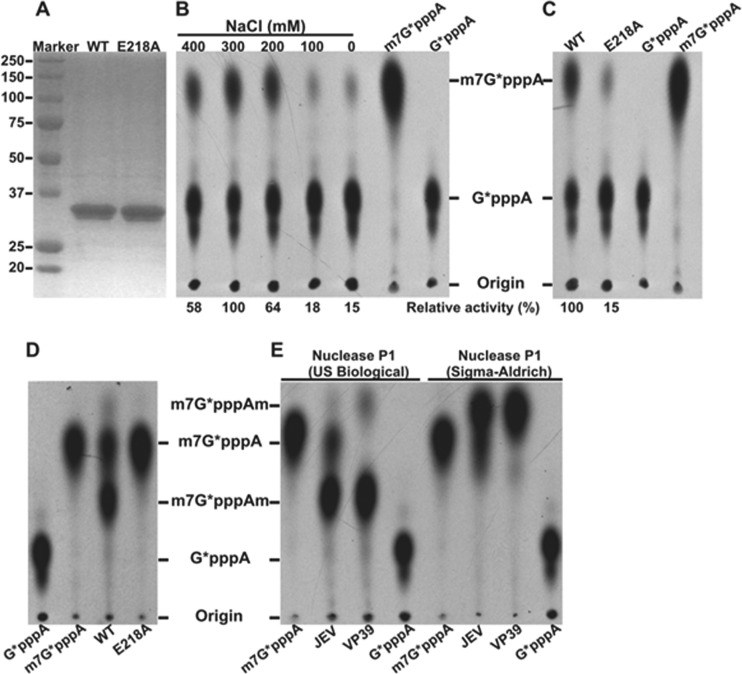
Although the MTase catalytic K-D-K-E tetrad is highly conserved among flaviviruses, the methylation of JEV MTase has not been experimentally demonstrated. Sequence alignment among various members of flavivirus (data not shown) indicates that amino acid E218 represents the glutamic acid within the K-D-K-E tetrad of the JEV MTase domain. We prepared recombinant WT and E218A mutant MTase proteins of JEV (representing the N-terminal 300 amino acids of NS5). SDS-PAGE analysis showed that the purified proteins were >95% pure with the expected molecular mass of 35 kDa (Fig. 1A). For detection of N-7 methylation (GpppA→m7GpppA), substrate G*pppA-RNA (representing the first 190 nucleotides of the 5′ end of the JEV genome) was incubated with the recombinant MTases in the presence of SAM. The reaction products were digested with nuclease P1 and analyzed on a TLC plate. We found the optimal conditions for N-7 methylation to be pH 6.0 (data not shown) and 300 mM NaCl (Fig. 1B). The E218A substitution reduced N-7 methylation to 15% of the WT level (Fig. 1C). For detection of 2′-O methylation, we measured the m7GpppA→m7GpppAm conversion. The E218A substitution completely knocked out the 2′-O methylation (Fig. 1D), demonstrating that E218 of the K-D-K-E tetrad is essential for the 2′-O methylation.
Fig 1.
N-7 and 2′-O cap methylations of the viral RNA by JEV MTase. (A) Analysis of recombinant JEV MTase proteins on 12% SDS-PAGE gels; the sizes (kilodaltons) of protein markers are labeled on the left. (B) N-7 methylation. N-7 methylation assays were performed using substrate G*pppA-RNA; the reaction mixtures were digested with nuclease P1 (purchased from US Biological) and analyzed on TLC plates. The conversion of G*pppA-RNA to m7G*pppA-RNA was quantified using a PhosphorImager. The 33P-labeled m7G*pppA or G*pppA marker is indicated on top of the TLC plate. The optimal concentration of NaCl was determined by titration. Relative activities are presented using the optimal level as 100%. (C) N-7 activity analysis of mutant E218A MTase. The methylation efficiency of the E218A MTase was compared with that of the WT MTase (set at 100%). Average results from three independent experiments are shown. (D) Analysis of 2′-O methylation activity of the E218A MTase. 2′-O methylation assays were performed using m7G*pppA-RNA substrate. The reaction mixtures were incubated at 23°C for 15 min, digested with nuclease P1 (US Biological), and analyzed on TLC plates. 33P-labeled cap structures (m7G*pppA or G*pppA) are indicated on the bottom of the TLC plate. The position of the origin and the migration positions of the G*pppA, m7G*pppA, and m7G*pppAm molecules are shown to the right of the TLC plate. The methylation efficiency of the E218A MTase was compared with that of the WT MTase (set at 100%). Average results from three independent experiments are shown. (E) Effect of nuclease P1 from two different vendors (indicated) on the migration of m7G*pppAm. 2′-O methylation assays were the same as those in panel D except that the reaction was done at 23°C for 30 min (JEV MTase) or 37°C for 30 min (VP39). Half of the samples were digested with nuclease P1 in 20 mM Tris-HCl (pH 7.5) buffer, and the other half of the samples were digested with nuclease P1 in the same buffer and then analyzed on TLC plates. 33P-labeled cap structures (m7G*pppA or G*pppA) are indicated on the bottom of the TLC plate. The position of the origin and the migration positions of the G*pppA, m7G*pppA, and m7G*pppAm molecules are shown to the left of the TLC plate.
Nuclease P1 from different vendors affects the migration of m7GpppAm molecule on a TLC plate.
Contradictory results were previously reported on the migration position of m7GpppAm on TLC plates. Some groups found that m7GpppAm migrated faster than m7GpppA (6, 18, 19), whereas other groups showed that m7GpppAm migrated slower than m7GpppA (20). As shown in Fig. 1E, after the 2′-O methylation reaction (m7GpppA→m7GpppAm) was cleaved with the nuclease P1 purchased from Sigma-Aldrich (catalog no. N8630), the m7GpppAm product migrated faster than m7GpppA. In contrast, after the reaction was cleaved with the nuclease P1 purchased from US Biological (catalog no. N7000), the m7GpppAm product migrated slower than m7GpppA. For this experiment, we used the WT JEV MTase as well as vaccinia virus VP39 (a well-characterized 2′-O MTase used as a positive control). We currently do not know what component(s) in the nuclease P1 reaction mixtures from the two vendors caused the difference in migration.
Recovery and characterization of the 2′-O MTase mutant JEV.
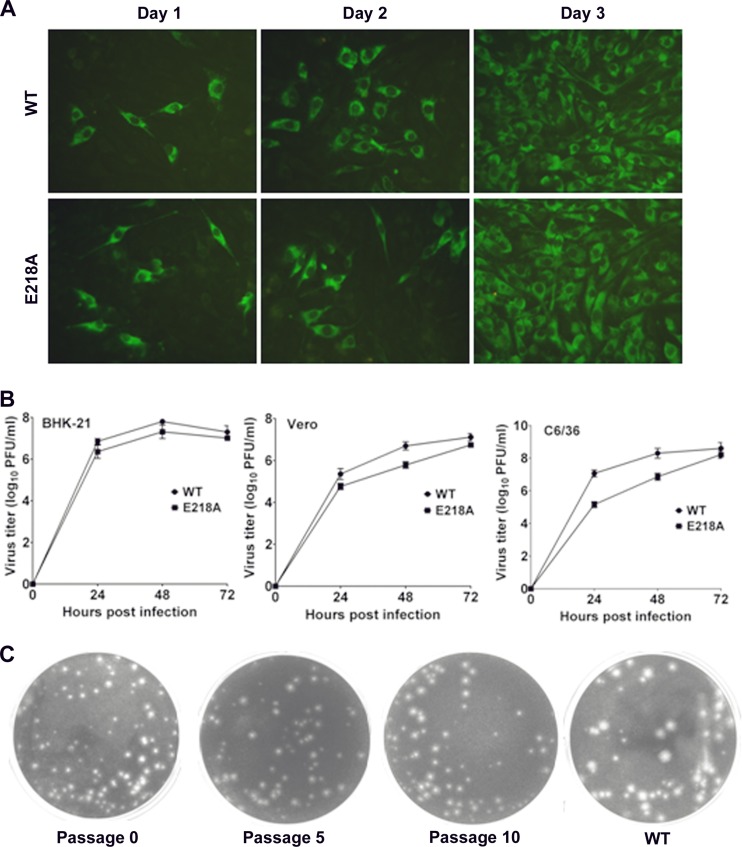
To generate a 2′-O MTase mutant JEV, the E218A mutation was introduced into the full-length infectious clone of JEV pAJE70 (13). Transfection of BHK-21 cells with genome-length RNAs generated numbers of viral E protein-positive cells equivalent between the WT and E218A RNAs (Fig. 2A). Genomic sequencing confirmed that the designed mutation E218A was retained in the recovered virus and that no additional mutations were introduced (data not shown). One-step growth curves in mammalian (BHK-21 and Vero) and mosquito (C6/36) cells showed that the mutant virus replicated slightly slower than the WT virus at 24 and 48 h but peaked at similar levels at 72 h postinfection (Fig. 2B). Interestingly, the E218A virus exhibited smaller plaque morphology than did the WT virus (Fig. 2C). Continuous culturing of the mutant virus on Vero cells for 10 rounds (3 to 4 days per round) did not change its small-plaque morphology (Fig. 2C). Genomic sequencing of the passage 10 virus showed that the engineered E218A change was retained without any extra mutation (data not shown). The results indicate that the 2′-O MTase mutant JEV is highly replicative and genetically stable in cell culture.
Fig 2.
Recovery and characterization of JEV MTase mutant virus. (A) BHK-21 cells were transfected with equal amounts of RNA transcripts from pAJE70 and pAJE70-E218A and subjected to IFA using anti-JEV E protein MAb 4D5 at the indicated time posttransfection. (B) Growth kinetics in different cell lines. BHK-21, Vero, and C6/36 cells were infected with WT and E218A viruses at an MOI of 1. Viral titers were measured at indicated times using standard plaque assays in BHK-21 cells. (C) Plaque morphology and genetic stability of E218A mutant. E218A virus recovered from RNA-transfected cells (passage 0), as well as from passaging on Vero cells for 5 and 10 rounds (passages 5 and 10), was analyzed by standard plaque assays and compared with the WT virus.
2′-O MTase mutant JEV is highly attenuated in mice.
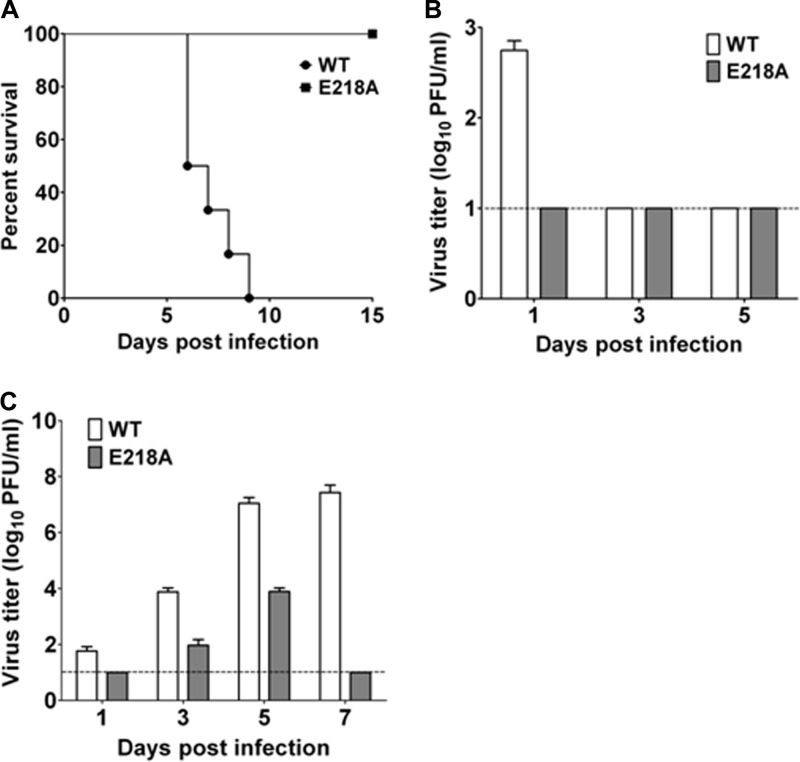
We compared the neuroinvasiveness of the WT and E218A viruses in mice. Groups of 3-week-old BALB/c mice were i.p. injected with 4 × 107 PFU of WT or E218A JEV; the mortality of the infected mice was monitored for 2 weeks. The WT-infected mice all died within 9 days, whereas the E218A virus-infected mice all survived with no neurological symptoms (Fig. 3A). Viremia was detected at day 1 postinfection in the WT-infected mice, whereas no viremia (i.e., below the detection limit of plaque assay) was observed in the E218A virus-infected mice (Fig. 3B). The WT virus was detected in mouse brains at days 1, 3, 5, and 7 postinfection; in contrast, the mutant virus could be detected in brains only at days 3 and 5 postinfection, and the titers were much lower than those from the WT group (Fig. 3C). These results demonstrate that the 2′-O MTase mutant JEV has significantly reduced neuroinvasiveness in mice.
Fig 3.
JEV 2′-O MTase mutant is highly attenuated in mice. (A) Neuroinvasiveness tests for WT and E218A mutant virus in mice. Groups of 3-week-old BALB/c mice (n = 6) were i.p. injected with 4 × 107 PFU of WT and E218A viruses. The mortality was monitored for 15 days or till death. The significance between survival curves from each group was analyzed by Kaplan-Meier survival analysis with log-rank tests. (B and C) Viral loads in mouse sera (B) and brain tissue (C) were measured by plaque assay at the indicated time postinfection. Dotted lines represent limits of detection.
We also compared the neurovirulence levels of the WT and E218A viruses in mice. Mice were i.c. inoculated with serial dilutions of the WT or mutant viruses. As shown in Table 1, the E218A mutant virus was significantly attenuated: the LD50 of the E218A virus was about 24-fold higher than that of the WT virus. The AST of the mutant virus-inoculated mice for each dose was significantly longer than that of the WT virus-infected mice. Collectively, these results demonstrate that the 2′-O MTase mutant of JEV is attenuated in mice. In addition, we sequenced the mutant viruses recovered from the dead mice after i.c. inoculation of high doses (≥1 × 101.9 PFU); the engineered E218A mutation was retained from all samples with no other nucleotide changes detected. This result demonstrates the in vivo stability of the E218A mutant virus.
Table 1.
Comparison of neurovirulence levels between the WT and MTase E218A mutant viruses in micea
| Virus | Dose (log10 PFU) | No. dead/no. inoculated (% mortality) | LD50 (PFU) | AST (range; days) |
|---|---|---|---|---|
| WT | 3.9 | 8/8 (100) | 7.9 | 4.6 (4–6) |
| 2.9 | 8/8 (100) | 4.9 (4–6) | ||
| 1.9 | 8/8 (100) | 8.1 (7–9) | ||
| 0.9 | 4/8 (50) | 12.0 (7–15) | ||
| E218A | 3.9 | 8/8 (100) | 188.4 | 8.0.0 (6–10) |
| 2.9 | 8/8 (100) | 8.8 (7–11) | ||
| 1.9 | 2/10 (20) | 12.75 (10–15) | ||
| 0.9 | 0/8 (0) | 15 (15) |
Groups of 3-week-old BALB/c mice were i.c. injected with various doses of WT and E218A mutant viruses. The number of surviving mice was recorded daily.
The 2′-O MTase mutant virus induces a protective immune response in mice.
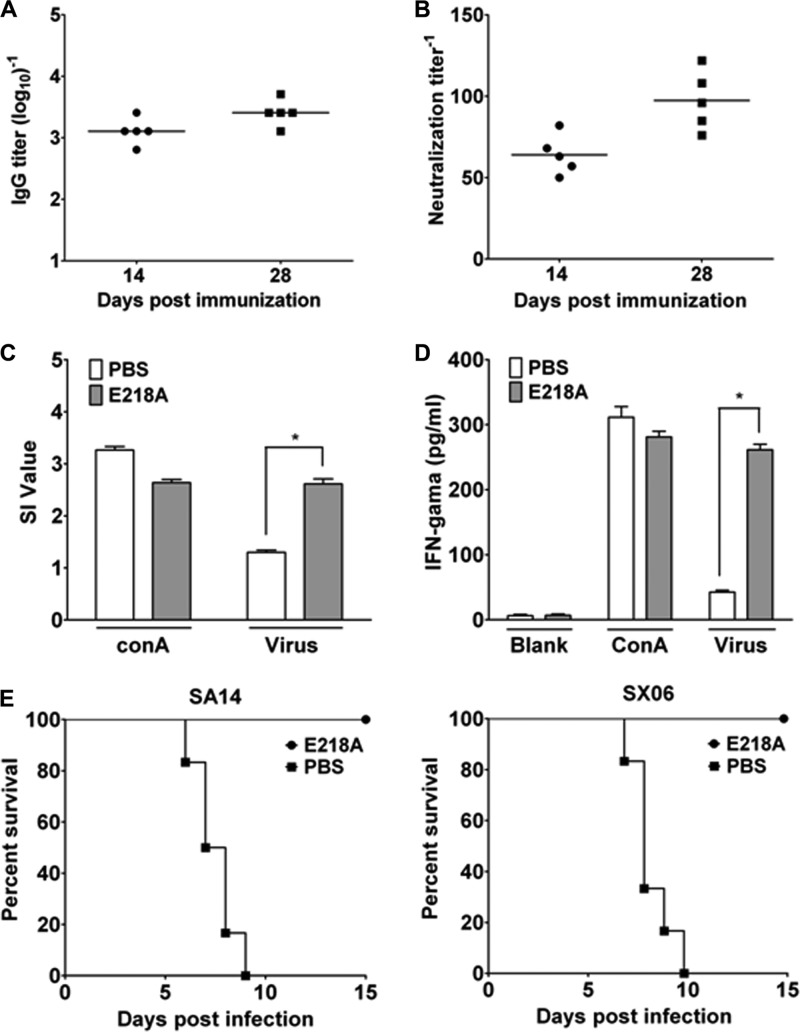
The immunogenicity of the 2′-O MTase mutant virus was evaluated in mice. JEV-specific IgG and neutralizing antibodies were assayed by IFA and PRNT at 14 and 28 days postimmunization. A single-dose immunization with 1 × 104 PFU of E218A virus induced robust IgG and neutralizing antibodies (Fig. 4A); the geometric mean titer (GMT) of neutralizing antibody against JEV reached 97.4 at 28 days postimmunization (Fig. 4B). As expected, no JEV-specific antibodies were detected in the sera collected prior to the immunization or from the PBS-immunized animals (data not shown).
Fig 4.
JEV 2′-O MTase mutant is immunogenic and protective in mice. Mice immunized with 1 × 104 PFU of E218A or in the control group were bled at 14 and 28 days postimmunization. The titers of IgG antibody (A) and neutralization antibody (B) were determined by IFA and PRNT, respectively. JEV-specific cellular immune response induced by E218A virus immunization was assayed by splenocyte proliferation assay (C) and IFN-γ production assay (D). On day 28 postimmunization, the spleens from immunized mice of various experimental groups were separated and the proliferation was tested in the presence of inactivated JEV or the positive control ConA. The culture supernatants were collected after 72 h of stimulation with inactivated JEV and ConA for IFN-γ assay. Challenge experiments were performed with JEV strains SA14 and SX-06 (E). Groups of mice (n = 10) immunized with E218A virus or PBS were challenged with 1 × 108 PFU of SA14 and SX-06. The mortality was recorded daily, and the survival curve from each group was analyzed by Kaplan-Meier survival analysis with log-rank tests.
Cellular immunity to JEV was determined by splenocyte proliferative response and IFN-γ production in splenocytes from the vaccinated mice on day 28 postimmunization. MTS assay showed that the SI value (2.61 ± 0.21) of the E218A virus-immunized mice was significantly higher than that of the control groups (1.30 ± 0.09) (Fig. 4C). IFN-γ production in splenocytes from the E218A virus-immunized mice was significantly higher than that of the control groups (261.43 ± 16.94 versus 42.62 ± 5.47) under the stimulation with JEV (Fig. 4D). Together, these results clearly demonstrate that a single-dose vaccination with the E218A virus induces robust humoral and cellular immune responses in mice.
Next, we challenged the immunized mice with lethal doses of JEV strains SX-06 and SA14, representative of genotypes I and III of JEV, respectively (Fig. 4E). All the PBS-immunized mice died within 6 to 10 days after the challenge, whereas all the E218A virus-immunized mice survived the lethal challenges, and none of them developed any clinical manifestations. Thus, a single dose of JEV 2′-O MTase mutant immunization provides full protection against lethal challenges with different JEV genotypes.
2′-O MTase mutant virus shows enhanced sensitivity to IFN and IFITs.
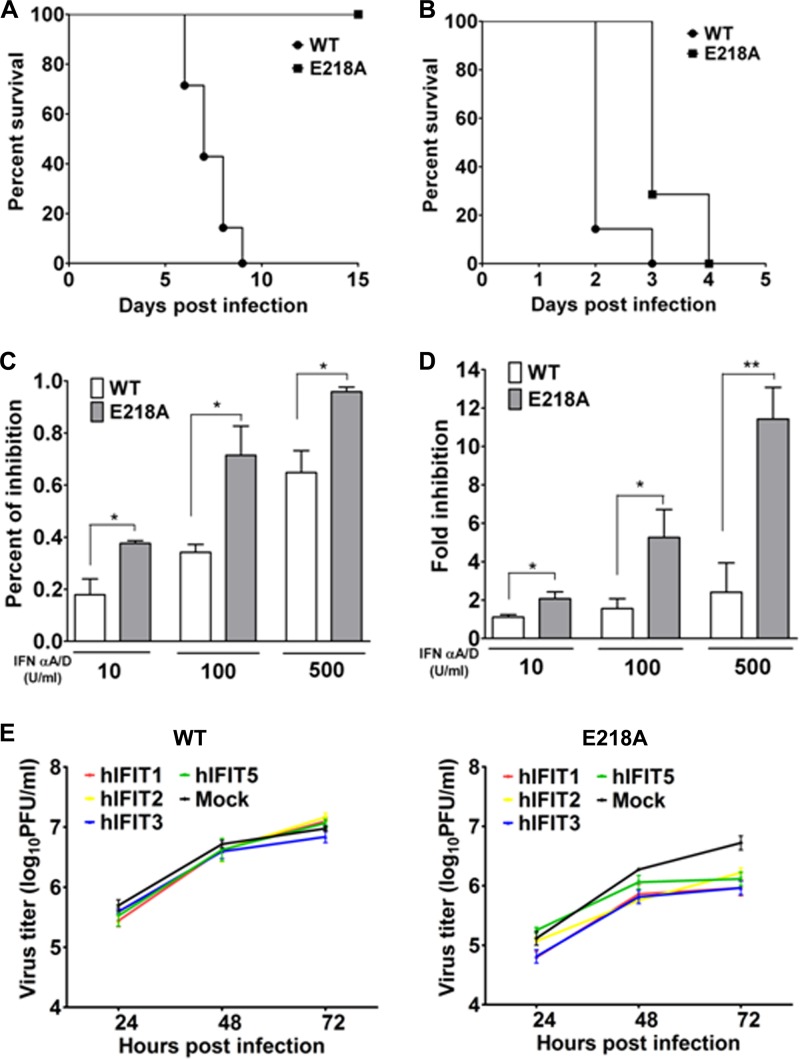
To explore the molecular mechanism of attenuation for 2′-O MTase mutant JEV, we compared the virulence levels of the WT and mutant viruses in mice deficient in type I IFN receptors (A129 mice) and their parental 129 mice. A129 and 129 mice were i.p. challenged with the WT and mutant JEVs. The results showed that even a high dose of 5 × 107 PFU of the E218A virus failed to kill any 129 mice (Fig. 5A), which was in agreement with the results from BALB/c mice (Fig. 3A). However, both the WT and E218A viruses showed 100% mortality within 4 days in the A129 mice; only a slight delay of death was observed in the E218A virus-infected mice compared with that for the WT virus (Fig. 5B), indicating the critical role of IFN signaling for the attenuation phenotype of the mutant virus.
Fig 5.
JEV 2′-O MTase mutant shows enhanced sensitivity to IFN and IFITs. (A and B) Groups of 3-week-old 129 mice (A) and A129 mice (B) were i.p. injected with 5 × 107 PFU of WT or E218A virus. The mortality was monitored for 15 days or till death. BHK-21 cells in a 96-well plate were infected with the WT or E218A mutant virus at an MOI of 5 for 1 h, followed by treatment with various doses of IFN-α A/D (10, 100, or 500 U/ml). (C and D) MTS assay (C) and plaque assay (D) were performed 48 h postinfection. (E) HEK293 cells were transfected with equal amount of plasmids IFIT-1, -2, -3, and -5 using Lipo2000 followed by infection with WT and E218A viruses at an MOI of 1. The viral titers in the supernatant were then assayed at the indicated times postinfection by plaque assay.
Next, we compared the sensitivities of the WT and mutant viruses to IFN-α pretreatment in cell culture. The result showed that the mutant virus was more sensitive to the antiviral effect of various concentrations of IFN-α than was the WT (Fig. 5C). Moreover, IFN-α treatment strongly inhibited the yield of E218A in BHK-21 cells in a dose-dependent manner, whereas the WT virus was only slightly impaired by the same dose of IFN-α treatment (Fig. 5D).
Finally, we compared the antiviral effects of human IFIT-1, -2, -3, or -5 on the WT and mutant viruses. HEK293 cells transgenically expressing individual IFITs were infected with the WT or mutant virus; the viral titers in supernatants were determined at the indicated times postinfection. The overexpression of IFIT-1, -2, -3, and -5 significantly inhibited the production of the mutant virus at 72 h postinfection, whereas no IFITs suppressed the production of the WT virus (Fig. 5E). As a control, Western blotting showed equal levels of IFIT-1, -2, -3, and -5 protein expression in the transfected HEK293 cells (data not shown). Taken together, we conclude that the in vivo attenuation of the 2′-O MTase mutant JEV is attributable to the enhanced sensitivity to the antiviral action of IFN and IFIT proteins.
DISCUSSION
This study aimed to test the hypothesis of using 2′-O methylation-defective virus as a vaccine approach. Using JEV as a model, we functionally analyzed the E218A mutation of the viral NS5 protein in cap methylation, viral replication, mouse virulence, and evasion of innate immune response. We chose JEV as the model to test the 2′-O MTase vaccine approach because of the availability of a good mouse model for this virus. In agreement with other MTase results, the E218A mutation of the JEV MTase K-D-K-E tetrad completely abolished the 2′-O methylation activity while the N-7 methylation activity was retained. However, the effect of the E218A mutation on N-7 methylation is more significant in JEV MTase (15% of the WT activity) than in WNV MTase (76 to 92% of the WT activity [7]) and DENV MTase (59% of the WT activity [10]). These results indicate the plasticity of the role of the MTase K-D-K-E tetrad in the methylation activities among various flaviviruses.
We showed that recombinant JEV lacking 2′-O MTase activity was replicative but was highly sensitive to the antiviral effects of IFN and IFITs. Compared with cells infected with the WT JEV, cells infected with the mutant virus did not induce higher levels of IFN production (data not shown). The latter result is similar to that observed for the 2′-O MTase mutant WNV (3, 4) but different from that observed for the 2′-O MTase mutant MHV (which showed higher IFN production than did the WT virus [4]). However, the 2′-O MTase mutant JEV was sensitively inhibited by all four tested human IFITs, whereas the 2′-O MTase mutant WNV was inhibited only by mouse IFIT-2, not IFIT-1, -3, or -5 (3). This difference in sensitivity to IFITs between the mutant JEV and WNV could be due to IFITs from different species being used in the two studies. Human IFITs are critical for the inhibition of viral infections (21–23), but the functions of these IFITs are only partially understood. Nevertheless, the mutant 2′-O MTase JEV showed reduced virulence in mice. Furthermore, mice immunized with a single dose of the mutant JEV were completely protected against lethal challenge with virulent JEV strains. These results demonstrate the potential of this rationally designed 2′-O MTase mutant virus for further development as a vaccine approach.
Our results support the concept that an attenuated virus could be rationally designed by altering the ability of virus to antagonize innate immunity (1). Previously, Talon et al. proposed and tested this concept by reducing the ability of influenza virus to antagonize the type I IFN response (1). Nonstructural protein 1 (NS1) of influenza A and B viruses antagonizes the IFN response following infection and contributes to the virulence (24); viruses with partial deletions of NS1 are attenuated and yet induce a protective immune response in mice (1).
A live JE vaccine strain, SA14-14-2, derived from continual passaging in cells and animals, has been widely used in Asia and has saved millions of lives. Currently, the live SA14-14-2 vaccine has been licensed in 10 Asian countries, and primary hamster kidney cells were approved for vaccine production by the World Health Organization (16). However, the SA14-14-2 vaccine has not yet been approved for use in the markets of developed countries. With the development of reverse genetics of flavivirus (25–27), rational design of recombinant live attenuated JEV vaccine has become possible. A novel live attenuated JEV vaccine, IMOJEV, using the yellow fever 17D virus as a backbone, was recently licensed in Australia and Thailand (28, 29). The results presented in this study demonstrate that the JEV 2′-O MTase mutant E218A virus could potentially serve as a novel live JEV vaccine candidate.
The 2′-O MTase mutant vaccine has several advantages over the licensed JEV vaccines. First, the E218A virus showed an ideal attenuation profile in cells and animals. It completely lost its neuroinvasiveness (Fig. 3A and B). The E218A mutant virus was stable; no second-site mutation was detected even after 10 passages (>30 days) in Vero cells. The engineered mutation was also retained in the virus recovered from the mice that died of high-dose i.c. inoculation of the E218A virus, demonstrating the in vivo stability of the mutant virus. Second, the E218A virus exhibited an ideal immunogenicity; a single immunization with the E218A virus induced high titers of neutralizing antibodies (GMT = 97.4) as well as robust cellular immune responses in mice (Fig. 4). Previous clinical trials and animal experiments with JEV vaccine showed that a neutralization antibody titer of more than 10 was protective (30). Third, the 2′-O MTase mutant JEV was attenuated due to the single E218A mutation in NS5 protein via a defined mechanism in association with the host IFN response. Previous studies based on SA14-14-2 or other attenuated strains indicate the critical role of several mutations in the E protein in governing the attenuation phenotype (13, 25, 31–34). However, these mutations are within the major protective antigen E protein and could alter the native antibody repertoire, leading to a high risk of emergence of neutralization escape mutants. For the current 2′-O MTase approach, we could improve the safety of the 2′-O MTase mutant vaccine by engineering double or triple mutations within the K-D-K-E active site of the JEV MTase. This would minimize the reversion of the mutant MTase to the WT sequence.
Overall, by in vitro and in vivo characterization of the 2′-O methylation of JEV MTase, we provide various lines of evidence for the highly attenuated recombinant JEV mutant E218A virus as a potential JEV live vaccine approach. This vaccine approach warrants further development, especially considering the great need for a low-cost but high-efficacy vaccine for a neglected disease in developing countries. Moreover, this rational design approach can be readily applicable to other viruses with defined 2′-O MTase activity for potential vaccine development.
ACKNOWLEDGMENTS
We thank Andreas Pichlmair (CEMM, Austria) for providing the human IFIT plasmids and Yongxin Yu (NIFDC, Beijing, China) for helpful discussions.
This work was supported by the Basic Research Project of China (grant 2012CB518904) and the National Natural Science Foundation of China (grants 81101243 and 31270974). The P.-Y.S. group is partially supported by the TCR flagship STOP Dengue program from the National Medical Research Council in Singapore. C.-F.Q. was supported by the Beijing Nova Program of Science and Technology (grant 2010B041).
Footnotes
Published ahead of print 13 March 2013
REFERENCES
- 1. Talon J, Salvatore M, O'Neill RE, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 97:4309–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furuichi Y, Shatkin AJ. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55:135–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81:3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2012. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 8:e1002642 doi:10.1371/journal.ppat.1002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chung KY, Dong H, Chao AT, Shi PY, Lescar J, Lim SP. 2010. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2′-O methyltransferase activity in dengue virus. Virology 402:52–60 [DOI] [PubMed] [Google Scholar]
- 10. Dong H, Chang DC, Xie X, Toh YX, Chung KY, Zou G, Lescar J, Lim SP, Shi PY. 2010. Biochemical and genetic characterization of dengue virus methyltransferase. Virology 405:568–578 [DOI] [PubMed] [Google Scholar]
- 11. Pan XL, Liu H, Wang HY, Fu SH, Liu HZ, Zhang HL, Li MH, Gao XY, Wang JL, Sun XH, Lu XJ, Zhai YG, Meng WS, He Y, Wang HQ, Han N, Wei B, Wu YG, Feng Y, Yang DJ, Wang LH, Tang Q, Xia G, Kurane I, Rayner S, Liang GD. 2011. Emergence of genotype I of Japanese encephalitis virus as the dominant genotype in Asia. J. Virol. 85:9847–9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng YQ, Dai JX, Ji GH, Jiang T, Wang HJ, Yang HO, Tan WL, Liu R, Yu M, Ge BX, Zhu QY, Qin ED, Guo YJ, Qin CF. 2011. A broadly flavivirus cross-neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of E protein. PLoS One 6:e16059 doi:10.1371/journal.pone.0016059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye Q, Li XF, Zhao H, Li SH, Deng YQ, Cao RY, Song KY, Wang HJ, Hua RH, Yu YX, Zhou X, Qin ED, Qin CF. 2012. A single nucleotide mutation in NS2A of Japanese encephalitis-live vaccine virus (SA14-14-2) ablates NS1′ formation and contributes to attenuation. J. Gen. Virol. 93:1959–1964 [DOI] [PubMed] [Google Scholar]
- 14. Kang X, Li Y, Fan L, Lin F, Wei J, Zhu X, Hu Y, Li J, Chang G, Zhu Q, Liu H, Yang Y. 2012. Development of an ELISA-array for simultaneous detection of five encephalitis viruses. Virol. J. 9:56 doi:10.1186/1743-422X-9-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang T, Zhao H, Li XF, Deng YQ, Liu J, Xu LJ, Han JF, Cao RY, Qin ED, Qin CF. 2011. CpG oligodeoxynucleotides protect against the 2009 H1N1 pandemic influenza virus infection in a murine model. Antiviral Res. 89:124–126 [DOI] [PubMed] [Google Scholar]
- 16. Yu Y. 2010. Phenotypic and genotypic characteristics of Japanese encephalitis attenuated live vaccine virus SA14-14-2 and their stabilities. Vaccine 28:3635–3641 [DOI] [PubMed] [Google Scholar]
- 17. Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. 2011. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12:624–630 [DOI] [PubMed] [Google Scholar]
- 18. Alcaraz-Estrada SL, Manzano MI, Del Angel RM, Levis R, Padmanabhan R. 2010. Construction of a dengue virus type 4 reporter replicon and analysis of temperature-sensitive mutations in non-structural proteins 3 and 5. J. Gen. Virol. 91:2713–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bhattacharya D, Hoover S, Falk SP, Weisblum B, Vestling M, Striker R. 2008. Phosphorylation of yellow fever virus NS5 alters methyltransferase activity. Virology 380:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J, Wang JT, Whelan SP. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 103:8493–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fensterl V, Sen GC. 2011. The ISG56/IFIT1 gene family. J. Interferon Cytokine Res. 31:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 8:e1002712 doi:10.1371/journal.ppat.1002712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Z, Liang H, Zhou Q, Li Y, Chen H, Ye W, Chen D, Fleming J, Shu H, Liu Y. 2012. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 22:1328–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330 [DOI] [PubMed] [Google Scholar]
- 25. Liang JJ, Liao CL, Liao JT, Lee YL, Lin YL. 2009. A Japanese encephalitis virus vaccine candidate strain is attenuated by decreasing its interferon antagonistic ability. Vaccine 27:2746–2754 [DOI] [PubMed] [Google Scholar]
- 26. Pu SY, Wu RH, Yang CC, Jao TM, Tsai MH, Wang JC, Lin HM, Chao YS, Yueh A. 2011. Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. J. Virol. 85:2927–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yun SI, Kim SY, Rice CM, Lee YM. 2003. Development and application of a reverse genetics system for Japanese encephalitis virus. J. Virol. 77:6450–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dean CH, Alarcon JB, Waterston AM, Draper K, Early R, Guirakhoo F, Monath TP, Mikszta JA. 2005. Cutaneous delivery of a live, attenuated chimeric flavivirus vaccine against Japanese encephalitis (ChimeriVax)-JE) in non-human primates. Hum. Vaccin. 1:106–111 [DOI] [PubMed] [Google Scholar]
- 29. Halstead SB, Thomas SJ. 2011. New Japanese encephalitis vaccines: alternatives to production in mouse brain. Expert Rev. Vaccines 10:355–364 [DOI] [PubMed] [Google Scholar]
- 30. Van Gessel Y, Klade CS, Putnak R, Formica A, Krasaesub S, Spruth M, Cena B, Tungtaeng A, Gettayacamin M, Dewasthaly S. 2011. Correlation of protection against Japanese encephalitis virus and JE vaccine (IXIARO) induced neutralizing antibody titers. Vaccine 29:5925–5931 [DOI] [PubMed] [Google Scholar]
- 31. Arroyo J, Guirakhoo F, Fenner S, Zhang ZX, Monath TP, Chambers TJ. 2001. Molecular basis for attenuation of neurovirulence of a yellow fever virus/Japanese encephalitis virus chimera vaccine (ChimeriVax-JE). J. Virol. 75:934–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee E, Hall RA, Lobigs M. 2004. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J. Virol. 78:8271–8280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Monath TP, Arroyo J, Levenbook I, Zhang ZX, Catalan J, Draper K, Guirakhoo F. 2002. Single mutation in the flavivirus envelope protein hinge region increases neurovirulence for mice and monkeys but decreases viscerotropism for monkeys: relevance to development and safety testing of live, attenuated vaccines. J. Virol. 76:1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Z, Date T, Li Y, Kato T, Miyamoto M, Yasui K, Wakita T. 2005. Characterization of the E-138 (Glu/Lys) mutation in Japanese encephalitis virus by using a stable, full-length, infectious cDNA clone. J. Gen. Virol. 86:2209–2220 [DOI] [PubMed] [Google Scholar]