Abstract
Cache Valley virus (CVV)-induced malformations have been previously reproduced in ovine fetuses. To evaluate the development of the antiviral response by the early, infected fetus, before the development of immunocompetency, ovine fetuses at 35 days of gestation were inoculated in utero with CVV and euthanized at 7, 10, 14, 21, and 28 days postinfection. The antiviral immune response in immature fetuses infected with CVV was evaluated. Gene expression associated with an innate, immune response was quantified by real-time quantitative PCR. The upregulated genes in infected fetuses included ISG15, Mx1, Mx2, IL-1, IL-6, TNF-α, TLR-7, and TLR-8. The amount of Mx1 protein, an interferon-stimulated GTPase capable of restricting growth of bunyaviruses, was elevated in the allantoic and amniotic fluid in infected fetuses. ISG15 protein expression was significantly increased in target tissues of infected animals. B lymphocytes and immunoglobulin-positive cells were detected in lymphoid tissues and in the meninges of infected animals. These results demonstrated that the infected ovine fetus is able to initiate an innate and adaptive immune response much earlier than previously known, which presumably contributes to viral clearance in infected animals.
INTRODUCTION
Cache Valley virus (CVV) is a mosquito-borne bunyavirus of the family Bunyaviridae, genus Orthobunyavirus, Bunyamwera group (1), and endemic in North America (2). Serologic studies have shown that unlike most members of the family Bunyaviridae, the vectors of CVV also infect larger mammals, with virus being isolated from sheep, cattle, and horses and causing reproductive losses in small ruminants (2–8), similar to the disease caused by Akabane virus (9–11) and the newly identified Schmallenberg virus occurring in Europe (12). The virus has rarely been associated with meningoencephalitis in humans (13, 14). In utero ovine fetal infection causes abortion and fetal malformations, mainly affecting the musculoskeletal system (MSS) and the central nervous system (CNS) (2–5, 7, 15).
Previous studies describing experimental CVV-induced malformations in ovine fetuses showed that the development of fetal lesions depends on the fetal age when infected. If the virus is inoculated between 28 and 48 days of gestation (dg), fetal death and abortion or MSS and CNS malformations occur. No apparent malformations are observed if the virus is inoculated after 48 dg (2, 3). Virus isolation can only be made from infected fetuses in early development, and viral recovery from tissues of term abortions and malformed lambs is uniformly unsuccessful. The virus is cleared from infected tissues within a few weeks after infection (2, 15) and before the presumed age of fetal immunocompetency, at approximately 70 to 75 dg (16, 17).
The gestation period of the ewe is approximately 147 days. Ovine fetuses develop erythropoiesis, myelopoiesis, and megakaryopoiesis in the yolk sac and liver at approximately 17 dg (18). At approximately 20 to 25 dg, lymphocyte production begins in the thymus, and lymphocytes are in the bloodstream at 48 to 50 dg (18). At 45 to 50 dg, T and B lymphocytes and cells with surface immunoglobulins are present in the spleen and lymph nodes (19, 20). The lymph nodes become integrated into a lymphatic system around 65 dg (18). Establishing the time when fetuses are able to respond to antigens is difficult. Available data on adaptive immune response of ovine fetuses have been based on serum neutralization assays, which were established for use in mature animals. In addition, gestation assay points used in experiments are somewhat arbitrary. Ovine fetal antibody response to viral infection at titers greater than 1:2 has been detected after 76 to 78 dg (11). Because the syndesmochorial placenta of ruminants prevents passage of immunoglobulins from the ewe to the fetus (21), antibodies in fetuses and precolostral newborns are those produced by the fetus. For this reason, in utero viral infections can be diagnosed in aborted fetuses and stillborn ruminants using serum neutralization tests. Such testing is necessary with CVV because the fetus clears the virus long before the end of gestation (2, 5).
Previously, we have demonstrated that fetuses infected with CVV early in gestation (∼35 dg) have low viral antigen and RNA signal in tissues around 56 dg and are able to clear the infection before development of an adaptive immune system at 75 dg (15). Similarly, age-based findings have been described in Akabane virus-infected fetuses (9, 11, 16). Because no effective serum neutralization antibody (an adaptive immune response) has been detected in ovine fetuses at the time when CVV is cleared from fetuses (18), it may be that the ovine fetus mounts an innate immune response for early viral clearance. To test this hypothesis, the expression of selected genes associated with the innate immune response was determined in tissues of CVV-infected and noninfected ovine fetuses, and CVV mRNA was quantified in selected tissues of these fetuses to correlate the measured innate response with viral clearance. In addition, fetal Mx protein, an interferon (IFN)-stimulated GTPase previously associated with antiviral activity against bunyaviruses (22–26), was quantified in the fetal allantoic and amniotic fluids. Expression of the IFN-stimulated gene 15 (ISG15), which has been shown to be produced by fetuses in response to viral infection in utero (27–29), was evaluated in CVV-infected fetal tissues. Finally, the distribution of B and T lymphocytes and immunoglobulin-positive cells was evaluated in infected and noninfected, ovine fetal tissues in early gestation.
MATERIALS AND METHODS
Virus inoculation and sample harvesting.
A group of 15 seronegative, pregnant Rambouillet ewes was housed in BSL2 confinement buildings according to protocols approved by the Institution Animal Care and Use and the Institutional Biosafety Committee. At 35 dg, ewes were inoculated in utero with a 1-ml inoculum containing 105 50% tissue culture infectious doses of CVV (infected group) or 1 ml of minimum essential medium (mock-infected/control group), as previously described (15). The viral inoculum was derived from the second passage of an isolate from allantoic membrane from an experimentally infected fetus (3). At 7, 10, 14, 21, and 28 days postinfection (dpi), three ewes (one mock infected and two virus infected) were humanely euthanized. Selected fetal tissues and their fluids were harvested and immediately frozen at −80°C or placed in RNA later (Ambion Life Technologies, Carlsbad, CA) and frozen at −80°C. The remaining fetal tissues and placenta were fixed in 10% buffered formalin diluted in deionized water or in Davidson's AFA fixative (glacial acetic acid, 37% formaldehyde, 95% ethanol).
Real-time qPCR.
To quantify relative numbers of CVV RNA copies, and abundance of genes of interest, real-time quantitative PCR (qPCR) was performed on samples of brain and muscle harvested from CVV-infected or mock-infected ovine fetuses according to a previously published protocol with minor modifications (30). Briefly, RNA was isolated from harvested samples and homogenized in TRIzol reagent (Gibco-BRL, Bethesda, MD) according to the manufacturer's recommendation. In order to eliminate contamination with genomic DNA, the extracted RNA was treated with the DNase 1 amplification reagent (Invitrogen, Carlsbad, CA), and the RNA concentration was quantified by spectrophotometry. RNA quality was determined by denaturing samples in a 1.5% agarose gel electrophoresis stained with ethidium bromide and visualized on a UV transilluminator. The cDNA was synthesized from 500 ng of total RNA combined with primer mix containing oligo(dT) primer (0.2 μg/ml), random hexamer primer (300 μg/ml; Invitrogen), and deoxynucleoside triphosphate mix (10 mM each), followed by incubation at 65°C for 5 min. After incubation, SuperScript II reverse transcriptase (Invitrogen) was added to the reaction according to manufacturer's recommendations and reverse transcribed under the following conditions: 25°C for 10 min, 42°C for 60 min, and 70°C for 5 min. To test for genomic DNA contamination, control reactions were prepared without reverse transcriptase.
Specific oligonucleotide primers (Table 1) were generated in Oligo 5 program (Molecular Biology Insights, Inc.) for the following genes: the M segment of glycoprotein 1 of CVV, alpha interferon (IFN-α), IFN-β, TNF-α, IL-1, IL-6, Mx1, Mx2, ISG15, TLR7, and TLR8. The real-time qPCRs was performed using the ABI prism 7900HT system (Applied Biosystems, Foster, CA) with power SYBR green PCR Master Mix (Applied Biosystems). Primer specificity and efficiency (−3.6 > slope > −3.1) were confirmed by using a test amplification run. The data were normalized using cycle threshold (CT) values for the ovine glyceraldehyde 3-phosphate dehydrogenase gene (GADPH) in each sample. Semiquantitative analysis was calculated with the ΔΔCT method, and results are expressed as the relative fold changes compared to the lowest value for mock-infected samples. To calculate the relative amounts of viral mRNA, the relative fold changes were compared to the infected sample with the lowest ΔΔCT.
Table 1.
Primer sequences used for RT-qPCR
| Gene | Accession no. | Primer sequence (5′–3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| CVV | AF082576.1 | CAC CAG CGA AAT CCC AAT CAC CA | CTC CAG ACA TAG CAC CCA CCA |
| TNF-αα | X55152.1 | CGG CGT GGA GCT GAA AGA CAA | CTG CGA GTA GAT GAG GTA AA |
| IL-1 | NM_001009465.2 | AGT GAT GGC TTG CTA CAG T | CCG AGG TCC AGG TGT T |
| IL-6 | X68723.1 | GAG GGA AAT CAG GAA ACT GT | CTC GTT TGA GGA CTG CAT CT |
| TLR7 | HQ529279.1 | ACT CCT TGG GGC TAG ATG GT | GCT GGA GAG ATG CCT GCT AT |
| TLR8 | FJ905847.1 | TCC ACA TCC CAG ACT TTC TA | GTT CTT GTC CTC ACT CTC TT |
| Mx1 | JN377734.1 | GTA CGA GCC GAG TTC TCC AA | ATG TCC ACA GCA GGC TCT TC |
| Mx2 | NM_001078652.1 | CAT CCA TAA ATC GCT CCC CTT GT | GCT CCT CTG TCG CCC TCT GGT |
| ISG15 | FJ844480.1 | TGA CGG TGA AGA TGC TAG GG | ACT GCT TCA GCT CGG ATA CC |
| IFN-α | X55152.1 | ACC CAG CAC ACC TTC CAG CTC TT | CCT CGC AGC CCC TCC TC |
| IFN-β | EU276065.1 | TGG TTC TCC TGC TGT GTT TCT C | CGT TGT TGG AAT CGA AGC AA |
Protein slot blot.
A protein slot blot assay was used to determine and compare relative concentrations of the Mx1 protein in the fetal fluids of infected versus noninfected fetuses. The starting concentration of protein in allantoic and amniotic fluids was determined by using the Bradford assay as recommended by the manufacturer (Bio-Rad Laboratories). According to the protein concentration, samples were prepared and diluted to 12 μg of protein for the amniotic fluid and 50 μg of protein for the allantoic fluid in a final volume of 200 μl of Tris-buffered saline (TBS). The volume was deposited onto a nitrocellulose membrane (Bio-Rad Laboratories, Richmond, CA) in a Bio-Dot SF microfiltration apparatus (Bio-Rad Laboratories) according to a modified protocol, as previously published (31). Briefly, nonspecific binding sites were blocked by immersing the membrane in 5% skim milk in TBS-Tween (TBST) for 1 h at room temperature on an orbital shaker. The membranes were incubated overnight at 4°C in 2.5% milk-TBST containing a rabbit anti-ovine Mx1 polyclonal antibody (a gift from Troy Ott, Penn State University) (32) at a concentration of 1:10,000. The membrane was washed three times and then incubated for 1 h with goat anti-rabbit IgG (1:10,000 dilution in 2.5% milk-TBST). After incubation with the secondary antibody, the membrane was washed three times and further incubated for 1 min with Bio-Rad chemiluminescence reagent. The relative concentration of the Mx1 protein in triplicate samples was determined quantitatively in relative light units using a Bio-Rad Chemidoc imager and assessed by using QuantityOne Image analysis software (31).
Immunohistochemistry.
Immunohistochemistry was performed on 5-μm, deparaffinized sections of paraformaldehyde-fixed tissues mounted onto positively charged, silanized slides using an automated system for immunohistochemistry (DakoCytomation Autostainer; Dako, Carpinteria, CA) with minor modifications to a previously described immunohistochemistry protocol (15). Antigen retrieval was performed in a decloaker chamber (Biocare Medical, Concord, CA) with slides soaked in citrate buffer (pH 6.0). Sections were incubated with anti-ISG15 polyclonal antibody (diluted 1:500) for 30 min (the antibody was a gift from Thomas Hansen, Colorado State University) (33), CD3 (diluted 1:400) polyclonal antibody for 35 min (Dako), anti-CD79a polyclonal antibody (diluted 1:200) for 30 min (Dako), anti-ovine IgG (diluted 1:500) for 35 min, and anti-ovine IgM (diluted 1:2,000) for 30 min (KPL, Inc., Gaithersburg, MD). The primary antibody was followed by incubation with the secondary antibody (MACH 2 anti-rabbit polymer-HRP from Biocare Medical [CD3, IgG, and IgM] or ImmPress anti-mouse immunoglobulin (peroxidase) polymer from Vector Laboratories [ISG15, CD79a]) for 20 min. A chromogen complex, 3,3-diaminobenzidine tetrachloride (DAB; Dako), was used to detect the targeted antigens, and sections were counterstained with hematoxylin. Slides were coverslipped with Permount mounting solution. Tissues were evaluated to determine percentage of infected cells in each examined organ and graded as follows: mild, <3% of cells positive; moderate, between 3 and 15% of cells positive; and marked, >15% of cells positive.
Statistical analysis.
The quantitative data for mRNA expression and protein levels on fetal fluids were subjected to the Wilcoxon signed-rank test to determine differences between infected and noninfected groups. Differences between groups at different days postinfection were subjected to a one-pair Student t test. In order to reduce heterogeneity among samples, the data were log transformed if necessary, and P values below 0.05% were considered statistically significant. The data are presented as means and overall standard errors.
RESULTS
Real-time qPCR.
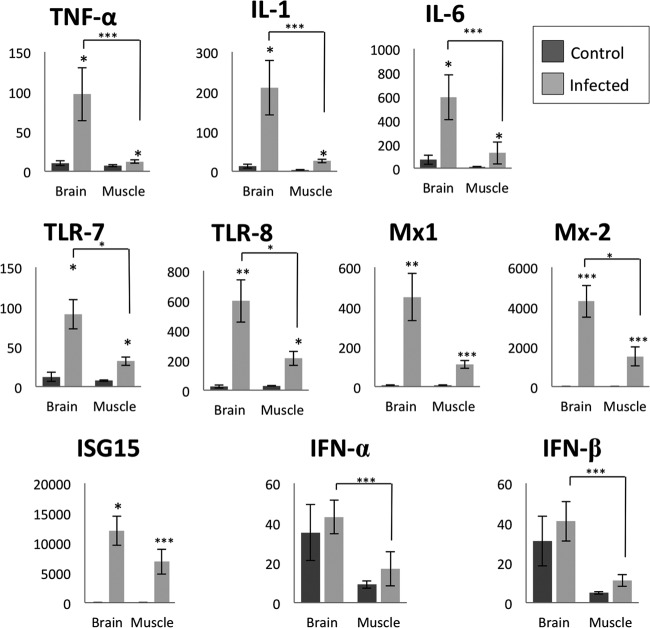
No significant difference in gene expression was observed either between infected animals at each different days postinfection or between control animals at different days of gestation. Therefore, the data for each gene for each evaluated tissue were pooled into two groups: control and infected animals (Fig. 1). qPCR analysis revealed that the IL-1, IL-6, Mx2, ISG15, TLR7, and TLR8 genes were upregulated (P < 0.05) in the brain and skeletal muscle (SKM) of CVV-infected fetuses compared to noninfected fetuses. Only in the brain was the TNF-α gene significantly upregulated. All examined genes, but not ISG15 and Mx1, had significantly higher upregulation in the brain compared to the skeletal muscle (P < 0.05). Similarly, the relative number of copies of CVV was significantly higher (P < 0.05) in the brain than in skeletal muscle (Fig. 2). Progressively lower means of relative number of copies of CVV were observed in both brain and SKM with the progression of the infection. A significant difference (P < 0.05) was observed in the brains of fetuses at between 14 and 28 dpi (CVV in the brains of 10-dpi fetuses was excluded from analysis due to high variability among these samples) and in SKM when comparing 10 to 14 dpi, 10 to 21 dpi, and 10 to 28 dpi.
Fig 1.
Relative levels of genes expression in brain and skeletal muscle of control and Cache Valley virus-infected animals. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Fig 2.
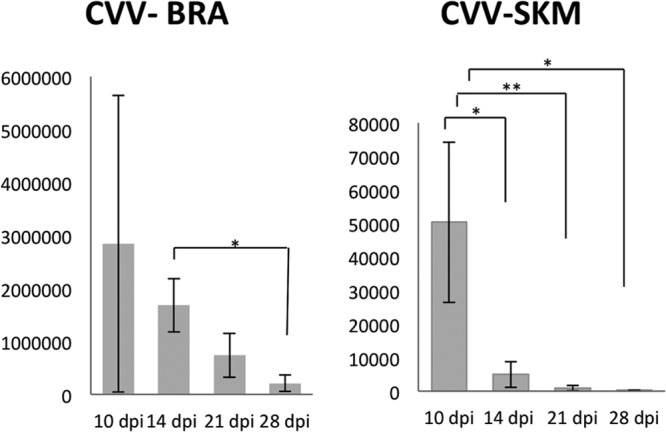
Relative number of Cache Valley virus RNA copies in main target tissues, brain (BRA) and skeletal muscle (SKM), at 10, 14, 21, and 28 dpi. *, P < 0.05; **, P < 0.01.
Protein slot blot.
A significant (P < 0.01) increase in the amount of Mx1 protein in the allantoic and amniotic fluid occurred in infected fetuses versus noninfected fetuses at each time point examined (Fig. 3). Similar to the quantitative data collected with the real-time qPCR, no effect of day or interaction of day versus infection was detected in evaluated samples.
Fig 3.
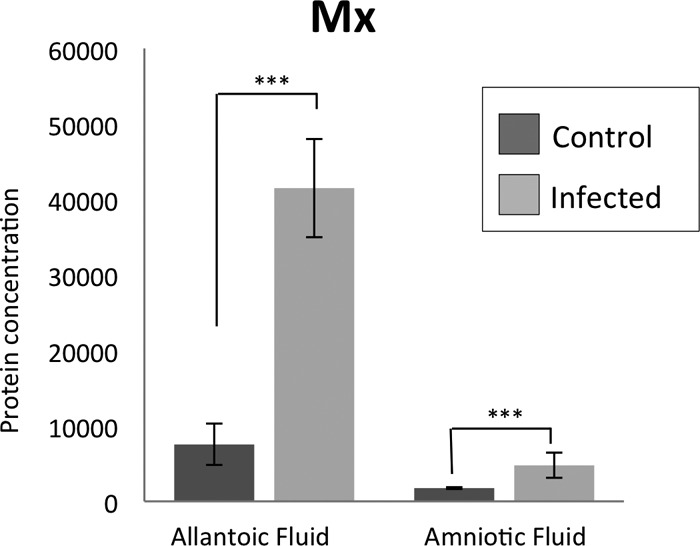
Relative amounts of ovine Mx1 protein in allantoic and amniotic fluid in control and infected animals. The presented values (y axis) are the relative light units using the Bio-Rad Chemidoc imager and assessed using QuantityOne Image analysis software. The starting protein concentration for the samples from the allantoic fluid was 50 μg and for the amniotic fluid was 12 μg. The data was log transformed for normalization. The infected fetuses had relatively higher amounts of Mx1 protein in the allantoic and amniotic fluid compared to the control fetuses. ***, P < 0.005.
Immunohistochemistry for ISG15.
ISG15 was detected in several tissues of all infected fetuses, but no ISG15 immunolabeling was observed in control fetuses (Fig. 4). In infected fetuses, a strong signal was observed in numerous cells of the cerebral parenchyma and meninges, and fewer cells were positive for ISG15 in the spinal cord. Most fetuses at 7 to 21 dpi had marked ISG15 antigen signal in the brain, with moderate signal observed at 28 dpi. The ISG15 antigen signal in the spinal cord of infected fetuses varied between mild to moderate at all test points. Multifocal clusters of cells with positive ISG15 immunolabeling were observed in the SKM of infected fetuses through 21 dpi with only rare cells having a positive signal at 28 dpi. Other tissues with clusters of cells with positive signal included: heart, smooth muscle of intestine, tongue, fibroblasts in the subcutaneous tissue, wall of large arteries, cells around small blood vessels, spleen (one fetus at 21 dpi), and rarely in the lungs, tonsils, and thymus of fetuses infected earlier.
Fig 4.
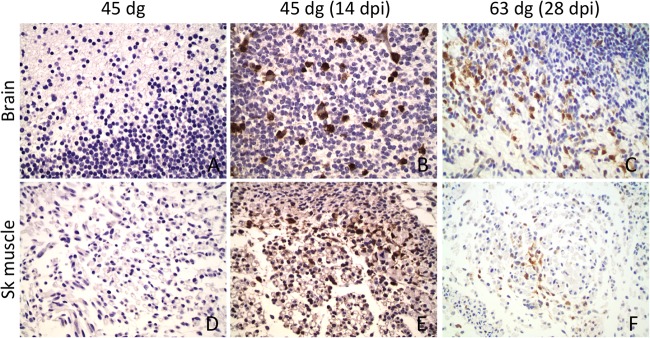
Immunohistochemistry for ISG15 in brain and skeletal muscle. A rabbit polyclonal antibody against ovine ISG15 protein was stained with DAB chromogen (brown) on the brains (A, B, and C) and skeletal muscle (D, E, and F) from 45-dg control fetuses (A and D) and from infected fetuses at 14 dpi (45 dg) (B and E) and 28 dpi (63 dg) (C and F). Numerous cells in the brains and skeletal muscle of infected fetuses are positive for ISG15. No positive cells were observed in the tissues of control fetuses.
Immunohistochemistry of B and T lymphocytes, IgG, and IgM.
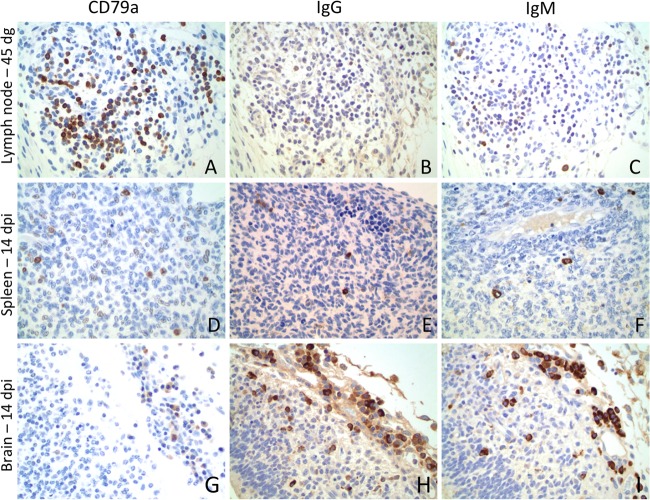
At 7 dpi/42 dg, scattered CD79a-positive cells were observed in the dorsal abdominal cellular lymphoid aggregates around the abdominal aorta, which correspond to sites of development of rudimentary renal or mesenteric lymph nodes, and rarely in the hepatic sinusoids in both control and infected animals. At 10 dpi/45 dg, slightly increased numbers of CD79a-, IgG-, and IgM-positive cells were observed in the same locations in infected and control animals (Fig. 5A to C). Rare IgM-positive cells were observed in the meninges of one infected animal. At 14 dpi/49 dg, slightly increased numbers of CD79a-, IgG-, and IgM-positive cells were observed in the sites described above, and at this time point, CD79a- and IgM-positive cells were in the spleen of one of the infected animals (Fig. 5D to F). Surprisingly, another infected animal also had a marked infiltration of CD79a-, IgG-, and IgM-positive cells in the meninges of the brain and spinal cord and in rare cells within the parenchyma (Fig. 5G to I). Increased numbers of CD79a-, IgG-, and IgM-positive cells were arranged around splenic arterioles in both infected and control animals at 21 dpi/56 dg and 28 dpi/63 dg, besides also being in the previously described sites. Numerous IgG- and IgM-positive cells were observed in the meninges of the spinal cord of two infected fetuses at 21 dpi, and rare immunoglobulin-positive cells were observed in one fetus at 28 dpi. Moderate to marked numbers of CD3-positive cells, evaluated only in sections of brain and transverse sections of the abdomen, including the lumbar spinal cord of control and infected fetuses at 21 and 28 dpi, were detected in the spleen and in renal and mesenteric lymph nodes in all fetuses. Scattered CD3-positive cells were in the meninges of the brain and spinal cord of one infected fetus at 14 dpi and in the meninges of the spinal cord in one fetus at 21 dpi.
Fig 5.
Immunohistochemistry for B cells, plasma cells, and ovine IgG and IgM in the lymph node, spleen, and brain. A polyclonal antibody against B cells and plasma cells (CD79a), ovine IgG, and ovine IgM were stained with DAB chromogen (brown). CD79a (A), IgG (B) and IgM (C) positive cells were observed in rudimentary lymph nodes at 42 to 45dg in control fetuses. CD79a (D)-, IgG (E)-, and IgM (F)-positive cells were observed in the spleens of infected fetuses. A remarkable number of CD79a (G)-, IgG (H)-, and IgM (I)-positive cells were seen in the leptomeninges of an infected fetus at 14 dpi.
DISCUSSION
Ovine fetuses are able to mount an immune response associated with CVV infection earlier in gestation than previously described (10, 11, 16, 17). Although the fetal immune response was not able to completely clear virus by the end of the study at 63 dg (15), infected ovine fetuses were able to reduce the viral load within the CNS and MSS. Viral signal was also markedly reduced or absent in the ganglion, retina, kidney, and heart.
CVV targets mainly cells within the CNS and MSS, and viral signal was markedly reduced in the brain and almost completely cleared in the skeletal muscle after 21 to 28 dpi in fetuses infected at 35 dg (15). Similarly, a progressive reduction in viral load, as shown by mean relative amounts of CVV RNA copies, was observed in the brain and SKM in the present study, with significant differences observed between most days of infection in the SKM, and in the brain at between 14 and 28 dpi. In addition, in all groups, the relative numbers of CVV RNA copies in the brain were significantly higher than in the skeletal muscle.
The Mx protein, an IFN-stimulated GTPase involved in intracellular trafficking, membrane remodeling, and fusion processes, is capable of restricting growth of viruses, including influenza virus, measles virus, and bunyaviruses (including the Orthobunyavirus, Phlebovirus, and Hantavirus genera) (22–24, 26, 34–38). In viral infections, the Mx protein acts by interfering with transport of viral components in infected cells. In a model proposed by Haller and Kochs (35), MxA forms large, membrane-associated self-assemblies that store monomers of this protein. With infection, monomers bind to viral target structures, forming new assemblies involving viral intermediates that lead to mislocalization of viral components and consequent inhibition of viral replication (35). Human MxA has been shown to inhibit replication of La Crosse virus by binding to the viral nucleocapsid protein and forming large copolymers that accumulate in the perinuclear region (24). Previous studies have demonstrated antiviral activity of bovine Mx2 in cattle infected with bovine viral diarrhea virus (BVDV) (28, 39). Because Mx has a demonstrated antiviral effect against bunyaviruses, the present study quantified expression of the Mx molecule in infected tissues. A significant upregulation of ovine Mx1 and Mx2 was observed in evaluated tissues from infected fetuses compared to controls. Increased secretion of Mx1 was identified in fetal fluids in infected ovine fetuses. Thus, this protein likely contributes to the clearance of CVV infection.
The ISG15 gene was markedly upregulated in the brain and in the skeletal muscle and highly expressed in the tissues from infected animals. Similar to the Mx protein, ISG15 is induced by type I IFN and has been associated with antiviral activity (29). In recent studies, ISG15 has been shown to conjugate to proteins in a manner similar to ubiquitin (40, 41). Modifications and interference of antiviral signaling pathways involving ISG15 are mechanisms used by Crimean-Congo hemorrhagic fever virus, a bunyavirus, to evade the innate immune response (40). Upregulation of numerous ISG genes, including ISG15, and continuous stimulation of the innate antiviral response have been demonstrated in blood and tissues of bovine fetuses and cattle infected with BVDV (28, 39).
Although the ISG15, Mx1, and Mx2 genes were abundantly expressed in all infected fetuses, the expression of other selected IFN-stimulated genes was not altered. Previous studies have demonstrated that ovine fetuses within the second and third trimester of gestation are able to produce levels of interferon similar or greater to those produced by adult animals in response to a viral infection (42). Since type I IFN-induced genes were upregulated, it is presumed that the expression of type I IFN genes were also upregulated at the same or similar times and that, due to the short half-life of type I IFNs, the timing of upregulation of the type I IFN(s) was not detected at the time points evaluated. Even though less likely, one must consider the possibility of the upregulation of both ISG15 and ovine Mx by an IFN-independent pathway.
CD79a-positive cells (B lymphocytes and plasma cells) were observed in the spleens of CVV-infected fetuses at 49 dg, a finding which is in agreement with previous studies that identified B lymphocytes in ovine fetal lymph nodes and the spleen as early as 47 to 48 dg (19, 20, 43). Both infected and noninfected fetuses had scattered CD79a-, IgM-, and IgG-positive cells in lymphoid aggregates in the dorsal aspect of the abdominal cavity, sites of development of rudimentary renal and/or mesenteric lymph nodes, as early as 42 and 45 dg, respectively. The identification of CD79a-positive cells in regional lymph nodes earlier than observed in the fetal spleen supports the previously proposed idea that extrasplenic sites have the potential of producing B lymphocytes (43, 44) and potentially explains the fact that splenectomized fetuses are also capable of colonizing Peyer's patches with B lymphocytes (43, 45).
A marked infiltration of CD79a-, IgM-, and IgG-positive cells was observed within the meninges with fewer cells within the parenchyma of the brains and spinal cords of infected fetuses at 49 dg. It appears that the ovine fetus not only mounts an innate immune response to a viral infection but is also able to stimulate an adaptive immune response. Unfortunately, blood samples were not available from the fetuses during these experiments; therefore, fetal neutralizing antibodies could not be assayed. In ovine fetuses infected with Akabane virus, the earliest day of gestation where IgM- and IgG-positive cells were observed in tissues was at 59 to 60 dg. Titers of >1:4 were detected only after 100 dg (10, 46). Two other studies detected Akabane virus neutralizing antibodies after 76 to 78 dg, with titers ranging from 8 to 64 in one of these studies (11, 16).
CVV is a viral pathogen with a tropism for the ovine fetal CNS, skeletal muscle, and fetal membranes. The study of early infection demonstrated that a tropism correlated well with the CNS and musculoskeletal malformations observed in spontaneous CVV disease. With CVV, the development of arthrogryposis probably has a multifactorial pathogenesis involving effects on developing neurons, myocytes, and fetal membranes. CVV is the only source of viral infection shown to cause oligohydramnios. The virus is partially cleared from most fetal tissues by ∼65 dg (28 dpi), before the presently accepted onset of ovine immunocompetence.
The ovine fetus is able to mount an immune response associated with CVV infection earlier in gestation by upregulation of genes that participate in the innate immune response. The infected fetus upregulates Mx genes, which have been shown to restrict growth of bunyaviruses. In addition, cells of the adaptive immune response—CD79a-, IgM-, and IgG-positive cells—are found in large numbers in infected tissues. Whether the immunoglobulin in these cells is actually CVV specific is still unclear. Further studies should be conducted for better understanding of the immune system development in the ovine fetuses exposed to CVV and other viral infections.
ACKNOWLEDGMENTS
This study was supported by the USDA Animal Formula Health Research grant AH-9249.
We thank Andy Ambrus, the staff of the histology laboratory, Hoai Jaclyn Li from the Department of Veterinary Pathobiology, Andrew Steelman from the Department of Veterinary Integrative Biosciences at Texas A&M University, and Pamela J. Ferro from the Texas Veterinary Medical Diagnostic Laboratory for their excellent technical support.
Footnotes
Published ahead of print 6 March 2013
REFERENCES
- 1. Schmaljohn CS, Nichol ST. 2007. Bunyaviridae, p 1741–1790 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology. Lippincott/Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Edwards JF. 1994. Cache Valley virus. Vet. Clin. N. Am. Food Anim. Pract. 10:515–524 [DOI] [PubMed] [Google Scholar]
- 3. Chung SI, Livingston CW, Jr, Edwards JF, Gauer BB, Collisson EW. 1990. Congenital malformations in sheep resulting from in utero inoculation of Cache Valley virus. Am. J. Vet. Res. 51:1645–1648 [PubMed] [Google Scholar]
- 4. Chung SI, Livingston CW, Jr, Jones CW, Collisson EW. 1991. Cache Valley virus infection in Texas sheep flocks. J. Am. Vet. Med. Assoc. 199:337–340 [PubMed] [Google Scholar]
- 5. Concha-Bermejillo Ad. l. 2003. Cache Valley Virus is a cause of fetal malformation and pregnancy loss in sheep. Small Ruminant Res. 49:1–9 [Google Scholar]
- 6. Edwards JF, Angulo A, Pannell E. 2003. Theriogenology question of the month. JAMA 222:1361–1362 [DOI] [PubMed] [Google Scholar]
- 7. Edwards JF, Livingston CW, Chung SI, Collisson EC. 1989. Ovine arthrogryposis and central nervous system malformations associated with in utero Cache Valley virus infection: spontaneous disease. Vet. Pathol. 26:33–39 [DOI] [PubMed] [Google Scholar]
- 8. Erwin P. 2010. First case of Cache Valley fever found in Kansas, p 3 In Bryant B., Domer K. (ed), Kansas Animal Health News; Topeka, KS [Google Scholar]
- 9. Charles JA. 1994. Akabane virus. Vet. Clin. N. Am. Food Anim. Pract. 10:525–546 [DOI] [PubMed] [Google Scholar]
- 10. Hashiguchi Y, Nanba K, Kumagai T. 1979. Congenital abnormalities in newborn lambs following Akabane virus infection in pregnant ewes. Natl. Inst. Anim. Health Q. (Tokyo) 19:1–11 [PubMed] [Google Scholar]
- 11. Parsonson IM, Della-Porta AJ, Ohalloran ML, Snowdon W, Fahey KJ, Standfast HA. 1981. Akabane virus infection in the pregnant ewe. 1. Growth of virus in the fetus and the development of the fetal immune response. Vet. Microbiol. 6:197–207 [Google Scholar]
- 12. Gibbens N. 2012. Schmallenberg virus: a novel viral disease in northern Europe. Vet. Rec. 170:58. [DOI] [PubMed] [Google Scholar]
- 13. Campbell GL, Mataczynski JD, Reisdorf ES, Powell JW, Martin DA, Lambert AJ, Haupt TE, Davis JP, Lanciotti RS. 2006. Second human case of Cache Valley virus disease. Emerg. Infect. Dis. 12:854–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sexton DJ, Rollin PE, Breitschwerdt EB, Corey GR, Myers SA, Dumais MR, Bowen MD, Goldsmith CS, Zaki SR, Nichol ST, Peters CJ, Ksiazek TG. 1997. Life-threatening Cache Valley virus infection. N. Engl. J. Med. 336:547–549 [DOI] [PubMed] [Google Scholar]
- 15. Rodrigues Hoffmann A, Welsh CJ, Wilcox Varner P, de la Concha-Bermejillo A, Marchand Ball J, Ambrus A, Edwards JF. 2012. Identification of the target cells and sequence of infection during experimental infection of ovine fetuses with Cache Valley virus. J. Virol. 86:4793–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McClure S, McCullagh P, Parsonson IM, McPhee DA, Della-Porta AJ, Orsini A. 1988. Maturation of immunological reactivity in the fetal lamb infected with Akabane virus. J. Comp. Pathol. 99:133–143 [DOI] [PubMed] [Google Scholar]
- 17. Popp SK, Mann DA, Milburn PJ, Gibbs AJ, McCullagh PJ, Wilson JD, Tonjes RR, Simeonovic CJ. 2007. Transient transmission of porcine endogenous retrovirus to fetal lambs after pig islet tissue xenotransplantation. Immunol. Cell Biol. 85:238–248 [DOI] [PubMed] [Google Scholar]
- 18. Miyasaka M, Morris B. 1988. The ontogeny of the lymphoid system and immune responsiveness in sheep. Prog. Vet. Microbiol. Immunol. 4:21–55 [PubMed] [Google Scholar]
- 19. Maddox JF, Mackay CR, Brandon MR. 1987. Ontogeny of ovine lymphocytes. II. An immunohistological study on the development of T lymphocytes in the sheep fetal spleen. Immunology 62:107–112 [PMC free article] [PubMed] [Google Scholar]
- 20. Maddox JF, Mackay CR, Brandon MR. 1987. Ontogeny of ovine lymphocytes. III. An immunohistological study on the development of T lymphocytes in sheep fetal lymph nodes. Immunology 62:113–118 [PMC free article] [PubMed] [Google Scholar]
- 21. Tizard IR. 1996. Immunity in the fetus and newborn, p 237–250 In Veterinary immunology: an introduction, 5th ed WB Saunders, Philadelphia, PA [Google Scholar]
- 22. Frese M, Kochs G, Feldmann H, Hertkorn C, Haller O. 1996. Inhibition of bunyaviruses, phleboviruses, and hantaviruses by human MxA protein. J. Virol. 70:915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hefti HP, Frese M, Landis H, Di Paolo C, Aguzzi A, Haller O, Pavlovic J. 1999. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La Crosse virus and other lethal viral infections. J. Virol. 73:6984–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kochs G, Janzen C, Hohenberg H, Haller O. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc. Natl. Acad. Sci. U. S. A. 99:3153–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sandrock M, Frese M, Haller O, Kochs G. 2001. Interferon-induced rat Mx proteins confer resistance to Rift Valley fever virus and other arthropod-borne viruses. J. Interferon Cytokine Res. 21:663–668 [DOI] [PubMed] [Google Scholar]
- 26. Stertz S, Dittmann J, Blanco JC, Pletneva LM, Haller O, Kochs G. 2007. The antiviral potential of interferon-induced cotton rat Mx proteins against orthomyxovirus (influenza), rhabdovirus, and bunyavirus. J. Interferon Cytokine Res. 27:847–855 [DOI] [PubMed] [Google Scholar]
- 27. Chen S, Short JA, Young DF, Killip MJ, Schneider M, Goodbourn S, Randall RE. 2010. Heterocellular induction of interferon by negative-sense RNA viruses. Virology 407:247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shoemaker ML, Smirnova NP, Bielefeldt-Ohmann H, Austin KJ, van Olphen A, Clapper JA, Hansen TR. 2009. Differential expression of the type I interferon pathway during persistent and transient bovine viral diarrhea virus infection. J. Interferon Cytokine Res. 29:23–35 [DOI] [PubMed] [Google Scholar]
- 29. Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dorniak P, Bazer FW, Spencer TE. 2011. Prostaglandins regulate conceptus elongation and mediate effects of interferon tau on the ovine uterine endometrium. Biol. Reprod. 84:1119–1127 [DOI] [PubMed] [Google Scholar]
- 31. Satterfield MC, Bazer FW, Spencer TE. 2006. Progesterone regulation of preimplantation conceptus growth and galectin 15 (LGALS15) in the ovine uterus. Biol. Reprod. 75:289–296 [DOI] [PubMed] [Google Scholar]
- 32. Johnson GA, Joyce MM, Yankey SJ, Hansen TR, Ott TL. 2002. The Interferon Stimulated Genes (ISG) 17 and Mx have different temporal and spatial expression in the ovine uterus, suggesting more complex regulation of the Mx gene. J. Endocrinol. 174:R7–R11 [DOI] [PubMed] [Google Scholar]
- 33. Austin KJ, Carr AL, Pru JK, Hearne CE, George EL, Belden EL, Hansen TR. 2004. Localization of ISG15 and conjugated proteins in bovine endometrium using immunohistochemistry and electron microscopy. Endocrinology 145:967–975 [DOI] [PubMed] [Google Scholar]
- 34. Haller O, Frese M, Kochs G. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. 17:220–230 [DOI] [PubMed] [Google Scholar]
- 35. Haller O, Kochs G. 2002. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710–717 [DOI] [PubMed] [Google Scholar]
- 36. Haller O, Staeheli P, Kochs G. 2007. Interferon-induced Mx proteins in antiviral host defense. Biochimie 89:812–818 [DOI] [PubMed] [Google Scholar]
- 37. Haller O, Stertz S, Kochs G. 2007. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 9:1636–1643 [DOI] [PubMed] [Google Scholar]
- 38. Kochs G, Reichelt M, Danino D, Hinshaw JE, Haller O. 2005. Assay and functional analysis of dynamin-like Mx proteins. Methods Enzymol. 404:632–643 [DOI] [PubMed] [Google Scholar]
- 39. Hansen TR, Smirnova NP, Van Campen H, Shoemaker ML, Ptitsyn AA, Bielefeldt-Ohmann H. 2010. Maternal and fetal response to fetal persistent infection with bovine viral diarrhea virus. Am. J. Reprod. Immunol. 64:295–306 [DOI] [PubMed] [Google Scholar]
- 40. Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. 2011. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U. S. A. 108:2228–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang D, Zhang DE. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J. Interferon Cytokine Res. 31:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rinaldo CR, Jr, Overall JC, Jr, Glasgow LA. 1975. Viral replication and interferon production in fetal and adult ovine leukocytes and spleen cells. Infect. Immun. 12:1070–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Press CM, Hein WR, Landsverk T. 1993. Ontogeny of leucocyte populations in the spleen of fetal lambs with emphasis on the early prominence of B cells. Immunology 80:598–604 [PMC free article] [PubMed] [Google Scholar]
- 44. Alitheen N, McClure S, McCullagh P. 2007. Detection and quantification of IgM(+) lymphocytes in fetal lamb spleen, liver, and lymph nodes by flow cytometry. Immunol. Cell Biol. 85:391–393 [DOI] [PubMed] [Google Scholar]
- 45. Press CM, McCullagh P, Landsverk T. 2001. Effect of early fetal splenectomy on prenatal B-cell development in sheep. Immunology 102:131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Narita M, Kawashima K. 1993. Detection of Akabane viral antigen and immunoglobulin-containing cells in ovine fetuses by use of immunoperoxidase staining. Am. J. Vet. Res. 54:420–424 [PubMed] [Google Scholar]