Abstract
Recent genome-wide association studies (GWASs) have identified genetic variants associated with blood pressure (BP). We investigated whether genetic risk scores (GRSs) constructed of these variants would predict incident cardiovascular disease (CVD) events. We genotyped 32 common single nucleotide polymorphisms (SNPs) in several Finnish cohorts, with up to 32,669 individuals after exclusion of prevalent CVD cases. The median follow-up was 9.8 years, during which 2,295 incident CVD events occurred. We created GRSs separately for systolic (SBP) and diastolic BP (DBP) by multiplying the risk allele count of each SNP by the effect size estimated in published GWASs. We performed Cox regression analyses with and without adjustment for clinical factors including BP at baseline in each cohort. The results were combined by inverse variance-weighted fixed-effects meta-analysis. The GRSs were strongly associated with SBP and DBP and baseline hypertension (all p<10−62). Hazard ratios comparing the highest quintiles of SBP and DBP genetic risk scores with the lowest quintiles after adjustment for age, age squared and sex, were 1.25 (1.07–1.46, p = 0.006) and 1.23 (1.05–1.43, p = 0.01), respectively, for incident coronary heart disease; 1.24 (1.01–1.53, p = 0.04) and 1.35 (1.09–1.66, p = 0.005) for incident stroke; and 1.23 (1.08–1.40, p = 2×10−6) and 1.26 (1.11–1.44, p = 5×10−4) for composite CVD. In conclusion, BP findings from GWASs are strongly replicated. GRSs comprised of bona fide BP SNPs predicted cardiovascular disease risk, consistent with a life-long effect on BP of these variants collectively.
Keywords: Hypertension, blood pressure, genetics, cardiovascular disease, prospective cohort study, genetic risk score
Introduction
Elevated blood pressure (BP) is a strong, independent and modifiable risk factor for stroke and heart disease.1,2BP is a heritable trait with estimated heritability of 0.4-0.5,3 and recent well-powered genome-wide association studies (GWAS) have identified several genetic loci which are associated with systolic (SBP), diastolic blood pressure (DBP) or commonly both.4-8 While the variants have modest effects on BP, their presence may act over the entire life course and therefore lead to substantial increases in risk of cardiovascular and cerebrovascular disease. For example, it was recently found that common genetic variants are associated with preclinical blood pressure traits even in childhood.9The intra-individual and measurement variability of BP is high10 and therefore several measurements are optimally needed over time to reliably determine a person’s BP level. In principle, genetic background is stable and could, in borderline cases, help clinicians decide whether BP treatment is needed or alter the intensity of BP treatment.
We genotyped 32 genetic variants which have been previously reported to be associated with BP at genome-wide significance and investigated whether genetic risk scores (GRSs) constructed of these variants would be significant predictors of incident cardiovascular (CVD) events in prospective, population-based cohorts from Finland.
Methods
An expanded description of the Methods section is available in the online-only Data Supplement.
Study Populations
FINRISK surveys are cross-sectional, population-based studies conducted every 5 years since 1972 to monitor the risk of chronic diseases. For each survey, a representative random sample was selected from 25–74 year old inhabitants of different regions in Finland. The survey included a questionnaire and a clinical examination, at which a blood sample was drawn, with linkage to national registers of cardiovascular and other health outcomes. The study protocol has been described elsewhere.11 Study participants were followed up through 31 December 2010. The current study included eligible individuals from FINRISK surveys conducted in 1992, 1997, 2002, and 2007 (total n=27,838).
Health 2000 was based on a stratified two-stage cluster sampling from the National Population Register to represent the total Finnish population aged 30 years and over. The Mini-Finland Health Survey was originally conducted between 1978 and 1980 in similar manner to the Health 2000 Study. Of the Mini-Finland participants, 985 living in seven large cities participated in a follow-up study in 2001 at which DNA was collected. Health 2000 and Mini-Finland cohorts were analyzed pooled, adjusting for study cohort. The survey included an interview about medical history, health-related lifestyle habits, and a clinical examination at which a blood sample was drawn. A detailed description of the study protocol is available at: http://www.terveys2000.fi/doc/methodologyrep.pdf. Study participants were followed up through 31 December 2010.After restricting the study to participants aged ≤80 years at baseline, there were 6,731 individuals eligible for the current study.
The Helsinki Birth Cohort Study (HBCS) is composed of 8,760 individuals born between the years 1934-44 in one of the two main maternity hospitals in Helsinki, Finland. Between 2001 and 2004, a randomly selected sample of 928 males and 1,075 females participated in a clinical follow-up study with a focus on cardiovascular, metabolic and reproductive health, cognitive function and depressive symptoms. The participants were followed up through 31 December 2010, and 1,676 participants were eligible for the present study.12
The Oulu Project Elucidating Risk of Atherosclerosis (OPERA) is a population-based, epidemiological study examining risk factors and disease end points of atherosclerotic cardiovascular diseases. The hypertensive cohort (cases) consisted of 600 subjects (300 men and 300 women, aged 40 to 59 years at the time of selection) from the town of Oulu randomly selected from the national register of medication reimbursements for moderate or severe hypertension. For each year of birth (1931 to 1950), 15 hypertensive men and 15 hypertensive women were selected. For each hypertensive subject, an age- and sex-matched control was randomly selected from all inhabitants of Oulu excluding subjects with reimbursement for hypertension medication.13 Study participants were followed up through 31 December 2009. For the present study, 1,000 participants were eligible.
Blood pressure measurement methods for each of the cohorts are described in the online-only Data Supplement.
Follow-up
During follow-up, hospitalization and mortality data were obtained from the Finnish National Hospital Discharge Register and the National Causes-of-Death Register. These registers cover all cardiovascular events that have led either to hospitalization or death in Finland. The cardiovascular diagnoses in these registers have been validated.14,15Coronary heart disease (CHD) was defined as non-fatal myocardial infarction, unstable angina pectoris, coronary revascularization (coronary artery bypass graft surgery or percutaneous transluminal coronary angioplasty), or death due to CHD. Cardiovascular disease (CVD) included CHD and ischemic stroke events. The clinical outcomes were linked to study subjects using their unique national social security ID, which is assigned to every permanent resident of Finland. Events prior to baseline were traced back to 1970 when computerized records first became available in Finland. In the OPERA study, however, register data was available only from the baseline onwards, and events prior to baseline were determined by a detailed interview of a physician. All study subjects provided written informed consent. The local institutional ethical review boards approved each study.
SNP Selection, Genotyping and Genetic Risk Scores
We selected 32 single nucleotide polymorphisms (SNP; Table S1) which have been associated with SBP or DBP in GWAS.4-8 Details of the genotyping are provided in the online-only Data Supplement. We calculated the GRS using the reported effect sizes from the reference studies as weights per copy of the coded allele for each individual SNP. Different SNPs thus contribute different weights, as opposed to an alternate approach in which no weighting of effects is used and each SNP allele counts equally in the score. The coded allele is the allele coded 0, 1 or 2 according to the number of copies of the allele. A one unit increase in the GRS corresponds to a 1 mm Hg increase in the predicted SBP or DBP as a result of the aggregated predicted effects of all 32 SNPs. Missing genotype data for each SNP was imputed using the average coded allele frequency within each study cohort. However, if more than 60% of the SNP genotypes were missing for a given individual, the GRS was set as missing for that individual. Two GRSs were calculated, one for SBP and one for DBP.
Statistical Analyses
Associations of the GRSs and, as a secondary analysis, individual SNPs with SBP or DBP with imputation for use of antihypertensive therapy (+15mmHg for SBP; +10mmHg for DBP as an estimate of the expected BP off antihypertensive therapy)16 was performed using linear regression, adjusting for baseline age and its square, sex, and geographic region (eastern vs western Finland).The analysis was repeated with further adjustment for BMI, alcohol consumption (grams/week), and leisure time physical activity (moderate to high vs low). Association of the GRSs and individual SNPs with baseline hypertension (HTN, defined as SBP ≥140 mmHg or DBP ≥ 90 mmHg or use of antihypertensive therapy) was analyzed using logistic regression adjusted for the same covariates as above.
Associations of the GRSs and secondarily individual SNPs with incident CVD outcomes were analyzed using Cox proportional hazards regression with age as the time scale. All models were stratified by sex and adjusted for geographic region. As a secondary analysis we included also prevalent CVD cases, i.e., using the first event since birth. In these analyses we used time from birth as the time scale except in HBCS, which used elapsed time from the first date of eligibility, 1 January 1970. Power analysis for detecting a single SNP effect in the Cox regression for incident CVD is provided in Figure S1.
The incident outcome associations were then analyzed using two further adjustments. First, we adjusted for the following Framingham Risk Score risk factors: total cholesterol, high-density lipoprotein cholesterol, current smoking and baseline diabetes, plus lipid-lowering treatment. The last covariate is not part of the Framingham Score, but it was significant in most models, improving model fit and validity of the proportional hazards assumption. Second, we adjusted additionally for SBP or DBP (in the case of the DBP GRS) and for antihypertensive treatment. In addition to the analyses of the primary CVD endpoint, we performed secondary Cox regression analyses using ischemic stroke and CHD events as the outcomes with the same covariate adjustments as in the primary analyses.
In order to examine the possibility that the GRS derived for SBP or DBP would not be the optimal GRS for predicting incident cardiovascular events we derived another GRS specifically for CVD, using the Cox regression results of the individual SNPs with incident CVD in the present studies. This CVD GRS used betas or hazard ratios from the meta-analyses (Table S3) as weights for the risk allele counts.
We confirmed that Cox proportional hazards assumptions were met using scaled Schoenfeld residuals (R function cox.zph). The results from individual studies were combined using inverse variance-weighted fixed effect meta-analysis, checking for heterogeneity using the I2 measure; I2> 0.5 is considered evidence for significant heterogeneity.17 This limit was not exceeded in any of the analyses, beyond chance expectation.
Using the Health 2000 and FINRISK 92 and 97 cohorts which have 10 year follow-up, we further assessed the utility of the GRS for 10-year CVD risk prediction by estimating the net reclassification improvement (NRI), clinical NRI for prospective data,18integrated discrimination improvement (IDI)19,and explained relative risk for the GRS. The statistical significance of AUC change between models with and without GRS was tested with the correlated C-index approach.200 Model calibration was assessed with the Hosmer-Lemeshow goodness-of-fit test.21All statistical analyses were performed using R version 2.15.22In general, we considered two-sided p<0.05 statistically significant. For the association tests of individual SNPs with BP/HTN and cardiovascular events, we used Bonferroni correction to account for 32 independent tests.
Results
The baseline characteristics of the study cohorts are given in Table 1. Individually, 23 of the 32 SNPs were associated with SBP or DBP in the same direction as previously reported, after accounting for multiple testing (P<0.0016 = P<0.05/32, Table S1). Directions of effect were consistent for all SNPs but two. Effects on SBP or DBP were highly correlated in the Finnish studies with the original estimated effects reported in the literature (SBP r=0.75, DBP r=0.71). GRSs were strong predictors for SBP, DBP and HTN (Table 2, all p<10−62). The average proportion of variance in SBP or DBP explained by each respective score was generally greater than that estimated previously8: 1.20% in SBP and 1.18% in DBP using weighted averages across all cohorts. The results for the individual cohorts are provided in Table S4.
Table 1.
Characteristics of the study cohorts
| Study | FR 19921 (N=5,465) |
FR 1997 (N=6,692) |
FR 2002 (N=7,951) |
FR 2007 (N=4,334) |
Health 2000 (N=5,797) |
OPERA (N=856) |
HBCS (N=1,574) |
|---|---|---|---|---|---|---|---|
|
Follow-up time from
baseline, yrs; Median (IQR) |
18.8 (0.1) | 13.8 (0.1) | 8.8 (0.1) | 3.9 (0.1) | 10.0 (0.3) | 17.6 (1.1) | 8.0(1.4) |
| Sex | |||||||
| Men | 2,483 (45.4%) |
3,104 (46.4%) |
3,611 (45.4%) |
1,929 (44.5%) |
2,601(44.9 %) |
419 (48.9%) |
639(40.6% ) |
| Women | 2,982 (54.6%) |
3,588 (53.6%) |
4,340 (54.6%) |
2,405 (55.5%) |
3,196(55.1 %) |
437 (51.1%) |
935(59.4% ) |
| Age at baseline, yrs | 44.2± 11.3 | 47.3± 13.0 | 47.4± 13.0 | 51.6± 13.5 | 52.7± 12.8 | 50.8 ± 5.8 | 61.4 ± 2.9 |
| Blood pressure | |||||||
| Systolic (mm Hg) | 135.4 ± 19.4 |
135.4± 19.7 |
134.7 ± 19.8 |
136.7 ± 20.5 |
134.3 ± 20.5 | 147.9 ± 21.9 |
145.4± 20.4 |
| Diastolic (mm Hg) | 81.2 ± 11.9 |
82.2± 11.3 | 79.0 ± 11.4 |
79.4± 11.2 | 82.3± 10.9 | 89.0 ± 12.2 |
89.1± 10.3 |
| HTN | 2,626(48.1 %) |
3,256(48.7 %) |
3,330 (41.9%) |
2,335 (53.9%) |
2,857 (49.3%) |
655 (76.5%) |
1,121(71.2 %) |
| Antihypertensive medication |
481 (8.8%) |
802 (12.0%) |
1,055 (13.3%) |
873 (20.1%) |
1,241 (21.4%) |
409 (47.8%) |
475(30.2% ) |
| Cholesterol(mmol/l) | |||||||
| Total | 5.6 ± 1.1 | 5.5 ± 1.1 | 5.6 ± 1.1 | 5.3 ± 1.1 | 6.0 ± 1.1 | 5.7 ± 1.0 | 6.0 ± 1.1 |
| LDL (Friedewald) | 3.6 ± 1.0 | 3.5 ± 0.9 | 3.4 ±1.0 | 3.2 ± 0.9 | 3.8 ± 1.2 | 3.5 ± 0.9 | 3.7 ± 0.9 |
| HDL | 1.4 ± 0.3 | 1.4 ± 0.4 | 1.5 ± 0.4 | 1.4 ± 0.4 | 1.4 ± 0.4 | 1.4 ± 0.4 | 1.6 ± 0.4 |
| Body-mass index(kg/m2) | 26.1 ± 4.4 | 26.6 ± 4.6 | 26.8 ± 4.7 | 27.1 ± 4.9 | 27.0± 4.6 | 27.6 ± 4.7 | 27.5± 4.7 |
| Current smoker | 1,514(27.7 %) |
1,621(24.2 %) |
2,073 (26.1%) |
772(17.2% ) |
1,655(28.5 %) |
247 (28.9%) |
376(23.9% ) |
| Diabetes mellitus | 191(3.5%) | 342(5.1%) | 383(4.8%) | 349(8.1%) | 392(6.8%) | 68 (8.0%) | 217(13.8% ) |
| Prevalent cases * | |||||||
| Coronary heart disease |
72(1.3%) | 147(2.1%) | 210(2.6%) | 130(2.9%) | 179(3.0%) | 117 (11.7%) |
79(4.7%) |
| Ischemic stroke | 27 (0.5%) | 69(1.0%) | 87(1.1%) | 68(1.5%) | 100(1.6%) | 37 (3.7%) | 28(1.7%) |
| Cardiovascular disease |
97(1.7%) | 204 (3.0%) | 284(3.4%) | 188(4.2%) | 265(4.4%) | 144 (14.4%) |
102(6.1%) |
| Incident cases * | |||||||
| Coronary heart disease |
391(7.2%) | 411(6.1%) | 229(2.9%) | 67(1.5%) | 360(6.2%) | 104 (12.1%) |
66(4.2%) |
| Ischemic stroke | 210(3.8%) | 231(3.5%) | 115(1.4%) | 26(0.6%) | 154 (2.7%) | 49 (5.7%) | 30(1.9%) |
| Cardiovascular disease |
555 (10.2%) |
599(9.0%) | 326(4.1%) | 88(2.0%) | 495(8.5%) | 141 (16.5%) |
91(5.8%) |
The numbers refer to participants for whom the GRSs are available. Unless otherwise noted, continuous measures are mean±SD and dichotomous measures are N (%). Prevalent CVD cases are excluded from everything else except the prevalent case numbers. Health 2000 was restricted to participants aged ≤80 years. Abbreviations: FR, FINRISK; IQR, Interquartile range; N, number of individuals; LDL, low-density lipoprotein; HDL, high-density lipoprotein; HTN, hypertension. HTN was defined as SBP ≥140 mmHg or DBP ≥ 90 mmHg or use of antihypertensive therapy.
In the case counts some persons might have both coronary heart disease and ischemic stroke, hence they do not necessary add up to number of persons with cardiovascular disease.
Table 2.
Association of the GRSs with BP and incident cardiovascular disease events.
| Trait | Systolic BP (N=37,176) | Diastolic BP (N=37,168) | HTN (N=37,245) | |||
|---|---|---|---|---|---|---|
| Analysis | mm Hg (95% CI) | P | mm Hg (95% CI) | P | OR (95% CI) | P |
| SBP GRS * | 1.1 (1.0;1.2) | 1×10−87 | 0.6 (0.6;0.7) | 4×10−77 | 1.12 (1.11;1.14) | 1×10−65 |
| SBP GRS † | 1.1 (1.0;1.2) | 4×10−90 | 0.6 (0.6;0.7) | 2×10−81 | 1.13 (1.12;1.15) | 3×10−67 |
| DBP GRS * | 1.7 (1.5;1.9) | 9×10−78 | 1.0 (0.9;1.2) | 3×10−82 | 1.20 (1.17;1.22) | 1×10−62 |
| DBP GRS † | 1.7 (1.5;1.8) | 1×10−79 | 1.0 (0.9;1.2) | 9×10−87 | 1.21 (1.18;1.24) | 3×10−64 |
| Trait |
CHD (N=32,953; 2,408 events)
‡ (N=32,669; 1,628events) §,∥,¶ |
STR (N=32,953; 1,222events)
‡ (N=32,669; 815events) §,∥,¶ |
CVD (N=32,953; 3,294events)
‡ (N=32,669; 2,295events)§,∥,¶ |
|||
| Analysis | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P |
|
| ||||||
| SBP GRS ‡ | 1.05 (1.03;1.08) | 3×10−6 | 1.05 (1.02;1.09) | 0.001 | 1.05 (1.03;1.07) | 4×10−8 |
| SBP GRS § | 1.04 (1.01;1.07) | 0.003 | 1.03 (1.00;1.07) | 0.07 | 1.04 (1.02;1.06) | 5×10−4 |
| SBP GRS ∥ | 1.04 (1.01;1.07) | 0.005 | 1.04 (1.00;1.07) | 0.05 | 1.04 (1.02;1.06) | 6×10−4 |
| SBP GRS ¶ | 1.03 (1.00;1.05) | 0.06 | 1.02 (0.98;1.06) | 0.24 | 1.03 (1.00;1.05) | 0.02 |
| DBP GRS ‡ | 1.08 (1.04;1.11) | 4×10−5 | 1.08 (1.02;1.13) | 0.002 | 1.08 (1.05;1.11) | 8×10−7 |
| DBP GRS § | 1.06 (1.01;1.10) | 0.01 | 1.06 (1.00;1.12) | 0.06 | 1.06 (1.02;1.10) | 0.001 |
| DBP GRS ∥ | 1.05 (1.01;1.10) | 0.01 | 1.06 (1.00;1.12) | 0.05 | 1.06 (1.02;1.10) | 0.002 |
| DBP GRS ¶ | 1.04 (1.00;1.08) | 0.08 | 1.04 (0.98;1.10) | 0.24 | 1.04 (1.01;1.08) | 0.02 |
Linear regression, logistic regression and Cox regression (age as the time scale) were used as appropriate. Studies were combined using inverse variance-weighted fixed-effects meta-analysis. HTN was defined as SBP ≥140 mmHg or DBP ≥ 90 mmHg or use of antihypertensive therapy. Effect estimates are shown as increase in blood pressure on the mm Hg scale or hazard ratio for dichotomous outcomes for a one unit increase in GRS. One unit for the SBP GRS is one mm Hg increase in predicted SBP and for the DBP GRS one mm Hg increase in predicted DBP, based on effect estimates derived from the published GWAS. Abbreviations: BP, blood pressure; DBP, diastolic blood pressure; SBP, systolic blood pressure; HTN, hypertension; GRS, genetic risk score; CHD, coronary heart disease; CI, confidence interval; CVD, cardiovascular disease; HR, hazard ratio; N, number of individuals; OR, odds ratio; STR, stroke, including ischemic stroke and intracerebral hemorrhages.
Models adjusted for baseline age and its square, sex and study region. In the linear regressions BP measurements were corrected for the use of antihypertensive therapy (+15mmHg for SBP; +10mmHg for DBP).
Additionally adjusted for BMI, leisure time physical activity (high/moderate vs. none) and average alcohol consumption (7-day abs. gr. or high vs. low or none depending on the study).
Prevalent cases were also included. Models stratified for sex and adjusted for study region. OPERA was excluded from the analyses.
Only incident cases. Models stratified for sex and adjusted for study region.
Only incident cases. Additionally adjusted for log(total cholesterol), log(HDL cholesterol), baseline diabetes and current smoking status.
Only incident cases. Additionally adjusted for all above and BP treatment and SBP or DBP depending on the trait.
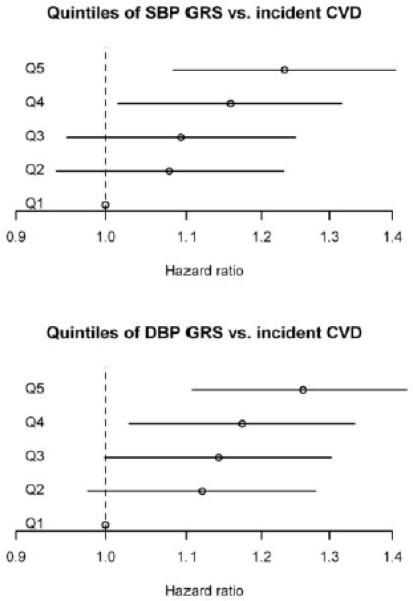
After excluding 1,284 individuals with prevalent CVD at baseline, 2,295 incident CVD events occurred during a median follow-up time of 9.8 (IQR 5.1) years. In total, the study participants contributed 347,955 person-years of follow-up. GRSs showed significant, independent and roughly linear associations with CVD risk (Table 2, Table 3, Figure 1). As expected, the observed effects on SBP or DBP of a predicted 1 mm Hg increase in GRS, based on the previously published per-SNP effect estimates, were 1.1 and 1.0 mm Hg, for the SBP and DBP GRSs, respectively (Table 2). A 4% increased hazard for coronary heart disease, stroke or cardiovascular disease was observed for each predicted 1 mm Hg increase in SBP GRS, and 6-8% for each predicted 1 mm Hg increase in DBP GRS, in models adjusting for non-BP clinical risk factors. After further adjustment for antihypertensive treatment and baseline SBP or DBP for their respective GRSs, the hazard ratios were reduced only slightly and for the most part remained statistically significant (Table 2). In models, adjusting for age, age squared and sex, increasing quintile of SBP or DBP GRS was associated with roughly linear increases in BP, HTN prevalence and risk of incident cardiovascular disease, with or without inclusion of prevalent cases (Table 3). For example, the highest compared to lowest quintile of SBP GRS was associated with a hazard ratio for CVD of 1.30 (95% CI 1.17–1.45, p=1×10−6) including prevalent and incident events and 1.23 (95% CI 1.08-1.40, p=2×10-6) including only incident events and the highest quintile of DBP GRS was associated with a hazard ratio of 1.30 (95% CI 1.17–1.45, p=2×10−6) and 1.26 (95% CI 1.11-1.44, p=5×10−4), respectively (Table 3). No individual SNPs were statistically significantly associated with CVD risk after correcting for 32 tests, although some were nominally associated (p<0.05, Table S3).
Table 3.
Blood pressure and cardiovascular outcome risk by quintile of blood pressure GRS.
| BP Quintile | SBP (mm Hg)* |
DBP (mm Hg)* |
HTN OR (95% CI)* |
CHD prev + inc HR (95% CI)‡ |
CHD inc only HR (95% CI)§ |
STR prev + inc HR (95% CI)‡ |
STR inc only HR (95% CI)§ |
CVD prev + inc HR (95% CI)‡ |
CVD inc only HR (95% CI)§ |
|---|---|---|---|---|---|---|---|---|---|
| SBP GRS Q2 | 1.4 (0.8;2.1) P = 1×10−5 |
0.9 (0.5;1.3) P = 5×10−6 |
1.18 (1.09;1.27) P = 2×10−5 |
1.09 (0.96;1.24) P = 0.19 |
1.07 (0.92;1.26) P = 0.39 |
1.16 (0.97;1.39) P = 0.10 |
1.22 (0.99;1.50) P = 0.07 |
1.09 (0.97;1.22) P = 0.14 |
1.08 (0.94;1.23) P = 0.03 |
| SBP GRS Q3 | 2.9 (2.2;3.5) P = 3×10−16 |
1.7 (1.3;2.1) P = 4×10−18 |
1.37 (1.27;1.48) P = 2×10−16 |
1.13 (0.99;1.28) P = 0.08 |
1.15 (0.98;1.34) P = 0.09 |
1.05 (0.87;1.26) P = 0.63 |
1.04 (0.84;1.30) P = 0.70 |
1.10 (0.99;1.23) P = 0.09 |
1.09 (0.96;1.25) P = 0.05 |
| SBP GRS Q4 | 3.7 (3.0;4.3) P = 3×10−29 |
2.1 (1.8;2.5) P = 5×10−28 |
1.50 (1.39;1.61) P = 2×10−25 |
1.15 (1.01;1.31) P = 0.03 |
1.16 (1.00;1.36) P = 0.06 |
1.16 (0.97;1.39) P = 0.11 |
1.20 (0.97;1.48) P = 0.09 |
1.14 (1.02;1.28) P = 0.02 |
1.16 (1.02;1.32) P = 0.003 |
| SBP GRS Q5 | 5.8 (5.1;6.4) 1×10−69 |
3.3 (2.9;3.6) P = 3×10−62 |
1.82 (1.69;1.96) P = 2×10−53 |
1.32 (1.16;1.50) P = 2×10−5 |
1.25 (1.07;1.46) P = 0.006 |
1.29 (1.08;1.55) P = 0.004 |
1.24 (1.01;1.53) P = 0.04 |
1.30 (1.17;1.45) P = 1×10−6 |
1.23 (1.08;1.40) P = 2×10−6 |
|
DBP GRS
Q2 |
1.3 (0.7;2.0) P = 7×10−5 |
1.0 (0.6;1.4) P = 1×10−7 |
1.18 (1.10;1.28) P = 1×10−5 |
1.13 (0.99;1.29) P = 0.07 |
1.09 (0.93;1.28) P = 0.30 |
1.20 (1.00;1.44) P = 0.05 |
1.29 (1.05;1.60) P = 0.02 |
1.13 (1.01;1.27) P = 0.03 |
1.12 (0.98;1.28) P = 0.10 |
|
DBP GRS
Q3 |
3.1 (2.4;3.7) P = 1×10−20 |
1.8 (1.4;2.2) P = 6×10−20 |
1.36 (1.26;1.46) P = 3×10−15 |
1.15 (1.01;1.31) P = 0.03 |
1.20 (1.03;1.41) P = 0.02 |
1.11 (0.92;1.33) P = 0.28 |
1.14 (0.920;1.42) P = 0.23 |
1.12 (1.00;1.25) P = 0.05 |
1.14 (1.00;1.30) P = 0.05 |
|
DBP GRS
Q4 |
3.63 (3.0;4.3) P = 2×10−28 |
2.3 (1.9;2.7) P = 3×10−31 |
1.49 (1.38;1.61) P = 5×10−25 |
1.21 (1.06;1.37) P = 0.004 |
1.20 (1.03;1.41) P = 0.02 |
1.14 (0.95;1.36) P = 0.18 |
1.15 (0.925;1.429) P = 0.21 |
1.18 (1.06;1.32) P = 0.003 |
1.17 (1.03;1.34) P = 0.02 |
|
DBP GRS
Q5 |
5.2 (4.6;5.9) P = 2×10−57 |
3.3 (2.9;3.7) P = 4×10−65 |
1.76 (1.63;1.89) P = 1×10−47 |
1.30 (1.14;1.47) P = 6×10−5 |
1.23 (1.05;1.43) P = 0.01 |
1.32 (1.11;1.58) P = 0.002 |
1.35 (1.09;1.66) P = 0.005 |
1.30 (1.17;1.45) P = 2×10−6 |
1.26 (1.11;1.44) P = 5×10−4 |
Linear regression, logistic regression and Cox regression (age as the time scale) were used as appropriate. Studies were combined using inverse variance-weighted fixed-effects meta-analysis. HTN was defined as SBP ≥140 mmHg or DBP ≥ 90 mmHg or use of antihypertensive therapy. Effect estimates are shown as increase in blood pressure on the mm Hg scale or hazard ratio for dichotomous outcomes for quintiles 2-4 for the SBP and DBP GRS compared to the bottom quintile. Abbreviations: DBP, diastolic blood pressure; SBP, systolic blood pressure; HTN, hypertension; GRS, genetic risk score; CHD, coronary heart disease; CI, confidence interval; CVD, cardiovascular disease; HR, hazard ratio; OR, odds ratio; STR, stroke, including ischemic stroke and intracerebral hemorrhages.
Models adjusted for baseline age and its square, sex and study region. In the linear regressions BP measurements were corrected for the use of antihypertensive therapy (+15mmHg for SBP; +10mmHg for DBP).
Prevalent and incident cases were included. Models stratified for sex and adjusted for study region. OPERA was excluded from the analyses.
Only incident cases. Models stratified for sex and adjusted for study region.
Figure 1.
Association of the SBP and DBP GRS quintiles with incident CVD. Cox regression models were adjusted for sex and study region. Age was used as the time scale and results were combined using inverse variance-weighted fixed-effects meta-analysis.
The associations of a CVD GRS, created using CVD effects estimated in the Finnish cohorts, with cardiovascular events were stronger than the associations with the BP GRSs with less attenuation after adjustment for Framingham Risk Score components (HR=2.40,p=6×10−5 for incident CHD; HR=2.07, p=0.013 for incident stroke; and HR=2.40,P=2×10−6 for incident CVD; Table S5). However, because the CVD GRS was derived from the same studies in which its association with CVD events was tested, the strength of the association might have been overestimated.
Reclassification analyses in Health 2000, FINRISK 1992 and 1997 showed that the SBP GRS did not improve CVD risk discrimination over and above the standard Framingham risk score (which includes SBP) when assessed using ROC curves and C-statistics, or IDI (Tables S6,S7). There was no net improvement in NRI, either (Tables S6, S7). However, the clinical NRI, i.e., reclassification in the intermediate risk group of 5-20%, was 3.8% and statistically significant (p=5×10−5). Model calibration was good in both models with and without the GRS (FigureS2). We found no significant interaction of sex, baseline age, BMI or antihypertensive treatment with the GRS effect on risk of incident CVD.
Discussion
We tested previously established SNPs associated with BP in a large collection of population-based studies in Finland. We replicated 23 SNPs at a stringent Bonferroni-corrected significance threshold and found directional consistency for 30 out of 32 SNPs. Genetic risk scores weighted according to previously reported effect estimates were highly associated with BP and hypertension. An important novel finding of our study was that both SBP- and DBP-based genetic risk scores were strongly associated with risk of incident CVD among those free of CVD at baseline. These associations were largely independent of standard CVD risk factors, including BP measured at baseline.
The field of genetic associations has historically been riddled with irreproducible results, largely due to overly permissive significance thresholds and inadequate power from limited sample sizes.23 The widespread adoption of stringent p-value thresholds and the development of genotyping platforms allowing the efficient genotyping of scores to millions of variants in tens of thousands of individuals have enabled identification of reproducible associations. We have replicated many of the BP variants identified in GWAS meta-analyses some of which included six times the sample size examined here.
Our findings are consistent with a causal effect of BP on cardiovascular disease. This is well accepted given the broad epidemiologic support for this relationship, the existence of Mendelian syndromes of hypertension and premature cardiovascular disease and the modification of risk by lowering blood pressure through behavioral or pharmacologic means.24 Most of the SNPs that have been identified lie in chromosomal regions with a heterogeneous set of genes without a single dominant pathway apparent. Despite these heterogeneous effects, when the weak effects of the individual SNPs are considered jointly, a strong and consistent effect on cardiovascular risk is observed.
BP is a highly variable, dynamic measure, with minute-to-minute variation influenced by activity, posture, mental stress, medication, etc. We used the mean of two BP measurements. However, we were limited to the examination of BP at a single time point, which is clearly an imprecise surrogate for the lifetime of exposure to higher BP that contributes to the pathogenesis of cardiovascular disease and could lead to regression dilution of the true impact of BP and other time-varying factors. This, in fact, highlights the potential value of the study of precisely measured genotypes. Genetic variants are fixed and when set against the background of a large dynamic range may capture a fixed component to lifetime BP exposure. Thus, small genetic effects on BP may translate into comparatively large effects when compounded over a lifetime. In the present study, we found no net improvement in NRI due to SBP or DBP GRS over and above the Framingham equation. It should be noted, however, that the Framingham equation estimates ten-year risk only, as risk factors change over time, whereas for BP GRS the estimation of lifetime risk might be more appropriate as genotype is invariant over time.
For decades, investigators have sought to resolve essential hypertension into well-defined subtypes that might benefit from specific therapies, such as low-renin, high-aldosterone hypertension. However, these have not led to widespread adoption or recommendation for specific therapies by most public health guidelines.24 Given the limited power to detect the weak SNP effects that have so far been found, it is estimated that hundreds of common variants of similarly modest effect will ultimately be found to exist.8 Assuming as yet unidentified effects are as modest as those identified to date, it seems unlikely that common genetic variation will ultimately accomplish this task.
Some SNPs that fall in targets of antihypertensives are potential candidates to modulate the response to antihypertensive therapy. For example, a common BP-associated missense polymorphism lies in ADRB1 which encodes the β1 adrenergic receptor, a target of beta adrenergic receptor antagonists. Whether such BP-associated variants also influence response to antihypertensive therapy, awaits results from large clinical trials.
Strengths of the current study include the large sample size, the population-based sampling of the cohorts examined, the measurement of BP precisely and in a uniform manner, the availability of relevant covariates, the large number of CVD outcomes available for prospective analyses and the single country of origin for all samples. Limitations include the inability to generalize to non-European ancestry groups. We lacked the ability to adjust for time-varying clinical factors that influence BP. The proportion of variation explained by the SNPs remained low, and the level of prediction for events was also relatively small. We lacked power to demonstrate replication even at nominal p<0.05 of some of the modest BP effects reported in prior GWAS.
Perspectives
We found that genetic risk scores comprised of 32 SNPs identified in GWAS of BP were strongly associated with risk of incident cardiovascular disease, even after adjustment for baseline BP. While the genetic risk scores were not associated with significant reclassification of CVD risk when added to the Framingham Risk Score, a more complete compendium of genetic variation that reproducibly influences BP will ultimately need to be tested.
Supplementary Material
Novelty and Significance.
What Is New?
Recent GWAS studies have identified several genetic variants associated with BP.
The predictive power of BP-associated genetic variants for incident CVD events has not been established
What Is Relevant?
GRSs comprised of 32 SNPs identified in GWAS of BP were strongly associated with risk of incident CVD, even after adjustment for baseline BP and antihypertensive treatment.
A more complete compendium of genetic variation that reproducibly influences BP will ultimately need to be tested.
Summary
These findings are consistent with a lifelong effect on BP of these variants and a causal effect of BP on CVD risk.
Acknowledgments
We wish to thank Ms. Annukka M. Lahtinen from the Research Programs Unit, Molecular Medicine and Department of Medicine, University of Helsinki, who participated in the Sequenom genotyping, Mr. Antti-Pekka Sarin from the Institute for Molecular Medicine Finland (FIMM), University of Helsinki, who did the genotyping QC and Dr. Merja Santaniemi from the Institute of Clinical Medicine, Department of Medicine, University of Oulu, who helped in coding the follow-up events in the OPERA cohort.
Sources of Funding: FINRISK was mainly funded by the National Institute for Health and Welfare. Additional support was obtained from the Academy of Finland (grant numbers 129 494 and 139 635) to Dr Salomaa. The Health 2000 Study is funded by the National Institute for Health and Welfare (THL), the Finnish Centre for Pensions (ETK), The Social Insurance Institution of Finland (KELA), The Local Government Pensions Institution (KEVA) and other organizations listed on the website of the survey (http://www.terveys2000.fi). The OPERA study was supported by the Finnish Foundation for Cardiovascular Research. The Helsinki Birth Cohort Study has been supported by grants to Dr. Eriksson from the Academy of Finland (Grant numbers 129 255, 126 775, 135 072) the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, Ahokas Foundation and Juho Vainio Foundation. Dr Kontula was supported by the Sigrid Juselius Foundation and the Finnish Foundation for Cardiovascular Research. Dr Newton-Cheh and the SNP genotyping were supported by the Burroughs Wellcome Fund and the National Institutes of Health (HL098283).
Footnotes
Conflicts of Interest/Disclosures: Dr Newton-Cheh reports that he is a member of a scientific advisory board for hypertension and heart failure at Merck. No other disclosures were reported.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.MacMahon S, Peto R, Cutler J, Collins R, Sorlie P, Neaton J, Abbott R, Godwin J, Dyer A, Stamler J. Blood pressure, stroke, and coronary heart disease. Part 1, prolonged differences in blood pressure: Prospective observational studies corrected for the regression dilution bias. Lancet. 1990;335:765–774. doi: 10.1016/0140-6736(90)90878-9. [DOI] [PubMed] [Google Scholar]
- 2.Collins R, Peto R, MacMahon S, Hebert P, Fiebach NH, Eberlein KA, Godwin J, Qizilbash N, Taylor JO, Hennekens CH. Blood pressure, stroke, and coronary heart disease. Part 2, short-term reductions in blood pressure: Overview of randomised drug trials in their epidemiological context. Lancet. 1990;335:827–838. doi: 10.1016/0140-6736(90)90944-z. [DOI] [PubMed] [Google Scholar]
- 3.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 4.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nature genetics. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nature genetics. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, Laing S, Corso B, Navis G, Kwakernaak AJ, van der Harst P, Bochud M, Maillard M, Burnier M, Hedner T, Kjeldsen S, Wahlstrand B, Sjogren M, Fava C, Montagnana M, Danese E, Torffvit O, Hedblad B, Snieder H, Connell JM, Brown M, Samani NJ, Farrall M, Cesana G, Mancia G, Signorini S, Grassi G, Eyheramendy S, Wichmann HE, Laan M, Strachan DP, Sever P, Shields DC, Stanton A, Vollenweider P, Teumer A, Volzke H, Rettig R, Newton-Cheh C, Arora P, Zhang F, Soranzo N, Spector TD, Lucas G, Kathiresan S, Siscovick DS, Luan J, Loos RJ, Wareham NJ, Penninx BW, Nolte IM, McBride M, Miller WH, Nicklin SA, Baker AH, Graham D, McDonald RA, Pell JP, Sattar N, Welsh P, Munroe P, Caulfield MJ, Zanchetti A, Dominiczak AF. Genome-wide association study of blood pressure extremes identifies variant near umod associated with hypertension. PLoS genetics. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, Rice K, Verwoert GC, Launer LJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, Caulfield M, van Duijn CM, Ridker PM, Munroe PB, Levy D. Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension. 2011;57:903–910. doi: 10.1161/HYPERTENSIONAHA.110.158667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Artigas MS, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr., Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oikonen M, Tikkanen E, Juhola J, Tuovinen T, Seppala I, Juonala M, Taittonen L, Mikkila V, Kahonen M, Ripatti S, Viikari J, Lehtimaki T, Havulinna AS, Kee F, Newton-Cheh C, Peltonen L, Schork NJ, Murray SS, Berenson GS, Chen W, Srinivasan SR, Salomaa V, Raitakari OT. Genetic variants and blood pressure in a population-based cohort: The cardiovascular risk in young finns study. Hypertension. 2011;58:1079–1085. doi: 10.1161/HYPERTENSIONAHA.111.179291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothwell PM, Howard SC, Dolan E, O’Brien E, Dobson JE, Dahlof B, Sever PS, Poulter NR. Prognostic significance of visit-to-visit variability, maximum systolic blood pressure, and episodic hypertension. Lancet. 2010;375:895–905. doi: 10.1016/S0140-6736(10)60308-X. [DOI] [PubMed] [Google Scholar]
- 11.Vartiainen E, Laatikainen T, Peltonen M, Juolevi A, Mannisto S, Sundvall J, Jousilahti P, Salomaa V, Valsta L, Puska P. Thirty-five-year trends in cardiovascular risk factors in finland. International journal of epidemiology. 2010;39:504–518. doi: 10.1093/ije/dyp330. [DOI] [PubMed] [Google Scholar]
- 12.Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. The New England journal of medicine. 2005;353:1802–1809. doi: 10.1056/NEJMoa044160. [DOI] [PubMed] [Google Scholar]
- 13.Kiema TR, Kauma H, Rantala AO, Lilja M, Reunanen A, Kesaniemi YA, Savolainen MJ. Variation at the angiotensin-converting enzyme gene and angiotensinogen gene loci in relation to blood pressure. Hypertension. 1996;28:1070–1075. doi: 10.1161/01.hyp.28.6.1070. [DOI] [PubMed] [Google Scholar]
- 14.Pajunen P, Koukkunen H, Ketonen M, Jerkkola T, Immonen-Raiha P, Karja-Koskenkari P, Mahonen M, Niemela M, Kuulasmaa K, Palomaki P, Mustonen J, Lehtonen A, Arstila M, Vuorenmaa T, Lehto S, Miettinen H, Torppa J, Tuomilehto J, Kesaniemi YA, Pyorala K, Salomaa V. The validity of the finnish hospital discharge register and causes of death register data on coronary heart disease. European journal of cardiovascular prevention and rehabilitation : official journal of the European Society of Cardiology, Working Groups on Epidemiology & Prevention and Cardiac Rehabilitation and Exercise Physiology. 2005;12:132–137. doi: 10.1097/00149831-200504000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Tolonen H, Salomaa V, Torppa J, Sivenius J, Immonen-Raiha P, Lehtonen A. The validation of the finnish hospital discharge register and causes of death register data on stroke diagnoses. European journal of cardiovascular prevention and rehabilitation : official journal of the European Society of Cardiology, Working Groups on Epidemiology & Prevention and Cardiac Rehabilitation and Exercise Physiology. 2007;14:380–385. doi: 10.1097/01.hjr.0000239466.26132.f2. [DOI] [PubMed] [Google Scholar]
- 16.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: Antihypertensive therapy and systolic blood pressure. Statistics in medicine. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 17.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 18.Pencina MJ, D’Agostino RB, Sr., Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Statistics in medicine. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pencina MJ, D’Agostino RB, Sr., D’Agostino RB, Jr., Vasan RS. Evaluating the added predictive ability of a new marker: From area under the roc curve to reclassification and beyond. Statistics in medicine. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207-112. [DOI] [PubMed] [Google Scholar]
- 20.Antolini L, Nam BH, D’Agostico RB. Inference on correlated discrimination measures in survival analysis: A nonparametric approach. Communications in statistics. Theory and methods. 2004;33:2117–2135. [Google Scholar]
- 21.Hosmer DW, Hosmer T, Le Cessie S, Lemeshow S. A comparison of goodness-of-fit tests for the logistic regression model. Statistics in medicine. 1997;16:965–980. doi: 10.1002/(sici)1097-0258(19970515)16:9<965::aid-sim509>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 22.Team RDC. R: A language and environment for statistical computing. 2005 [Google Scholar]
- 23.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nature genetics. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 24.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr., Jones DW, Materson BJ, Oparil S, Wright JT, Jr., Roccella EJ. The seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure: The JNC 7 report. JAMA : the journal of the American Medical Association. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.