Abstract
53BP1 contributes to activation of the G2/M checkpoint downstream of ATM and MDC1 in response to ionizing radiation and promotes nonhomologous end-joining (NHEJ) in mammalian cells. In order to determine whether the cellular activities of 53BP1 are conserved in the model organism C. elegans, we analyzed the function of its homolog, HSR-9 in response to DNA damage. Deletion or Mos1-insertion in hsr-9 did not affect the sensitivity of worms to double strand DNA breaks (DSBs), as reflected in embryonic survival and larval development. Nevertheless, the hsr-9 mutations, as well as a lig-4 deletion, reversed the hypersensitivity of rad-54-deficient worms to DSBs. In addition, oocyte chromosomal aberrations, which were increased by rad-54 knockdown in response to DSBs, were also reduced by the hsr-9 mutations. The hsr-9 mutations did not prevent the cell cycle arrest induced by DSBs in mitotically proliferating germ cells. However, they attenuated apoptosis induced by DSBs, but not when CEP-1 (a p53 ortholog) was absent, suggesting that HSR-9 functions in the same pathway as CEP-1. We concluded that the 53BP1 homolog in C. elegans is not directly involved in cell cycle arrest in response to DSBs, but that it promotes apoptosis and also a form of NHEJ that occurs only when rad-54 is deficient.
Introduction
53BP1 (p53 binding protein 1) was discovered as a p53-binding protein and its primary role was initially thought to be stabilizing p53 in response to ionizing radiation [1], [2]. However, more important roles in cell cycle checkpoints and DNA repair were identified later. When double-strand DNA breaks (DSBs) are formed, ATM is activated with the help of the Mre11/Rad50/NBS1 (MRN) complex and phosphorylates histone H2AX, MDC1, and 53BP1 [3], [4]. MDC1 recruits RNF8 (RING-finger protein 8), which ubiquitinates histones H2A and H2AX, and then 53BP1 [5]–[9]. 53BP1 activates CHK2 and SMC1, and leads to activation of the G2/M and S phase checkpoint [2], [10]–[13]. 53BP1 and MDC1 were found to mediate checkpoint activation only at low doses of ionizing radiation, but were later observed to participate in maintaining G2/M arrest even at high doses of ionizing radiation [14]. Although 53BP1 is a downstream target of ATM, it can also stimulate the kinase activity of ATM [15].
In addition to its role in cell cycle arrest, 53BP1 promotes nonhomologous end-joining (NHEJ) and suppresses homologous recombination (HR) [7]. On the other hand, the upstream checkpoint protein, MDC1, mediates homologous recombination [7]. Brca1-deficiency (Brca1 Δ11/Δ11) induces the senescence and death of MEF cells, but this is rescued by deletion of 53BP1 [16]. It has been suggested that this rescue occurs because 53BP1 inhibits DNA-end resection by CtIP in Brca1-deficient cells, thus suppressing homologous recombination and permitting NHEJ. Similarly, the 53BP1 ortholog in S. cerevisiae, Rad9, inhibits DNA resection at DSBs and uncapped telomeres [17]. The unprotected telomeres present in TRF2 knockout cells have greater mobility in the presence of 53BP1, and this leads to telomere fusions via NHEJ [18]. Similarly, 53BP1 is required for class-switch recombination and facilitates long-range DNA end-joining in V(D)J recombination in lymphocytes [13], [19], [20].
53BP1 has a number of phosphorylation sites [21] and contains dimerization, tandem Tudor, and BRCT domains. The dimerization and BRCT domains contribute to oligomerization of 53BP1, and the BRCT domain mediates interaction with other DNA damage response proteins such as the MRN complex [15]. The oligomerized tandem Tudor domain is sufficient for recognition of DSBs and binds to di-methylated histones at H3K79 and H4K20 [22], [23]. DOT1L, an H3K79 methyltransferase, is needed to recruit 53BP1 to DSBs in human cell lines, and Dot1 plays the same role in nuclear localization of Rad9 (a 53BP1 ortholog) in S. cerevisiae [24]. The histone H4K20 methyltransferases, SUV4-20, SET8 (PR-SET7/8), and MMSET (WHSC1) are reported to affect 53BP1 accumulation at DSBs in human cell lines [25]–[27]. Likewise, histone methylation at H4Lys20 mediated by Set9 controls the recruitment of Crb2 (a 53BP1 ortholog) to DSBs in S. pombe [28], [29].
DNA damage checkpoint proteins, such as ATM, ATR, CHK1, CHK2, MRE11/RAD50, the 9-1-1 complex, and RPA, are functionally conserved in C. elegans [30]–[33]. In order to assess which 53BP1 activities related to DSBs are conserved in the C. elegans homolog HSR-9, we investigated the roles of HSR-9 in cell cycle control, DNA repair, and apoptosis after DSB formation.
Results
The C. elegans 53BP1 homolog, HSR-9 and its nuclear localization
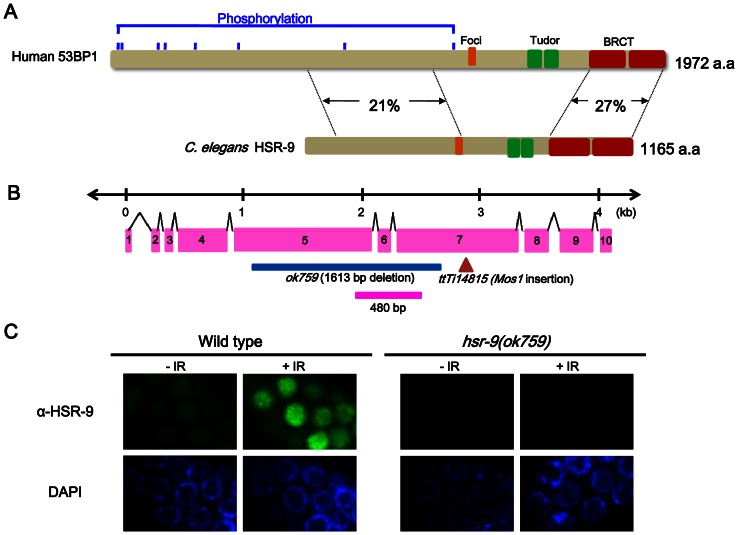
By carrying out a BLAST search of the C. elegans “orfeome”, we identified a protein called HSR-9 that had 27% amino acid identity with 53BP1 in the C-terminal BRCT region (Hs 53BP1: amino acids 1678∼1965, Ce HSR-9: 871∼1154), and 21% identity in the N-terminal domain (53BP1: 704∼1142, HSR-9: 111∼562) (Figure 1A). Moreover, the regions needed for nuclear focus formation (53BP1, 1251∼1271; HSR-9, 564∼582) and the tandem Tudor folds (53BP1, 1486∼1602; HSR-9, 734∼854), which bind to methylated histones, were conserved [22], [34]. To assess whether C. elegans HSR-9 is a functional homolog of 53BP1, we examined its intracellular location using antibodies against a polypeptide fragment of HSR-9 (496∼655 a.a.) (Figure 1B). Immuno-stained HSR-9 was observed in the nuclei of proliferating germ cells under normal conditions and the localization was much more prominent after γ-irradiation (Figure 1C). There was no signal in the hsr-9(ok759) germ cells either before or after γ-irradiation (Figure 1C), confirming that the antibody was specific for HSR-9. The nuclear level of HSR-9 protein also increased after UV treatment and DNA replication inhibition using hydroxyurea (Figure S1 in File S1), which suggests a role in response to various types of DNA damage.
Figure 1. The 53BP1 homolog in C. elegans, HSR-9 protein, accumulates in the nuclei of C. elegans germ cells in response to γ-rays.
(A) HSR-9 is the most similar of the predicted open reading frames in C. elegans to human 53BP1 in its C-terminal BRCT domains (27% amino acid identity) and N-terminus (21% amino acid identity). The regions controlling nuclear focus formation (Foci) and the Tudor domains are also conserved [22], [34]. (B) The deletion in hsr-9(ok759) is shown in blue in this representation of the gene, and the Mos1 insertion is in brown. The polypeptide fragment used for antibody production is indicated by the pink bar. (C) HSR-9 was immuno-localized to the nuclei of wild-type germ cells in the mitotically proliferating region of gonads 3 h after γ-ray (75 Gy) treatment. Scale bar, 10 µm.
Mutations of hsr-9 do not affect sensitivity to DSBs in the germ line and noncycling somatic cells
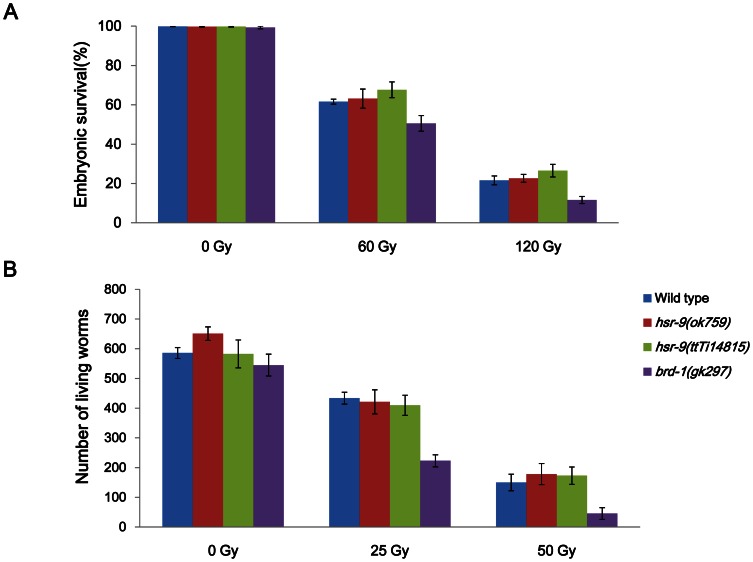
In order to test whether HSR-9 has a role in the DNA damage response as expected, we obtained an hsr-9(ok759) mutant with a deletion from Exon 5 to Exon 7 (Figure 1B). We also used another knockout mutant hsr-9(ttTi14815), which has a Mos1 insertion in the open reading frame (Figure 1B). The genetic backgrounds of the mutants were cleaned up by outcrossing with the wild-type N2 strain. We then irradiated L4 stage worms with γ-rays and scored the hatching of progeny embryos, to detect any effects of the hsr-9 mutations on sensitivity to DSBs (Figure 2A). The mutation gk297 of brd-1 which encodes the C. elegans BARD1 homolog, decreased embryonic hatching after γ-irradiation, as expected from its role in HR [35]. In contrast, hatching of the hsr-9(ok759) deletion mutant was not significantly affected at 60 Gy (Student’s t test, p value = 0.48) or at 120 Gy (p = 0.99) (Figure 2A). Nor did the Mos1 insertion mutant, hsr-9(ttTi14815), differ from the wild-type N2 strain in its sensitivity to γ-rays (at 60 Gy, p = 0.13; at 120 Gy, p = 0.36). Since hsr-9 mutations did not significantly affect the survival of L4-stage germ cells at either the mitotically proliferating or meiotic pachytene stage, worms were instead γ-irradiated at the L1 stage (Figure 2B). In L1 survival assays, the radiation-sensitivity of only mitotically proliferating germ cells can be measured by scoring adult fertility as the number of surviving larvae in the next generation [36]. The number of living larvae was significantly reduced compared with the wild type in the positive control, brd-1(gk297). However, neither of the hsr-9 mutations, ok759 and ttTi14815, significantly decreased the number of worms produced after 25 and 50 Gy irradiation (N2 vs. ok759, p = 0.05 at 0 Gy, p>0.5 at 25 and 50 Gy; N2 vs. ttTi14815, p>0.5 at 0, 25, and 50 Gy). Therefore, it can be concluded that the hsr-9 mutations affect neither the survival of proliferating nor of meiotic germ cells after DSB formation.
Figure 2. HSR-9 does not affect the survival of L4 stage germ cells or the proliferation of L1 stage germ cells.
(A) Embryonic hatching was scored after exposing L4 stage worms of wild-type N2 and hsr-9 mutants to γ-rays and collecting embryos between 24 and 48 h after irradiation. Hatching rates of wild-type N2, hsr-9(ok759), and hsr-9(ttTi14815) mutant embryos were not significantly different at 60 Gy (Student’s t test: N2 vs. ok759, p = 0.48; N2 vs. ttTi14815, p = 0.13) or at 120 Gy (N2 vs. ok759, p = 0.99; N2 vs. ttTi14815, p = 0.36). (B) Five P0 worms of each strain were exposed to γ-rays (25 and 50 Gy) at the L1 stage and grown for 2 days. F1 embryos were collected over the following 24 h, and live larvae (L1-L3 stage) that hatched from the embryos were counted 24 later. p values between N2 and hsr-9(ok759 or ttTi14815) at all radiation doses were larger than 0.5, except for p = 0.05 at 0 Gy between N2 and hsr-9(ok759).
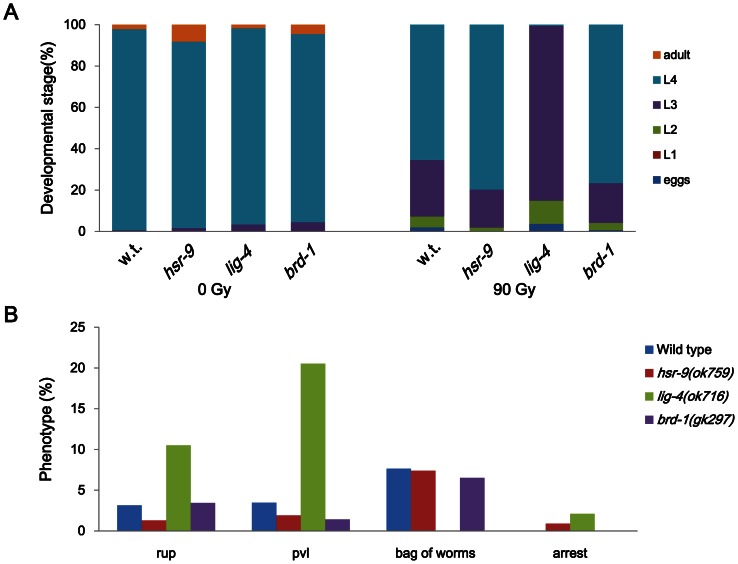
Since NHEJ is important for DSB repair in non-cycling somatic cells of C. elegans and 53BP1 promotes NHEJ in mammalian cells [7], [37], we presumed that HSR-9, the 53BP1 homolog in C. elegans, would influence larval growth after DSB formation. When late embryos were treated with γ-rays (90 Gy) and cultivated to adulthood, larvae of the lig-4(ok716) mutant, which has a deletion in the gene for ligase 4, developed more slowly than wild-type larvae, as shown in Figure 3A. This result is very similar to the previous observations on C. elegans mutants deficient in NHEJ proteins such as CKU-80 (Ku80 ortholog), CKU-70 (Ku70 ortholog), and LIG-4 [38]. In contrast, brd-1(gk297) worms, defective in HR, did not show a significant retardation in growth rate, and neither did the hsr-9 mutants. lig-4(ok716) worms developed a large number of morphological abnormalities as larvae and adults, such as ruptured body and protruding vulva, whereas wild-type N2, brd-1(gk297) and hsr-9(ok759) did not (Figure 3B). These results imply that contrary to our expectation, HSR-9 does not play a significant role in NHEJ during postembryonic development.
Figure 3. hsr-9 deletion does not affect post-embryonic development after γ-ray treatment.
Late embryos of wild-type N2, hsr-9(ok759), lig-4(ok716), and brd-1(gk297) were treated with γ-rays (90 Gy). (A) Larvae hatched from treated and untreated embryos were examined every day to estimate their growth rates, and the percentages of worms at different larval stages at 48 h after the irradiation were calculated. (B) Developmental abnormalities of treated wild-type (w.t.) and mutant worms were scored on the 3rd day of adulthood (120 h after the irradiation). Phenotypes scored were ruptured body (rup), protruding vulva (pvl), bag of worms (hatched larvae inside the mothers) and growth arrest (arrest).
hsr-9 and lig-4 mutations rescue the hypersensitivity to DSBs of rad-54-deficient germ cells
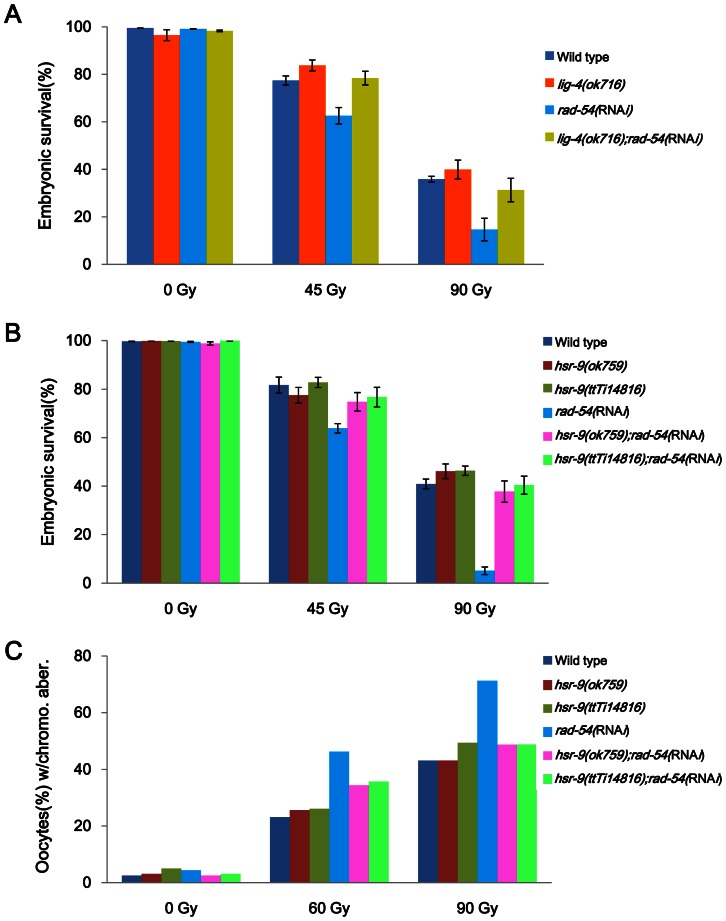
Homologous recombination (HR) is the major DSB repair pathway in the germ cells and cycling somatic cells of C. elegans, whereas NHEJ is the major pathway in noncycling somatic cells [38]. Nevertheless, NHEJ influences the sensitivity of germ cells to DNA damage in particular genetic backgrounds. For example, lig-4 mutation suppresses the hypersensitivity of fcd-2 (a FANCD2 ortholog) mutant germ cells to interstrand DNA crosslinks (ICLs) and also suppresses the association of oocyte chromosomes seen in the double-deficiency strain of fcd-2 and a meiotic recombination gene [39]. Therefore, we examined the possibility that lig-4 mutation might affect the hypersensitivity to DSBs of worms defective in homologous recombination. Knockdown of the C. elegans ortholog of RAD54 which participates in strand invasion and branch migration during HR [38], [40]–[43], decreased embryonic survival after γ-irradiation (Figure 4), as reported previously [38]. The deletion mutation ok716 of LIG-4, which is involved in the NHEJ pathway, expectedly rescued the embryonic lethality in rad-54(RNAi) worms after γ-irradiation (Figure 4A).
Figure 4. hsr-9 mutations rescue the hypersensitivity of rad-54-deficient worms to γ-rays as lig-4 mutations do.
Wild-type N2, hsr-9(ok759), hsr-9(ttTi14815), and lig-4(ok716) worms were fed E. coli cells containing double-stranded RNA of rad-54 from the L1 stage. (A) L4 stage worms deficient in either lig-4 or rad-54, or both were treated with γ-rays (45 Gy and 90 Gy), and embryos were collected between 24 and 48 h after irradiation. The p values for hatching rates were: rad-54 vs. lig-4;rad-54, p = 0.01 at 45 Gy and p = 0.07 at 90 Gy; lig-4 vs. lig-4;rad-54, p = 0.24 at 45Gy and p = 0.26 at 90 Gy. (B) L4 stage worms deficient in either hsr-9 or rad-54, or both were treated with γ-rays as in (A). The p values for hatching rates at 45 Gy were: hsr-9(ok759 or ttTi14815) vs. rad-54, p<0.01; rad-54 vs. hsr-9;rad-54, p<0.03; hsr-9 vs. hsr-9;rad-54, p>0.20. The hatching rates of hsr-9 and hsr-9;rad-54 did not differ significantly at 90 Gy either (p>0.14). (C) L4 stage worms were irradiated with 60 and 90 Gy of γ-rays and fixed after 24 h. After staining worms with DAPI, the number of chromosomes in each oocyte was counted under a fluorescence microscopy (80 oocytes per strain). The percentage of chromosomal aberrations was calculated from the number of oocytes containing abnormal number of chromosomes (as opposed to 6 bivalent chromosomes) divided by the total number of oocytes observed.
We then tested whether HSR-9, whose homolog promotes NHEJ in mammalian cells [7], [16], [18], also influences DSB repair in worms deficient in RAD-54. Indeed, both hsr-9 mutations, ok759 and ttTi14815, rescued the embryonic lethality of rad-54(RNAi) at 45 and 90 Gy (Figure 4B), as did the lig-4(ok716) mutation. Rescue by the hsr-9 mutations was also observed with respect to oocyte chromosomal aberrations (defined by abnormal numbers of chromosomes) after γ-ray treatment (60 and 90 Gy) (Figure 4C): the percentage of abnormal oocytes was about 2 fold greater (2.0 fold at 60 Gy, 1.7 fold at 90 Gy) in rad-54(RNAi) than in the wild type, whereas the increase by rad-54 knockdown was significantly less in the hsr-9 mutant backgrounds (<1.4 fold at 60 Gy, <1.1 fold at 90 Gy).
hsr-9 mutations do not influence normal cell cycle arrest in response to DSBs
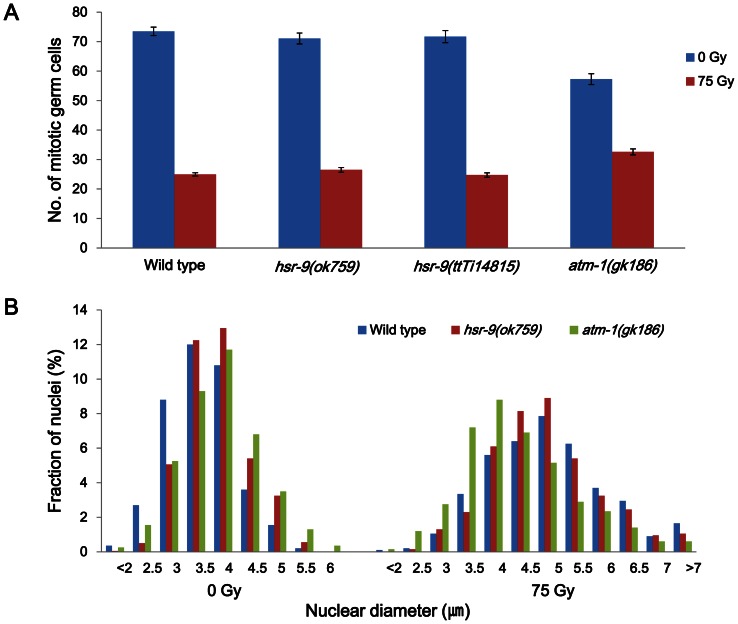
To detect a role of HSR-9 in the DNA damage checkpoint, gonads were stained with DAPI at 12 h after γ-ray treatment (75 Gy) and the number of nuclei in the mitotic region of each gonad arm was counted. Numbers of germ cells were decreased to the same extent in the two hsr-9 mutants as in wild-type N2 (N2, Δn = 49±2(SEM); hsr-9(ok759), Δn = 45±2; hsr-9(ttTi14816), Δn = 47±2; p>0.5 between N2 and hsr-9 mutants), whereas the reduction was less in atm-1(ok186) (N2, Δn = 49±2(SEM); atm-1(gk186), Δn = 25±2; p<0.03) (Figure 5A). This suggests that unlike ATM-1, HSR-9 is not significantly involved in cell cycle arrest. We also quantified the extent of cell cycle arrest by measuring the diameters of germ cell nuclei in the mitotically proliferating regions of gonads. Germ cells normally enlarge after γ-irradiation due to G2 arrest and subsequent cell growth [44]–[46]. Before γ-ray treatment, the nuclear diameters of the hsr-9 and atm-1 mutants were slightly larger than those of wild type N2 (N2, average nuclear diameter (d) = 3.60±0.02(SEM)µm; hsr-9(ok759), 3.87±0.02 µm; atm-1(gk186), 3.92±0.02 µm). As expected, after γ-ray treatment, their average values (d) increased in all three strains (N2, 5.01±0.04(SEM) µm; hsr-9(ok759), 4.94±0.04 µm; atm-1(gk186), 4.42±0.04 µm). However, the distribution of nuclear sizes in the hsr-9(ok759) worms was much closer to that in wild-type worms than to that in atm-1(ok186) worms, and consequently the same was true for averages of nuclear diameters (Figure 5B). This result agrees with that in Figure 5A, supporting the view that HSR-9 does not play a significant role in cell cycle arrest after DSB formation, at least not one as important as that of ATM-1.
Figure 5. Cell cycle arrest in response to γ-rays is normal in the hsr-9(ok7599) mutants.
Wild-type N2, hsr-9(ok759), hsr-9(ttTi14815), and atm-1(gk186) worms were treated with 75 Gy of γ-irradiation as L4 stage larvae. Twelve hours later, gonads were isolated and stained with DAPI. (A) Germ cells in the mitotically proliferating regions of the gonads (within 75 µm of the distal tip cell) were counted under a fluorescence microscope (30 gonad arms per data point). (B) The diameters of 40 nuclei in the mitotically proliferating region of each gonad arm were measured (number of nuclei = 800 for each strain before and after irradiation) for wild-type N2, hsr-9(ok759), and atm-1(gk186) worms. The percentages of the total nuclei of different diameters are plotted. Very small (<2 µm) and very large (>7 µm) nuclei were grouped together.
hsr-9 mutations attenuate apoptosis induced by DSBs
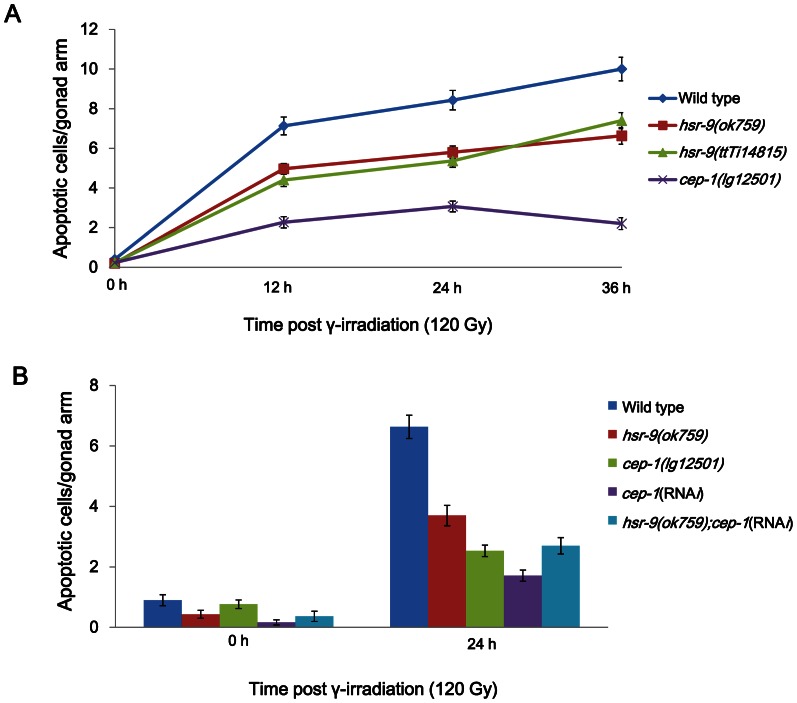
Although we did not detect a role of HSR-9 in cell cycle arrest in response to DSBs, we examined whether the deletion mutation affects apoptosis induced by DSBs. In the C. elegans germline, more than half of the germ cells spontaneously undergo apoptosis at the pachytene stage, and this can be detected by either observing cell corpses under Nomarski optics or staining for condensed chromosomes [47]. L4 stage hermaphrodites were treated with γ-rays (120 Gy), and dying germ cells at the pachytene stage were scored at 12 h intervals after staining with SYTO-12. hsr-9(ok759) and hsr-9(ttTi14815) mutants contained fewer apoptotic cells than wild-type worms (Figure 6A). However, the reductions of apoptotic cell numbers in hsr-9(ok759) and hsr-9(ttTi14815) were less than in cep-1(lg12501), which has a deletion in the gene coding for a P53 ortholog [48], [49]. In order to determine whether HSR-9 influences apoptosis via the same pathway as CEP-1, we measured apoptotic cells of hsr-9(ok759);cep-1(RNAi) after γ-ray treatment (120 Gy, 24 h later). The apoptotic cell number in this strain was similar to that in cep-1(lg12501) and slightly higher than in cep-1(RNAi) (hsr-9(ok759);cep-1(RNAi) vs. cep-1(lg12501), p = 0.62; hsr-9(ok759);cep-1(RNAi) vs. cep-1(RNAi) p<0.01) (Figure 6B). The fact that the reductions in apoptotic cell numbers in the two single deficiency strains did not add up to the reduction in the double deficiency strain, suggests that HSR-9 functions in the same pathway as CEP-1. To confirm this, transcripts of egl-1, a cell-death activator downstream of cep-1 [50], were measured by gene-specific real-time PCR after reverse transcribing total RNA (Figure S2 in File S1). The transcripts were significantly lower in hsr-9(ok759) worms than in wild-type worms (p = 0.01), but not as low as in cep-1(lg12501) worms. The reduced level of egl-1 transcripts in hsr-9(ok759) is likely to have contributed to the reduction in apoptosis.
Figure 6. hsr-9 mutations attenuate apoptosis induced by γ-rays.
(A) Wild type, hsr-9(ok759), hsr-9(ttTi14815), and cep-1(lg12501) L4 stage worms were irradiated with γ-rays (120 Gy), and stained with SYTO-12 dye at 0, 12, 24, and 36 h after γ-ray treatment. Intensely stained pachytene stage apoptotic cells were scored in each gonad arm. (B) Wild type and hsr-9(ok759) worms were fed E. coli cells containing double-stranded RNA of cep-1 from the L1 stage. Wild type, hsr-9(ok759), cep-1(lg12501), cep-1(RNAi), and hsr-9(ok759);cep-1(RNAi) L4 stage worms were treated with γ-rays (120 Gy), and apoptotic germ cells were counted as in (A).
Discussion
53BP1-deficient mice are growth-retarded, immuno-deficient, radiation-sensitive, and cancer prone [51]. In contrast, we found that C. elegans mutants of the 53BP1 homolog, HSR-9, were not hypersensitive to DSBs as measured by embryonic survival and larval growth after γ-ray treatment (Figures 1 and 2). However, the hsr-9 mutations unexpectedly reversed the hypersensitivity to DSBs in rad-54-deficient worms, suggesting that it has a role in DSB repair (Figure 4). The observation is very similar to the finding that the phenotypes of BRCA1-deficient (Brca1 Δ11/Δ11) mice, such as embryonic lethality, senescence, and cell death, are rescued by 53BP1 knockout [16]. The suppression of the BRCA1-deficiency phenotypes by 53BP1 mutation has been explained by the mechanism that 53BP1 inhibits end-resection at DSBs, thereby shifting the DSB repair towards NHEJ rather than HR [37]. Thus, although HR is compromised in BRCA1-deficient cells and leads to error-prone repair by NHEJ, the absence of 53BP1 allows DSBs to be repaired by error-free HR. To test whether this genetic relationship was conserved in C. elegans, we generated a double mutant of brc-1 (BRCA1 homolog) and hsr-9. However, the brc-1(tm1145);hsr-9(ok759) mutant was as hypersensitive to DSBs as brc-1(tm1145), in contrast to the mammalian situation (Figure S3 in File S1, brc-1;hsr-9 vs. brc-1, p>0.3). This difference is probably related to the fact that the CTIP homolog in C. elegans, COM-1, is not required for RAD-51 loading at exogenously-induced DSBs, but is essential at meiotic DSBs generated by SPO-11 [52]. Therefore, C. elegans CTIP does not appear contribute to the processing of exogenously-induced DSBs and so is unlikely to interact with BRC-1 during end-resection. Instead, the C. elegans homolog of RAD54, which is involved in strand invasion and branch migration during homologous recombination, interacts with the 53BP1 homolog, HSR-9 (Figures 4B and 4C). A deletion mutation of lig-4, which is involved in NHEJ pathway, also rescued the embryonic lethality due to rad-54 knockdown after γ-ray treatment (Figure 4A). The fact that both HSR-9 and LIG-4 are involved in similar genetic interactions with RAD-54 suggests that HSR-9 probably promotes NHEJ in the context of the HR defect associated with RAD-54 deficiency. Although it is clear that hsr-9 mutation rescues rad-54 knockdown phenotypes, the mechanism responsible is not evident. One possibility is that RAD-54 interacts with the MRE-11 or EXO-1 nucleases during the end-resection of DSBs, as BRCA1 does with CTIP in mammalian cells. Another possibility is that chromatin modulation by RAD-54, a SNF2 family protein, is normally counterbalanced by binding of HSR-9 to di-methylated histones near DSBs [22], [23]: in the absence of RAD-54 at DSBs, abnormal chromatin structures may be formed as a result of HSR-9 activity, and HR cannot proceed.
The pan-nuclear level of HSR-9 was significantly increased in germ cells after DSB formation (Figure 1C), but there were no clear foci of HSR-9 in contrast to the case of mammalian 53BP1. Similarly, C. elegans ATM-1 did not form nuclear foci unlike its mammalian homolog [53], implying that the homologs of mammalian proteins that form nuclear foci in response to DNA damage do not always form foci in worms. 53BP1 has several S/TQ sites that can be phosphorylated by ionizing or UV radiation, which is consistent with the idea that ATM and ATR catalyze its phosphorylation [21]. Indeed, we observed that nuclear HSR-9 increased after inhibiting DNA replication with hydroxyurea and after exposure to UV–radiation (Figure S1 in File S1). Despite the nuclear accumulation of HSR-9, hsr-9 mutations did not affect the arrest of germ cell proliferation induced by DSBs, whereas the atm-1 mutation did partially prevent the arrest (Figure 5). Thus, HSR-9 does not seem to participate in checkpoint activation, unlike its homolog in mammalian cells. One reason for this difference could be the fact that the C. elegans homolog of CHK2, a downstream target of 53BP1, does not play a significant role in the response to DSBs [54], [55]. Another reason could be the high dose of γ-rays that we used, because a need for 53BP1 in checkpoint activation was observed only at low doses of ionizing radiation [2], [12].
Previously, hsr-9 knockdown was found to result in reduced progeny survival, defective cell cycle arrest, and attenuated apoptosis in response to DSBs in a high-throughput protein interaction analysis of DNA damage responses in C. elegans [56]. In contrast to these effects, we did not observe any effects of hsr-9 mutations on progeny survival or cell cycle arrest. Nevertheless, hsr-9 mutations did attenuate germline apoptosis after DSB formation (Figure 6), like hsr-9 knockdown in the previous report [56]. Double deficiency for hsr-9 and cep-1 (the p53 ortholog in C. elegans) did not further decrease apoptotic cell numbers in the germline compared to single deficiency for cep-1, suggesting that HSR-9 promotes apoptosis induced by DSBs via the same pathway as CEP-1. The fact that egl-1 mRNA expression, which is induced by CEP-1, is decreased by hsr-9 mutation further confirms that the two proteins function in the same pathway (Figure S2 in File S1). HSR-9 probably mediates the activation of CEP-1 in response to DSBs, as 53BP1 contributes to the accumulation of p53 in mammalian cells [2]. It is intriguing that RAD-54 and MRE-11, acting in the homologous recombination pathway, promote apoptosis in response to UV-C radiation in C. elegans [41], as HSR-9 does in response to DSBs.
We conclude that HSR-9 mediates apoptosis induced by DSBs in the same pathway as CEP-1. However, there is no clear evidence that HSR-9 functions in cell cycle arrest. Similarly, normal cell cycle arrest was observed in response to DSBs after knocking down CEP-1 [49]. In spite of the fact that HSR-9 appears not to function in DSB repair in the wild type genetic background, HSR-9 mutations reverse the hypersensitivity of RAD-54-deficient cells to DSBs like LIG-4 deficiency. This suggests that HSR-9 can shift the choice of DSB repair pathway from HR to NHEJ, just as 53BP1 promotes NHEJ in BRCA1-deficient cells. Therefore, the roles of 53BP1 in mammalian cells, mediating apoptosis and shunting DSB repair to NHEJ, are to a degree conserved in the 53BP1 ortholog of C. elegans [16], [37].
Materials and Methods
C. elegans strains
The standard wild-type Bristol N2 strain, as well as the hsr-9(ok759), atm-1(gk186), brd-1(gk297), lig-4(ok716), cep-1(lg12501), and brc-1(tm1145) mutants were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN, USA). We obtained a validated hsr-9(ttTi14815) strain (with a Mos1 transposon inserted in the 7th exon of hsr-9) from Drs. Laurent Ségalat (Université Claude Bernard Lyon 1, France) and Maïté Carre-Pierrat (Plateforme “Biologie de Caenorhabditis elegans”, France). To generate uniform genetic backgrounds, hsr-9(ok759), hsr-9(ttTi14815), brd-1(gk297), brc-1(tm1145), and lig-4(ok716) were outcrossed with wild type N2 worms 4, 3, 3, 6, and 2 times, respectively. The double mutant brc-1(tm1145);hsr-9(ok759) was generated by crossing brc-1(tm1145) with hsr-9(ok759). The atm-1(gk186) mutant had been previously outcrossed 6 times [53]. All strains were maintained by feeding them E. coli OP50-1 cells at 20°C. To confirm that the Mos1 transposon inserted in hsr-9(ttTi14815) was not spontaneously excised, the genomic DNA next to the inserted transposon was amplified from hsr-9(ttTi14815) (Figure S4 in File S1). When the genomic DNA fragment was analyzed by agarose gel electrophoresis, the Mos1 insertion was found to be present.
Bacteria-mediated RNAi
E. coli HT115(DE3) cells harboring a plasmid containing the cDNA of cep-1 and rad-54 were obtained from the C. elegans RNAi v1.1 feeding library (Open Biosystems) and Ahringer’s RNAi feeding library, respectively. The E. coli cells were cultured in liquid LB medium containing ampicillin (50 µg/me) and tetracycline (5 µg/me) for 12 h at 37°C. Aliquots of the cultures were spread on NGM plates containing 1 mM IPTG and carbenicillin (25 µg/me), and left at room temperature for 24 h. For rad-54 RNAi and cep-1 RNAi, C. elegans worms were grown on the plates from the L1 stage.
L4-stage gem cell survival and oocyte chromosomal aberration after γ-ray treatment
Twenty wild type, hsr-9(ok759), hsr-9(ttTi14815), brd-1(gk297), lig-4(ok716), rad-54(RNAi), hsr-9;rad-54(RNAi), and lig-4(ok716);rad-54(RNAi) worms at the L4 stage were exposed to 60 Gy and 120 Gy (or 45 Gy and 90 Gy) of γ-rays using a 137Cs source (IBL 437C, CIS Biointernational). Embryos were collected between 24 and 48 h after irradiation, and their hatching was scored 24 h later. To score oocytes containing abnormal numbers of chromosomes, worms were fixed 24 h after irradiation (60 and 90 Gy), stained with DAPI, and observed with a fluorescence microscope (DMRHC, Leica).
Survival of L1-stage germ cells after γ-ray treatment
The survival of L1-stage germ cells was measured according to the protocol of Craig et al. [36] with slight modification. Wild type, hsr-9(ok759), hsr-9(ttTi14815) and brd-1(gk297) worms at the L1 stage were treated with 25 Gy and 50 Gy of γ-rays. After 48 h, five worms of the P0 generation were plated onto each of 3 NGM plates containing E. coli OP50-1 lawn, and embryos were collected for 24 h. The number of living worms (F1 generation) was counted 24 h later.
Measurement of developmental abnormalities after γ-ray treatment
Twenty adult worms were allowed to lay eggs for 1 h. After further incubation for 4 h, late-stage embryos were irradiated with γ-rays (90 Gy). Hatched worms were examined with a stereomicroscope once per day to estimate their growth rates until the control (untreated wild-type) worms reached the L4 stage. Developmental abnormalities were scored when the controls reached the 3rd adult day. Phenotypes scored were ruptured body (rup), protruding vulva (pvl), bag of worms (hatched larvae inside the mothers) and growth arrest (arrest).
Assessment of cell cycle arrest after γ-ray treatment
To measure the effect of γ-rays on cell cycle arrest, wild type, hsr-9(ok759), hsr-9(ttTi14815), and atm-1(gk186) larvae were treated at the L4 stage with γ-rays (75 Gy). After 12 h, their gonads were dissected, stained with DAPI, and observed with a fluorescence microscope (DMRHC, Leica). Cell cycle stages were determined by two methods. First, we counted the number of nuclei within 75 µmof the distal tip cells of each gonad, that is, in the mitotically proliferating region. Second, we measured the diameters of about 40 nuclei in the mitotically proliferating region of each gonad.
SYTO-12 staining for apoptotic germ cells
To identify apoptotic cells induced by double-strand DNA breaks, wild type, hsr-9(ok759), hsr-9(ttTi14815), cep-1(lg12501), and hsr-9(ok759);cep-1(RNAi) L4 stage worms were irradiated with γ-rays (120 Gy). Worms were harvested at 0, 12, 24, and 36 h after γ-ray treatment, and incubated in 33 µM SYTO-12 for 2 h at 25°C. After 3 washes in 1× PTw, they were allowed to recover for 1 h on NGM plates with bacterial lawn, and then mounted on glass slides with agar pads. Apoptotic cells at the pachytene germ stage were observed with a fluorescence microscope.
Preparation of anti-HSR-9 antibody
Total RNA was isolated from mixed-stage C. elegans worms using an easy-BLUE Total RNA Extraction kit (Intron Biotechnology). A cDNA pool was synthesized from the total RNA using M-MLV reverse transcriptase (Intron Biotechnology) and oligo(dT) primer, and the full-length cDNA of HSR-9 was amplified by gene-specific PCR. PCR was also used to amplify a cDNA fragment (nucleotides 1486∼1965) of the open reading frame, which was cloned into pGEM-T vector (Promega). The forward and reverse primers were 5′-GGATCCCGTAGATCTACAAGGGCTAAG and 5′-GAATTCCGAAGTTCCTACAGACGATAC, respectively. The recombinant plasmid DNA was digested at the BamHI and EcoRI sites (underlined), and the cDNA product was recloned into pGEX4T-1 vector. The resulting plasmid pGEX4T-1/HSR-9 was transferred into E. coli BL21(DE3) cells, and protein expression was induced by incubation with 1 mM IPTG for 4 h at 37°C. The fusion protein was isolated and used to raise antibodies in SD rats.
Immunostaining of gonads
The heads of adult C. elegans worms were cut open, and the gonads and intestines pushed out. These were fixed in 3% paraformaldehyde and 0.1 M K2HPO4 (pH 7.2) for 1 h at room temperature. They were then washed 5 times with 1× PTw and placed in methanol at −20°C overnight for post-fixation. After 5 washes in 1× PTw, the samples were left at room temperature for 1 h in blocking solution (a mixture of equal volumes of goat serum (Gibco-BRL) and 1× PTw). The fixed specimens were reacted with HSR-9 antiserum diluted in blocking solution (1∶50) at 4°C overnight. After five washes in 1× PTw, specimens were incubated with FITC-conjugated goat anti-rat immunoglobulin (1∶1000 dilution, Molecular Probes) at room temperature for 1 h. They were washed 3 times, stained with DAPI for 15 min, and washed twice with 1× PTw. They were then placed on glass slides coated with 2% agarose, followed by addition of antifade reagent (Invitrogen), and observed with a fluorescence microscope (DMRHC, Leica).
Quantitative RT-PCR of egl-1 mRNA
Adult wild type, hsr-9(ok759), and cep-1(lg12501) worms were irradiated with 120 Gy of γ-rays and incubated for 6 h. Total RNA was isolated using an easy-BLUE Total RNA Extraction kit (Intron Biotechnology). Total RNA (2 µg) was reverse-transcribed using M-MLV reverse transcriptase (Intron Biotechnology) and oligo(dT) primer. Relative amounts of egl-1 cDNA were measured by real-time PCR using IQ SYBR Green Supermix (BIORAD) in a CFX96 Detection System (BIORAD). Primer pairs were designed as suggested by Hofmann et al [30], and γ-tubulin was used as a standard to calculate the relative abundance of egl-1 mRNA. The primer pairs were for egl-1, 5′-CAGGACTTCTCCTCGTGTGAAGATTC and 5′-GAAGTCATCGCACATTGCTGCTA, and for γ-tubulin, 5′-AAGATCTATTGTTCTACCAGGC and 5′-CTTGAA CTTCTTGTCCTTGAC.
Identification of Mos1 insertion by PCR amplification of genomic DNA
To detect the Mos1 insertion in the 7th exon of the hsr-9 gene, single wild type and hsr-9(ttTi14815) worms were lysed, and a genomic DNA fragment containing the insertion site was amplified. PCR was carried out in a 2720 Thermal Cycler (Applied Biosystems) using primers, 5′-TCTCCTGCTGCTAAGAATCG and 5′-ACGAGTAGCCTCCGATATTGT. Amplified DNA fragments were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and visualized with a UV transilluminator.
Supporting Information
Figure S1, HSR-9 protein accumulates in the nuclei of C. elegans germ cells in response to various types of DNA damage. (A) HSR-9 was immuno-localized to the nuclei of wild-type germ cells in the mitotically proliferating region of gonads in response to hydroxyurea (HU, 60 mM) treatment for 16 h. (B) Nuclear levels of HSR-9 at 3 h after UV (150 J/m2) irradiation. Scale bar, 10 µm. Figure S2, hsr-9 deletion decreases egl-1 mRNA expression after γ-ray treatment. Adult wild type, hsr-9(ok759), and cep-1(lg12501) worms were irradiated with 120 Gy of γ-rays, and total RNA was isolated 6 h later. cDNA pools were prepared by reverse transcription, and relative amounts of egl-1 cDNA were measured by real-time PCR. Figure S3, hsr-9 mutation cannot relieve the hypersensitivity of brc-1 mutants to γ-rays. Wild-type N2, hsr-9(ok759), brc-1(tm1145), and brc-1(tm1145);hsr-9(ok759) worms were irradiated with γ-rays (45 Gy and 90 Gy) at the L4 stage. Embryos were collected between 24 and 48 h after the irradiation, and their hatching was scored 24 h later. brc-1(tm1145) mutation but not hsr-9(ok759) resulted in hypersensitivity to DSBs. The double mutant was very similar to the single brc-1 mutant in its sensitivity to DSBs (at both 45 Gy and 90 Gy, brc-1 vs. brc-1;hsr-9, p>0.3). Figure S4, The presence of Mos1 insertion in the hsr-9(ttTi14815) mutant is confirmed by PCR amplification of a genomic DNA fragment. Single worms were picked and lysed, followed by PCR amplification of the genomic DNA fragment flanking the insertion site. A 0.75 kb DNA fragment was produced from wild-type worms, and a much longer DNA fragment (2.0 kb) from the hsr-9(ttTi14815) mutant.
(PDF)
Acknowledgments
C. elegans N2, hsr-9(ok759), atm-1(gk186), brd-1(gk297), lig-4(ok716), cep-1(lg12501), and brc-1(tm1145) were provided by the C. elegans Genetics Center (St. Paul, MN), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The hsr-9(ttTi14815) in UMS3421 was provided by Drs. Laurent Ségalat (Université Claude Bernard Lyon 1, France) and Maïté Carre-Pierrat (Plateforme “Biologie de Caenorhabditis elegans”, France). We thank Mi So Park (Yonsei Univ.) for assistance with rad-54 experiments.
Funding Statement
This work was supported by the Nuclear Research & Development Program (grant code: 2010-0017583) and a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. 2012-029610) to HSK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S (1994) Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci U S A 91: 6098–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang B, Matsuoka S, Carpenter PB, Elledge SJ (2002) 53BP1, a mediator of the DNA damage checkpoint. Science 298: 1435–1438. [DOI] [PubMed] [Google Scholar]
- 3. Anderson L, Henderson C, Adachi Y (2001) Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol 21: 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rappold I, Iwabuchi K, Date T, Chen J (2001) Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol 153: 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huen MS, Grant R, Manke I, Minn K, Yu X, et al. (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, et al. (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900. [DOI] [PubMed] [Google Scholar]
- 7. Xie A, Hartlerode A, Stucki M, Odate S, Puget N, et al. (2007) Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell 28: 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Minter-Dykhouse K, Ward I, Huen MS, Chen J, Lou Z (2008) Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. J Cell Biol 181: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eliezer Y, Argaman L, Rhie A, Doherty AJ, Goldberg M (2009) The direct interaction between 53BP1 and MDC1 is required for the recruitment of 53BP1 to sites of damage. J Biol Chem 284: 426–435. [DOI] [PubMed] [Google Scholar]
- 10. DiTullio RA Jr, Mochan TA, Venere M, Bartkova J, Sehested M, et al. (2002) 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 4: 998–1002. [DOI] [PubMed] [Google Scholar]
- 11. Wu J, Prindle MJ, Dressler GR, Yu X (2009) PTIP regulates 53BP1 and SMC1 at the DNA damage sites. J Biol Chem 284: 18078–18084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, et al. (2002) DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol 4: 993–997. [DOI] [PubMed] [Google Scholar]
- 13. Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, et al. (2004) 53BP1 is required for class switch recombination. J Cell Biol 165: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shibata A, Barton O, Noon AT, Dahm K, Deckbar D, et al. (2010) Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G(2)/M checkpoint arrest. Mol Cell Biol 30: 3371–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee JH, Goodarzi AA, Jeggo PA, Paull TT (2010) 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J 29: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao L, Xu X, Bunting SF, Liu J, Wang RH, et al. (2009) A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell 35: 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, et al. (2008) Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 27: 1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, et al. (2004) 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol 5: 481–487. [DOI] [PubMed] [Google Scholar]
- 20. Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, et al. (2008) 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jowsey P, Morrice NA, Hastie CJ, McLauchlan H, Toth R, et al. (2007) Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair (Amst) 6: 1536–1544. [DOI] [PubMed] [Google Scholar]
- 22. Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, et al. (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411. [DOI] [PubMed] [Google Scholar]
- 23. Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, et al. (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wysocki R, Javaheri A, Allard S, Sha F, Cote J, et al. (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25: 8430–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, et al. (2008) Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem 283: 12085–12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, et al. (2010) Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40: 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pei H, Zhang L, Luo K, Qin Y, Chesi M, et al. (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, et al. (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614. [DOI] [PubMed] [Google Scholar]
- 29. Greeson NT, Sengupta R, Arida AR, Jenuwein T, Sanders SL (2008) Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J Biol Chem 283: 33168–33174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, et al. (2002) Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol 12: 1908–1918. [DOI] [PubMed] [Google Scholar]
- 31. Stergiou L, Hengartner MO (2004) Death and more: DNA damage response pathways in the nematode C. elegans. Cell Death Differ 11: 21–28. [DOI] [PubMed] [Google Scholar]
- 32. Garcia-Muse T, Boulton SJ (2005) Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J 24: 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayashi M, Chin GM, Villeneuve AM (2007) C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet 3: e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zgheib O, Pataky K, Brugger J, Halazonetis TD (2009) An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Mol Cell Biol 29: 1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, et al. (2004) BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol 14: 33–39. [DOI] [PubMed] [Google Scholar]
- 36. Craig AL, Moser SC, Bailly AP, Gartner A (2012) Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line. Methods Cell Biol 107: 321–352. [DOI] [PubMed] [Google Scholar]
- 37. Bunting SF, Callen E, Wong N, Chen HT, Polato F, et al. (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clejan I, Boerckel J, Ahmed S (2006) Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics 173: 1301–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, et al. (2010) Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35. [DOI] [PubMed] [Google Scholar]
- 40. Nimonkar AV, Dombrowski CC, Siino JS, Stasiak AZ, Stasiak A, et al. (2012) Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem 287: 28727–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stergiou L, Eberhard R, Doukoumetzidis K, Hengartner MO (2011) NER and HR pathways act sequentially to promote UV-C-induced germ cell apoptosis in Caenorhabditis elegans. Cell Death Differ 18: 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazina OM, Rossi MJ, Deakyne JS, Huang F, Mazin AV (2012) Polarity and bypass of DNA heterology during branch migration of Holliday junctions by human RAD54, BLM, and RECQ1 proteins. J Biol Chem 287: 11820–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ceballos SJ, Heyer WD (2011) Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta 1809: 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO (2000) A conserved checkpoint pathway mediates DNA damage—induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- 45. Crittenden SL, Leonhard KA, Byrd DT, Kimble J (2006) Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell 17: 3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moser SC, von Elsner S, Bussing I, Alpi A, Schnabel R, et al. (2009) Functional dissection of Caenorhabditis elegans CLK-2/TEL2 cell cycle defects during embryogenesis and germline development. PLoS Genet 5: e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO (1999) Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- 48. Derry WB, Putzke AP, Rothman JH (2001) Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. [DOI] [PubMed] [Google Scholar]
- 49. Schumacher B, Hofmann K, Boulton S, Gartner A (2001) The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol 11: 1722–1727. [DOI] [PubMed] [Google Scholar]
- 50. Conradt B, Horvitz HR (1998) The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93: 519–529. [DOI] [PubMed] [Google Scholar]
- 51. Ward IM, Minn K, van Deursen J, Chen J (2003) p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23: 2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Penkner A, Portik-Dobos Z, Tang L, Schnabel R, Novatchkova M, et al. (2007) A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J 26: 5071–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee SJ, Gartner A, Hyun M, Ahn B, Koo HS (2010) The Caenorhabditis elegans Werner syndrome protein functions upstream of ATR and ATM in response to DNA replication inhibition and double-strand DNA breaks. PLoS Genet 6: e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Higashitani A, Aoki H, Mori A, Sasagawa Y, Takanami T, et al. (2000) Caenorhabditis elegans Chk2-like gene is essential for meiosis but dispensable for DNA repair. FEBS Lett 485: 35–39. [DOI] [PubMed] [Google Scholar]
- 55. MacQueen AJ, Villeneuve AM (2001) Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev 15: 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, et al. (2002) Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127–131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, HSR-9 protein accumulates in the nuclei of C. elegans germ cells in response to various types of DNA damage. (A) HSR-9 was immuno-localized to the nuclei of wild-type germ cells in the mitotically proliferating region of gonads in response to hydroxyurea (HU, 60 mM) treatment for 16 h. (B) Nuclear levels of HSR-9 at 3 h after UV (150 J/m2) irradiation. Scale bar, 10 µm. Figure S2, hsr-9 deletion decreases egl-1 mRNA expression after γ-ray treatment. Adult wild type, hsr-9(ok759), and cep-1(lg12501) worms were irradiated with 120 Gy of γ-rays, and total RNA was isolated 6 h later. cDNA pools were prepared by reverse transcription, and relative amounts of egl-1 cDNA were measured by real-time PCR. Figure S3, hsr-9 mutation cannot relieve the hypersensitivity of brc-1 mutants to γ-rays. Wild-type N2, hsr-9(ok759), brc-1(tm1145), and brc-1(tm1145);hsr-9(ok759) worms were irradiated with γ-rays (45 Gy and 90 Gy) at the L4 stage. Embryos were collected between 24 and 48 h after the irradiation, and their hatching was scored 24 h later. brc-1(tm1145) mutation but not hsr-9(ok759) resulted in hypersensitivity to DSBs. The double mutant was very similar to the single brc-1 mutant in its sensitivity to DSBs (at both 45 Gy and 90 Gy, brc-1 vs. brc-1;hsr-9, p>0.3). Figure S4, The presence of Mos1 insertion in the hsr-9(ttTi14815) mutant is confirmed by PCR amplification of a genomic DNA fragment. Single worms were picked and lysed, followed by PCR amplification of the genomic DNA fragment flanking the insertion site. A 0.75 kb DNA fragment was produced from wild-type worms, and a much longer DNA fragment (2.0 kb) from the hsr-9(ttTi14815) mutant.
(PDF)