Abstract
Background
The saprophytic pathogen Listeria monocytogenes has to cope with a variety of acidic habitats during its life cycle. The impact of low-temperature coupled with pH decrease for global gene expression and subsequent virulence properties, however, has not been elucidated.
Results
qRT-PCR revealed for the first time a transient, acid triggered prfA induction of approximately 4-fold, 5.7-fold, 7-fold and 9.3-fold 60 to 90 min after acid shock of L. monocytogenes at 37°C, 25°C, 18°C, and 10°C, respectively. Comparable data were obtained for seven different L. monocytogenes strains, demonstrating that prfA induction under these conditions is a general response of L. monocytogenes. Transcriptome analysis revealed that the in vivo-relevant genes bsh, clpP, glpD, hfq, inlA, inlB, inlE, lisR, and lplA1 as well as many other genes with a putative role during infection are transiently induced upon acid shock conducted at 25°C and 37°C. Twenty-five genes repressed upon acid shock are known to be down regulated during intracellular growth or by virulence regulators. These data were confirmed by qRT-PCR of twelve differentially regulated genes and by the identification of acid shock-induced genes influenced by σB. To test if up regulation of virulence genes at temperatures below 37°C correlates with pathogenicity, the capacity of L. monocytogenes to invade epithelial cells after acid shock at 25°C was measured. A 12-fold increased number of intracellular bacteria was observed (acid shock, t = 60 min) that was reduced after adaptation to the level of the unshocked control. This increased invasiveness was shown to be in line with the induction of inlAB. Using a nematode infection assay, we demonstrated that Caenorhabditis elegans fed with acid-shocked L. monocytogenes exhibits a shorter time to death of 50% (TD50) of the worms (6.4 days) compared to infection with unshocked bacteria (TD50 = 10.2 days).
Conclusions
PrfA and other listerial virulence genes are induced by an inorganic acid in a temperature-dependent manner. The data presented here suggest that low pH serves as a trigger for listerial pathogenicity at environmental temperatures.
Keywords: Listeria monocytogenes, Virulence genes, Acidic pH, prfA, Invertebrates, Transcriptome, Global response, Invasion
Background
The foodborne pathogen L. monocytogenes possesses a broad range of growth temperature (4°C to 45°C) and has been isolated from a variety of habitats including soil, decaying plants, water and animals [1]. This facultatively intracellular Gram-positive bacterium can cause systemic infections especially in immuno-compromised people with symptoms such as septicaemia, encephalomeningitis, placentitis and stillbirth. A main challenge for L. monocytogenes is the switch from a saprophytic lifestyle to a successful infection of mammals. Its capability to survive in low pH habitats such as fermented food including silage as well as in acidic host compartments like the stomach, the small intestine and phagosomes is an adaptation strategy common to both stages [2]. This aciduric capacity in turn raises persistent safety problems for the food industry [3]. On the other hand, stresses like acid are known clues for virulence gene regulation in many pathogenic organisms that may use the decrease in pH during stomach transit as signal to up regulate virulence genes [4,5]. However, the response of listerial virulence genes to acidic conditions under environmental temperatures, and a possible impact for the life cycle of this saprophyte, remains to be elucidated.
The response of L. monocytogenes to low pH involves the alternative sigma factor SigB (σB) and is characterized by the synthesis of a number of proteins to mount a significant acid tolerance response (ATR). The ATR is capable of protecting cells against otherwise lethal acid stress conditions [6]. Several lines of evidence indicated that an adaptive acid tolerance response is required for pathogenicity of L. monocytogenes, and that the ability of this pathogen to survive gastric acid fluid and to invade host cells is directly linked to the activation of the ATR [7-9]. This is supported by the finding that the glutamate decarboxylase (GAD) system as the most important ATR component is required for listerial survival in the gastric environment [10], and that the deletion of LisRK, a two-component system (TCS) for signal transduction involved in the regulation of acid resistance, resulted in growth phase-dependent sensitivity to acid stress and a significant reduction in virulence [11]. A further protein linking a low pH-response to listerial virulence is the cysteine transport associated protein CtaP, which is necessary for growth in acidified BHI (pH 5.5) and for virulence after intragastric infection of mice [12].
The translation of PrfA, the master virulence gene regulator of L. monocytogenes, is regulated by post transcriptional mRNA-folding and blocked at temperatures below 30°C [13,14]. In accordance, it has been reported that prfA is not induced by low pH [15,16]. Interestingly, L. monocytogenes grown at 4°C or 37°C exhibited similar infection phenotypes upon intragastrical inoculation [17]. Furthermore, the production of PrfA and PrfA-dependent gene products was observed at low-temperature and within cells from the fruit fly Drosophila melanogaster[18,19]. This is in line with the demonstration that L. monocytogenes is pathogenic towards invertebrates such as D. melanogaster, the greater wax moth Galleria mellonella and the nematode Caenorhabditis elegans as model organisms for listerial pathogenicity [20-23]. These findings prompted us to investigate the putative connectivity between acid stress, low temperature gene induction, and virulence properties of L. monocytogenes.
Here we demonstrate by transcriptional analysis after treatment of L. monocytogenes with the inorganic acid HCl that prfA and other virulence factors are transiently induced by acidification at lower (10°C, 18°C, and 25°C), as well as at mammalian body temperature (37°C). We further show that σB-influenced genes participate in the ATR also at 25°C, and that low pH increases epithelial cell entry of L. monocytogenes as well as its virulence towards C. elegans.
Results
Temperature-dependent induction of prfA after acid shock
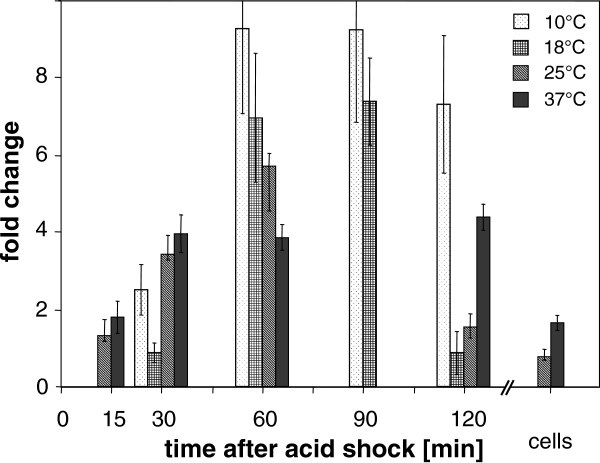
L. monocytogenes cultures grown to middle exponential phase were subjected to acid shock at different temperatures, and prfA transcript levels were examined with qRT-PCR. At 37°C, prfA showed approximately 4-fold higher transcript levels between 30 to 120 min after exposure to pH 5.0 with respect to the control. At 25°C, 18°C and 10°C, prfA was up regulated approximately 5.7-fold, 7.0-fold and 9.3-fold at 60 min after acid shock, respectively. prfA mRNA decreased after acid adaptation at 25°C and 37°C to nearly the same transcriptional level obtained before acid shock (Figure 1). Remarkably, the lower the tested temperature, the higher was the fold induction in comparison with untreated cells at the same temperature. The relative amount of prfA transcript at 25°C with respect to 37°C was 2.6 before acid shock (data not shown). Thus, the induction of prfA at 25°C and 60 min after acid shock compared to 37°C without acid shock was 2.2. Similarly, for 18°C and time point 60 min, a 3.7-fold increase compared to 37°C under pre-shock conditions was observed.
Figure 1.
Transcriptional induction of prfA upon acid shock at different time points. Samples were taken before (t = 0) and at five time points until two hours after acid shock. Mean values of fold expression ± standard deviation calculated as ratio between prfA transcription after and before HCl treatment are presented. To obtain a sample of adapted cells, acid-shocked L. monocytogenes (120 min pH 5) were diluted and grown at pH 5.2 to OD600nm = 0.5. Expression of prfA at 10°C and 18°C was measured by real time qRT-PCR in replicates of three and at 25°C and 37°C in two biologically independent experiments, respectively. The prfA induction was not determined at some time points (e.g., for 10 and 18°C at 15 min, 25 and 37°C at 90 min, and 10 and 18°C for the adaptation sample).
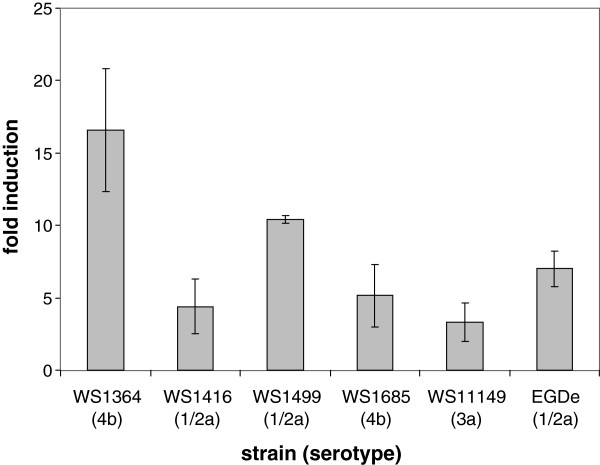
To exclude that this temperature-dependent prfA response to low pH is restricted to strain EGDe (serovar 1/2a), we examined six additional L. monocytogenes strains belonging to the serovars 1/2a, 3a and 4b for prfA transcription. For this purpose, the cultures were incubated for 1 h at pH 5.0 and 18°C, and their RNA was analyzed by quantitative real-time PCR (qRT-PCR). For five strains we received prfA induction levels of between threefold (WS11149) and 17-fold (WS1364) compared to unshocked bacteria (Figure 2). A correlation between serovar and induction levels could not be found. In one serovar 3a strain, WSLC 1211, no prfA transcription above basal level could be measured at neutral pH. However, the threshold cycle (CT) of RNA amplification with RNA from 18°C samples was the lowest in comparison with the other strains, suggesting a strong induction of prfA in WSLC 1211 after acid shock at low temperature. The 7-fold induction of prfA in strain EGDe corresponds to the mean fold induction value of 8.0 measured for the five additional L. monocytogenes strains. Taken together, these data demonstrate that the strong transcriptional increase found for prfA in strain EGDe under the described pH and temperature conditions is not a strain-specific phenomenon, but is generally observed in L. monocytogenes.
Figure 2.
prfA induction levels in L. monocytogenes strains. The transcriptional change of the prfA mRNA from five additional L. monocytogenes strains belonging to serotypes 1/2a, 3a and 4b at 18°C and 60 min after acid shock to pH 5.0 was measured by qRT-PCR in three independent experiments. The value for strain EGDe from Figure 1 is given in the last column as reference.
Global transcriptional response upon acidification at 25°C and 37°C
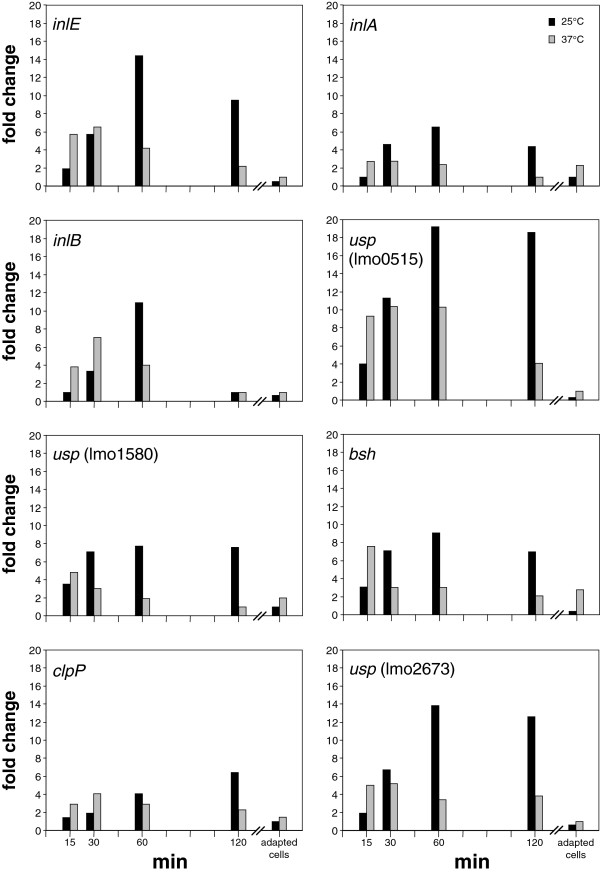
To assess the global impact of acid shock in gene induction, transcription profiling of L. monocytogenes was performed at 15, 30, 60, and 120 min post acid shock using microarray analysis. At 60 min after acid shock, 338 genes were found to be significantly up regulated at 25°C, and 383 genes at 37°C (Additional file 1). To attribute these experimental results to virulence properties, we conducted a comprehensive literature search for L. monocytogenes genes that are required for full virulence in vivo. We considered a gene to play a role during infection if its mutation showed attenuation in a mouse model (Additional file 2). Of these genes, we found 15 to be up regulated after acid shock at both 25°C and 37°C as summarized in Table 1. The genes mainly belong to the categories metabolism, regulation, stress and adhesins, and encode the following factors: bile salt hydrolase (Bsh), an ABC transporter (lmo0137), lipoate protein ligase A (lplA1), the proteolytic subunit of Clp protease (clpP), the RNA-binding protein Hfq, the TCS response regulator LisR, a PerR-like regulator (lmo1683), the response regulator DegU, three homologs of universal stress proteins (Usps; lmo0515, lmo1580, lmo2673), a hypothetical protein (SepA) and three internalins (InlA, InlB, InlE). Some of these genes, namely inlA, inlB, inlE, bsh, clpP and the three usp genes showed a significantly higher transcription rate at 25°C in comparison to 37°C at most time points after acid shock (Figure 3). Interestingly, their transcription ratios reverted to pre-shock levels after adaptation to low pH. Additionally, we noted induction of frvA, btlB and purB to be up regulated upon acid shock at 25°C, but not at 37°C (Table 1). These genes encode a Fur-regulated virulence factor, a factor similar to bile acid dehydratase, and an adenylosuccinate lyase. We also found zinA and fbpA to be up regulated at 37°C, but not at lower temperature.
Table 1.
List of genes required for full virulencein vivothat are induced after acid shock at 25 and 37°C
|
Locus |
Gene |
Functional annotation or prediction |
Commentsa |
T |
Min after acid shock |
Adaptationb |
|||
|---|---|---|---|---|---|---|---|---|---|
| 15 | 30 | 60 | 120 | ||||||
|
Induced after acid shock at both 25°C and 37°C. | |||||||||
| lmo0137 |
|
ABC transporter |
Required for full virulence in mice
[24] |
25°C |
4.0 |
5.0 |
5.8 |
9.5 |
0.8 |
| 37°C |
19.2 |
5.7 |
7.9 |
6.3 |
n.a. |
||||
| lmo0264 |
inlE |
InlE, internalin E |
Required for full virulence in mice
[25] |
25°C |
1.9 |
5.7 |
14.4 |
9.5 |
0.5 |
| 37°C |
5.7 |
6.5 |
4.2 |
2.2 |
1* |
||||
| lmo0433 |
inlA |
InlA, internalin A |
Required for host cell entry in vivo[26], PrfA-regulated
[27]and σB[28] |
25°C |
1* |
4.6 |
6.5 |
4.4 |
1* |
| 37°C |
2.7 |
2.8 |
2.4 |
1* |
2.3 |
||||
| lmo0434 |
inlB |
InlB, internalin B |
Required for host cell entry in vivo[26], σB-regulated
[28] |
25°C |
1* |
3.3 |
10.9 |
1* |
0.7 |
| 37°C |
3.8 |
7.1 |
4.0 |
1* |
1* |
||||
| lmo0515 |
|
Universal stress protein |
Important for growth in vivo[29] |
25°C |
4.0 |
11.3 |
19.2 |
18.6 |
0.3 |
| 37°C |
9.3 |
10.4 |
10.3 |
4.1 |
1* |
||||
| lmo0931 |
lplA1 |
Lipoate protein ligase A |
Aborted replication in macrophages and in vivo[30] |
25°C |
1* |
1* |
1.7 |
2.3 |
1.5 |
| 37°C |
1.9 |
2.1 |
2.4 |
2.5 |
1* |
||||
| lmo1295 |
hfq |
RNA-binding protein |
Contributes to virulence
[31] |
25°C |
1* |
2.1 |
3.4 |
2.8 |
0.6 |
| 37°C |
1* |
3.7 |
2.8 |
1.7 |
1* |
||||
| lmo1377 |
lisR |
TCS response regulator |
Contributes to virulence
[11,32] |
25°C |
1* |
0.9 |
1.7 |
2.9 |
1* |
| 37°C |
1* |
1* |
2.0 |
2.0 |
1.3 |
||||
| lmo1580 |
|
Universal stress protein |
Important for growth in vivo[29] |
25°C |
3.5 |
7.1 |
7.7 |
7.6 |
1* |
| 37°C |
4.8 |
3.0 |
1.9 |
1* |
2.0 |
||||
| lmo1683 |
perR |
Transcription regulator (Fur family), PerR in B. subtilis |
Mutant is attenuated in mice
[33] |
25°C |
1* |
2.6 |
3.5 |
5.4 |
1* |
| 37°C |
1.6 |
4.0 |
5.1 |
4.6 |
2.3 |
||||
| lmo2067 |
bshc |
Bile salt hydrolase |
PrfA-
[34] and σB-regulated
[35], necessary for gastrointestinal persistence
[36] and virulence
[37] |
25°C |
3.1 |
7.1 |
9.1 |
7.0 |
0.4 |
| 37°C |
(7.6) |
(3.0) |
(3.0) |
(2.1) |
(2.8) |
||||
| lmo2157 |
sepA |
Hypothetical protein |
Intracellularly up regulated
[38,39], deletion decreases virulence
[40] |
25°C |
1* |
2.8 |
5.9 |
5.4 |
1* |
| 37°C |
3.2 |
5.0 |
4.3 |
1.6 |
1* |
||||
| lmo2468 |
clpP |
ATP-dependent Clp protease, proteolytic subunit |
Required for full virulence
[41] and induced intracellularly
[38] |
25°C |
1.4 |
1.9 |
4.1 |
6.4 |
1* |
| 37°C |
2.9 |
4.1 |
2.9 |
2.3 |
1.5 |
||||
| lmo2515 |
degU |
Response regulator |
Contributes to virulence
[42] |
25°C |
0.9 |
0.9 |
2.0 |
3.0 |
1* |
| 37°C |
1* |
2.3 |
2.9 |
1.9 |
1* |
||||
| lmo2673 |
|
Universal stress protein |
Important for growth in vivo[29] |
25°C |
1.9 |
6.7 |
13.8 |
12.6 |
0.6 |
| 37°C |
5.0 |
5.2 |
3.4 |
3.8 |
1* |
||||
|
Induced after acid shock at 25°C, but not at 37°C | |||||||||
| lmo0641 |
frvA |
Fur-regulated virulence factor |
A putative P-type ATPase required for virulence
[43] |
25°C |
2.5 |
5.9 |
6.2 |
4.9 |
2.4 |
| lmo0754 |
btlB |
Bile acid 7-α-dehydratase |
Important for intestinal persistence
[36] |
25°C |
1.9 |
2.0 |
1.9 |
1.5 |
1* |
| lmo1773 |
purB |
Adenylosuccinate lyase |
Mutants attenuated for systemic infection in mice
[44] |
25°C |
1.0 |
0.9 |
2.4 |
1* |
1* |
|
Induced after acid shock at 37°C, but not at 25°C | |||||||||
| lmo0153 |
zinA |
Zinc ABC transporter, Zn-binding |
Contributes to full virulence in vivo[45] |
37°C |
1* |
1* |
1.85 |
2.28 |
1.38 |
| lmo1829 | fbpA | Homologies to atypical fibronectin-binding proteins | Increases adherence, required for intestinal and liver colonization [46] | 37°C | 1* | 2.0 | 1.8 | 2.3 | 1* |
Fold induction values: The integer digits (up regulated genes) or the first post decimal digit (down regulated genes) is significant (p < 0.05).
n.a.: data not available; T, temperature.
1*: No significant difference (p > 0.05) between experiment and control; the value was therefore set to 1.
aComments are based on the given reference(s).
bThe pH for adaptation was 5.2 instead of 5.0 as for all other time points, to allow for significant growth at low pH.
cThe 37°C values for bsh did not pass the quality threshold due to insufficient number of independent data points; the data available are in brackets.
Figure 3.
Up regulation of internalin and other in vivo-relevant genes upon acid shock. Transcriptional induction of selected genes transcriptionally induced after acid shock at 25°C (black columns) compared to acid shock at 37°C (grey columns) as deduced by microarray analysis. All genes have experimentally been demonstrated to play a role in listerial virulence (Additional file 2). Samples were taken 15, 30, 60 and 120 min after acid shock, and from acid-adapted cells. Two biological and two technical replicates were performed on microarray for each time point measured.
We identified 33 additional genes by our approach that have not been demonstrated to play a role in vivo, but are required for epithelial cell invasion or intracellular replication, are induced during intracellular replication, or are influenced by σB, VirR or PrfA (Table 2). Two of these genes code for internalin-homologs.
Table 2.
List of genes identified in this study that are influenced by PrfA, σB, or VirR, are required for epithelial cell invasion or intracellular replication, are induced during intracellular replication, or are homologs of internalin
|
Locus |
Gene |
Functional annotation or prediction |
Comments from literature |
T |
min after acid shock |
Adaptation1 |
|||
|---|---|---|---|---|---|---|---|---|---|
| 15 | 30 | 60 | 120 | ||||||
|
Induced after acid shock at both 25°C and 37°C | |||||||||
| lmo0206 |
|
Hypothetical protein |
PrfA-influenced, up regulated intracellularly
[38] |
25°C |
1* |
2.0 |
3.2 |
3.1 |
0.9 |
| 37°C |
1.4 |
4.1 |
2.4 |
1* |
1* |
||||
| lmo0223 |
cysK |
CysK, highly similar to cysteine synthase |
PrfA-box upstream, cell wall proteome
[47] |
25°C |
2.3 |
6.2 |
6.6 |
4.1 |
n.a. |
| 37°C |
2.5 |
3.9 |
2.5 |
2.9 |
2.0 |
||||
| lmo0593 |
|
Nitrate transporter |
Up regulated intracellularly
[38] |
25°C |
0.6 |
1* |
1.9 |
2.2 |
0.4 |
| 37°C |
6.2 |
5.7 |
6.6 |
2.9 |
1* |
||||
| lmo0596 |
|
Hypothetical protein |
Contains PrfA- and RpoS-boxes
[48], σB-dependent
[35] |
25°C |
0.8 |
1.7 |
2.0 |
1.7 |
0.5 |
| 37°C |
10.1 |
5.9 |
4.6 |
3.3 |
5.7 |
||||
| lmo0610 |
inl |
Peptidoglycan bound protein (LPXTG motif) |
Internalin-homolog
[49] |
25°C |
1.5 |
3.7 |
6.4 |
5.7 |
0.4 |
| 37°C |
9.1 |
5.9 |
5.4 |
1* |
1* |
||||
| lmo0669 |
|
Oxidoreductase |
σB[50] and PrfA-regulated
[48], required for intracellular survival
[51] |
25°C |
1.8 |
4.4 |
8.3 |
7.7 |
0.7 |
| 37°C |
3.6 |
5.7 |
5.0 |
3.7 |
3.8 |
||||
| lmo0759 |
|
Lactoylglutathione lyase |
Mutant is intracellularly attenuated
[39] |
25°C |
2.6 |
4.9 |
8.2 |
11.0 |
0.8 |
| 37°C |
4.8 |
4.9 |
4.5 |
2.9 |
1* |
||||
| lmo0783 |
|
Mannose-specific PTS component IIB |
σB-dependent
[52] |
25°C |
0.7 |
1* |
2.2 |
2.0 |
0.8 |
| lmo0880 |
|
Cell wall associated protein precursor (LPXTG motif) |
Up regulated intracellularly
[38], σB-regulated
[53] |
25°C |
2.4 |
6.9 |
9.1 |
8.8 |
1* |
| 37°C |
50.1 |
18.8 |
12.5 |
23.9 |
17.4 |
||||
| lmo1138 |
clpP |
Proteolytic component of Clp protease |
Up regulated intracellularly
[38] |
25°C |
n.a. |
1.8 |
3.4 |
4.5 |
n.a. |
| 37°C |
2.0 |
4.5 |
2.8 |
1.4 |
n.a. |
||||
| lmo1293 |
glpD |
Glycerol-3-phosphate dehydrogenase |
Up regulated intracellularly
[38] |
25°C |
n.a. |
4.9 |
7.7 |
5.9 |
n.a. |
| |
|
|
Required for intracellular growth
[39] |
37°C |
5.7 |
4.4 |
3.3 |
5.3 |
7.0 |
| lmo1302 |
lexA |
SOS response regulator |
Up regulated intracellularly
[38] |
25°C |
1.8 |
2.8 |
3.5 |
5.0 |
1* |
| 37°C |
2.0 |
2.4 |
3.5 |
2.2 |
1* |
||||
| lmo1416 |
|
Hypothetical protein |
Deletion renders L. monocytogenes bile sensitive
[36] |
25°C |
1* |
1.7 |
7.8 |
11.5 |
4.7 |
| 37°C |
1* |
8.9 |
7.7 |
9.4 |
5.9 |
||||
| lmo1734 |
gltS |
Glutamate synthase (large subunit) |
Intracellularly up regulated
[39] |
25°C |
1.7 |
2.0 |
5.2 |
1* |
1* |
| 37°C |
1* |
6.9 |
6.0 |
1* |
2.4 |
||||
| lmo1991 |
ilvA |
Threonine dehydratase |
Involved in intravacuolar survival
[38], intracellularly up regulated
[39] |
25°C |
1.7 |
2.1 |
2.9 |
3.1 |
0.9 |
| 37°C |
2.7 |
3.3 |
3.8 |
2.9 |
1* |
||||
| lmo2085 |
|
Peptidoglycan bound protein (LPXTG motif) |
Up regulated intracellularly
[38], σB-dependent
[53] |
25°C |
1.7 |
6.3 |
9.4 |
7.3 |
0.7 |
| 37°C |
6.1 |
5.8 |
3.9 |
4.2 |
1* |
||||
| lmo2156 |
|
Hypothetical protein |
VirR regulated
[54] |
25°C |
8.3 |
18.0 |
11.1 |
4.6 |
2.0 |
| 37°C |
6.2 |
9.9 |
4.6 |
4.6 |
1.7 |
||||
| lmo2191 |
spxA |
Hypothetical protein |
Up regulated intracellularly
[38] |
25°C |
1* |
1.9 |
3.6 |
3.6 |
0.8 |
| 37°C |
6.2 |
4.7 |
4.1 |
4.1 |
2.8 |
||||
| lmo2199 |
ohrA |
Hypothetical protein |
Involved in hydroxyperoxide resistance
[55], up regulated intracellularly
[38] |
25°C |
1* |
1.7 |
3.4 |
7.4 |
1.5 |
| 37°C |
1.5 |
2.1 |
3.8 |
3.9 |
1* |
||||
| lmo2200 |
ohrR |
Transcription regulator |
Involved in hydroxyperoxide resistance
[55], up regulated intracellularly
[38] |
25°C |
1* |
2.5 |
5.2 |
10.2 |
2.0 |
| 37°C |
9.8 |
11.2 |
11.2 |
14.0 |
3.4 |
||||
| lmo2206 |
clpB |
similar to endopeptidase Clp ATP-binding chain B |
chaperone, up regulated intracellularly
[38,39] |
25°C |
2.9 |
5.8 |
5.8 |
4.1 |
n.a. |
| 37°C |
7.2 |
6.9 |
3.5 |
1.5 |
n.a. |
||||
| lmo2434 |
gadD3 /gadD |
Glutamate decarboxylase |
Up regulated during acid stress
[15] and intracellularly
[39], σB-dependent
[56] |
25°C |
1* |
5.7 |
10.1 |
11.1 |
0.4 |
| 37°C |
1.8 |
2.5 |
2.4 |
1* |
1.5 |
||||
| lmo2570 |
|
Integral membrane protein |
PrfA-regulated
[48] |
25°C |
0.7 |
2.5 |
5.8 |
7.6 |
0.8 |
| 37°C |
2.1 |
4.1 |
3.4 |
2.7 |
1* |
||||
| lmo2571 |
|
Nicotinamidase |
PrfA-regulated
[48] |
25 °C |
0.5 |
4.1 |
3.4 |
2.7 |
1* |
| 37°C |
12.6 |
3.6 |
8.2 |
10.1 |
1* |
||||
| lmo2572 |
|
Dihydrofolate reductase, chain A |
PrfA-regulated
[48] |
25°C |
1* |
3.9 |
8.6 |
9.1 |
1* |
| 37°C |
5.5 |
3.9 |
n.a. |
3.7 |
2.3 |
||||
| lmo2573 |
|
Zinc-binding dehydrogenase |
PrfA-regulated
[48] |
25 °C |
2.3 |
6.8 |
10.6 |
10.0 |
0.5 |
| 37°C |
14.3 |
6.1 |
6.2 |
4.3 |
6.5 |
||||
| lmo2695 |
|
Dihydroxyaceton kinase |
Mutant attenuated in macrophages (Fuchs et al., unpublished data) |
25°C |
1* |
5.5 |
8.2 |
5.9 |
0.8 |
| 37°C |
2.8 |
2.6 |
2.6 |
2.8 |
2.1 |
||||
| lmo2696 |
|
Dihydroxyaceton kinase |
Mutant attenuated in macrophages (Fuchs et al., unpublished data) |
25°C |
n.a. |
5.3 |
13.5 |
18.6 |
n.a. |
| 37°C |
1.8 |
5.5 |
3.2 |
2.3 |
1.7 |
||||
| lmo2713 |
|
Secreted protein with GW repeat |
Up regulated intracellularly
[38], σB-regulated
[48] |
25°C |
1.5 |
3.0 |
5.0 |
3.9 |
1* |
| 37°C |
3.1 |
2.5 |
4.0 |
2.8 |
1.5 |
||||
|
Induced after acid shock at 25°C, but not at 37°C | |||||||||
| lmo1538 |
glpK1 |
Glycerol kinase |
Up regulated intracellularly
[39] |
25°C |
4.3 |
3.5 |
2.5 |
1.5 |
4.6 |
|
Induced after acid shock at 37°C, but not at 25°C | |||||||||
| lmo0232 |
clpC |
Endopeptidase Clp, ATP-binding chain C |
Plays a role in adhesion and invasion, and modulates the expression of InlA, InlB and ActA
[57] |
37°C |
2.7 |
9.6 |
6.0 |
2.0 |
1.8 |
| lmo1439 |
sod |
Superoxide dismutase |
Mutant attenuated in survival within macrophages
[58] |
37°C |
1* |
1* |
2.5 |
2.7 |
2.0 |
| lmo2250 | arpJ | ABC-transporter for arginine | Mutant attenuated in intracellular growth [59] | 37°C | 1* | 2.3 | 2.0 | 2.0 | 1* |
See legend of Table 1.
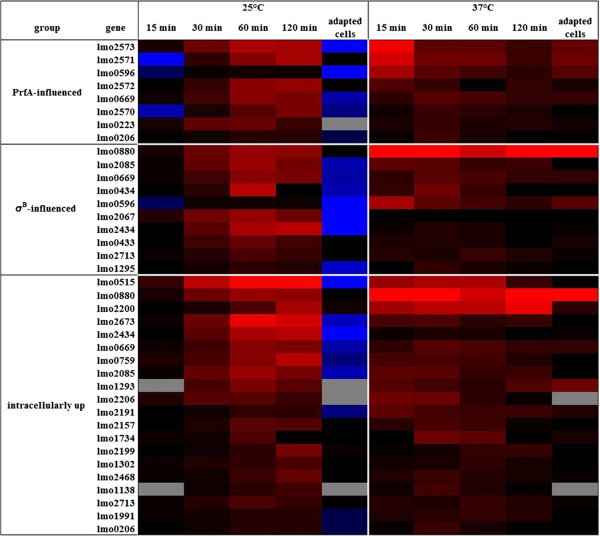
Acid shock is characterized by a mainly posttranslational activation of σB in Gram positive bacteria [60]. Accordingly, no induction of sigB transcription was observed in the microarray experiments. However, as demonstrated by the induction of σB-influenced genes (lmo0433, lmo0434, lmo0596, lmo0669, lmo0783, lmo0880, lmo0913, lmo1295, lmo2067, lmo2085, lmo2434, lmo2713; Tables 1 and 2), σB seems to be activated after acid shock. Furthermore, we observed down regulation of several genes belonging to the rsb-operon after acid shock, namely rsbR, rsbS, rsbT, and rsbU. The rsbRSTU-gene products are involved in the upstream switch module of the σB -regulation cascade [61]. Sixty min after acid shock at 25°C, a 5.9-fold, 3.6-fold, 5.3-fold, and 4.8-fold down regulation of these genes was recorded. Their expression levels were restored to pre-shock values when acid adaptation had taken place (data not shown).
To summarize these findings, we grouped those genes into the categories of PrfA- or σB-influenced or intracellularly up regulated genes. Genes in all three groups are up regulated to various degrees not only at body temperature, but also at lower environmental temperature (Figure 4). In contrast to the experiments performed at 37°C, most of the genes analyzed at 25°C reach their maximal expression 60 min or later post acid shock.
Figure 4.
Heat map of genes influenced by PrfA or σB, or of intracellularly induced genes. Average expression of in vivo-relevant expression after acid shock at 25°C and at 37°C. In both experimental settings, samples were taken 15, 30, 60 and 120 min after acid shock, and from acid-adapted cells. Genes have been grouped according to their putative regulation by PrfA and σB, as well as to up regulation during intracellular growth. Up regulation is colored in shades of red, down regulation in shades of blue. “No change” is indicated by black and missing values by grey coloring.
Data validation
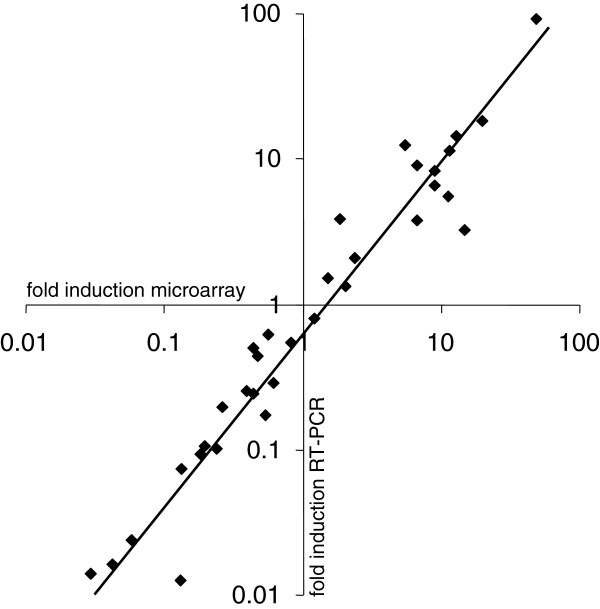
Although two biological and two technical replicates on each array were performed, twelve up and down regulated genes representing virulence and metabolic factors as well as regulatory genes and those involved in motility were chosen for qRT-PCR to further validate the microarray results (Additional file 3). This approach confirmed induced transcription of prfA, hly, flhB, and cheA, and a set of genes encoding the following predicted functions: a transcriptional regulator (lmo0109), glutamine ABC transporter (lmo0847 and lmo1740), a subunit of a sugar ABC transporter (lmo1389), the IIA component of a mannose-specific phosphotransferase system (PTS, lmo1997), a protein similar to glutamate decarboxylases (lmo2434), and a stress protein (lmo2784). Down regulation of rsbR (lmo0889) encoding a positive regulator of σB-activity was also approved by qRT-PCR. 16S rRNA transcription was used for normalization. The values obtained correlated well with the respective microarray data (Figure 5). Calculation revealed a high level of concordance (r = 0.966), demonstrating a significant correlation between the induction or repression levels determined by microarrays and by qRT-PCR. The qRT-PCR analysis thus confirmed the microarray-derived expression data of the genes investigated.
Figure 5.
Correlation of microarray data and qRT-PCR analysis of selected genes. The following genes were selected for microarray validation by qRT-PCR: prfA, hly, flhB, cheA, lmo0109, lmo0847, lmo1740, rsbR, lmo1389, lmo1997, lmo2434, and lmo2784 (see Additional file 3 for more functional details). The validated genes checked at various time points (♦) displayed a significant correlation (level of correlation = 0.966) between the data obtained by microarray analysis and by qRT-PCR. Three independent qRT-PCR experiments were performed.
Listerial genes with lower transcript levels at 37°C and 25°C following acid shock
Twenty-five genes observed to be down regulated at both temperatures after acid shock have been mentioned in the literature to be intracellularly down regulated and/or to be negatively controlled by the virulence regulators PrfA, DegU, MogR or PerR (Additional file 4). Some of these genes code for proteins that are important for cell division and replication (dnaA, lmo0394, minD, minC, divIVA, ftsZ, pbpB, ftsX, ftsE, murA/namA, cydA). They all have in common to be down regulated during intracellular growth, a response that is also reflected by slower growth rates after acid shock (data not shown). Another group of genes (lmo0178, lmo0644, lmo0675, lmo1604, plsX, dra) are known to be repressed by PrfA, PerR, DegU, MogR, and Fri (Additional file 4), suggesting that these and other down regulated genes (listed in Additional file 4; iap, lmo0847, lmo0888, lmo0970, pycA, lmo1081, lmo1084, lmo1254, fabD, pbpB, oppA, lmo2202, lgt) are not necessary within acidic host compartments. Another set of in vivo-relevant genes was observed in our study to be down regulated upon acid shock at 25°, but not at 37°C. These genes include purA, inlH, lmo0540/lmo2229/lmo2754 encoding penicillin-binding proteins, flaA, lmo0848 encoding an ABC transporter, dal, clpE, racE, lpd, bilEA, zurR, zurM, lapB, mprF, gtcA, and ami (Additional files 1 and 2). Their negative transcriptional response to low-temperature may indicate a predominant virulence role towards mammals rather than invertebrates. In contrary, the metabolic genes pgl, lap and gap, as well as brtA encoding a bile sensor (Additional files 1 and 2) are down regulated only at 37°C and might therefore contribute to pathogenicity towards invertebrates.
Increased invasiveness of listerial cells acid-shocked at 25°C
To investigate the biological relevance of acid shock at low temperature on virulence gene induction in L. monocytogenes, bacteria were cultivated at 25°C in neutral BHI medium, well below the reported 30°C-threshold for prfA gene induction [13], and then transferred to BHI medium at pH 5.0 for acid shock as described. The efficiency of unshocked, acid-shocked and acid-adapted EGDe cells to enter Caco-2 cells was determined. For this purpose, the epithelial cell layer was incubated for one hour at a multiplicity of infection (MOI) of 10. Extracellular bacteria were then eliminated, and the number of intracellular cells was determined by plating host cell lysates. In comparison to bacteria grown at pH 7.2, a strong 6.2-fold and 11.7-fold increased invasiveness of listerial cells 30 and 60 min after acid shock was observed (Figure 6). L. monocytogenes cells adapted to low pH behaved similar to those grown in BHI medium at pH 7.2 as control. These data clearly indicate that the exposure to acidic pH at 25°C enhances transiently the ability of L. monocytogenes to invade colon epithelial cells.
Figure 6.
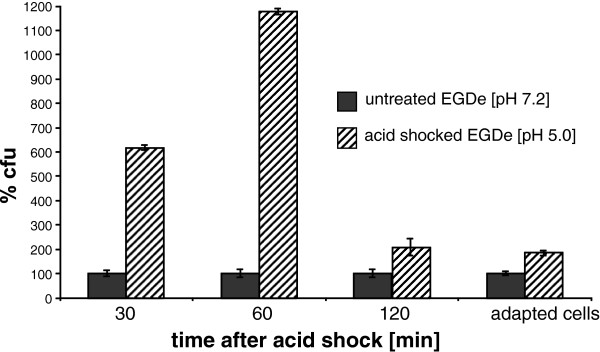
Invasiveness of L. monocytogenes after acid shock at 25°C. Invasion assays were performed with Caco-2 cells. Untreated and acid-shocked EGDe cells were used for infection at an MOI of 10. The invasion process was blocked after 1 h by cell washing and gentamycin treatment, and the epithelial cells were lysed to enumerate intracellular listeriae. The percentage of colony forming units (cfu) derived from acid treated inoculate as normalized to the control using untreated cells. Standard deviations of three independent invasion assays, performed at least as triplicates, are shown. In each experiment, the significance value was below 0.05 (p < 0.05) as calculated by the Student’s t-test. The fold difference in invasion is approximately 6-fold at 30 min and 12-fold at 60 min after acid shock.
Acid-shocked L. monocytogenes reduce the lifespan of C. elegans
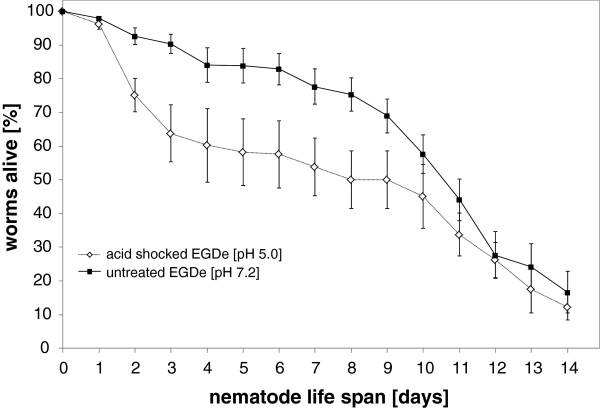
The induction of bsh and the internalin genes inlA, inlB and inlE at acidic pH in combination with low temperature might point to a role of this listerial response in the environment. To further test this hypothesis, acid-shocked L. monocytogenes were fed to C. elegans. The nematode has already been demonstrated to be an established model to study pathogenicity mechanisms of L. monocytogenes[23]. L. monocytogenes were acid-shocked for 1 h at pH 5.0 and washed three times in fresh medium. Untreated EGDe cells served as control. Listerial cells were freshly prepared every day to serve as feed for the nematodes. The nematodes were kept at room temperature (22°C), and the number of viable and dead worms was monitored each day. The time point at which 50% of worms were dead (TD50) was calculated as 6.4 ± 0.5 days upon infection with acid-shocked L. monocytogenes and 10.2 ± 0.3 days when using unshocked cells (Figure 7). The significant reduction (p < 0.05) of the nematode’s life span by 3.8 days due to low pH shocked L. monocytogenes demonstrates the biological relevance of differential gene induction at environmental temperatures below 37°C as a trigger for listerial pathogenicity towards invertebrates.
Figure 7.
Nematode infection assays. Death curves of nematodes fed with acid-shocked L. monocytogenes (◊) and unshocked control cells (■). Surviving worms were transferred daily to freshly prepared acid-shocked or unshocked L. monocytogenes for up to 14 d. The error bars are from four independent experiments using 20 worms in each condition, totaling 160 worms.
Discussion
Environmental conditions which L. monocytogenes is exposed to prior to host invasion are decisive for its infective potential as virulence gene expression is generally down regulated after cold shock or in rich medium [62,63]. In contrast, exposure to high temperature (42°C), oxidants, entry into stationary phase induces the transcription of virulence genes, or oxygen limitation increases invasiveness 100-fold [64]. Stimuli for prfA induction include body temperature, low iron concentration, high osmolarity, activated charcoal, oxidative stress and conditions simulating the gastrointestinal system [13,14,65-67], whereas fermentable sugars repress PrfA-dependent virulence genes [34,68].
Our finding of prfA induction upon acid shock contrasts the outcome of previous studies [15,68] whose experimental conditions, however, are not comparable to those used in our work. Cells had been taken from stationary phase or from cultures grown to exponential phase in the presence of low pH, thus allowing the cells to adapt to acid. Likewise, Garner et al.[69] reported an attenuated invasiveness of L. monocytogenes for Caco-2 cells, when the bacteria were grown at pH 5.5 in the presence of organic acids, and Rieu et al. [70] demonstrated a decrease in virulence gene transcription after 5 h at pH 4.0 achieved with acetic acid. In both studies, listerial cells had been adapted to low pH previous to the experiment. These results are in line with our data showing prfA transcription levels and listerial invasiveness of epithelial cells to decrease to nearly pre-shock levels after adaptation to low pH (Figures 1, 4 and 6). However, conducting a short time experiment, Werbrouck et al.[16] failed to detect prfA induction after 1 h acid shock to pH 5.5. This conflicting finding might be explained by their use of organic acids that are much more harmful to the bacteria. To mimic gastric conditions, we, in contrast, added inorganic HCl to acid shock L. monocytogenes. The observed changes do not reflect growth-phase dependent physiological changes because listerial growth after acid shock was not observed for at least 1 hour (data not shown); therefore, the transcriptional response appears acid-shock specific. The prfA inducibility by HCl at 37°C as demonstrated here might be considered as an imitation of conditions during the stomach passage where acid is an early signal for a possible host entry, thus preparing bacteria for infection [71].
We assume that in contrast to a temporary acid shock, prolonged mild acid conditions are often encountered by saprophytic bacteria in the environment, namely upon geological processes, fermentation of plant material, activity of plant roots or food processing [7,17,72]. As a result of this acid adaptation process, virulence gene expression probably decreases in L. monocytogenes. Due to the ATR, the acid adapted listeriae also show an enhanced capacity to survive the gastric barrier as reflected by higher survival rates 15 min after intragastric infection of mice [73]. In this case, a further signal for virulence gene induction occurs after stomach exit, namely exposure to short chain fatty acids accompanied with pH neutralization [74].
An unexpected result of this study was the induction of prfA and other in vivo-relevant genes at temperatures lower than 37°C after acid shock (Figures 1, 4). Johansson et al. reported that prfA mRNA is stable at 30°C, but PrfA is only formed at 37°C [13]. According to Loh et al. [75], PrfA expression is approximately 16-times lower at 30°C compared to 37°C and not detectable in L. monocytogenes cultivated at 20°C in BHI medium. However, using a prfA-promoter fusion to gfp, substantial expression of the reporter protein at 30°C in the heterologous system of E. coli was visible [75]. This suggests PrfA expression to be generally possible at temperatures below 37°C. Accordingly, it was argued that prfA upregulation occurs most likely at the σB-dependent P2 promoter and not via the σA-dependent P1 promoter, producing monocistronic prfA mRNA which is not blocked at temperatures below 30°C [34].
A remarkable high number of genes involved in virulence, controlled by one of the main virulence regulators or induced during intracellular growth, also positively responded to acid shock at both temperatures, indicating that this signal contributes to the adaptation of L. monocytogenes during infection. A differential temperature response was observed for few genes only (Tables 1 and 2), and we hypothesize that this finding points to their requirement in specific hosts. While zinA, fbpA, clpC, sod and arpJ at 37 °C might specifically be induced in mammalian hosts, the low temperature-induced genes frvA, btlB, purB and glpK1 may indicate a role in invertebrates. A set of up regulated genes identified here is influenced by σB. σB is not only responsible for listerial survival under acid stress conditions [76], but for the transcription of several in vivo-relevant genes such as bsh, inlAB and opuCA[28,35,51,77]. Remarkably, σB contributes to virulence and gastrointestinal spread, but not systemic infection [78].
Uptake of L. monocytogenes into non-phagocytotic cells is mainly mediated by InlAB [27,28]. In agreement with the above reported increased invasiveness at 25°C, we observed a transient induction of inlA and inlB (e.g., at 60 min 6.5-fold for inlA and 10.9-fold for inlB; Figure 3), despite a normally lesser transcription at lower temperatures [19,79]. Enough InlAB seems to be produced for invasion after acid shock [57]. We detected a 12-fold increase in cell culture invasiveness one hour after acid shock at 25°C, compared to a modest 3.1-fold increase after acid shock at 37°C or at 30°C [9,80]. In contrast to acidic stress at 37°C [81], listerial cells adapted to acid lost their higher potential to invade Caco-2 cells. σB, which is active at low environmental pH, contributes to invasion by controlling inlAB[28]. This is in accordance with our finding that σB-dependent genes are induced upon acid shock.
The above described data prompted us to investigate acid shock induced prfA transcription at even lower temperatures of 18°C and 10°C. Unexpectedly, lower temperatures corresponded with higher folds of prfA induction when compared to unshocked cells at the same temperature (Figure 1). A similar response was observed for six other L. monocytogenes strains. We wondered whether the increase of virulence factor transcription due to acidic conditions at temperatures below 37°C confers increased pathogenicity of L. monocytogenes under yet unknown conditions. To answer this question, we applied a nematode killing assay. When we fed acid-shocked L. monocytogenes to C. elegans we could observe that the nematodes died faster compared to nematodes fed with unshocked bacteria. These observations clearly showed the biological relevance of virulence gene induction after acid shock at room temperature. D. melanogaster and its cells grown at 30°C have also successfully been introduced as a model for listerial infection [18,20]. Therefore, it could be hypothesized that other poikilothermal animals are hosts of L. monocytogenes, although virulence genes appear generally down regulated at low temperature [63].
Indeed, Listeria spp. have been isolated from reptiles, amphibians, fish, as well as snails, crustaceans, diptera and leeches in the past [82-85], but to our knowledge no acute illness of such hosts upon listerial infection has been reported. Interestingly, the above mentioned poikilothermal animals acidify their stomach or gut content during digestion [86-91], and the resulting low pH may serve as a signal of host entry and virulence gene induction at environmental temperatures.
Conclusions
Environmental conditions have been identified here as a clue for L. monocytogenes virulence. A persistent low pH appears to signal that L. monocytogenes has entered a low pH habitat outside animals, resulting in the down regulation of virulence genes and the development of a full ATR. In contrast, a transient low pH at environmental temperature is a signal for this pathogen to potentially have entered a host as evidenced by the increase in cell culture invasion capabilities and faster killing of nematodes. This phenotype is paralleled by an unexpected induction of prfA transcription, which is transient and increases with temperature decrease.
Methods
Culture conditions and acid shock
If not mentioned otherwise, L. monocytogenes EGDe (serovar 1/2a) was used for the experiments. The following L. monocytogenes strains were taken from the Weihenstephan ListeriaCollection (WSLC, ZIEL, Abteilung Mikrobiologie, Technische Universität München, Freising, Germany): WSLC1211 (serovar 3a), WSLC1364 (4b), WSLC1416 (1/2a), WSLC1499 (1/2a), WSLC1685 (4b), WSLC11149 (3a). Cells were grown with shaking (180 rpm) in near neutral BHI (pH 7.2) media at 10°C, 18°C, 25°C or 37°C until an optical density at 600 nm (OD600nm) of 0.5 and then exposed to sub lethal acid stress (pH 5.0, BHI adjusted with 5 M HCl). Samples were taken immediately before acid shock as a control (0 min), or 15, 30, 60, 90 and 120 min after acid shock. In each experiment, the control samples were used as reference. Two hours after exposure to pH 5.0, cells were diluted 1:100 into fresh BHI broth at pH 5.2. A sample of cells adapted in this way was taken from this culture when it had reached the middle logarithmic growth phase (OD600 = 0.5). A pH of 5.2 was chosen to allow significant growth compared to pH 5.
RNA isolation
Ten ml cell suspension was added to 20 ml of bacterial RNA Protect (QIAGEN, Hilden, Germany) and sedimented by centrifugation at 5,000 × g for 5 min. The pellets were immediately frozen in liquid nitrogen. Total RNA was isolated using an RNeasy tissue kit (QIAGEN) according to the manufacturer’s recommendations with the following modifications. The frozen cells were resuspended in 700 μl RTL buffer containing 1% β-mercaptoethanol and disrupted thrice in a Ribolyzer (Hybaid, Heidelberg, Germany) using Lysing Matrix B beads (Qbiogene, Heidelberg, Germany) for 45 s at a speed of 6.5 m/s. The cells were cooled on ice between runs. Finally, the tubes were centrifuged for 2 min at 16,100 × g, and the supernatant was removed and processed as described by the manufacturer. DNase digestion was performed using Ambion’s DNA-free (Ambion, Cambridgeshire, United Kingdom) according to the manufacturer’s instructions.
Real time quantitative qRT-PCR
One μg of isolated total RNA was transcribed with random primers to cDNA by Superscript™ III (Invitrogen, Karlsruhe, Germany). The cDNA was 10-fold serially diluted and real time PCR was performed in an iCycler (BioRad, München, Germany) using ABsolute™ QPCR SYBR® Green Fluorescein mix (ABgene, Hamburg, Germany). Primer for prfA-analysis are lmo0200F3 (5′-GTATCACAAAGCTCACGAGT-3′) and lmo0200R3 (5′- TGTATCAATAAAGCCAGACAT-3′). Primer binding to different genetic regions for microarray confirmation are listed in Additional file 3. 16S rRNA was used to normalize the qRT-PCR data as described previously [92].
Epithelial cell invasion assay
Human colon epithelial cells (Caco-2 cells, ATCC HTB-37), 2.5 × 105 per well, were seeded in a 24-well culture plate and cultivated until infection at 37°C and 5% CO2 in RPMI 1640 supplemented with 10% fetal calf serum. Caco-2 cells were not differentiated, and passages 7–13 were used. L. monocytogenes was grown in BHI (pH 7.2) at 25°C until OD600 = 0.5 and acid-shocked to pH 5.0 using HCl for 30 and 60 min as described above. A sample of bacteria adapted to low pH was obtained as described above. Epithelial cells were washed thrice with PBS/Mg2+Ca2+ and finally covered for 1 h at 37°C with 500 μl RPMI 1640 containing bacterial cells to a multiplicity of infection of 10. Subsequently, the Caco-2 cells were washed thrice with PBS/Mg2+Ca2+, incubated for 60 min in 500 μl RPMI 1640 containing 10 μg ml-1 gentamycin, and washed with PBS/Mg2+Ca2+. The infected Caco-2 cells were lysed in 1 ml cold Triton X-100 (0.1%), and intracellular bacteria were quantified by plating appropriate dilutions.
C. elegans infection assays
The infection of C. elegans with EGDe was essentially performed as described recently [93]. Briefly, L. monocytogenes was grown in BHI to OD600nm = 0.5 at 30°C. An aliquot of this culture was removed as a control, and the remaining cell suspension was acidified to pH 5.0 for 1 h with HCl as described. Ten ml of the acid-shocked and the control culture were centrifuged. The sedimented cells were washed three times in fresh BHI, and the final pellet was spread on nematode growth medium (NGM) agar plates of 8.5 cm diameter. Plates were equilibrated to room temperature (22°C) before use. Twenty C. elegans L4 larvae were then transferred onto the listerial lawn. The worms were transferred daily to freshly prepared L. monocytogenes as described above, and their survival was monitored for 14 d. Worms were considered dead if they did not respond to touching. The time for 50% of the worms to die (TD50) was calculated using the dose–response curve (drc) of the software package R and the function LL.2. The package drc calculates the time at which the inflection point locates at 50% deaths and gives a standard error within a 95% confidence interval estimated by using all given data points [94].
Microarray analysis
Forty μg of total RNA were reversely transcribed using 9 μg random hexamer primer (Invitrogen), 200U Superscript III RNase H- Reverse Transcriptase (Gibco, Life Technologies), and 2 μl of either Cy3- or Cy5-conjugated dCTP (Amersham Biosciences, Freiburg, Germany). The labeled cDNA was applied to microarray analysis with 70mer oligos (Operon Biotechnologies, Cologne, Germany) spotted on epoxy-slides [39]. Competitive microarray hybridization was performed over night at 50°C in duplicate from two independent experiments, with dye swap arrangements for each experiment. The spots were identified and measured using the software ImaGene (Biodiscovery, El Segundo, CA, USA). After median local background correction, the data were normalized globally using the lowess method [95] implemented in the program. Technical replicates on the same array were combined. Only genes exhibiting substantial changes of RNA levels upon acid treatment (at least threefold signal intensities above local background, expression ratios ≥2 for at least at one time point, more than three valid ratio values) were considered, and the final data were analyzed using GeneSight (Biodiscovery).
Abbreviations
PTS: Phosphotransferase system; TCS: Two-component system; ATR: Acid tolerance response.
Competing interests
The authors declare that they have no competing financial interests.
Authors’ contributions
SS supervised the study, and KN and TMF drafted the manuscript. KN performed RT-PCR and supervised the worm assays, PS conducted the microarray analysis, KS was responsible for cell culture assays. KN and TMF analyzed the data. All authors read and approved the final manuscript.
Supplementary Material
Transcriptomic dynamic of L. monocytogenes upon acid shock at two different temperatures.
List of genes from L. monocytogenes whose knockout led in all cases to attenuation in mouse infection experiments.
List of the genes used for the qRT-PCR validation of the L. monocytogenes microarray experiments.
List of genes significantly (p ≤ 0.05) down regulated after acid shock at both 25°C and 37°C.
Contributor Information
Klaus Neuhaus, Email: neuhaus@wzw.tum.de.
Peter Satorhelyi, Email: peter.satorhelyi@fermentia.hu.
Kristina Schauer, Email: kristina.schauer@mh.vetmed.uni-muenchen.de.
Siegfried Scherer, Email: siegfried.scherer@wzw.tum.de.
Thilo M Fuchs, Email: thilo.fuchs@wzw.tum.de.
Acknowledgements
The authors thank Jan Blohberger for performing the nematode infection assays. Parts of this work were supported by the Competence Centre PathoGenoMik funded by the Federal Ministry of Education and Research (BMBF) of Germany and by the European Union’s (EU) 6th Framework Programme TRUEFOOD (Traditional United European Food), which is an Integrated Project financed by the EU for RTD (contract number FOOD-CT-2006-016264). The information in this document reflects only the author’s views and the EU is not liable for any use that may be made of the information contained therein.
References
- Fenlon DR. In: Listeria, Listeriosis, and Food Safety. Volume 1. 2. Ryser ET, Marth EH, editor. New York: Marcel Dekker Inc; 1999. Listeria monocytogenes in the natural environment; pp. 21–38. [Google Scholar]
- Gahan CGM, Hill C. In: Microbial Stress Adaptation and Food Safety. Yousef AE, Juneja VK, editor. CRC Press; 2002. Relationship between stress adaptation and virulence in foodborne pathogenic bacteria. [Google Scholar]
- Bowman JP, Lee Chang KJ, Pinfold T, Ross T. Transcriptomic and phenotypic responses of Listeria monocytogenes strains possessing different growth efficiencies under acidic conditions. Appl Environ Microbiol. 2010;76:4836–4850. doi: 10.1128/AEM.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack HM, Gahan CG, Hill C. A novel promoter trap identifies Listeria monocytogenes promoters expressed at a low pH within the macrophage phagosome. FEMS Microbiol Lett. 2007;274:139–147. doi: 10.1111/j.1574-6968.2007.00832.x. [DOI] [PubMed] [Google Scholar]
- van der Veen S, van Schalkwijk S, Molenaar D, de Vos WM, Abee T, Wells-Bennik MH. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology. 2010;156:374–384. doi: 10.1099/mic.0.035196-0. [DOI] [PubMed] [Google Scholar]
- Ryan S, Hill C, Gahan CG. Acid stress responses in Listeria monocytogenes. Adv Appl Microbiol. 2008;65:67–91. doi: 10.1016/S0065-2164(08)00603-5. [DOI] [PubMed] [Google Scholar]
- Gandhi M, Chikindas ML. Listeria: A foodborne pathogen that knows how to survive. Int J Food Microbiol. 2007;113:1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- O’Driscoll B, Gahan CG, Hill C. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl Environ Microbiol. 1996;62:1693–1698. doi: 10.1128/aem.62.5.1693-1698.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MP, Petrone G, Di Biase AM, Ammendolia MG, Superti F, Seganti L. Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophage-like cells. Microb Pathog. 2000;29:137–144. doi: 10.1006/mpat.2000.0379. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Gahan CG, Hill C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol. 2001;40:465–475. doi: 10.1046/j.1365-2958.2001.02398.x. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Emerson N, Gahan CG, Hill C. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J Bacteriol. 1999;181:6840–6843. doi: 10.1128/jb.181.21.6840-6843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xayarath B, Marquis H, Port GC, Freitag NE. Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol Microbiol. 2009;74:956–973. doi: 10.1111/j.1365-2958.2009.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/S0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Freitag NE, Boor KJ. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect Immun. 2006;74:2505–2512. doi: 10.1128/IAI.74.5.2505-2512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MP, Petrone G, Di Biase AM, Longhi C, Penta M, Tinari A, Superti F, Fabozzi G, Visca P, Seganti L. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect Immun. 2002;70:4369–4378. doi: 10.1128/IAI.70.8.4369-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbrouck H, Vermeulen A, Van Coillie E, Messens W, Herman L, Devlieghere F, Uyttendaele M. Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int J Food Microbiol. 2009;134:140–146. doi: 10.1016/j.ijfoodmicro.2009.03.022. [DOI] [PubMed] [Google Scholar]
- Czuprynski CJ. Listeria monocytogenes: silage, sandwiches and science. Anim Health Res Rev. 2005;6:211–217. doi: 10.1079/AHR2005111. [DOI] [PubMed] [Google Scholar]
- Cheng LW, Portnoy DA. Drosophila S2 cells: an alternative infection model for Listeria monocytogenes. Cell Microbiol. 2003;5:875–885. doi: 10.1046/j.1462-5822.2003.00327.x. [DOI] [PubMed] [Google Scholar]
- McGann P, Ivanek R, Wiedmann M, Boor KJ. Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity of these proteins among the listeriae. Appl Environ Microbiol. 2007;73:2806–2814. doi: 10.1128/AEM.02923-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Altincicek B, Hain T, Domann E, Vilcinskas A, Chakraborty T. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl Environ Microbiol. 2010;76:310–317. doi: 10.1128/AEM.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce SA, Gahan CG. Molecular pathogenesis of Listeria monocytogenes in the alternative model host Galleria mellonella. Microbiology. 2010;156:3456–3468. doi: 10.1099/mic.0.040782-0. [DOI] [PubMed] [Google Scholar]
- Thomsen LE, Slutz SS, Tan MW, Ingmer H. Caenorhabditis elegans is a model host for Listeria monocytogenes. Appl Environ Microbiol. 2006;72:1700–1701. doi: 10.1128/AEM.72.2.1700-1701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Geginat G, Liang C, Goebel W, Dandekar T, Fuchs TM. Deciphering the intracellular metabolism of Listeria monocytogenes by mutant screening and modelling. BMC Genomics. 2010;11:573. doi: 10.1186/1471-2164-11-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffelsbauer D, Bubert A, Engelbrecht F, Scheinpflug J, Simm A, Hess J, Kaufmann SH, Goebel W. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol Gen Genet. 1998;260:144–158. doi: 10.1007/s004380050880. [DOI] [PubMed] [Google Scholar]
- Gaillard JL, Jaubert F, Berche P. The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J Exp Med. 1996;183:359–369. doi: 10.1084/jem.183.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingnau A, Domann E, Hudel M, Bock M, Nichterlein T, Wehland J, Chakraborty T. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect Immun. 1995;63:3896–3903. doi: 10.1128/iai.63.10.3896-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Marquis H, Boor KJ. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology. 2005;151:3215–3222. doi: 10.1099/mic.0.28070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifart Gomes C, Izar B, Pazan F, Mohamed W, Mraheil MA, Mukherjee K, Billion A, Aharonowitz Y, Chakraborty T, Hain T. Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e in vitro and in vivo. PLoS One. 2011;6:e24965. doi: 10.1371/journal.pone.0024965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan M, Moors MA, Portnoy DA. Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- Christiansen JK, Larsen MH, Ingmer H, Søgaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, Bauer S, Beier D, Kuhn M. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect Immun. 2005;73:3152–3159. doi: 10.1128/IAI.73.5.3152-3159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea RB, Gahan CG, Hill C. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect Immun. 2004;72:717–727. doi: 10.1128/IAI.72.2.717-727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortti M, Monzo HJ, Lacharme-Lora L, Lewis DA, Vázquez-Boland JA. The PrfA virulence regulon. Microbes Infect. 2007;9:1196–1207. doi: 10.1016/j.micinf.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Rauch M, Luo Q, Müller-Altrock S, Goebel W. SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J Bacteriol. 2005;187:800–804. doi: 10.1128/JB.187.2.800-804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M, Sleator RD, Gahan CG, Hill C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun. 2005;73:894–904. doi: 10.1128/IAI.73.2.894-904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, Cossart P. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Przybilla K, Stühler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskirch LM, Paterson Y. Listeria monocytogenes: a potent vaccine vector for neoplastic and infectious disease. Immunol Rev. 1997;158:159–169. doi: 10.1111/j.1600-065X.1997.tb01002.x. [DOI] [PubMed] [Google Scholar]
- Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol Microbiol. 2000;35:1286–1294. doi: 10.1046/j.1365-2958.2000.01773.x. [DOI] [PubMed] [Google Scholar]
- Knudsen GM, Olsen JE, Dons L. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol Lett. 2004;240:171–179. doi: 10.1016/j.femsle.2004.09.039. [DOI] [PubMed] [Google Scholar]
- McLaughlin HP, Xiao Q, Rea RB, Pi H, Casey PG, Darby T, Charbit A, Sleator RD, Joyce SA, Cowart RE. A putative P-type ATPase required for virulence and resistance to haem toxicity in Listeria monocytogenes. PLoS One. 2012;7:e30928. doi: 10.1371/journal.pone.0030928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith NG, Kim JW, Azizoglu R, Kathariou S, Czuprynski C. Purine biosynthesis mutants (purA and purB) of serotype 4b Listeria monocytogenes are severely attenuated for systemic infection in intragastrically inoculated A/J mice. Foodborne Pathog Dis. 2012;9:480–486. doi: 10.1089/fpd.2011.1013. [DOI] [PubMed] [Google Scholar]
- Corbett D, Wang J, Schuler S, Lopez-Castejon G, Glenn S, Brough D, Andrew PW, Cavet JS, Roberts IS. Two zinc uptake systems contribute to the full virulence of Listeria monocytogenes during growth in vitro and in vivo. Infect Immun. 2012;80:14–21. doi: 10.1128/IAI.05904-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Bourdichon F, Cabanes D, Lecuit M, Fsihi H, Cossart P. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol Microbiol. 2004;53:639–649. doi: 10.1111/j.1365-2958.2004.04138.x. [DOI] [PubMed] [Google Scholar]
- Schaumburg J, Diekmann O, Hagendorff P, Bergmann S, Rohde M, Hammerschmidt S, Jansch L, Wehland J, Karst U. The cell wall subproteome of Listeria monocytogenes. Proteomics. 2004;4:2991–3006. doi: 10.1002/pmic.200400928. [DOI] [PubMed] [Google Scholar]
- Milohanic E, Glaser P, Coppée JY, Frangeul L, Vega Y, Vázquez-Boland JA, Kunst F, Cossart P, Buchrieser C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- Pucciarelli MG, Calvo E, Sabet C, Bierne H, Cossart P. Garcia-del Portillo F: Identification of substrates of the Listeria monocytogenes sortases A and B by a non-gel proteomic analysis. Proteomics. 2005;5:4808–4817. doi: 10.1002/pmic.200402075. [DOI] [PubMed] [Google Scholar]
- Homerova D, Bischoff M, Dumolin A, Kormanec J. Optimization of a two-plasmid system for the identification of promoters recognized by RNA polymerase containing Staphylococcus aureus alternative sigma factor σB. FEMS Microbiol Lett. 2004;232:173–179. doi: 10.1016/S0378-1097(04)00063-1. [DOI] [PubMed] [Google Scholar]
- Sue D, Fink D, Wiedmann M, Boor KJ. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology. 2004;150:3843–3855. doi: 10.1099/mic.0.27257-0. [DOI] [PubMed] [Google Scholar]
- Abram F, Starr E, Karatzas KA, Matlawska-Wasowska K, Boyd A, Wiedmann M, Boor KJ, Connally D, O’Byrne CP. Identification of components of the Sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl Environ Microbiol. 2008;74:6848–6858. doi: 10.1128/AEM.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes σB regulates stress response and virulence functions. J Bacteriol. 2003;185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol. 2005;57:1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x. [DOI] [PubMed] [Google Scholar]
- Dussurget O, Dumas E, Archambaud C, Chafsey I, Chambon C, Hébraud M, Cossart P. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol Lett. 2005;250:253–261. doi: 10.1016/j.femsle.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Wemekamp-Kamphuis HH, Wouters JA, de Leeuw PP, Hain T, Chakraborty T, Abee T. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl Environ Microbiol. 2004;70:3457–3466. doi: 10.1128/AEM.70.6.3457-3466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Milohanic E, Berche P. ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect Immun. 2000;68:7061–7068. doi: 10.1128/IAI.68.12.7061-7068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambaud C, Nahori MA, Pizarro-Cerda J, Cossart P, Dussurget O. Control of Listeria superoxide dismutase by phosphorylation. J Biol Chem. 2006;281:31812–31822. doi: 10.1074/jbc.M606249200. [DOI] [PubMed] [Google Scholar]
- Klarsfeld AD, Goossens PL, Cossart P. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol Microbiol. 1994;13:585–597. doi: 10.1111/j.1365-2958.1994.tb00453.x. [DOI] [PubMed] [Google Scholar]
- van Schaik W, Abee T. The role of σB in the stress response of Gram-positive bacteria – targets for food preservation and safety. Curr Opin Biotechnol. 2005;16:218–224. doi: 10.1016/j.copbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Chaturongakul S, Boor KJ. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl Environ Microbiol. 2004;70:5349–5356. doi: 10.1128/AEM.70.9.5349-5356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YC, Raengpradub S, Boor KJ, Wiedmann M. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl Environ Microbiol. 2007;73:6484–6498. doi: 10.1128/AEM.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JB, Roldgaard BB, Christensen BB, Licht TR. Oxygen restriction increases the infective potential of Listeria monocytogenes in vitro in Caco-2 cells and in vivo in guinea pigs. BMC Microbiol. 2007;7:55. doi: 10.1186/1471-2180-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MP, Longhi C, Petrone G, Polidoro M, Valenti P, Seganti L. Modulation of actA gene expression in Listeria monocytogenes by iron. J Med Microbiol. 2000;49:681–683. doi: 10.1099/0022-1317-49-8-681. [DOI] [PubMed] [Google Scholar]
- Myers ER, Dallmier AW, Martin SE. Sodium chloride, potassium chloride, and virulence in Listeria monocytogenes. Appl Environ Microbiol. 1993;59:2082–2086. doi: 10.1128/aem.59.7.2082-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Olesen I, Andersen T, Fang W, Jespersen L. Survival of Listeria monocytogenes in simulated gastrointestinal system and transcriptional profiling of stress- and adhesion-related genes. Foodborne Pathog Dis. 2010;7:267–274. doi: 10.1089/fpd.2009.0361. [DOI] [PubMed] [Google Scholar]
- Behari J, Youngman P. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect Immun. 1998;66:3635–3642. doi: 10.1128/iai.66.8.3635-3642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner MR, James KE, Callahan MC, Wiedmann M, Boor KJ. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl Environ Microbiol. 2006;72:5384–5395. doi: 10.1128/AEM.00764-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu A, Guzzo J, Piveteau P. Sensitivity to acetic acid, ability to colonize abiotic surfaces and virulence potential of Listeria monocytogenes EGD-e after incubation on parsley leaves. J Appl Microbiol. 2010;108:560–570. doi: 10.1111/j.1365-2672.2009.04463.x. [DOI] [PubMed] [Google Scholar]
- Clifford T. Ability of Listeria monocytogenes to survive the stresses encountered during gastrointestinal transit. Microbiologist. 2006;7:45. [Google Scholar]
- Dakora FD, Phillips DA. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil. 2002;245:35–47. doi: 10.1023/A:1020809400075. [DOI] [Google Scholar]
- Saklani-Jusforgues H, Fontan E, Goossens PL. Effect of acid-adaptation on Listeria monocytogenes survival and translocation in a murine intragastric infection model. FEMS Microbiol Lett. 2000;193:155–159. doi: 10.1111/j.1574-6968.2000.tb09418.x. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Ferreira A, O’Byrne CP, Boor KJ. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl Environ Microbiol. 2001;67:4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain T, Hossain H, Chatterjee SS, Machata S, Volk U, Wagner S, Brors B, Haas S, Kuenne CT, Billion A. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e σB regulon. BMC Microbiol. 2008;8:20. doi: 10.1186/1471-2180-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner MR, Njaa BL, Wiedmann M, Boor KJ. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun. 2006;74:876–886. doi: 10.1128/IAI.74.2.876-886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann P, Wiedmann M, Boor KJ. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl Environ Microbiol. 2007;73:2919–2930. doi: 10.1128/AEM.02664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy RA, Chan YC, Bowen BM, Boor KJ, Wiedmann M. Growth temperature-dependent contributions of response regulators, σB, PrfA, and motility factors to Listeria monocytogenes invasion of Caco-2 cells. Foodborne Pathog Dis. 2010;7:1337–1349. doi: 10.1089/fpd.2010.0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen I, Vogensen FK, Jespersen L. Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress. Foodborne Pathog Dis. 2009;6:669–680. doi: 10.1089/fpd.2008.0243. [DOI] [PubMed] [Google Scholar]
- Weber A, Prell A, Potel J, Schäfer R. Vorkommen von Listeria monocytogenes bei Schlangen, Schildkröten, Echsen und Amphibien in der Heimtierhaltung. Berl Munch Tierarztl Wochenschr. 1993;106:293–295. [PubMed] [Google Scholar]
- Botzler RG, Wetzler TF, Cowan AB. Listeria in aquatic animals. J Wildl Dis. 1973;9:163–170. doi: 10.7589/0090-3558-9.2.163. [DOI] [PubMed] [Google Scholar]
- Kuzina LV, Peloquin JJ, Vacek DC, Miller TA. Isolation and identification of bacteria associated with adult laboratory Mexican fruit flies, Anastrepha ludens (Diptera: Tephritidea) Curr Microbiol. 2001;42:290–294. doi: 10.1007/s002840110219. [DOI] [PubMed] [Google Scholar]
- Jallewar PK, Kalorey DR, Kurkure NV, Pande VV, Barbuddhe SB. Genotypic characterization of Listeria spp. isolated from fresh water fish. Int J Food Microbiol. 2007;114:120–123. doi: 10.1016/j.ijfoodmicro.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Andrade DV, De Toledo LF, Abe AS, Wang T. Ventilatory compensation of the alkaline tide during digestion in the snake Boa constrictor. J Exp Biol. 2004;207:1379–1385. doi: 10.1242/jeb.00896. [DOI] [PubMed] [Google Scholar]
- Wang T, Busk M, Overgaard J. The respiratory consequences of feeding in amphibians and reptiles. Comp Biochem Physiol A Mol Integr Physiol. 2001;128:535–549. [PubMed] [Google Scholar]
- Smith LS. (Ed): Digestion in teleost fishes. Rome: Food and Agriculture Organization of the United Nations; 1980. [Google Scholar]
- Charrier M, Brune A. The gut microenvironment of helicid snails (Gastropoda: Pulmonata): in-situ profiles of pH, oxygen, and hydrogen determined by microsensors. Can J Zool. 2003;81:928–935. doi: 10.1139/z03-071. [DOI] [Google Scholar]
- Ahearn GA, Clay LP. Kinetic analysis of electrogenic 2 Na + −1 H + antiport in crustacean hepatopancreas. Am J Physiol. 1989;257:R484–R493. doi: 10.1152/ajpregu.1989.257.3.R484. [DOI] [PubMed] [Google Scholar]
- Terra WR, Costa RH, Ferreira C. Plasma membranes from insect midgut cells. An Acad Bras Cienc. 2006;78:255–269. doi: 10.1590/S0001-37652006000200007. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier B, Starke M, Higel F, Scherer S, Fuchs TM. Yersinia enterocolitica infection and tcaA-dependent killing of Caenorhabditis elegans. Appl Environ Microbiol. 2010;76:6277–6285. doi: 10.1128/AEM.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz C, Streibig JC. Bioassay analysis using R. J Stat Softw. 2005;12 [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptomic dynamic of L. monocytogenes upon acid shock at two different temperatures.
List of genes from L. monocytogenes whose knockout led in all cases to attenuation in mouse infection experiments.
List of the genes used for the qRT-PCR validation of the L. monocytogenes microarray experiments.
List of genes significantly (p ≤ 0.05) down regulated after acid shock at both 25°C and 37°C.