Abstract
A recombinant sleeping disease virus (rSDV) was previously shown to be totally attenuated and provide long-term protection in trout (C. Moriette, M. Leberre, A. Lamoureux, T. L. Lai, M. Brémont, J. Virol. 80:4088–4098, 2006). Sequence comparison of the rSDV to wild-type genomes exhibited a number of nucleotide changes. In the current study, we demonstrate that the virulent phenotype of SDV was essentially associated with two amino acid changes, V8A and M136T, in the E2 glycoprotein, with the V8A change mostly being involved in the acquisition of the virulent phenotype.
TEXT
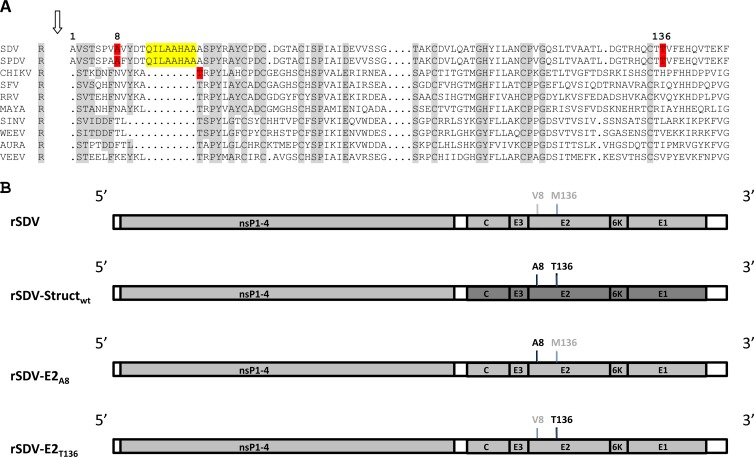
Sleeping disease in trout was first observed in France in 1985 (1). Disease in rainbow trout is characterized by the abnormal behavior of fish, which stay on their side at the bottom of the tanks, reminiscent of a sleeping state; thus, the name sleeping disease (2). Viral etiology of this disease has been suspected (3, 4), and this was further confirmed in our laboratory as being alphavirus-like (5). Genome sequences of the sleeping disease virus (SDV) and the closely related salmon pancreas disease virus (SPDV) have now been entirely determined (5, 6). SDV and SPDV genomes consist of a single positive-stranded RNA molecule of 11,900 and 11,919 nucleotides in length, respectively. As for all alphaviruses, their genomes encode two polyproteins, which, after processing through proteolytic cleavages, produce mature viral products: the nonstructural proteins nsP1, nsP2, nsP3, and nsP4, the structural proteins capsid (C), and the two external glycoproteins (E2 and E1). The close relationship between SDV and SPDV allowed suggesting the unofficial classification as salmonid alphavirus (SAV). This proposed classification is based on at least three main features: (i) SDV and SPDV are closely related but differ from the other alphaviruses, (ii) nonstructural and structural proteins are larger than those of mammalian alphaviruses, and (iii) arthropod-independent virus transmission to the host has been demonstrated in cohabitation experiments (7) and also through experimental infections by bath immersion, even for a single fish. To our knowledge, this phenomenon has never been documented for mammalian alphaviruses. An SDV-derived infectious cDNA has been established previously by constructing a pSDV plasmid, which contains a full-length SDV genome cDNA copy derived from a highly virulent S49P SDV strain (4), under the control of a cytomegalovirus (CMV) promoter (8). Following transfection of fish cells with pSDV, the recovered rSDV was shown to be totally attenuated and protective in trout. Nucleotide sequencing of wild-type SDV (wtSDV) and rSDV genomes showed a large number of nucleotide changes that could explain the pathogenic versus attenuated phenotype change. In the current study, we investigated if we could determine which nucleotide and, thus, which amino acid changes were responsible for the phenotype change. Starting from the initial pSDV construct, the complete cDNA fragment, of roughly 4.2 kb, encoding the structural proteins was removed by BlpI-EcoRV restriction enzyme digestion (Fig. 1) and replaced by that derived from the virulent wtSDV amplified by reverse transcription-PCR (RT-PCR), using the primers SDVcDNA [5′-TCTAGAGATATCT(17)-3′] and SDVPCR (5′-GAATTCGCTCAGCATGTTTCCCATGCAATTCACC-3′). (Restriction enzyme sites EcoRV and BlpI, respectively, are underlined.) Following digestion with BlpI and EcoRV, the PCR product was inserted in the digested pSDV, leading to pSDVStructwt. The chimeric rSDVStructwt was rescued following electroporation of the pSDVStructwt DNA plasmid construct into bluegill fry BF2 cells (Amaxa Biosystems). Since the viral titer is very low following DNA plasmid construct transfection into fish cells, all of the recovered SDV recombinants were amplified though two passages on fresh cells, and then their titers were determined. To evaluate whether the chimeric rSDVStructwt replicates as well as rSDV in cell culture, a comparative growth curve was established. Both the rSDV and rSDVStructwt viruses are indistinguishable in terms of replication kinetics (data not shown). To evaluate the pathogenicity of this chimeric rSDVStructwt in trouts, four tanks containing 50 juvenile trout each (mean weight of 4.4 g) were infected by bath immersion with 5 × 104 PFU/ml of wtSDV, rSDV, or rSDVStructwt or were mock infected. Mortalities were recorded over a period of 60 days. As illustrated in Fig. 2, the chimeric rSDVStructwt is as pathogenic as the wtSDV, and both viruses induce a similar rate of 57% of cumulative mortality at 2 months postinfection. In contrast, mock- or rSDV-infected trout all survived. This observation indicates that while the rSDV is totally attenuated, the amino acid changes present in the wtSDV structural polyprotein are sufficient to restore the wild-type phenotype of the chimeric rSDVStructwt. A comparison of the nucleotide sequences (GenBank accession numbers KC593282 and KC593283) of the whole structural region of rSDV and wtSDV from RNA genome extracted from viruses produced after 2 passages in cell culture (as described above) show that only 2 single-nucleotide substitutions (GCC versus GTC and ATG versus ACG), leading to two nonconservative amino acid changes, were present in the E2 glycoprotein. The amino acid change V8A (according to SDV E2 amino acid numbering; and attenuated versus virulent SDV) is located in the amino terminus of the E2 glycoprotein in a domain which is unique in E2 of the salmonid alphavirus compared to those of other alphaviruses sequenced so far, as shown in the multiple alignments of the E2 amino acid sequences of the main alphavirus prototypes (Fig. 3A). The second amino acid change, M to T, is in position 136 of E2. To correct each single mutation in the attenuated SDV genome, a BsrGI DNA fragment of 1,940 nucleotides (Fig. 1) was amplified by PCR using the following primers: BsrGIF, 5′-CATGATGGATGGAGTGTACAATT-3′, and BsrGIR, 5′-CTTGCAGCTCTGTACATACGCAA-3′. The PCR product was cloned in the pJET 1.2 vector (Fermentas) and served as a template to correct the E2 gene sequence. Mutagenesis of V8A and M136T was achieved through a site-directed mutagenesis assay using the QuikChange Multi site-directed mutagenesis kit (Stratagene) using the following mutagenesis primers: SDVMutVA, 5′-CGTCGCCTGTCGCCGCTTACGACACACAAATTCTCG-3′, and SDVMutMT, 5′-CGGCATCAATGCACCACGGTTTTCGAACATCAAG-3′. Following mutagenesis, a BsrGI DNA insert was excised from the pJET 1.2 vector and inserted back into the BsrGI-digested pSDV, leading to the final constructs pSDV-E2V8A and pSDV-E2M136T. Both plasmid constructs were separately electroporated (Amaxa Biosystems) into BF2 cells. The corresponding rSDV-E2A8 and rSDV-E2T136 mutants were recovered (Fig. 3B) and amplified, and their titers were determined after 2 passages in cell culture (Table 1). Part of the genome encoding the structural proteins has been sequenced after amplification in cell culture to ascertain that the E2 A8 and T136 mutations are still present and that no other mutations appeared. All of the other viruses were indistinguishable in cell culture, with the exception of the rSDV-E2A8 mutant, which shows slower growth kinetics (Fig. 4). For a drastic test of the in vivo pathogenicity of each rSDV, trout were infected by intraperitoneal injection. Thus, 25 trout per tank were infected by injection of 105 PFU/fish of each rSDV (after concentration of virus inoculum, if needed), and cumulative mortalities were recorded as described above over a period of 60 days. As shown in Fig. 5, the cumulative mortality rate recorded at 60 days postinfection was 40 and 8% for rSDV-E2A8 and rSDV-E2T136, respectively, whereas no mortality could be observed with rSDV. These data indicate that only a single amino acid change, A8V, in the SDV E2 glycoprotein is responsible for almost 90% of the rSDV attenuation. rSDV-E2T136, recovered from dead fish, was used to infect BF2 cells, and then, through RT-PCR and nucleotide sequencing, it was confirmed that the expected E2 amino acid changes were still present (data not shown). In the past, several studies on other alphaviruses, like Sindbis virus (SINV) and Venezuelan equine encephalitis virus, have shown that few residue changes in the E2 glycoprotein have a drastic impact on virulence (9, 10). For SINV, two changes have been identified in E2, S1R and S114R. The two amino acid changes identified in the current study in the E2 SDV were in good agreement with a very recent study published on another alphavirus, chikungunya virus, showing that two changes in the E2 glycoprotein are responsible for the attenuation in the vaccine strain (11). Interestingly, as for SDV, one of the amino acid changes, T12I, is also located in the extreme amino terminus of the chikungunya virus E2 glycoprotein. When the three-dimensional structure of the SDV p62 polyprotein is available, it will be interesting to visualize how this limited amino acid change induces such a drastic change in viral pathogenicity.
Fig 1.
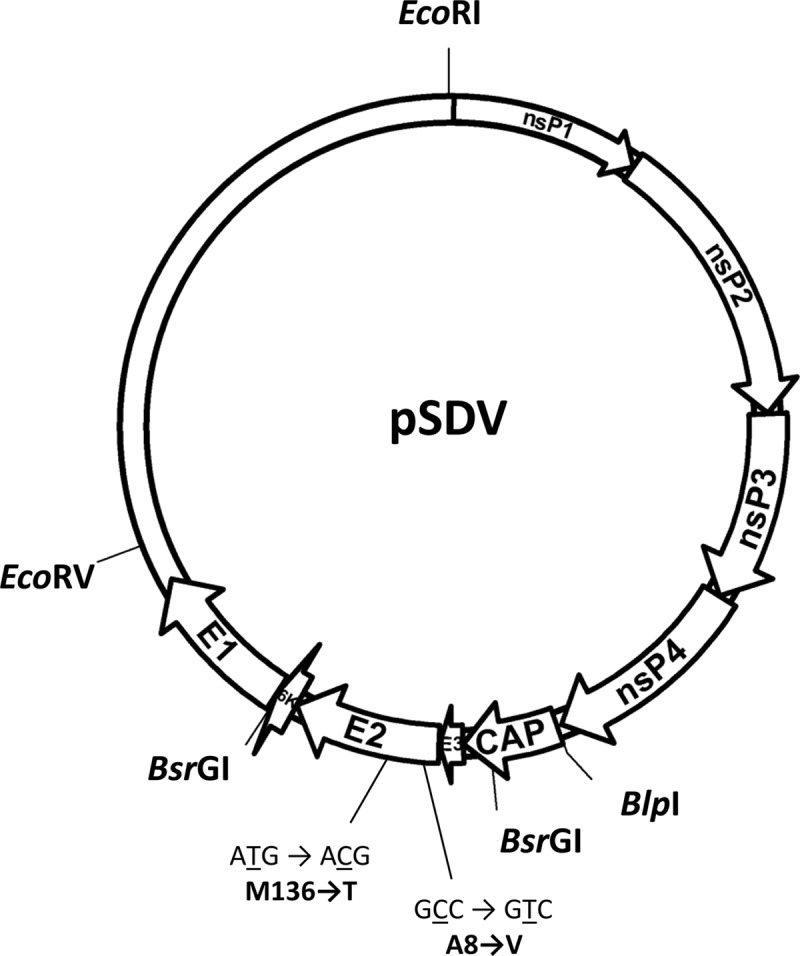
Schematic representation of the pSDV infectious cDNA. Restriction enzyme sites used in the study are indicated, as are the nucleotides and amino acid changes in the E2 glycoprotein.
Fig 2.
Pathogenicity of rSDVStructwt in trout. Fifty juvenile trout (mean weight, 4.4 g) were infected by bath immersion with 5 × 104 PFU/ml of wtSDV, rSDV, or rSDVStructwt or were mock infected. Mortalities were recorded every day postinfection over a 2-month period of time. A comparison of survival between groups was performed with the log rank χ2 test on the Kaplan-Meier survival data using GraphPad Prism (GraphPad, San Diego, CA).
Fig 3.
(A) Multiple-sequence alignment of alphavirus E2 glycoprotein amino acid sequences. E2 glycoprotein amino acid sequences from prototype alphaviruses are aligned with that of salmonid alphavirus (SAV), SDV, and SPDV. E2 SAV amino acids A8 and T136 are highlighted in red, as is T12 of E2 from chikungunya virus (CHIKV). The extra E2 domain for SAV is highlighted in yellow. Amino acids in gray shading are the residues conserved in all of the presented E2 alphavirus sequences. SFV, Semliki Forest virus; RRV, Ross River virus; MAYA, Mayaro virus; WEEV, Western equine encephalitis virus; AURA, Aura virus; VEEV, Venezuelan equine encephalitis virus. (B) Schematic representation of the genome of the various rSDV. Amino acid changes in the E2 glycoprotein are indicated at positions 8 and 136.
Table 1.
Titer of the viruses used in this study after 2 passages in cell culture
| Virus | Titer (PFU/ml) |
|---|---|
| rSDV | 6 × 107 |
| rSDV-E2A8 | 2 × 104 |
| rSDV-E2T136 | 1.45 × 108 |
| rSDVStructwt | 8 × 107 |
| wtSDVa | 5 × 108 |
wtSDV is reference strain S49P.
Fig 4.
Growth curve kinetics of the various rSDV. BF2 cells plated in 24-well plates were infected at a multiplicity of infection of 0.1 with each of the rSDV. At various days postinfection, aliquots of viral supernatant were taken and clarified. Viral titers were determined by plaque assay using serial 10-fold dilutions to infect (in duplicate) BF-2 cells plated in 96-well plates. Plaques were visualized 6 days later by indirect immunofluorescence assay with a monoclonal antibody, 17H23, directed against E2. The titers were expressed in PFU/ml. Viral titers were compared by one-way analysis of variance (ANOVA) with Tukey's multiple-comparison post test using GraphPad Prism (GraphPad, SanDiego, CA). Viruses that were assigned to statistically similar groups (and, thus, share a letter) are not significantly different from each other (P > 0.05), whereas those that were not assigned to statistically similar groups are significantly different (P < 0.05).
Fig 5.
Cumulative mortalities induced in trout by the various SDV. Twenty-five juvenile trout (mean weight, 3 g) were infected by intraperitoneal injection with 105 PFU/fish of rSDV, rSDVStructwt, rSDV-E2A8, or rSDV-E2T136 or were mock infected. Mortalities were recorded each day postinjection over a 2-month period. For statistical grouping, a comparison of survival between groups was performed with the log rank χ2 test on the Kaplan-Meier survival data using GraphPad Prism (GraphPad, San Diego, CA). Viruses that were assigned to statistically similar groups (and, thus, share a letter) are not significantly different from each other (P > 0.05), whereas those that were not assigned to statistically similar groups are significantly different (P < 0.01).
Nucleotide sequence accession numbers.
Sequences for the structural proteins of the attenuated and virulent form of SDV have been deposited in GenBank under the numbers KC593282 and KC593283, respectively.
ACKNOWLEDGMENTS
We thank members of the fish facilities (Infectiologie Expérimentale des Rongeurs et Poissons, INRA) for taking care of experimental fish. We thank Wendy Brand-Williams (INRA) for carefully reading the manuscript. We also thank M. C. Vaney (Pasteur Institute) for her helpful suggestions in alignment of multiple E2 sequences.
Footnotes
Published ahead of print 28 February 2013
REFERENCES
- 1. Anonymous 1985. Maladie du sommeil et pollutions expérimentales. Technical report no. 118. Laboratoire National de Pathologie des Animaux Aquatiques, CNEVA, Brest, France [Google Scholar]
- 2. Boucher P, Baudin-Laurencin F. 1994. Sleeping disease of salmonids. Bull. Eur. Assoc. Fish Pathol. 14:179–180 [Google Scholar]
- 3. Boucher P, Castric J, Baudin Laurencin F. 1994. Observation of virus-like particles in rainbow-trout Oncorhynchus mykiss infected with sleeping disease virulent material. Bull. Eur. Assoc. Fish Pathol. 14:215–216 [Google Scholar]
- 4. Castric J, Baudin Laurencin F, Brémont M, Jeffroy J, Le Ven A, Bearzotti M. 1997. Isolation of the virus responsible for sleeping disease in experimentally infected rainbow trout (Oncorhynchus mykiss). Bull. Eur. Assoc. Fish Pathol. 17:27–30 [Google Scholar]
- 5. Villoing S, Béarzotti M, Chilmonczyk S, Castric J, Brémont M. 2000. Rainbow trout sleeping disease virus is an atypical alphavirus. J. Virol. 74:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weston J, Villoing S, Brémont M, Castric J, Pfeffer M, Jewhurst V, McLoughlin M, Rødseth O, Christie KE, Koumans J, Todd D. 2002. Comparison of two aquatic alphaviruses, salmon pancreas disease virus and sleeping disease virus, by using genome sequence analysis, monoclonal reactivity, and cross-infection. J. Virol. 76:6155–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boucher P, Le Ven A, Baudin-Laurencin F. 1995. Transmission expérimentale de la maladie du sommeil: histologie et caractérisation de l'agent infectieux. Nouv. Sci. Technol. 13:115–117 [Google Scholar]
- 8. Moriette C, Leberre M, Lamoureux A, Lai TL, Brémont M. 2006. Recovery of a recombinant salmonid alphavirus fully attenuated and protective for rainbow trout. J. Virol. 80:4088–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kinney RM, Chang GJ, Tsuchiya KR, Sneider JM, Roehrig JT, Woodward TM, Trent DW. 1993. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J. Virol. 67:1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gorchakov R, Wang E, Leal G, Forrester NL, Plante K, Rossi SL, Partidos CD, Adams AP, Seymour R, Weger J, Borland EM, Sherman MB, Powers AM, Osorio JE, Weaver SC. 2012. Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J. Virol. 86:6084–6096 [DOI] [PMC free article] [PubMed] [Google Scholar]