Abstract
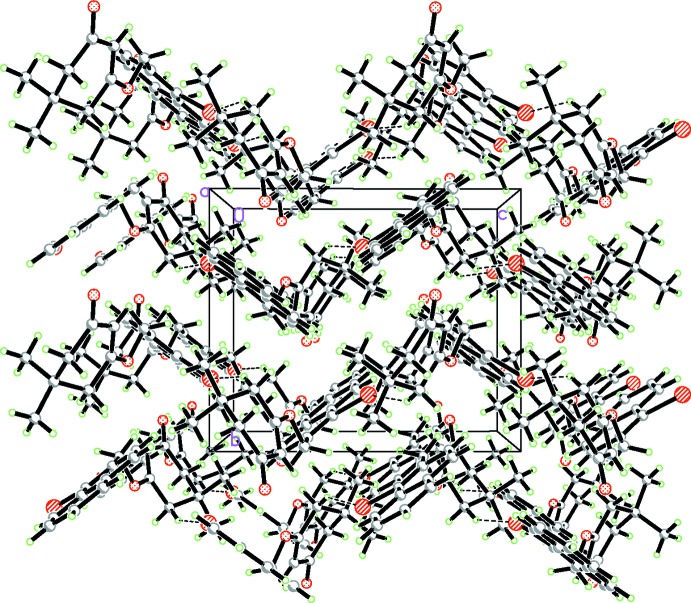
In the title compound, C19H19BrO2, the cyclohexenone ring adopts an envelope conformation with the C atom bearing the methyl substituents as the flap. In the crystal, weak π–π stacking is observed between parallel aromatic rings of adjacent molecules, the centroid–centroid distance being 3.694 (6) Å. The entire bromonaphthylmethyl unit is disordered over two orientations, with a site-occupancy ratio of 0.5214 (19):0.4786 (19).
Related literature
For the biological activity and applications of cyclohex-2-enone derivatives, see: Aghil et al. (1992 ▶); Correcia et al. (2001 ▶); Ghorab et al. (2011 ▶).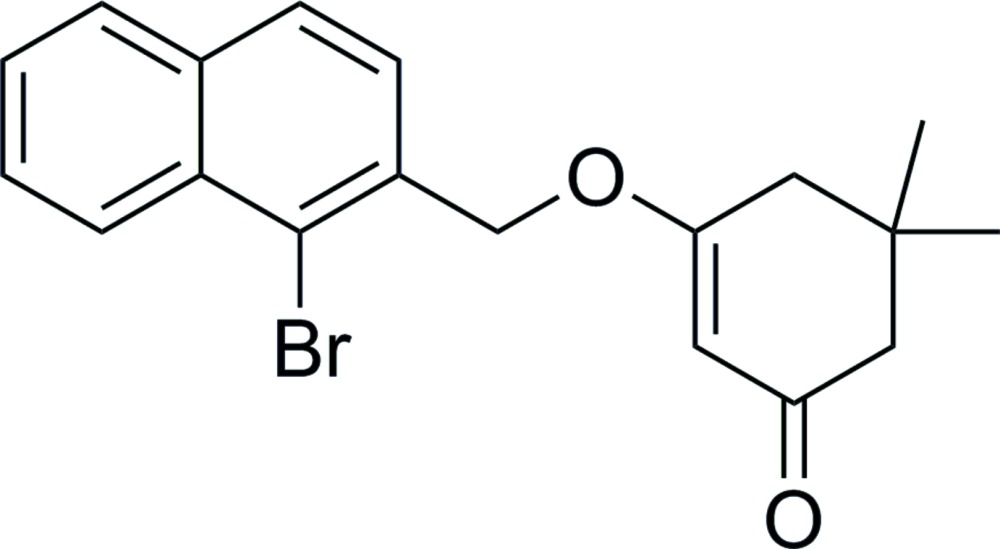
Experimental
Crystal data
C19H19BrO2
M r = 359.25
Monoclinic,
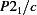
a = 13.986 (3) Å
b = 9.9970 (18) Å
c = 11.859 (2) Å
β = 91.169 (2)°
V = 1657.8 (5) Å3
Z = 4
Mo Kα radiation
μ = 2.48 mm−1
T = 296 K
0.32 × 0.29 × 0.27 mm
Data collection
Bruker SMART 1000 CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.504, T max = 0.554
11934 measured reflections
3075 independent reflections
1931 reflections with I > 2σ(I)
R int = 0.074
Refinement
R[F 2 > 2σ(F 2)] = 0.061
wR(F 2) = 0.199
S = 1.07
3075 reflections
222 parameters
72 restraints
H-atom parameters constrained
Δρmax = 0.26 e Å−3
Δρmin = −0.20 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681301026X/xu5694sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681301026X/xu5694Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681301026X/xu5694Isup3.cdx
Supplementary material file. DOI: 10.1107/S160053681301026X/xu5694Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 21172057).
supplementary crystallographic information
Comment
Cyclohex-2-enone derivatives display a wide range of biological activities (Aghil et al., 1992; Correcia et al., 2001). Moreover, they have been frequently used as precursors in the synthesis of heterocyclic compounds (Ghorab, et al., 2011). The title compound is the derivative of cyclohex-2-enones, and we report its crystal structure here.
In the title compound, C19H19BrO2, all the bond lengths and bond angles are within normal ranges. The cyclohexenone ring adopts an envelope conformation with the C atom bearing two methyl groups as the flap atom. All the atoms in the o-bromonaphthylmethyl group are disordered over two positions with site occupancy factors of 0.521 (2) and 0.479 (2).
In the crystal structure, weak π-π stacking is observed between parallel aromatic rings of adjacent molecules, the centrods distance being 3.694 (6) Å.
Experimental
To a solution of 1-bromo-2-(bromomethyl)naphthalene (0.15 g, 0.5 mmol) and 5,5-dimethylcyclohexane-1,3-dione (0.14 g, 1.0 mmol) in DMF (3 ml) were added K2CO3 (0.21 g, 1.5 mmol) and CuI (0.01 g, 0.05 mmol). The mixture was stirred at 373 K until a complete conversion. It was cooled to room temperature and added with water, then extracted with ethyl ether (5 ml × 3). The combined organic phases were concentrated under vacuum. The crude product was purified by column chromatography eluting with ethyl acetate/hexane (10–20%) to give the title compound with the yield of 32% (0.057 g, 0.16 mmol). Single crystals, suitable for X-ray diffraction analysis, were obtained by slow evaporation of the solvents from a chloroform-ethyl acetate (1:1 v/v) solution of the title compound.
Refinement
H atoms were positioned geometrically and refined using riding model with C—H = 0.93–0.97 Å, Uiso(H) = 1.2Ueq(C). The The bromonaphthalene moiety is disordered over two orientations, the site occupancies were refined to 0.5214 (19):0.4786 (19), the ADP of corresponding atoms in the disordered components were restrained as the same.
Figures
Fig. 1.
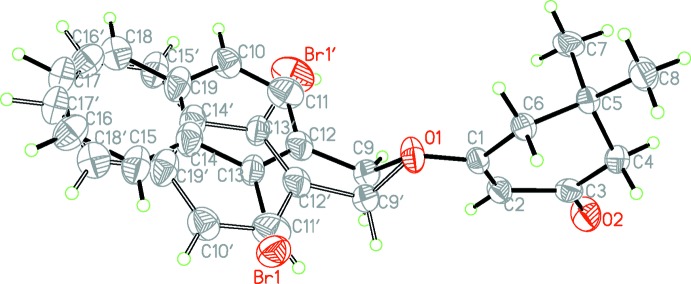
Molecular structure of the title compound with the disorder atoms, with displacement ellipsoids drawn at the 30% probability level.
Fig. 2.
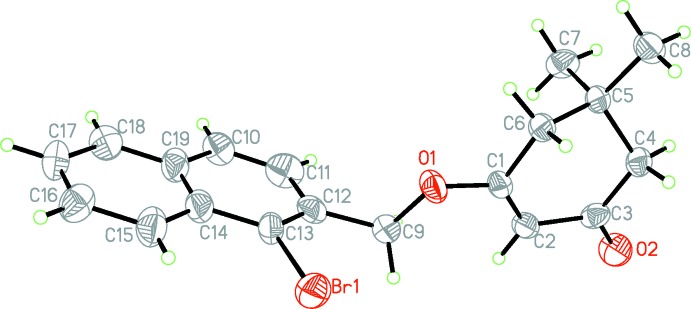
Molecular structure of the title compound without the disorder atoms, with displacement ellipsoids drawn at the 30% probability level.
Fig. 3.
Crystal structure of the title compound with view along the a axis (disorder atoms have been omitted for clarity).
Crystal data
| C19H19BrO2 | F(000) = 736 |
| Mr = 359.25 | Dx = 1.439 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2538 reflections |
| a = 13.986 (3) Å | θ = 2.5–20.6° |
| b = 9.9970 (18) Å | µ = 2.48 mm−1 |
| c = 11.859 (2) Å | T = 296 K |
| β = 91.169 (2)° | Block, colourless |
| V = 1657.8 (5) Å3 | 0.32 × 0.29 × 0.27 mm |
| Z = 4 |
Data collection
| Bruker SMART 1000 CCD area-detector diffractometer | 3075 independent reflections |
| Radiation source: fine-focus sealed tube | 1931 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.074 |
| phi and ω scans | θmax = 25.5°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −16→16 |
| Tmin = 0.504, Tmax = 0.554 | k = −12→12 |
| 11934 measured reflections | l = −14→14 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.061 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.199 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0635P)2 + 1.8223P] where P = (Fo2 + 2Fc2)/3 |
| 3075 reflections | (Δ/σ)max < 0.001 |
| 222 parameters | Δρmax = 0.26 e Å−3 |
| 72 restraints | Δρmin = −0.20 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Br1 | 0.93272 (9) | 0.21923 (16) | 0.49100 (14) | 0.0729 (5) | 0.5214 (19) |
| C9 | 0.8583 (5) | 0.0318 (10) | 0.6902 (8) | 0.0531 (17) | 0.5214 (19) |
| H9A | 0.8328 | 0.0122 | 0.6153 | 0.064* | 0.5214 (19) |
| H9B | 0.8464 | −0.0448 | 0.7381 | 0.064* | 0.5214 (19) |
| C10 | 1.1164 (5) | 0.0103 (9) | 0.7741 (7) | 0.0667 (19) | 0.5214 (19) |
| H10 | 1.1523 | −0.0316 | 0.8307 | 0.080* | 0.5214 (19) |
| C11 | 1.0182 (6) | −0.0061 (16) | 0.7684 (11) | 0.082 (4) | 0.5214 (19) |
| H11 | 0.9881 | −0.0599 | 0.8208 | 0.098* | 0.5214 (19) |
| C12 | 0.9649 (5) | 0.0574 (9) | 0.6848 (7) | 0.057 (2) | 0.5214 (19) |
| C13 | 1.0089 (4) | 0.1363 (8) | 0.6063 (6) | 0.0555 (17) | 0.5214 (19) |
| C14 | 1.1077 (4) | 0.1516 (9) | 0.6101 (7) | 0.068 (3) | 0.5214 (19) |
| C15 | 1.1529 (4) | 0.2292 (9) | 0.5302 (7) | 0.076 (3) | 0.5214 (19) |
| H15 | 1.1171 | 0.2707 | 0.4733 | 0.092* | 0.5214 (19) |
| C16 | 1.2513 (5) | 0.2448 (9) | 0.5352 (7) | 0.073 (3) | 0.5214 (19) |
| H16 | 1.2816 | 0.2968 | 0.4816 | 0.087* | 0.5214 (19) |
| C17 | 1.3047 (5) | 0.1829 (8) | 0.6201 (7) | 0.068 (3) | 0.5214 (19) |
| H17 | 1.3707 | 0.1934 | 0.6234 | 0.081* | 0.5214 (19) |
| C18 | 1.2595 (4) | 0.1054 (9) | 0.7000 (7) | 0.070 (2) | 0.5214 (19) |
| H18 | 1.2953 | 0.0638 | 0.7569 | 0.085* | 0.5214 (19) |
| C19 | 1.1611 (4) | 0.0897 (9) | 0.6950 (6) | 0.068 (3) | 0.5214 (19) |
| C9' | 0.8450 (6) | 0.0553 (11) | 0.6468 (9) | 0.0531 (17) | 0.4786 (19) |
| H9'1 | 0.8038 | 0.0595 | 0.5800 | 0.064* | 0.4786 (19) |
| H9'2 | 0.8458 | −0.0359 | 0.6748 | 0.064* | 0.4786 (19) |
| C10' | 1.0349 (5) | 0.2273 (10) | 0.4867 (7) | 0.0667 (19) | 0.4786 (19) |
| H10' | 1.0368 | 0.2777 | 0.4209 | 0.080* | 0.4786 (19) |
| C11' | 0.9485 (6) | 0.1796 (18) | 0.5240 (12) | 0.082 (4) | 0.4786 (19) |
| H11' | 0.8923 | 0.1996 | 0.4842 | 0.098* | 0.4786 (19) |
| C12' | 0.9454 (5) | 0.1022 (10) | 0.6205 (7) | 0.057 (2) | 0.4786 (19) |
| C13' | 1.0287 (5) | 0.0714 (9) | 0.6786 (7) | 0.0555 (17) | 0.4786 (19) |
| C14' | 1.1158 (4) | 0.1209 (10) | 0.6427 (7) | 0.068 (3) | 0.4786 (19) |
| C15' | 1.2003 (5) | 0.0901 (10) | 0.7016 (8) | 0.076 (3) | 0.4786 (19) |
| H15' | 1.1986 | 0.0369 | 0.7658 | 0.092* | 0.4786 (19) |
| C16' | 1.2875 (6) | 0.1393 (10) | 0.6641 (9) | 0.073 (3) | 0.4786 (19) |
| H16' | 1.3438 | 0.1188 | 0.7034 | 0.087* | 0.4786 (19) |
| C17' | 1.2901 (6) | 0.2191 (10) | 0.5678 (8) | 0.068 (3) | 0.4786 (19) |
| H17' | 1.3482 | 0.2519 | 0.5428 | 0.081* | 0.4786 (19) |
| C18' | 1.2056 (5) | 0.2498 (9) | 0.5089 (7) | 0.070 (2) | 0.4786 (19) |
| H18' | 1.2073 | 0.3031 | 0.4446 | 0.085* | 0.4786 (19) |
| C19' | 1.1184 (4) | 0.2007 (9) | 0.5463 (7) | 0.068 (3) | 0.4786 (19) |
| Br1' | 1.02132 (16) | −0.0384 (2) | 0.80852 (18) | 0.1011 (8) | 0.4786 (19) |
| C1 | 0.7221 (3) | 0.1315 (4) | 0.7734 (3) | 0.0451 (10) | |
| C2 | 0.6685 (3) | 0.0219 (5) | 0.7571 (4) | 0.0515 (11) | |
| H2 | 0.6915 | −0.0492 | 0.7149 | 0.062* | |
| C3 | 0.5733 (3) | 0.0146 (5) | 0.8065 (4) | 0.0525 (11) | |
| C4 | 0.5388 (3) | 0.1316 (5) | 0.8710 (4) | 0.0531 (11) | |
| H4A | 0.4962 | 0.1001 | 0.9288 | 0.064* | |
| H4B | 0.5020 | 0.1888 | 0.8204 | 0.064* | |
| C5 | 0.6191 (3) | 0.2159 (4) | 0.9280 (3) | 0.0471 (10) | |
| C6 | 0.6897 (3) | 0.2516 (4) | 0.8369 (4) | 0.0465 (10) | |
| H6A | 0.6598 | 0.3143 | 0.7846 | 0.056* | |
| H6B | 0.7448 | 0.2954 | 0.8714 | 0.056* | |
| C7 | 0.6693 (4) | 0.1348 (6) | 1.0219 (4) | 0.0652 (14) | |
| H7A | 0.7186 | 0.1882 | 1.0569 | 0.098* | |
| H7B | 0.6972 | 0.0558 | 0.9902 | 0.098* | |
| H7C | 0.6235 | 0.1095 | 1.0772 | 0.098* | |
| C8 | 0.5763 (4) | 0.3438 (6) | 0.9770 (5) | 0.0684 (14) | |
| H8A | 0.6263 | 0.3970 | 1.0107 | 0.103* | |
| H8B | 0.5307 | 0.3207 | 1.0332 | 0.103* | |
| H8C | 0.5450 | 0.3938 | 0.9178 | 0.103* | |
| O1 | 0.8111 (2) | 0.1500 (3) | 0.7355 (3) | 0.0642 (10) | |
| O2 | 0.5246 (3) | −0.0871 (4) | 0.7946 (3) | 0.0814 (12) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0660 (7) | 0.0816 (11) | 0.0705 (9) | 0.0125 (6) | −0.0091 (5) | −0.0032 (7) |
| C9 | 0.0528 (19) | 0.0533 (19) | 0.053 (2) | 0.0011 (10) | 0.0026 (10) | −0.0008 (10) |
| C10 | 0.073 (5) | 0.065 (5) | 0.061 (4) | 0.008 (4) | 0.007 (4) | 0.009 (4) |
| C11 | 0.102 (8) | 0.075 (9) | 0.068 (9) | 0.010 (6) | −0.003 (6) | 0.013 (6) |
| C12 | 0.063 (5) | 0.051 (5) | 0.060 (6) | 0.007 (4) | 0.011 (5) | −0.017 (4) |
| C13 | 0.040 (4) | 0.062 (5) | 0.065 (5) | 0.001 (3) | 0.007 (4) | −0.006 (3) |
| C14 | 0.052 (3) | 0.067 (6) | 0.084 (7) | −0.003 (3) | 0.014 (4) | −0.024 (5) |
| C15 | 0.055 (7) | 0.079 (6) | 0.096 (7) | −0.010 (5) | 0.012 (5) | −0.004 (5) |
| C16 | 0.059 (5) | 0.063 (6) | 0.096 (7) | 0.013 (4) | −0.012 (4) | −0.004 (5) |
| C17 | 0.044 (4) | 0.067 (6) | 0.093 (9) | −0.002 (4) | 0.009 (5) | 0.008 (6) |
| C18 | 0.075 (7) | 0.060 (5) | 0.077 (6) | −0.001 (5) | 0.008 (5) | 0.001 (4) |
| C19 | 0.050 (5) | 0.059 (5) | 0.097 (7) | 0.004 (4) | 0.009 (5) | −0.021 (5) |
| C9' | 0.0528 (19) | 0.0533 (19) | 0.053 (2) | 0.0011 (10) | 0.0026 (10) | −0.0008 (10) |
| C10' | 0.073 (5) | 0.065 (5) | 0.061 (4) | 0.008 (4) | 0.007 (4) | 0.009 (4) |
| C11' | 0.102 (8) | 0.075 (9) | 0.068 (9) | 0.010 (6) | −0.003 (6) | 0.013 (6) |
| C12' | 0.063 (5) | 0.051 (5) | 0.060 (6) | 0.007 (4) | 0.011 (5) | −0.017 (4) |
| C13' | 0.040 (4) | 0.062 (5) | 0.065 (5) | 0.001 (3) | 0.007 (4) | −0.006 (3) |
| C14' | 0.052 (3) | 0.067 (6) | 0.084 (7) | −0.003 (3) | 0.014 (4) | −0.024 (5) |
| C15' | 0.055 (7) | 0.079 (6) | 0.096 (7) | −0.010 (5) | 0.012 (5) | −0.004 (5) |
| C16' | 0.059 (5) | 0.063 (6) | 0.096 (7) | 0.013 (4) | −0.012 (4) | −0.004 (5) |
| C17' | 0.044 (4) | 0.067 (6) | 0.093 (9) | −0.002 (4) | 0.009 (5) | 0.008 (6) |
| C18' | 0.075 (7) | 0.060 (5) | 0.077 (6) | −0.001 (5) | 0.008 (5) | 0.001 (4) |
| C19' | 0.050 (5) | 0.059 (5) | 0.097 (7) | 0.004 (4) | 0.009 (5) | −0.021 (5) |
| Br1' | 0.1561 (17) | 0.0705 (11) | 0.0769 (14) | −0.0061 (9) | 0.0059 (10) | 0.0235 (9) |
| C1 | 0.043 (2) | 0.050 (3) | 0.042 (2) | 0.0009 (19) | 0.0091 (18) | 0.0007 (19) |
| C2 | 0.051 (3) | 0.054 (3) | 0.050 (2) | −0.008 (2) | 0.008 (2) | −0.009 (2) |
| C3 | 0.053 (3) | 0.062 (3) | 0.042 (2) | −0.017 (2) | −0.001 (2) | 0.002 (2) |
| C4 | 0.042 (2) | 0.064 (3) | 0.053 (3) | −0.006 (2) | 0.006 (2) | 0.001 (2) |
| C5 | 0.041 (2) | 0.057 (3) | 0.043 (2) | −0.003 (2) | 0.0087 (18) | −0.001 (2) |
| C6 | 0.045 (2) | 0.051 (3) | 0.044 (2) | −0.0027 (19) | 0.0065 (18) | 0.0026 (19) |
| C7 | 0.072 (3) | 0.087 (4) | 0.037 (2) | −0.014 (3) | 0.001 (2) | 0.010 (2) |
| C8 | 0.063 (3) | 0.073 (3) | 0.070 (3) | −0.004 (3) | 0.015 (3) | −0.017 (3) |
| O1 | 0.0533 (19) | 0.059 (2) | 0.081 (2) | −0.0126 (15) | 0.0272 (17) | −0.0207 (18) |
| O2 | 0.069 (2) | 0.083 (3) | 0.093 (3) | −0.035 (2) | 0.018 (2) | −0.018 (2) |
Geometric parameters (Å, º)
| Br1—C13 | 1.907 (6) | C13'—Br1' | 1.897 (7) |
| C9—O1 | 1.461 (10) | C14'—C15' | 1.395 (5) |
| C9—C12 | 1.515 (8) | C14'—C19' | 1.395 (5) |
| C9—H9A | 0.9700 | C15'—C16' | 1.395 (5) |
| C9—H9B | 0.9700 | C15'—H15' | 0.9300 |
| C10—C11 | 1.383 (6) | C16'—C17' | 1.395 (5) |
| C10—C19 | 1.387 (6) | C16'—H16' | 0.9300 |
| C10—H10 | 0.9300 | C17'—C18' | 1.395 (5) |
| C11—C12 | 1.382 (6) | C17'—H17' | 0.9300 |
| C11—H11 | 0.9300 | C18'—C19' | 1.395 (5) |
| C12—C13 | 1.376 (6) | C18'—H18' | 0.9300 |
| C13—C14 | 1.390 (6) | C1—C2 | 1.339 (6) |
| C14—C19 | 1.386 (5) | C1—O1 | 1.345 (5) |
| C14—C15 | 1.386 (5) | C1—C6 | 1.493 (6) |
| C15—C16 | 1.386 (5) | C2—C3 | 1.467 (6) |
| C15—H15 | 0.9300 | C2—H2 | 0.9300 |
| C16—C17 | 1.386 (5) | C3—O2 | 1.231 (6) |
| C16—H16 | 0.9300 | C3—C4 | 1.483 (7) |
| C17—C18 | 1.386 (5) | C4—C5 | 1.548 (6) |
| C17—H17 | 0.9300 | C4—H4A | 0.9700 |
| C18—C19 | 1.386 (5) | C4—H4B | 0.9700 |
| C18—H18 | 0.9300 | C5—C6 | 1.520 (6) |
| C9'—O1 | 1.499 (11) | C5—C8 | 1.531 (7) |
| C9'—C12' | 1.519 (9) | C5—C7 | 1.536 (6) |
| C9'—H9'1 | 0.9700 | C6—H6A | 0.9700 |
| C9'—H9'2 | 0.9700 | C6—H6B | 0.9700 |
| C10'—C19' | 1.379 (7) | C7—H7A | 0.9600 |
| C10'—C11' | 1.380 (6) | C7—H7B | 0.9600 |
| C10'—H10' | 0.9300 | C7—H7C | 0.9600 |
| C11'—C12' | 1.383 (6) | C8—H8A | 0.9600 |
| C11'—H11' | 0.9300 | C8—H8B | 0.9600 |
| C12'—C13' | 1.377 (6) | C8—H8C | 0.9600 |
| C13'—C14' | 1.389 (7) | ||
| O1—C9—C12 | 109.3 (7) | C14'—C15'—C16' | 120.0 |
| O1—C9—H9A | 109.8 | C14'—C15'—H15' | 120.0 |
| C12—C9—H9A | 109.8 | C16'—C15'—H15' | 120.0 |
| O1—C9—H9B | 109.8 | C15'—C16'—C17' | 120.0 |
| C12—C9—H9B | 109.8 | C15'—C16'—H16' | 120.0 |
| H9A—C9—H9B | 108.3 | C17'—C16'—H16' | 120.0 |
| C11—C10—C19 | 119.7 (4) | C18'—C17'—C16' | 120.0 |
| C11—C10—H10 | 120.2 | C18'—C17'—H17' | 120.0 |
| C19—C10—H10 | 120.2 | C16'—C17'—H17' | 120.0 |
| C12—C11—C10 | 120.1 (4) | C17'—C18'—C19' | 120.0 |
| C12—C11—H11 | 120.0 | C17'—C18'—H18' | 120.0 |
| C10—C11—H11 | 120.0 | C19'—C18'—H18' | 120.0 |
| C13—C12—C11 | 120.4 (4) | C10'—C19'—C18' | 120.4 (4) |
| C13—C12—C9 | 125.5 (6) | C10'—C19'—C14' | 119.6 (4) |
| C11—C12—C9 | 114.2 (6) | C18'—C19'—C14' | 120.0 |
| C12—C13—C14 | 120.0 (4) | C2—C1—O1 | 125.7 (4) |
| C12—C13—Br1 | 118.9 (4) | C2—C1—C6 | 123.7 (4) |
| C14—C13—Br1 | 121.1 (4) | O1—C1—C6 | 110.6 (4) |
| C19—C14—C15 | 120.0 | C1—C2—C3 | 119.5 (4) |
| C19—C14—C13 | 119.6 (3) | C1—C2—H2 | 120.2 |
| C15—C14—C13 | 120.4 (3) | C3—C2—H2 | 120.2 |
| C16—C15—C14 | 120.0 | O2—C3—C2 | 120.0 (4) |
| C16—C15—H15 | 120.0 | O2—C3—C4 | 121.7 (4) |
| C14—C15—H15 | 120.0 | C2—C3—C4 | 118.3 (4) |
| C17—C16—C15 | 120.0 | C3—C4—C5 | 114.4 (4) |
| C17—C16—H16 | 120.0 | C3—C4—H4A | 108.7 |
| C15—C16—H16 | 120.0 | C5—C4—H4A | 108.7 |
| C16—C17—C18 | 120.0 | C3—C4—H4B | 108.7 |
| C16—C17—H17 | 120.0 | C5—C4—H4B | 108.7 |
| C18—C17—H17 | 120.0 | H4A—C4—H4B | 107.6 |
| C17—C18—C19 | 120.0 | C6—C5—C8 | 109.8 (4) |
| C17—C18—H18 | 120.0 | C6—C5—C7 | 110.2 (4) |
| C19—C18—H18 | 120.0 | C8—C5—C7 | 110.0 (4) |
| C14—C19—C18 | 120.0 | C6—C5—C4 | 107.1 (3) |
| C14—C19—C10 | 120.2 (3) | C8—C5—C4 | 109.5 (4) |
| C18—C19—C10 | 119.8 (3) | C7—C5—C4 | 110.2 (4) |
| O1—C9'—C12' | 104.8 (7) | C1—C6—C5 | 112.2 (3) |
| O1—C9'—H9'1 | 110.8 | C1—C6—H6A | 109.2 |
| C12'—C9'—H9'1 | 110.8 | C5—C6—H6A | 109.2 |
| O1—C9'—H9'2 | 110.8 | C1—C6—H6B | 109.2 |
| C12'—C9'—H9'2 | 110.8 | C5—C6—H6B | 109.2 |
| H9'1—C9'—H9'2 | 108.9 | H6A—C6—H6B | 107.9 |
| C19'—C10'—C11' | 120.5 (4) | C5—C7—H7A | 109.5 |
| C19'—C10'—H10' | 119.8 | C5—C7—H7B | 109.5 |
| C11'—C10'—H10' | 119.8 | H7A—C7—H7B | 109.5 |
| C10'—C11'—C12' | 120.1 (4) | C5—C7—H7C | 109.5 |
| C10'—C11'—H11' | 120.0 | H7A—C7—H7C | 109.5 |
| C12'—C11'—H11' | 120.0 | H7B—C7—H7C | 109.5 |
| C13'—C12'—C11' | 119.9 (4) | C5—C8—H8A | 109.5 |
| C13'—C12'—C9' | 127.3 (7) | C5—C8—H8B | 109.5 |
| C11'—C12'—C9' | 112.8 (7) | H8A—C8—H8B | 109.5 |
| C12'—C13'—C14' | 120.4 (4) | C5—C8—H8C | 109.5 |
| C12'—C13'—Br1' | 118.4 (4) | H8A—C8—H8C | 109.5 |
| C14'—C13'—Br1' | 121.2 (4) | H8B—C8—H8C | 109.5 |
| C13'—C14'—C15' | 120.5 (4) | C1—O1—C9 | 116.1 (4) |
| C13'—C14'—C19' | 119.5 (4) | C1—O1—C9' | 117.1 (4) |
| C15'—C14'—C19' | 120.0 | C9—O1—C9' | 23.1 (5) |
| C19—C10—C11—C12 | −1 (2) | C12'—C13'—C14'—C19' | −0.8 (14) |
| C10—C11—C12—C13 | 1 (2) | Br1'—C13'—C14'—C19' | 179.6 (6) |
| C10—C11—C12—C9 | −179.1 (13) | C13'—C14'—C15'—C16' | 179.4 (11) |
| O1—C9—C12—C13 | −75.3 (11) | C19'—C14'—C15'—C16' | 0.0 |
| O1—C9—C12—C11 | 104.4 (12) | C14'—C15'—C16'—C17' | 0.0 |
| C11—C12—C13—C14 | 0.6 (17) | C15'—C16'—C17'—C18' | 0.0 |
| C9—C12—C13—C14 | −179.8 (9) | C16'—C17'—C18'—C19' | 0.0 |
| C11—C12—C13—Br1 | 178.9 (11) | C11'—C10'—C19'—C18' | −179.5 (12) |
| C9—C12—C13—Br1 | −1.5 (13) | C11'—C10'—C19'—C14' | 2.4 (17) |
| C12—C13—C14—C19 | −1.6 (12) | C17'—C18'—C19'—C10' | −178.1 (10) |
| Br1—C13—C14—C19 | −179.8 (5) | C17'—C18'—C19'—C14' | 0.0 |
| C12—C13—C14—C15 | 178.9 (7) | C13'—C14'—C19'—C10' | −1.3 (11) |
| Br1—C13—C14—C15 | 0.7 (11) | C15'—C14'—C19'—C10' | 178.1 (10) |
| C19—C14—C15—C16 | 0.0 | C13'—C14'—C19'—C18' | −179.4 (11) |
| C13—C14—C15—C16 | 179.5 (10) | C15'—C14'—C19'—C18' | 0.0 |
| C14—C15—C16—C17 | 0.0 | O1—C1—C2—C3 | 178.3 (4) |
| C15—C16—C17—C18 | 0.0 | C6—C1—C2—C3 | −2.0 (7) |
| C16—C17—C18—C19 | 0.0 | C1—C2—C3—O2 | −178.0 (5) |
| C15—C14—C19—C18 | 0.0 | C1—C2—C3—C4 | 1.4 (7) |
| C13—C14—C19—C18 | −179.5 (10) | O2—C3—C4—C5 | 151.0 (4) |
| C15—C14—C19—C10 | −179.0 (10) | C2—C3—C4—C5 | −28.4 (6) |
| C13—C14—C19—C10 | 1.5 (10) | C3—C4—C5—C6 | 52.9 (5) |
| C17—C18—C19—C14 | 0.0 | C3—C4—C5—C8 | 171.8 (4) |
| C17—C18—C19—C10 | 179.0 (9) | C3—C4—C5—C7 | −67.0 (5) |
| C11—C10—C19—C14 | −0.4 (15) | C2—C1—C6—C5 | 29.6 (6) |
| C11—C10—C19—C18 | −179.4 (11) | O1—C1—C6—C5 | −150.7 (4) |
| C19'—C10'—C11'—C12' | −1 (2) | C8—C5—C6—C1 | −170.9 (4) |
| C10'—C11'—C12'—C13' | −1 (2) | C7—C5—C6—C1 | 67.8 (5) |
| C10'—C11'—C12'—C9' | −178.3 (14) | C4—C5—C6—C1 | −52.1 (5) |
| O1—C9'—C12'—C13' | 83.4 (12) | C2—C1—O1—C9 | −11.2 (7) |
| O1—C9'—C12'—C11' | −99.3 (13) | C6—C1—O1—C9 | 169.1 (5) |
| C11'—C12'—C13'—C14' | 1.8 (18) | C2—C1—O1—C9' | 14.7 (8) |
| C9'—C12'—C13'—C14' | 178.9 (10) | C6—C1—O1—C9' | −165.1 (6) |
| C11'—C12'—C13'—Br1' | −178.6 (12) | C12—C9—O1—C1 | −162.9 (5) |
| C9'—C12'—C13'—Br1' | −1.5 (15) | C12—C9—O1—C9' | 98.9 (13) |
| C12'—C13'—C14'—C15' | 179.8 (8) | C12'—C9'—O1—C1 | 179.2 (6) |
| Br1'—C13'—C14'—C15' | 0.2 (12) | C12'—C9'—O1—C9 | −87.4 (13) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5694).
References
- Aghil, O., Bibby, M. C., Carrington, S. J., Douglas, K. T., Phillips, R. M. & Shing, T. K. M. (1992). Anti-Cancer Drug Des. 7, 67–82. [PubMed]
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Correcia, S. D., David, J. M., David, J. P., Chai, H. B., Pezzuto, J. M. & Cordell, G. A. (2001). Phytochemistry, 56, 781–784. [DOI] [PubMed]
- Ghorab, M. M., Al-Said, M. S. & El-Hossary, E. M. (2011). J. Heterocycl. Chem. 48, 563–571.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681301026X/xu5694sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681301026X/xu5694Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681301026X/xu5694Isup3.cdx
Supplementary material file. DOI: 10.1107/S160053681301026X/xu5694Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report