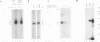Abstract
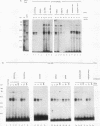
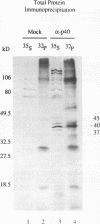
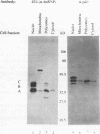
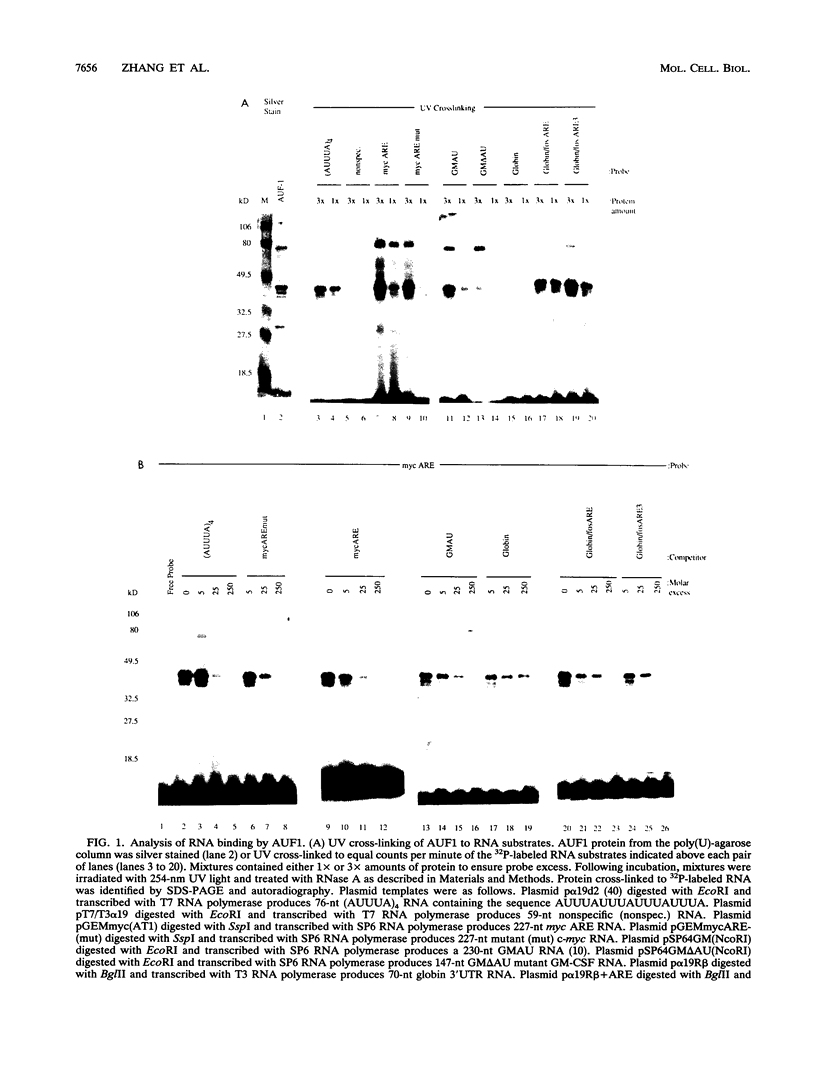
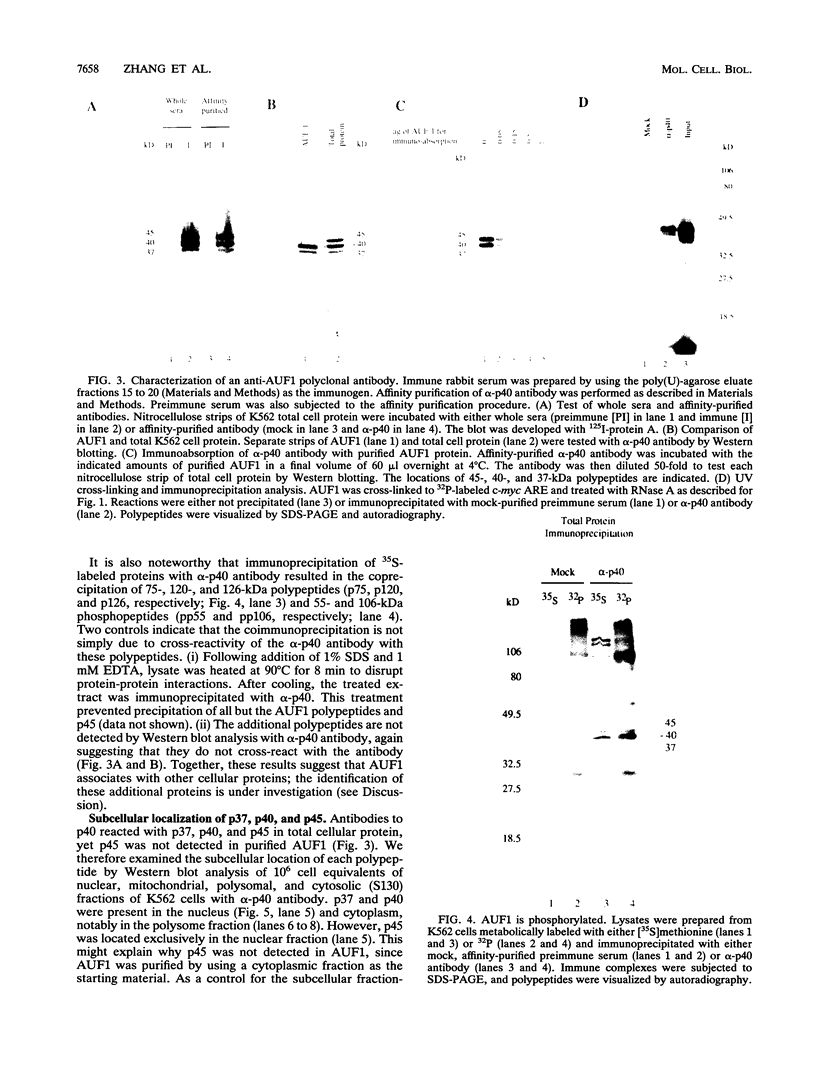
The degradation of some proto-oncogene and lymphokine mRNAs is controlled in part by an AU-rich element (ARE) in the 3' untranslated region. It was shown previously (G. Brewer, Mol. Cell. Biol. 11:2460-2466, 1991) that two polypeptides (37 and 40 kDa) copurified with fractions of a 130,000 x g postribosomal supernatant (S130) from K562 cells that selectively accelerated degradation of c-myc mRNA in a cell-free decay system. These polypeptides bound specifically to the c-myc and granulocyte-macrophage colony-stimulating factor 3' UTRs, suggesting they are in part responsible for selective mRNA degradation. In the present work, we have purified the RNA-binding component of this mRNA degradation activity, which we refer to as AUF1. Using antisera specific for these polypeptides, we demonstrate that the 37- and 40-kDa polypeptides are immunologically cross-reactive and that both polypeptides are phosphorylated and can be found in a complex(s) with other polypeptides. Immunologically related polypeptides are found in both the nucleus and the cytoplasm. The antibodies were also used to clone a cDNA for the 37-kDa polypeptide. This cDNA contains an open reading frame predicted to produce a protein with several features, including two RNA recognition motifs and domains that potentially mediate protein-protein interactions. These results provide further support for a role of this protein in mediating ARE-directed mRNA degradation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akashi M., Shaw G., Gross M., Saito M., Koeffler H. P. Role of AUUU sequences in stabilization of granulocyte-macrophage colony-stimulating factor RNA in stimulated cells. Blood. 1991 Oct 15;78(8):2005–2012. [PubMed] [Google Scholar]
- Aström J., Aström A., Virtanen A. In vitro deadenylation of mammalian mRNA by a HeLa cell 3' exonuclease. EMBO J. 1991 Oct;10(10):3067–3071. doi: 10.1002/j.1460-2075.1991.tb07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwater J. A., Wisdom R., Verma I. M. Regulated mRNA stability. Annu Rev Genet. 1990;24:519–541. doi: 10.1146/annurev.ge.24.120190.002511. [DOI] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bernstein P., Peltz S. W., Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989 Feb;9(2):659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel M., Iwai Y., Pluznik D. H., Cohen R. B. Binding of sequence-specific proteins to the adenosine- plus uridine-rich sequences of the murine granulocyte/macrophage colony-stimulating factor mRNA. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10001–10005. doi: 10.1073/pnas.89.21.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992 Mar 25;267(9):6302–6309. [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3' untranslated region of lymphokine mRNA. Mol Cell Biol. 1991 Jun;11(6):3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
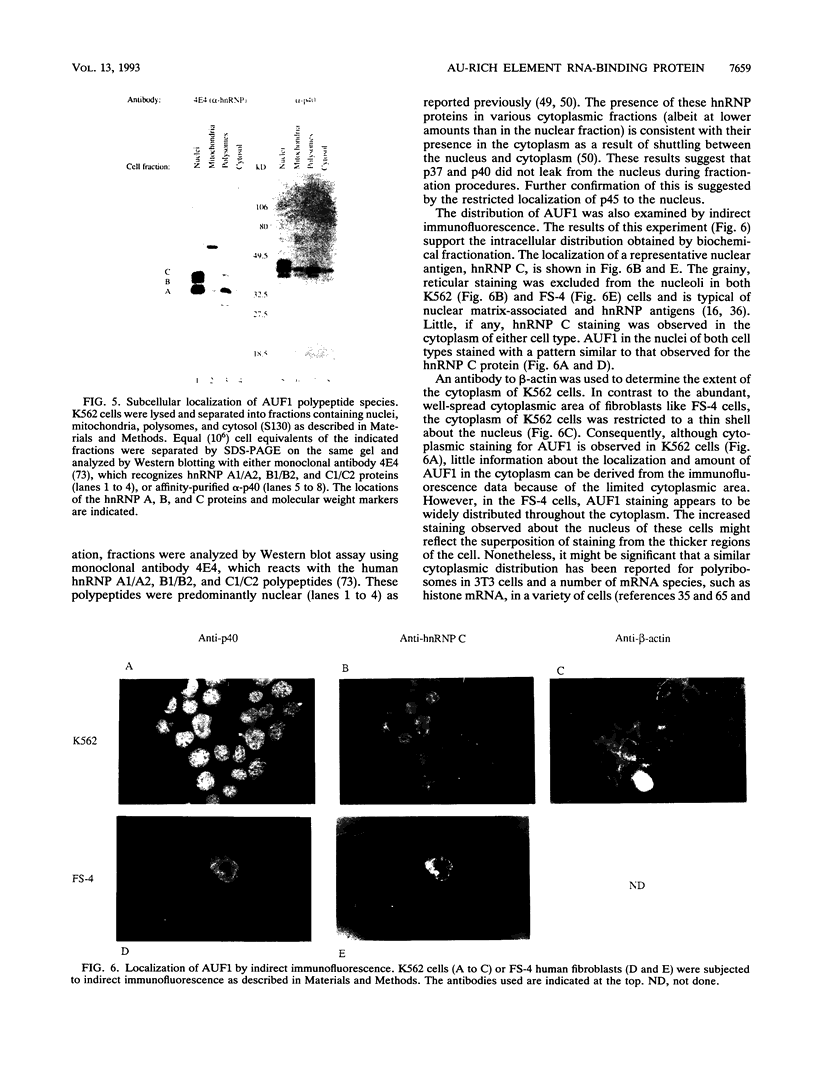
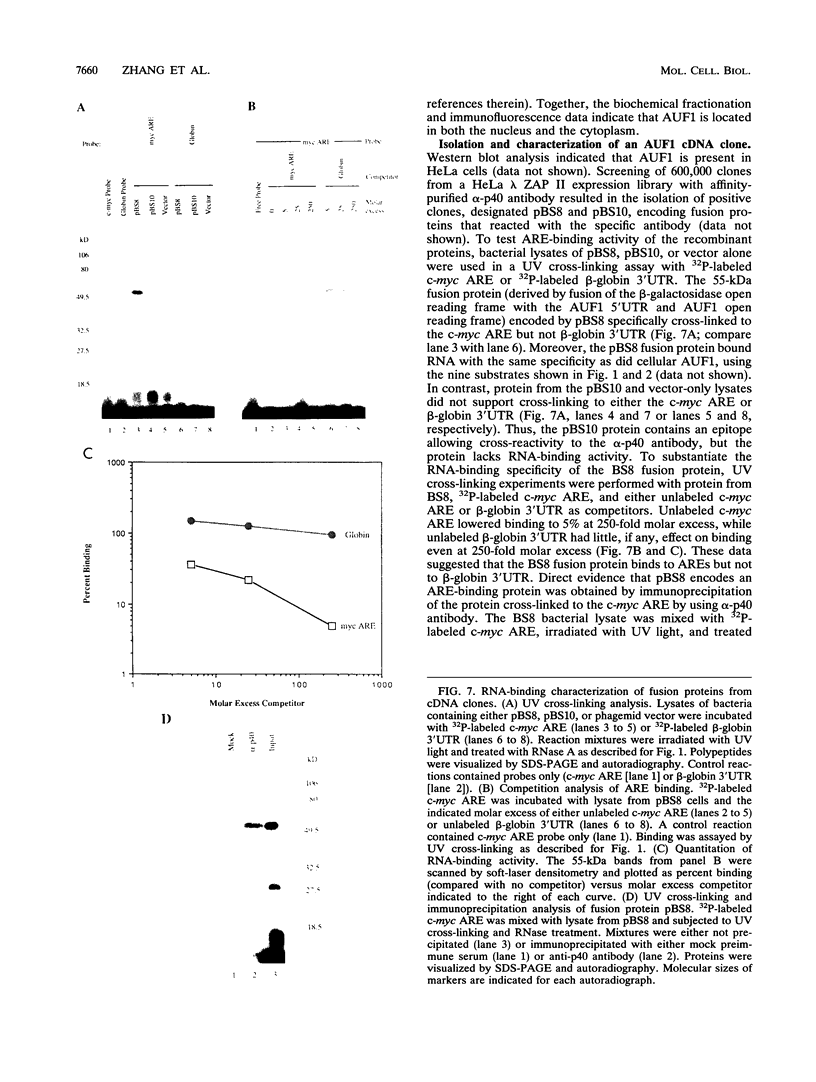
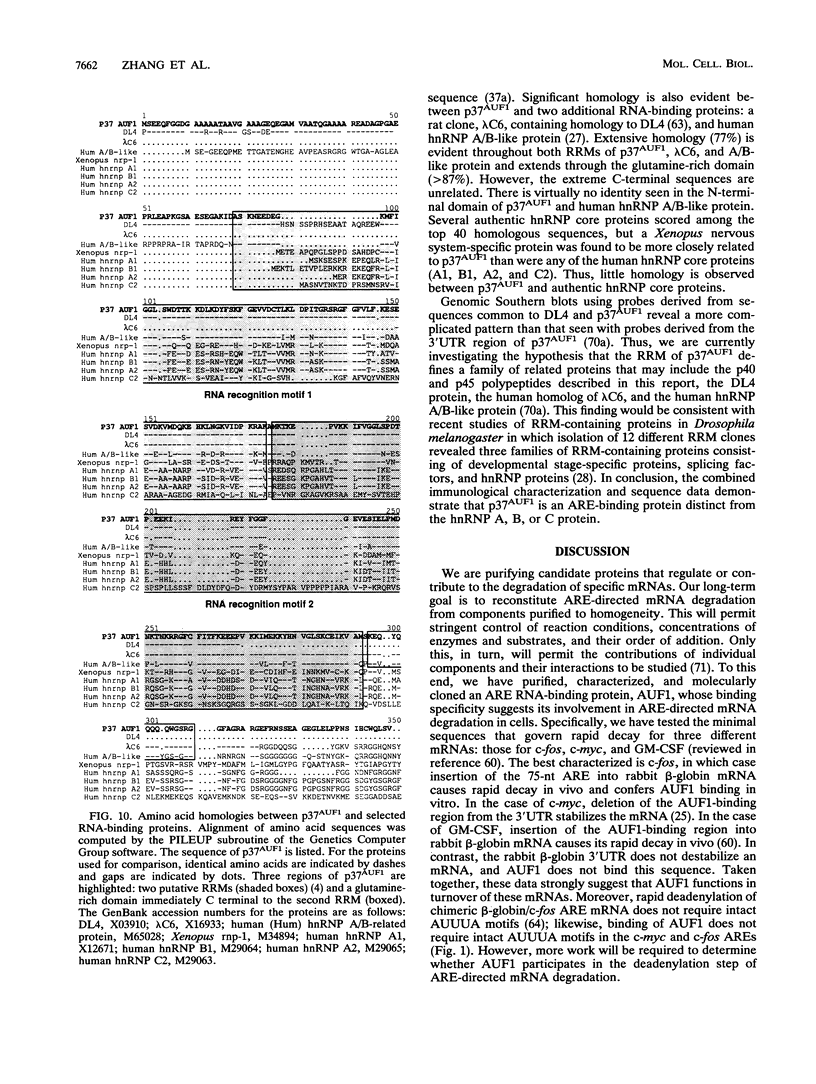
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Messenger RNA turnover in cell-free extracts. Methods Enzymol. 1990;181:202–209. doi: 10.1016/0076-6879(90)81122-b. [DOI] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989 May;9(5):1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Swanson M. S., Görlach M., Dreyfuss G. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9788–9792. doi: 10.1073/pnas.86.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M., Biamonti G., Tsoulfas P., Bassi M. T., Ghetti A., Riva S., Morandi C. cDNA cloning of human hnRNP protein A1 reveals the existence of multiple mRNA isoforms. Nucleic Acids Res. 1988 May 11;16(9):3751–3770. doi: 10.1093/nar/16.9.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaly N., Sabour M. P., Silver J. C., Aitchison W. A., Little J. E., Brown D. L. Monoclonal antibodies against nuclear matrix detect nuclear antigens in mammalian, insect and plant cells: an immunofluorescence study. Cell Biol Int Rep. 1986 Jun;10(6):421–428. doi: 10.1016/0309-1651(86)90037-8. [DOI] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A., Ley T. J., Humphries R. K., Fordis M., Schechter A. N. Inducible transcription of five globin genes in K562 human leukemia cells. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D., Mattaj I. W., Rio D. C. RNA-protein interactions. Cell. 1991 Dec 20;67(6):1041–1046. doi: 10.1016/0092-8674(91)90282-4. [DOI] [PubMed] [Google Scholar]
- Hamilton B. J., Nagy E., Malter J. S., Arrick B. A., Rigby W. F. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem. 1993 Apr 25;268(12):8881–8887. [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990 Sep;15(9):342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- Khan F. A., Jaiswal A. K., Szer W. Cloning and sequence analysis of a human type A/B hnRNP protein. FEBS Lett. 1991 Sep 23;290(1-2):159–161. doi: 10.1016/0014-5793(91)81249-8. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Baker B. S. Isolation of RRM-type RNA-binding protein genes and the analysis of their relatedness by using a numerical approach. Mol Cell Biol. 1993 Jan;13(1):174–183. doi: 10.1128/mcb.13.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991 Oct 25;266(30):19867–19870. [PubMed] [Google Scholar]
- Kruys V., Marinx O., Shaw G., Deschamps J., Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989 Aug 25;245(4920):852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- Kumar A., Williams K. R., Szer W. Purification and domain structure of core hnRNP proteins A1 and A2 and their relationship to single-stranded DNA-binding proteins. J Biol Chem. 1986 Aug 25;261(24):11266–11273. [PubMed] [Google Scholar]
- Lahiri D. K., Thomas J. O. A cDNA clone of the hnRNP C proteins and its homology with the single-stranded DNA binding protein UP2. Nucleic Acids Res. 1986 May 27;14(10):4077–4094. doi: 10.1093/nar/14.10.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A., Elfferich P., Knaken H. J., de Ruiter J., van der Eb A. J. Analysis of polyadenylation site usage of the c-myc oncogene. Nucleic Acids Res. 1989 Aug 25;17(16):6499–6514. doi: 10.1093/nar/17.16.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A. What determines the instability of c-myc proto-oncogene mRNA? Bioessays. 1992 Feb;14(2):119–124. doi: 10.1002/bies.950140209. [DOI] [PubMed] [Google Scholar]
- Lawrence J. B., Singer R. H., Marselle L. M. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989 May 5;57(3):493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lehner C. F., Eppenberger H. M., Fakan S., Nigg E. A. Nuclear substructure antigens. Monoclonal antibodies against components of nuclear matrix preparations. Exp Cell Res. 1986 Jan;162(1):205–219. doi: 10.1016/0014-4827(86)90439-8. [DOI] [PubMed] [Google Scholar]
- Lindstein T., June C. H., Ledbetter J. A., Stella G., Thompson C. B. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989 Apr 21;244(4902):339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- Lowell J. E., Rudner D. Z., Sachs A. B. 3'-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992 Nov;6(11):2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- Lozzio C. B., Lozzio B. B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975 Mar;45(3):321–334. [PubMed] [Google Scholar]
- Malter J. S. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989 Nov 3;246(4930):664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- Marcu K. B., Bossone S. A., Patel A. J. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. A binding consensus: RNA-protein interactions in splicing, snRNPs, and sex. Cell. 1989 Apr 7;57(1):1–3. doi: 10.1016/0092-8674(89)90164-5. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Gilmartin G. M., Reeves W. H., Nevins J. R. Multiple factors are required for poly(A) addition to a mRNA 3' end. Genes Dev. 1988 May;2(5):588–597. doi: 10.1101/gad.2.5.588. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992 Nov;6(11):2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- Myer V. E., Lee S. I., Steitz J. A. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1296–1300. doi: 10.1073/pnas.89.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E., Slor H., Pfeifer K., Hühn P., Bek A., Orsulic S., Ushijima H., Schröder H. C. Association of AUUUA-binding protein with A+U-rich mRNA during nucleo-cytoplasmic transport. J Mol Biol. 1992 Aug 5;226(3):721–733. doi: 10.1016/0022-2836(92)90628-w. [DOI] [PubMed] [Google Scholar]
- Pandey N. B., Sun J. H., Marzluff W. F. Different complexes are formed on the 3' end of histone mRNA with nuclear and polyribosomal proteins. Nucleic Acids Res. 1991 Oct 25;19(20):5653–5659. doi: 10.1093/nar/19.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Brewer G., Bernstein P., Hart P. A., Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryot Gene Expr. 1991;1(2):99–126. [PubMed] [Google Scholar]
- Piñol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992 Feb 20;355(6362):730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S., Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991 Jul 19;253(5017):312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Richter K., Good P. J., Dawid I. B. A developmentally regulated, nervous system-specific gene in Xenopus encodes a putative RNA-binding protein. New Biol. 1990 Jun;2(6):556–565. [PubMed] [Google Scholar]
- Ross J., Kobs G., Brewer G., Peltz S. W. Properties of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem. 1987 Jul 5;262(19):9374–9381. [PubMed] [Google Scholar]
- Sachs A. B., Bond M. W., Kornberg R. D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986 Jun 20;45(6):827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Deardorff J. A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992 Sep 18;70(6):961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- Savant-Bhonsale S., Cleveland D. W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a > 20S degradation complex. Genes Dev. 1992 Oct;6(10):1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- Schiavi S. C., Belasco J. G., Greenberg M. E. Regulation of proto-oncogene mRNA stability. Biochim Biophys Acta. 1992 Dec 16;1114(2-3):95–106. doi: 10.1016/0304-419x(92)90009-n. [DOI] [PubMed] [Google Scholar]
- Schröder H. C., Rottmann M., Wenger R., Bachmann M., Dorn A., Müller W. E. Studies on protein kinases involved in regulation of nucleocytoplasmic mRNA transport. Biochem J. 1988 Jun 15;252(3):777–790. doi: 10.1042/bj2520777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G. D., Cole M. D. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell. 1988 Dec 23;55(6):1115–1122. doi: 10.1016/0092-8674(88)90256-5. [DOI] [PubMed] [Google Scholar]
- Sharp Z. D., Smith K. P., Cao Z. D., Helsel S. Cloning of the nucleic acid-binding domain of the rat HnRNP C-type protein. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):306–309. doi: 10.1016/0167-4781(90)90073-b. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Singer R. H., Langevin G. L., Lawrence J. B. Ultrastructural visualization of cytoskeletal mRNAs and their associated proteins using double-label in situ hybridization. J Cell Biol. 1989 Jun;108(6):2343–2353. doi: 10.1083/jcb.108.6.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Green M. R. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993 Jan 15;259(5093):365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- Sun J., Pilch D. R., Marzluff W. F. The histone mRNA 3' end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 1992 Nov 25;20(22):6057–6066. doi: 10.1093/nar/20.22.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartwout S. G., Kinniburgh A. J. c-myc RNA degradation in growing and differentiating cells: possible alternate pathways. Mol Cell Biol. 1989 Jan;9(1):288–295. doi: 10.1128/mcb.9.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack J., Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3' untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991 Jun;11(6):3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriz S., Méchali M. Analysis of 3'-untranslated regions of seven c-myc genes reveals conserved elements prevalent in post-transcriptionally regulated genes. FEBS Lett. 1989 Jul 17;251(1-2):201–206. doi: 10.1016/0014-5793(89)81455-3. [DOI] [PubMed] [Google Scholar]
- Wahle E. The end of the message: 3'-end processing leading to polyadenylated messenger RNA. Bioessays. 1992 Feb;14(2):113–118. doi: 10.1002/bies.950140208. [DOI] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Wilusz J., Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol. 1990 Dec;10(12):6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Chen C. Y., Shyu A. B. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3' untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992 Jul;12(7):2931–2940. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Zapp M. L., Green M. R. Gene expression. RNA binding: beta s and basics. Nature. 1990 Dec 6;348(6301):485–486. doi: 10.1038/348485a0. [DOI] [PubMed] [Google Scholar]
- Zapp M. L., Green M. R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989 Dec 7;342(6250):714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]