Abstract
Despite improving understanding of glaucoma, key molecular players of neurodegeneration that can be targeted for treatment of glaucoma, or molecular biomarkers that can be useful for clinical testing, remain unclear. Proteomics technology offers a powerful toolbox to accomplish these important goals of the glaucoma research and is increasingly being applied to identify molecular mechanisms and biomarkers of glaucoma. Recent studies of glaucoma using proteomics analysis techniques have resulted in the lists of differentially expressed proteins in human glaucoma and animal models. The global analysis of protein expression in glaucoma has been followed by cell-specific proteome analysis of retinal ganglion cells and astrocytes. The proteomics data have also guided targeted studies to identify post-translational modifications and protein-protein interactions during glaucomatous neurodegeneration. In addition, recent applications of proteomics have provided a number of potential biomarker candidates. Proteomics technology holds great promise to move glaucoma research forward toward new treatment strategies and biomarker discovery. By reviewing the major proteomics approaches and their applications in the field of glaucoma, this article highlights the power of proteomics in translational and clinical research related to glaucoma and also provides a framework for future research to functionally test the importance of specific molecular pathways and validate candidate biomarkers.
1. Introduction
Glaucoma is commonly viewed as a neurodegenerative disease with multifactorial origin. It is increasingly evident that besides intraocular pressure-generated stress and aging, glaucomatous neurodegeneration involves genetic predispositions and epigenetic risk factors. Although recent experimental studies have achieved many advances in understanding of glaucomatous neurodegeneration, key molecular mechanisms that can serve as treatment targets remain unclear. In particular, the molecular mechanisms initiating and propagating the neuronal injury in different subcellular compartments of retinal ganglion cells (RGCs), the cross-talks between multiple molecular pathways, and the contribution of each pathway to structural and functional loss, are largely unknown (Libby et al., 2005; Whitmore et al., 2005; Almasieh et al., 2012; Tezel, 2013).
Uncovering the molecular mechanisms involved in neurodegeneration is a prerequisite for improved treatment strategies for neuroprotection, neurorescue, neuroregeneration, immunomodulation, and function gain in glaucoma (Tezel, 2009; Limb and Martin, 2011). As reviewed herein, this goal can be accomplished through the large-scale analysis of the entire complement of cellular proteins using the proteomics technology. A comprehensive picture of pathogenic mechanisms can be provided by proteomics, since proteins mediate the actions of genes and reflect important pathophysiological changes at the post-translational level. Particularly respecting the multiplicity of factors affecting the fate of RGCs and optic nerve axons in glaucoma, analysis of the altered protein expression and post-translational modifications that affect protein functions, and analysis of the protein interactions networks that determine the ultimate cell fate are crucial to identify pathogenic processes (Pandey and Mann, 2000; Anderson et al., 2001; Tyers and Mann, 2003). Proteomics is also essential for new drug development since many current drugs and their target molecules are proteins (Glish and Vachet, 2003).
In addition to defining the molecular mechanisms and new treatment strategies of glaucoma, another major goal of glaucoma research is biomarker discovery that can also be accomplished by the use of proteomics technology. As recently discussed at the ARVO/Pfizer Ophthalmic Research Institute Conference, 2011 (Bhattacharya et al., 2013), identification of reliable molecular biomarkers is strongly needed for clinical utility to detect the disease early, predict its prognosis, and monitor disease progression and treatment efficacy in patients with glaucoma.
Following a brief description of the main advantages and drawbacks of major proteomics approaches, this review will highlight the recent studies of glaucoma using proteomics analysis techniques that open up new avenues for glaucoma research aimed to better understand neurodegeneration and discover glaucoma-specific molecular biomarkers. By evaluating the use of proteomics in the field of glaucoma research, this review article is hoped to illustrate the potential of proteomics in translational and clinical research related to glaucoma and lay out a framework for future research in the field.
2. Proteomics technology
2.1. Overview of proteomics
The term “proteomics” was first introduced in mid-1990s for the aim of global characterization of a proteome (referring the PROTEins expressed by the genOME), including protein expression, structure, modifications, functions, and interactions (Domon and Aebersold, 2006). Proteomics is one of the most important post-genomic approaches to improve the understanding of gene function. However, compared to genome, proteome is a much more complex and dynamic system. Although proteins provide the most important clues to pathogenic mechanisms, their analysis is difficult due to large diversity in many properties, such as molecular size, dynamic range in quantity, and hydrophilicity or hydrophobicity. Considering that the human genome contains over 20,000 genes (Lander et al., 2001; Venter et al., 2001; Consortium, 2004) producing multiple proteins by alternative splicing, the human proteome yields millions of peptides to analyze by proteomics techniques. The proteome also differs from cell to cell, from time to time, and in response to external stimuli. Post-translational modifications of proteins, which can occur at multiple sites and multiple ways, multiply the challenges of proteomics analysis. Phosphorylation, oxidation, glycosylation, and proteolytic cleavage, are some of the roughly 200 forms of post-translational protein modifications (Krishna and Wold, 1993).
Proteomics has increasingly been used to study expression, structure, modifications, functions, and interactions of proteins for large-scale analysis of gene function directly at the protein level (Pandey and Mann, 2000; Anderson et al., 2001; Tyers and Mann, 2003). Classical techniques of biochemistry and cell biology, such as Western blot analysis, enzyme-linked immunosorbent assay (ELISA), or immunohistochemistry, aim at the analysis of expression or localization of selective proteins. Due to their hypothesis-driven nature, these analysis techniques are not only limited to the study of a few individual proteins but are also strongly biased to determine biological processes. Protein arrays or chips have been developed for rapid and high-throughput analysis of protein expression; however, development of a protein array large enough to investigate the function of the entire genome is also challenging (Jenkins and Pennington, 2001; Mitchell, 2002; Joos and Bachmann, 2009). Diverse proteomics approaches, including the use of mass spectrometry as a central analytical technique, have emerged for high-throughput protein studies. Compared to earlier protein identification techniques, mass spectrometry-based analysis offers an unbiased technique to handle complex protein mixtures with higher sensitivity (Domon and Aebersold, 2006; Kocher and Superti-Furga, 2007; Nilsson et al., 2010). Proteomics is a rapidly changing field because of extensive advances in the underlying technologies. Continuous improvements in ultrasensitive mass spectrometry techniques, along with the rapidly emerging analytical workflows and data analysis strategies, expand proteomics from mere protein profiling to high-throughput quantification of alterations in protein expression (Filiou et al., 2012), post-translational modifications (Jensen, 2006), and protein-protein interactions (Ho et al., 2002; Ping, 2003; Kocher and Superti-Furga, 2007). Proteomics can provide framework information about complex biological processes to build and test new hypotheses. When combined with complementary molecular, cellular, and pharmacological techniques, proteomics can reveal unknown pathogenic mechanisms and specific molecular targets for new treatments (Cravatt et al., 2007).
Tremendous technological progress has made the proteomics an important toolbox for answering biological questions in multiple fields. Comparative proteomics has provided the background for comprehension of pathogenic mechanisms in numerous diseases and complemented the interpretation of data arising from genomics studies. Besides powerful applications of proteomics in experimental research, identification of disease biomarkers has constituted a promising area of clinical proteomics. Parallel to growing applications in other fields of biomedical research, proteomics analysis techniques have also been increasingly used in ophthalmic research to advance molecular understanding and management of various ocular diseases (Steely and Clark, 2000; Lam et al., 2008; Mandal et al., 2009). In the past years, there has also been a major increase in studies using proteomics analysis techniques in the field of glaucoma research and provided bulk of new information about alterations in protein expression. Mass spectrometry-based analytical approaches also offer the opportunity for clinically relevant discovery of glaucoma-related molecular biomarkers. This review highlights how proteomics has been applied to uncover molecular mechanisms and identify candidate biomarkers of glaucoma.
2.2. Gel-based proteomics analysis
In earlier proteomics studies, gel-based analysis referring to separation approaches using protein electrophoresis has been the main approach for comparison of disease and control samples. This approach uses gel electrophoresis to simplify complex mixtures of protein samples for protein identification by mass spectrometry. In two-dimensional (2D) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), a mixture of proteins in biological samples is first separated on the basis of pI differences and then according to size, and the following gel staining allows spot visualization (O'Farrell, 1975). Fluorescent-based dyes, such as Sypro Ruby, have a greater dynamic range compared to Coomassie Brilliant Blue or silver-nitrate staining and do not impair extraction of peptides from the gel for following analysis by mass spectrometry (Harris et al., 2007). Using the gel-based technique, up to a few thousands of proteins can be separated from highly complex mixtures, and by comparison of the protein staining pattern, up-regulated and down-regulated proteins can be quantified (Patton, 2000; Rabilloud, 2002). Differential display analysis of large series of samples can be possible using special software to match, align, and compare the signal intensities of the gel spots (Wheelock and Buckpitt, 2005). This technique has been widely used in multiple fields for mapping the proteins and detecting their expression differences. In addition to information about variations in protein quantity, this type of analysis can also provide information about the occurrence of potential post-translational modifications, which are reflected by a shift of the protein spot on the gel (Patton, 2000; Rabilloud, 2002).
Although the gel-based analysis technique using 2D, or 1D, gel electrophoresis has been one of the most commonly used approaches for proteome analysis in many laboratories, it has many drawbacks and limitations, such as labor-intensive and time-consuming protocols, low sensitivity, poor quantification accuracy, and limited dynamic range. A major weakness hindering the standardized protein detection and quantification is related to inefficiency in resolving low abundant proteins, integral membrane proteins, and proteins at the extremes of pI or molecular weight. In order to address such problems inherent to SDS-PAGE, specific pre-treatments of the sample have been used to deplete most abundant proteins, enrich less abundant proteins, remove interfering substances, or add zwitterionic detergents. In addition, gels that cover limited pI ranges have been used to increase resolution. However, despite improvements leading to the generation of highly reliable equipment and protocols, the 2D-gel-based analysis of peptide mass maps has suffered from a number of shortcomings limiting the sensitivity and reproducibility (Herbert and Righetti, 2000; Zuo and Speicher, 2002; Gorg et al., 2004).
To solve the problems related to gel-to-gel variation, 2D-fluorescent difference gel electrophoresis (DIGE) has been introduced (Unlu et al., 1997). This approach employs a spectrally resolvable set of fluorescent dyes to label up to three protein mixture samples, including an internal standard. The labeled samples are then pooled for 2D-SDS-PAGE, and after excitation with the appropriate wavelength for each dye, expression differences between samples can be visualized by overlaying the captured images. Because proteins from different samples are separated in a single gel, this technique eliminates gel-to-gel variation, and reduces the reproducibility problems and difficulties in image analysis. A drawback is related to dependence of the technique on specific amino acids for tagging, since the absence of these amino acids may cause no labeling. In addition, co-migration of multiple proteins into a single spot is a problem intrinsic to gel-based quantification. Since relative quantification calculations are based on spot intensity, the intensity value may actually be related to one or multiple proteins present in the same spot (Marouga et al., 2005; Tannu and Hemby, 2006; Minden et al., 2009).
For protein identification through the gel-based technique, protein spots of interest are excised and subjected to analysis by mass spectrometry after in-gel digestion to peptides (Canas et al., 2007). Among the mass spectrometry-based analysis techniques, “peptide mass fingerprinting” refers to mass analysis of peptides to identify proteins by database analysis, “peptide sequencing” refers to determination of amino acid sequences by partial fragmentation of peptides through ionization. In common combinations of mass spectrometry instrumentation, different ionization sources, such as matrix-assisted laser desorption ionization (MALDI) or electrospray ionization (ESI), and different mass analyzers, such as time-of-flight (TOF), quadrupole, ion trap, high resolution orbitrap, and Fourier transform ion cyclotron resonance, are used. Each of these has their own advantages and weaknesses. A MALDI-TOF instrument is commonly employed for peptide mass fingerprinting/mapping, and a tandem mass spectrometry (MS/MS) instrument, such as MALDI-TOF/TOF or MALDI-Q-TOF, is used for peptide fragment fingerprinting or peptide sequencing (Aebersold and Mann, 2003).
Although introduction of highly sensitive mass spectrometers has initially seemed to help protein identification through the 2D-gel-based analysis, it has become clear that the thousands of spots seen in the gel maps are actually variants of a few hundred of the most abundant proteins (Fountoulakis et al., 2004), which along with the spot overlap, limit the accuracy of protein identification (Campostrini et al., 2005). Consequently, there has been a noticeable tendency toward the list of same proteins identified through the analysis of different systems (Petrak et al., 2008). Due to technical shortcomings, protein identification and differences detected in the protein expression level should be validated by complementary analysis techniques, such as Western blot analysis, ELISA, or immunohistochemistry using specific antibodies. As reviewed in section 3, differential display analysis of 2D-gels followed by mass spectrometry-based analysis of the excised gel spots for protein identification, as well as a similar 1D-gel-based approach, has been widely used for quantitative proteome profiling in glaucoma research, and provided valuable information.
2.3. Gel-free proteomics analysis
In recent years, increasing focus has been on the gel-free techniques of proteomics, and powerful instrumentation has been developed to yield improved data quality and higher protein identification throughput. In gel-free proteomics analysis, a complex protein mixture is first enzymatically (with trypsin or alternative enzymes) digested into peptides and the resulting peptides are separated by liquid chromatography (LC) before introduction into a mass spectrometer for fragmentation and sequencing to identify the parent proteins. The LC-MS/MS approach is now the preferred technique for “shotgun proteomics” that allows direct analysis of very complex peptide mixtures with high sensitivity and broad dynamic range. Since this technique uses separation mechanisms based on charge and hydrophobicity, as opposed to pI and molecular weight in the gel-based approach, it can facilitate the identification of low-abundant and integral membrane proteins and proteins with extreme pI or molecular weight. The current format in large-scale proteomics involves the analysis of tens of thousands of peptides with continuously enhancing dynamic range (Link et al., 1999; Aebersold and Mann, 2003; Powell et al., 2006; Mann and Kelleher, 2008).
Enzymatic digestion may generate 20–50 peptides from each protein depending on the size of the protein and specificity of the enzyme cleavage. It is currently not possible, and also not necessary, to achieve full protein sequence for every peptide digested from a protein. Consequently, the protein identification by direct LC-MS/MS analysis is based on sequencing of a rather few peptides from each protein, and the mass spectral data are then combined to compute a summarized value for the protein identity. Detecting and measuring multiple peptides, a minimum of two peptides, from one protein usually provides adequate information to trace back to the protein from which these peptides are generated. This strategy for proteomics analysis performed at the peptide level is called “bottom-up proteomics” (Link et al., 1999; Wolters et al., 2001).
Major challenges in bottom-up proteomics are sample complexity and large concentration differences of proteins. To overcome these challenges, abundant proteins may be removed from complex biological samples using affinity chromatography strategies. However, protein interactions may cause the depletion of low-abundant proteins along with the proteins being depleted from the sample. Although LC-based platforms reduce the sample complexity prior to mass spectrometry and allow detection of a greater number of peptides in tryptic digests, due to the complexity of proteome samples, traditional single dimensional LC could hardly meet the requirement of high resolution and high peak capacity for separation. To enhance an in-depth analysis for shotgun proteomics, evolving technologies for peptide separation have led the development of the multidimensional protein identification technology (MudPIT) (Washburn et al., 2001; Wolters et al., 2001). Through this technology digested samples are separated by two or more independent separation mechanisms in a consecutive manner. Among the various methodological combinations developed strong cation exchange (SCX) chromatography-reversed phase (RP) liquid chromatography is the most commonly used. Dramatically expanding new choices for multidimensional strategies include hydrophilic interaction chromatography (HILIC) and electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) (Zhang et al., 2010; Di Palma et al., 2012). The decision to use 1D- or 2D-LC is based on the complexity of protein sample. Although gel bands or spots containing less than 30 proteins can be analysed using 1D-LC, direct analysis of much more complex protein mixtures, such as whole cell lysates or serum, usually require 2D-LC for peptide separation.
In bottom-up proteomics, incomplete sequence coverage may result in losing the essential information necessary to distinguish different protein isoforms arising from alternative splicing, post-translational modifications, or proteolytic cleavage. Consequently, non-unique peptide sequences match to more than one protein candidate in a database, while unique sequences match only to one exclusive protein. This protein inference problem caused by presence of the same peptide sequence in multiple proteins or protein isoforms is a common challenge in bottom-up proteomics.
In an alternative approach, non-digested intact proteins are introduced into the mass spectrometer and subjected to gas-phase ion isolation and dissociation for protein fragmentation. This strategy, called “top-down proteomics”, has the potential to identify a larger fraction of protein sequences and the ability to also locate and characterize post-translational modification (Kelleher et al., 1999). In addition, the time-consuming protein digestion required for bottom-up methods is eliminated. This approach not only does increase the experimental efficiency, but also reduces the error rates for identification and quantification of proteins. However, despite detailed characterization, current top-down proteomics approaches are limited to the analysis of proteins with 500 or fewer amino acid residues (up to about 50 kDa) and have not yet proven useful for large-scale studies. The transition of this technique to broad applications will require further improvements in algorithms and software (Chait, 2006; Han et al., 2006; Armirotti and Damonte, 2010).
Similar to gel-based technique, the use of mass spectrometry plays a critical role in the workflow for gel-free analysis of tryptic digests. Although digested peptides can be analyzed by MALDI-TOF or MALDI-TOF/TOF, 1D-LC and 2D-LC are commonly coupled with ESI-MS/MS. Decision of which approach is most suitable for an analysis should depend on the analytical question, the existing samples, the available equipment, and the analysis time. MALDI-TOF and surface-enhanced laser desorption/ionization (SELDI)-TOF that is a modification of MALDI-TOF, are used for profiling, while MALDI-TOF/TOF and ESI-MS/MS can sequence the peptides to identify their parent proteins. The sensitivity of MALDI-TOF/MS is limited to characterizing the most abundant proteins in each sample; however, this method presents the benefit of a higher throughput platform (up to 96 individual protein samples). MALDI-TOF/MS is advantageous for analysis of gel-separated protein mixtures with lower complexity because each sample is analysed in minutes. SELDI-TOF data are given in mass-to-charge ratios (m/z), rather than peptide sequences for protein identification; however, it has been useful for rapid analysis of semi-complex samples by reducing the upfront separation, while preserving the fast analysis time of a MALDI platform. Although not desirable for discovery, the speed of analysis and data output, and low operation costs make the SELDI-TOF a suitable platform for clinical applications (Petricoin et al., 2002; Clarke et al., 2005; Zhang et al., 2010; Di Palma et al., 2012). Multidimensional separation allows the analysis of very complex samples, but greatly increases the analysis time to an hour for 1D-LC and 10 to 12 hours for 2D-LC-MS/MS.
Large sample requirements (50–200 μg of total protein per analysis), inadequate detection limits (0.1–10 ng protein per spot), and narrow dynamic range (less than 103) are major weaknesses of gel-based techniques. However, increasing sensitivity and accuracy of mass spectrometry enable direct (gel-free) analyses of as little as picogram amounts of protein samples. Although most commonly available LC-MS/MS systems require 10–50 μg of total protein sample to start, the modern nanoproteomics platform enables analyses of nanogram quantities of samples with individual protein identification sensitivity at the low zeptomole level. Dynamic range of protein measurements using nanoscale technologies can approach 106 for samples containing nanograms of total protein, while more abundant proteins can be detected from subpicogram amounts of samples (Shen and Smith, 2005; Xie et al., 2012). Advancing technologies for in-depth and high-throughput analysis of complex protein mixtures present the potential to eventually extend the analysis to a single mammalian cell.
2.3.1. Isotope labeling-based quantification techniques
With continuing progress in sensitivity, mass accuracy, peptide sequencing speed, and identification and scoring algorithms, along with the gradual shift from gel-based to gel-free techniques of mass spectrometry-based analysis, an entirely new toolbox has become available for quantitative comparisons that can be achieved by stable isotope labeling or label-free techniques (Filiou et al., 2012).
The principle of labeling-based quantitative proteomics relies on isotope incorporation at the protein or peptide level. The introduction of an isotope label (such as, 2H, 13C, 15N, or 18O) results in a predictable mass difference between a labeled peptide and the unlabeled counterpart, allowing the mass analyzer to discriminate between the two forms. Relative quantification can then be achieved by comparing the signal intensities of labeled and unlabeled samples by mass spectrometry. Two distinct techniques are commonly used for incorporation of stable isotopes into the proteome of interest, in vivo metabolically labeling (Ong et al., 2002) and in vitro stable chemical (Gygi et al., 1999; Ross et al., 2004) or enzymatic labeling (Yao et al., 2001).
In the first method, stable isotopes are incorporated metabolically into all proteins, in vivo. In vivo metabolic labeling methods include stable isotope labeling with amino acids in cell culture (SILAC) (Ong et al., 2002), and stable isotope labeling of mammals (SILAM) (Wu et al., 2004). In SILAC, two groups of cells are grown in culture media, one containing light isotopes and the other containing one or more heavy isotopes incorporated into the amino acids. After incorporation of these light and heavy amino acids into proteins during the process of cellular anabolism, equal numbers of cells in the two groups are combined, and the extracted proteins are digested for subsequent analysis by mass spectrometry. Thus, SILAC is the method of choice for labeling cultured cells. The major advantage of this technique is that labeling is performed at a very early stage of the experimental paradigm, and the labeled and reference samples are mixed prior to all subsequent procedures. This helps yielding accurate quantification by avoiding the differences that can arise from sample handling in later stages. Because the stable isotope tags are incorporated into the earliest possible stage of sample preparation, in vivo metabolic labeling ensures minimal variability among all quantitative methods. However, this technique cannot be applied to tissue samples or body fluids, it is costly and time-consuming requiring extended culture periods, and it may induce phenotypic alterations (Ong et al., 2003; Mann, 2006; Bantscheff et al., 2007). SILAM has been developed for metabolic labeling to quantify proteins from mammalian tissues but has not been widely used (Wu et al., 2004).
Heavy and light stable isotopes can also be introduced by chemically modifying the reactive functional groups in proteins or peptides. After chemical labeling, the labeled samples are combined for digestion, fractionation, and MS/MS analysis for protein identification and quantification (Ong and Mann, 2005). In vitro chemical labeling methods include isotope-coded affinity tag (ICAT) (Gygi et al., 1999), isobaric tag for relative and absolute quantification (iTRAQ) (Ross et al., 2004), and isotope-coded protein label (ICPL) (Schmidt et al., 2005). The ICAT relies on differential isotopic labeling of cysteine residues in proteins with biotin tags, thereby allowing the selective enrichment of modified peptides by avidin affinity chromatography. Proteins from two samples are labeled with isotopically light or heavy ICAT reagents. The ICAT has been a preferred method to analyze very complex protein mixtures where the dynamic range of protein concentration is an issue. This method has the advantage of labeling intact proteins prior to digestion, which can reduce variability. However, despite reducing the sample complexity, the cysteine-specific labeling also reduces the protein coverage. The iTRAQ uses a multiplexed tagging reagent that consists of a reporter group, including a tag with a specific mass in each individual reagent, a balance group, and a peptide reactive group. The balance group ranges in mass to ensure the constant combined mass for all reagents. Therefore, peptides labeled with different isotopes are isobaric and chromatographically indistinguishable. This is important for accurate quantification that is based on the comparison of reporter ion intensities. The most significant advantage of this technique is the capability to label up to eight different samples collected from multiple time-point experiments. However, fragment ion based-quantification strongly compromises the dynamic range of quantification. The ICPL method is based on isotopic labeling of lysine residues and available N-termini on intact proteins with light or heavy tags. This approach increases the level of labeling and provides multiplexed capability for simultaneous comparison of three samples, but also presents several disadvantages. Overall, the introduction of isotope labels during sample preparation is an advantage for chemical labeling techniques to lower the variability during sample handling. Despite technical difficulties and high costs, these techniques have been useful for large-scale or targeted analyses (Aggarwal et al., 2006; Bantscheff et al., 2007).
Regarding enzymatic 16O/18O labeling, this method relies on enzyme-catalyzed oxygen exchange in the presence of H218O. Two 16O atoms are typically replaced by two 18O atoms at the C-terminal carboxyl group of proteolytic fragments (Heller et al., 2003). The resulting mass shift between differentially labeled peptide ions permits identification, characterization, and quantification of proteins. Due to its ability to universally label almost all the carboxyl termini, enzymatic labeling is an ideal choice to quantitatively study mixtures of low molecular weight proteins and post-translationally modified peptides. In addition, isotopic 18O labeling does not alter the physical properties of peptides during analysis (Yao et al., 2001). Although the 16O/18O labeling is not yet a common technique compared to other quantitative proteomics techniques, recent advancements in the homogeneity of 18O incorporation and improvements made on the algorithms employed for calculation of 16O/18O ratios should result in its increased use. Its simplicity, specificity, affordability, and instantaneous applicability to even limited amount of samples, make this technique easily applicable for quantitative proteomics, particularly for proteome profiling of human tissue, plasma, or serum samples. When comparing the price of reagents needed, 18O labeling is far less expensive than all other stable isotope labeling techniques, approximately seven orders of magnitude less costly than iTRAQ labeling and two orders of magnitude less costly than ICAT and SILAC (Schnolzer et al., 1996; Sakai et al., 2005; Lopez-Ferrer et al., 2006; Miyagi and Rao, 2007).
An alternative method to labeling-based proteomics is the use of labeled whole proteomes as internal standards for quantitative comparisons (Ong, 2010). Although this approach is attractive, several issues remain to be addressed. For example, the labeled proteome may not accurately reflect the proteomes under investigation. A protein can be quantified only if it has been identified previously in the internal standard, as well as present in the two samples compared, which may not be always ensured even for high-abundant proteins. Consequently, a fraction of proteins may not be detectable, and valuable information may be lost (Filiou et al., 2012).
Thus, although there are a number of isotope labeling techniques employed for quantitative proteomics, there is no clear consensus in the literature for a best strategy, and further improvements are required to address existing limitations (Miyagi and Rao, 2007; Filiou et al., 2012). As reviewed in section 3, isotope labeling techniques have been used for quantitative analysis of differential protein expression in glaucoma. However, since isotopic labeling of proteins is not always practical and possesses several disadvantages, the label-free quantification has been an attractive alternative.
2.3.2. Label-free quantification techniques
The two major label-free quantification approaches are counting the number of mass spectra identifying a given peptide or protein (Liu et al., 2004), and area under the curve or signal intensity measurement based on precursor ion spectra (Bondarenko et al., 2002; Chelius and Bondarenko, 2002). Several studies have demonstrated that both spectral counting and extracted ion chromatograms of selected peptide ions correlate well with protein abundances in complex biological samples.
In label-free quantification, the time-consuming steps and costs of isotope labeling can be omitted. This approach also offers a wide dynamic range and allows for the comparison of an unlimited number of samples from any type of biological materials, thereby providing a higher degree of experimental flexibility. However, there is no sample multiplexing, and each sample has to be analyzed individually. Although the label-free quantification offering an inexpensive methodology are increasingly applied to quantitative proteomics, small numbers of spectral counts for low-abundant proteins may result in less robust measurements due to statistical limitations. In addition, data reproducibility may be an issue since sample processing and LC-MS/MS runs may vary across time on a given instrument or between different experiments. However, this can be overcome by running replicate samples and quality controls and by using software advances (Wiener et al., 2004; Old et al., 2005; Zhang et al., 2009; Lundgren et al., 2010; Neilson et al., 2011). Depending on the MS acquisition mode, there are two basic analytical methods. The first is termed data-dependent analysis where the mass spectrometer runs a parent ion scan to select the most abundant ions for fragmentation scans. Since data-dependent analysis results in compression of the dynamic range of quantifiable proteins, data-independent analysis has been developed as an alternative, which acquires tandem mass spectra at every m/z value without precursor selection. To increase the reproducibility and comprehensiveness of data collection, data-independent acquisition can also be applied for label-free quantification (Panchaud et al., 2009; Blackburn et al., 2010; Hengel et al., 2011; Krishnamurthy et al., 2011).
Number of studies have focused on the comparison of isotope labeling-based and label-free methods for quantitative proteomics (Filiou et al., 2012). A recent multicenter study conducted by The Association of Biomolecular Resource Facilities Proteomics Research Group was devoted to relative protein quantification (Turck et al., 2007). Fifty-two participating laboratories identified proteins and determined their relative quantities in the two samples containing the same proteins with different quantities, using gel-based methods, and isotope labeling-based or label-free direct analysis. Findings of this study indicated no superiority of labeling-based or label-free approaches for quantification and implied that there is no single gold standard method for quantitative analysis of the proteome, and scientists must choose the most appropriate method for particular purposes.
While the isotope labeling-based and label-free techniques are widely used for “relative protein quantification”, new mass spectrometry-based workflows have also been developed for “absolute protein quantification”. These methodologies relying on the isotope dilution principle use isotope-labeled analogue(s) of specific proteolytic peptide(s) (AQUA and QconCAT strategies) or protein(s) (PSAQ strategy) as quantification standards (Brun et al., 2009; Jaquinod et al., 2012). These strategies promise for powerful applications; however, they have been implemented in the proteomics field only recently and possess some problems. For example, limited number of proteotypic peptides eligible for quantification assays may adversely influence the quantification robustness (Schmidt et al., 2010).
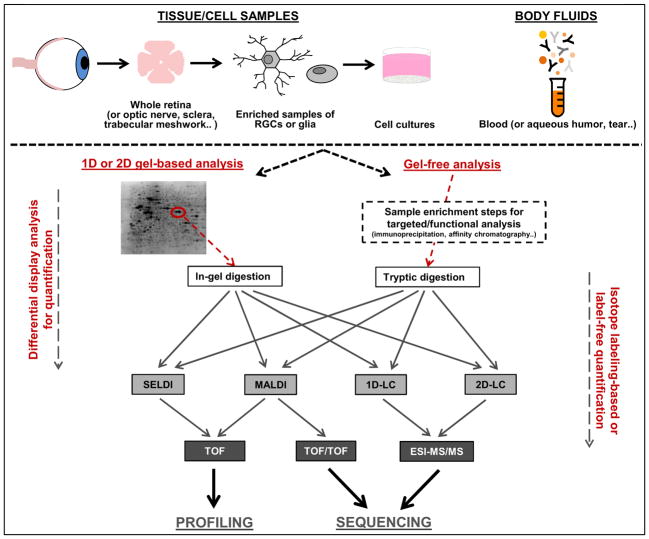
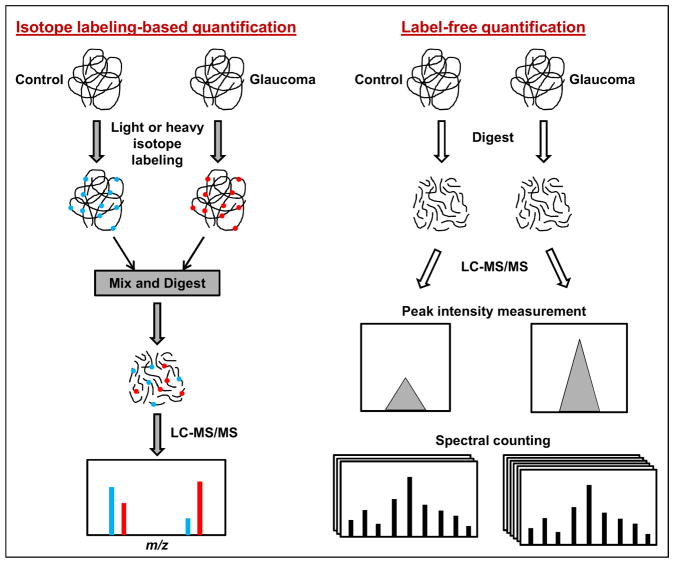
As reviewed in section 3, gel-based and gel-free techniques of protein analysis using mass spectrometry and isotope labeling-based or label-free techniques for relative quantification for protein expression between glaucomatous and control samples have been increasingly used to study molecular mechanisms and identify potential biomarker candidates of glaucoma. Figure 1 and Figure 2 present simplified workflows for commonly employed proteomics analysis and protein quantification techniques.
Figure 1.
Proteomics workflow. Both gel-based and gel-free proteomics techniques using mass spectrometry have been increasingly employed for high-throughput characterization of protein expression in glaucomatous human tissues and in vitro and in vivo experimental models. Proteomics analysis techniques also enable biomarker discovery in glaucoma.
Figure 2.
Quantitative proteomics analysis workflow. Both isotope labeling-based and label-free techniques using mass spectrometry have been applied for relative quantification of protein expression between glaucomatous and control samples.
2.4. Targeted and functional proteomics
The information gained from gel-based peptide mass fingerprinting/mapping or gel-free approaches of shotgun or “discovery proteomics” through peptide sequencing by LC-MS/MS analysis is roughly a list of identified proteins and their relative expression level. Analysis of protein functions requires additional methodology. In “functional proteomics”, selectively targeted protein samples can be prepared by a variety of affinity-based approaches to enrich modified proteins or isolate protein complexes. These enriched samples are then analyzed by LC-MS/MS for protein identification and quantification.
2.4.1. Identification of post-translational modifications
Regulation of protein expression is often affected by post-translational modifications. One of the main aims of current proteomics strategies is to identify proteins that carry post-translational modifications, map modified sites, and quantify the degree of modification. Phosphorylation, a dynamic process reversibly controlled by protein kinases and protein phosphatases, is one of the most prevalent post-translational protein modifications in eukaryotic cells. By determining the activity state, localization, turnover, and interactions of proteins with other proteins, phosphorylation plays a key role in regulation of different cellular processes, including signal transduction. Western blot analysis using phosphorylation site-specific antibodies is a commonly applied simple method for analysis of phosphorylated proteins. However, Western blot analysis relies on prior knowledge of the type and position of phosphorylation and the availability of antibodies. By contrast, mass spectrometry-based analysis can discover novel modifications as well as monitoring known modifications (Domon and Aebersold, 2006; Kocher and Superti-Furga, 2007; Nilsson et al., 2010).
2D-gel-based analysis with the inclusion of fluorescence-based stains, such as Pro-Q Diamond phosphoprotein stain, can be used for global analysis of phosphoproteins with high sensitivity and dynamic range (Steinberg et al., 2003; Wu et al., 2005; Agrawal and Thelen, 2009). When multiplexed with Sypro Ruby for total protein profiling, phosphoprotein gel staining provides a simple technique for detection and relative quantification of phosphoproteins. Since the stain non-covalently binds to phosphoproteins, it is also compatible with the following mass spectrometry-based protein identification. However, this technique suffers from the problems inherent to gel-based analysis.
Based on the prior knowledge of a protein sequence and the prediction of putative modified sites and corresponding peptide masses, tandem mass spectrometers can be programmed to detect and sequence the peptides that generate modification-specific signals. Mass spectrometry enables the site-specific assignment of post-translational modifications because the covalent addition of a chemical moiety to an amino acid leads to an increase in the molecular mass. For example, phosphorylation of a tyrosine residue (163 Da) increases its mass to 243 Da by the addition of an HPO3 group (+80 Da). In addition to peptide mass predictions that do not provide information on the exact location of the modified peptide, LC-MS/MS is also useful for mapping protein modifications. Since post-translational modifications are located at specific amino acid residues of proteins, usually in the context of a particular sequence motif, computational amino acid sequence analysis may reveal the potential sequence motifs for post-translational modifications in proteins (Mann and Jensen, 2003; Jensen, 2006). Continuously emerging workflows for quantitative analysis of protein phosphorylation can also provide site-specific data on the degree of phosphorylation by isotope-labeling or label-free quantification techniques or the addition of stable isotope labeled peptide/phosphopeptide pairs as internal standards (Olsen et al., 2006; Boehm et al., 2012a).
Despite many useful applications, phosphoproteomics approaches face many challenges to overcome (Mann and Jensen, 2003; Jensen, 2006). Besides the transient nature of phosphorylation, the same protein may be phosphorylated in different ways and at different sites, or a post-translational modification at a given site may be present in only a small fraction of the protein molecules. Since phosphoproteins usually have low abundance, more focused strategies to enrich phosphorylated proteins or peptides are a prerequisite to their analysis. Classical enrichment protocols involve immunoaffinity purification, such as immunoprecipitation with phosphorylation site-specific antibodies. However, it is important to consider that post-translational modifications might mask or alter the protein epitope normally recognized by the antibody, thereby leading to reduced recovery after immunoprecipitation. Phosphopeptides can also be enriched by immobilized metal ion-affinity chromatography (IMAC) (Chaga, 2001; Block et al., 2009; Ye et al., 2010) or metal oxide affinity chromatography (MOAC) (Rappsilber et al., 2007; Sugiyama et al., 2007). These methods are based on the affinity of phosphate moiety to metal ions (such as iron) or metal oxides (such as titanium dioxide) immobilized in a column. Such enrichment methods allow the separation of phosphopeptides from non-phosphorylated peptides for large-scale phosphoproteomics analysis by mass spectrometry.
Another post-translational protein modification, particularly relevant to glaucoma, is protein oxidation. Proteins are major targets of reactive oxygen and nitrogen species (ROS/RNS) and undergo both reversible redox reactions as part of their normal function and irreversible oxidative damage under disease conditions. Oxidative modifications of proteins may lead to a change in protein structure, function, and turnover, establish pathological hallmarks of disease, and contribute to disease development and progression. Similar to many other diseases and aging, oxidative processes are important for neuronal survival and inflammatory processes in glaucoma (Tezel, 2006; Tezel, 2011).
Oxidative protein modifications may result from oxidative cleavage of the protein backbone or amino acid side chains, direct oxidation of amino acid residues, or adduction by electrophilic species, such as lipid peroxidation products. Many of these oxidative modifications result in the introduction of carbonyl groups into the proteins. Further reaction of these adducts leads to formation of protein cross-linking and generation of advanced lipoxidation end products and advanced glycation end products (AGEs). Due to their chemical instability, instead of ROS/RNS, their stable metabolites and/or oxidation target products, such as nitrotyrosine, dityrosine, and protein carbonyls, are measured by targeted proteomics to study oxidized proteins (Dalle-Donne et al., 2005; Giustarini et al., 2009; Sultana and Butterfield, 2011). Redox proteomics is an increasingly emerging branch of proteomics to identify and quantify redox-based changes in the proteome. In the last few years, affinity chemistry-based methodologies combined with mass spectrometry have significantly contributed to better understanding of oxidative protein modifications (Butterfield and Dalle-Donne, 2012).
Despite developing new techniques for redox proteomics, many studies have investigated carbonylated proteins using a convenient immunochemical procedure detecting the dinitrophenyl derivatives of carbonyls. In this gel-based method, called 2D-oxyblot analysis, protein carbonyls react with 2,4-dinitrophenylhydrazine forming a hydrazone that is detectable by immunochemical labeling of gels, and sister gel pieces corresponding to reactive spots are then analyzed by mass spectrometry for protein identification (Butterfield, 2004; Tezel et al., 2005). Oxidative protein modifications, such as mass increase unique to the appended group, can also be distinguished by mass spectrometry. For example, a mass shift in the mass spectra corresponding to addition of the oxidation moiety to methionine (+16 Da) can be used for estimation of the methionine sulfoxide level of each protein (Stadtman et al., 2003; Davies, 2005; Houde et al., 2006; Fedorova et al., 2010). Evolving techniques of affinity chromatography and mass spectrometry allow enhanced isolation, identification, and chemical characterization of oxidative protein modifications. However, since different forms of oxidation occur simultaneously, differentiating various types of oxidative modifications may be difficult (Madian and Regnier, 2010). Section 3.2.1 reviews the studies aimed to identify glaucoma-related post-translational modifications.
2.4.2. Identification of protein interactions
Proteins control and execute the large majority of cellular functions; however, biological functions of proteins depend on the ability to interact with other biomolecules to form protein complexes. Intracellular signaling events often require transient or regulated interactions of proteins which play critical roles in cellular structure, regulation of gene expression, modulation of enzymatic substrate specificity, and cell fate. The study of protein interactions is an important post-genomics strategy to understand protein function. Characterizing protein complexes and mapping protein interactions can provide important insights into molecular mechanisms of specific cellular processes (Ho et al., 2002).
The complete description of protein interactions networks has long been one of the greatest challenges of proteomics. Although the yeast two-hybrid system has a real potential for protein interactome analysis, it presents several limitations in detecting the interactions of more than two proteins, interactions depending on post-translational modifications, or interactions involving membrane proteins. This technique also suffers from false positive and negative signals, thereby challenging the physiological relevance of detected interactions (Ito et al., 2002). Affinity-based purification techniques coupled with mass spectrometry have been proposed as an alternative to the yeast two-hybrid assay (Azarkan et al., 2007; Banks et al., 2012). The common theme of the affinity-based purification techniques is the use of an inherent interaction (affinity) of two biomolecules. If one of the molecules is immobilized on a solid support, the interacting molecule can be purified from a cell lysate along with associated proteins. There are many different affinity reagents proven useful for isolation of protein complexes, such as recombinant proteins, epitope-tagged proteins, antibodies, peptides, and nucleic acids. Immunoprecipitation, affinity tag (such as, GST) pull-down, epitope tagging, and tandem affinity purification are commonly used affinity approaches (Gavin et al., 2002; Bauer and Kuster, 2003).
The classical method to isolate a protein complex is co-immunoprecipitation that targets multi-protein complexes using an antibody against one of the components. This method isolates protein complexes without disrupting them; however depends on the availability, quality, and cross-reactivity of antibodies. The ability to isolate specific complexes may also be limited if the antibody-directed epitope is masked by interacting proteins. Additionally, antibody-binding may displace proteins that interact with the epitope region. To assure the success, several antibodies against different epitopes of a protein may be needed (Bauer and Kuster, 2003; Banks et al., 2012).
Another common technique, recombinant GST fusion protein-based affinity pull-down introduces exogenous full-length proteins into the cell lysate. In this assay, the bait protein competes with the endogenous protein to form a new multi-protein complex. This technique is robust, easy to use, and capable of capturing even weakly interacting and low-abundant proteins. However, since the technique is based on competition of the recombinant fusion protein with the corresponding endogenous component, the protein complex may not be isolated if all complex components are engaged in endogenous protein-protein interactions (Bauer and Kuster, 2003; Banks et al., 2012).
In epitope tagging, bait proteins can be fused to an epitope-tag and an antibody directed against the tag, instead of the bait protein, is used to isolate protein complexes. Despite high degree of reproducibility, this technique is largely limited to cell culture systems and the protein tag may influence protein function. The basic concept of tandem affinity purification is similar to epitope tagging, but the main difference is the sequential utilization of two tags, instead of one. This methodology combined with mass spectrometry is highly reproducible and applicable to large-scale studies, and represents a powerful tool for functional proteomics (Rigaut et al., 1999; Azarkan et al., 2007; Banks et al., 2012).
New technological advances for purification and analysis of protein complexes along with the emerging tools for downstream network analysis enable detailed understanding of the protein interactions networks. However, the proteomics techniques developed for analysis of protein interactions have not yet been broadly used in the field of glaucoma research. As reviewed in section 3.2.2, there is only one study focused on glaucoma-related protein interactions.
2.5. Computations and bioinformatics
As proteomics technologies improve to provide accurate qualitative and quantitative data on proteins, new computational techniques and bioinformatics databases are being developed to process and integrate the new information on proteins, their modifications, and interactions (Peri et al., 2003). The analysis of the raw datasets produced by mass spectrometry requires multiple steps for database-dependent search, statistical evaluation of search results, quantitative algorithms, and statistical analysis of the quantitative data. Multiple data analysis strategies and software tools have been developed for each of these steps, and the availability of appropriate software and algorithm solutions is critical to handle the complex and large amounts of mass spectral data obtained through isotope labeling-based or label-free quantitative approaches of proteomics (Mann and Kelleher, 2008; Mueller et al., 2008; Matthiesen et al., 2011). The integration of computational tools into detection and quantification of post-translational protein modifications is also a prerequisite for the interpretation of large-scale datasets to gain meaningful biological information. Database searching algorithms and bioinformatics prediction tools for detection of modified peptides by MS/MS analysis are continuously improving to allow post-translational modifications to be determined comprehensively. Computational methods have also been critical to generate protein interactions networks from proteomics datasets and provided a foundation for computational simulations of dynamic cell signaling processes (Bock and Gough, 2003; Rual et al., 2005; Pieroni et al., 2008; Sardiu and Washburn, 2011). While functional proteomics provides snapshot information about protein interactions wiring a protein network, the systems biology analyzes and quantifies perturbation-induced network changes and correlates dynamic networks with the cellular phenotype. Evolving data analysis strategies of the systems biology aim to integrate all of the information and build multidimensional and multidirectional networks of interacting molecules between genotypes and phenotypes to thereby provide novel pathophysiological perspectives (Bensimon et al., 2012).
Future studies will continue to seek improved approaches to identify and quantify proteins, their modifications and interactions by processing very small amounts of sample, and refined bioinformatics tools will aid researchers in better understanding, interpreting, and communicating the proteomics data.
3. Proteomics toward molecular mechanisms of glaucoma
3.1. Global proteomics analysis of glaucoma
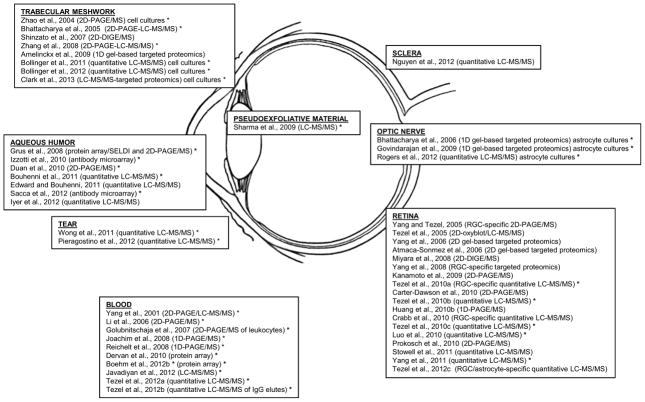
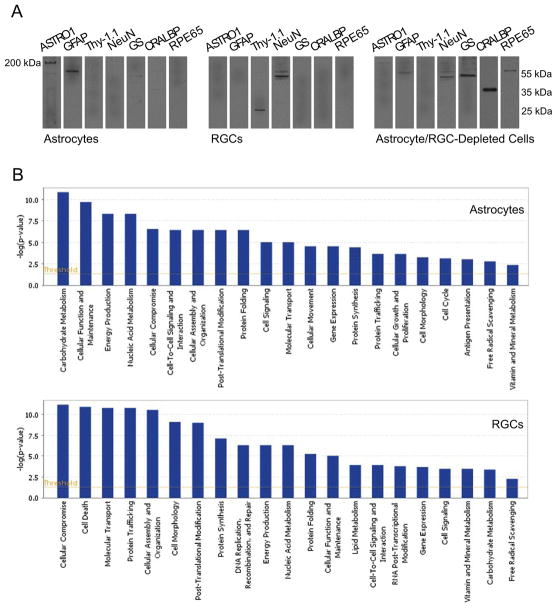
Mass spectrometry-based proteomics has been increasingly used for comprehensive understanding of the underlying molecular processes in glaucoma. Proteome analysis has provided both global information about alterations in protein expression and functional information about post-translational modifications and interactions of targeted proteins in glaucomatous samples. Besides animal tissues obtained from experimental models of glaucoma, a great deal of effort has been put into understanding of proteome changes in human glaucoma. Proteomics studies of human glaucoma have analysed diverse sample types, including retina, optic nerve, and trabecular meshwork tissues isolated from donor eyes, and aqueous humor and tear samples collected from patients. In addition to whole retinal proteins, enriched samples of RGCs and astrocytes have been analyzed to define cell-specific responses in animal models (Figure 1). This section will outline the contribution of proteomics studies to defining the molecular mechanisms in glaucoma. Figure 3 summarizes the related work in the field of glaucoma.
Figure 3.
Proteomics studies towards molecular mechanisms and biomarkers of glaucoma. A number of studies of human glaucoma and in vitro and in vivo experimental models have used proteomics analysis techniques. Distinct proteomics techniques have been applied to analyse diverse sample types, including optic nerve, retina, sclera, trabecular meshwork, aqueous humor, pseudoexfoliative material, tear, and blood. Ocular samples included tissue lysates unless indicated otherwise, such as cell cultures or enriched samples of RGCs or astrocytes. Blood samples included whole serum, IgG elutes, or isolated leukocytes. * indicates the studies including human samples. Proteomics analysis of glaucoma has resulted in the lists of differentially expressed proteins and contributed to current understanding of molecular mechanisms and biomarkers of glaucoma.
3.1.1. Proteomics findings in the retina and optic nerve
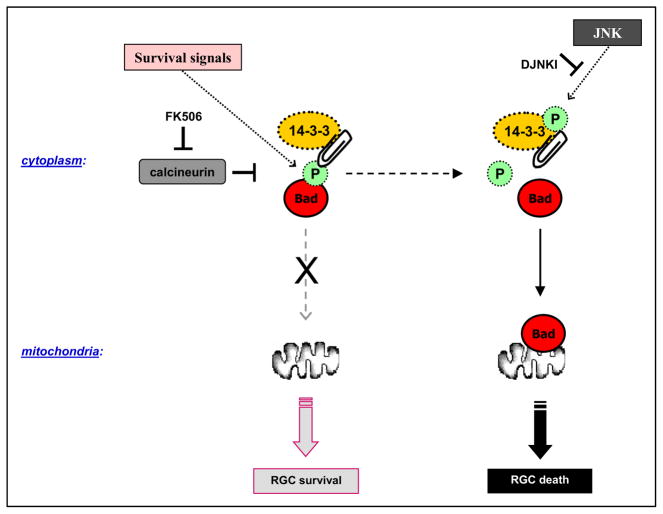
Our pioneering work for proteomics analysis of glaucomatous neurodegeneration has included the analysis of retinal proteome to characterize glaucoma-related alterations in human glaucoma and animal models. Our initial studies employed the conventional approach of 2D-SDS-PAGE followed by MALDI-TOF/MS or LC/MS/MS analysis to identify protein spots on 2D-gels. These studies complemented the proteomics analysis by other analysis techniques of biochemistry, cell biology, or molecular biology to validate and/or supplement the protein expression and relative quantification data. Through these studies using gel-based proteomics techniques, we identified oxidized (Tezel et al., 2005) and phosphorylated (Yang et al., 2008) retinal proteins in experimental rat glaucoma, and the analysis of 14-3-3 protein interactions presented a regulatory mechanism upstream of the mitochondrial cell death pathway in RGCs (reviewed in section 3.2).
Our following studies employed gel-free 2D-LC-MS/MS analysis for quantitative analysis of protein expression using 16O/18O labeling-based or label-free approaches. These comparative proteomics studies resulted in the identification of thousands of retinal proteins with high-confidence, including up-regulated or down-regulated proteins in human glaucoma and the rat model of ocular hypertensive glaucoma induced by hypertonic saline injections into episcleral veins. Findings of these studies led us to unexpected discoveries and molecular pathways to pursue for better understanding of pathogenic mechanisms. For example, we detected an erythrocyte protein, hemoglobin (Hb), in the human and rat retinal proteome, which exhibited up-regulation in glaucomatous samples (Tezel et al., 2010a). This observation motivated the studies focused on this important oxygen-binding protein to determine its relevance to glaucoma. First, RT-PCR and Western blot analyses of retinal protein samples, including the enriched samples of RGCs, along with the immunohistochemical analysis of histological sections from human and rat retinas, validated Hb expression in RGCs and astrocytes and a prominent up-regulation of this protein in glaucoma. Next, to determine whether Hb expression is regulated by ambient oxygen concentrations, we conducted in vitro experiments with primary cultures of RGCs and astrocytes incubated in the absence and presence of hypoxia. In addition, to determine the role of the HIF-1α/erythropoietin(EPO) signaling in Hb induction and cellular survival, we treated cell cultures with recombinant EPO and also used soluble EPO receptor to neutralize the EPO effects. These focused in vitro experiments revealed that hypoxia boosts glial Hb expression through the HIF-1α/EPO signaling in an autocrine manner and up-regulates Hb expression in RGCs in a paracrine manner, thereby providing an intrinsic mechanism to increase RGC survival. We also found that the up-regulated expression of Hb in glaucomatous human donor retinas and ocular hypertensive rat retinas is parallel to increased glial expression of EPO and up-regulation of EPO receptor on RGCs (Tezel et al., 2010a). Our earlier studies have also indicated up-regulation of HIF-1α in the glaucomatous human retina and optic nerve (Tezel and Wax, 2004). These findings provide new insights into how oxygen homeostasis is controlled in the inner retina through regulated expression of Hb in RGCs and astrocytes. Up-regulation of Hb in glaucoma appears to be an intrinsic compensatory mechanism facilitating the intracellular oxygen transport and perhaps also providing free radical scavenging.
Our proteomics studies of human glaucoma and animal models also identified various molecular components of the immune response pathways, which included complement activation, toll-like receptor (TLR) signaling, TNF-α/TNFR1 signaling, NF-κB activation, and inflammasome assembly. Regarding complement cascade, proteomics findings, also supported by retinal immunolabeling, revealed expression and differential regulation of different complement components, receptors, and regulators in human and rat glaucoma (Tezel et al., 2010c). Besides the classical complement pathway, differentially regulated complement components during glaucomatous neurodegeneration included those linked to the lectin pathway of complement activation. In addition, a major complement regulatory protein, complement factor H (CFH), exhibited a trend toward down-regulation in glaucomatous samples. When we focused on the regulation of CFH expression in accompanying in vitro experiments, we detected a prominent down-regulation of CFH expression in primary cultures of retinal cells exposed to H2O2-induced oxidative stress. This observation, along with the previous evidence of increased protein oxidation in the glaucomatous human retina (Tezel et al., 2007; Tezel et al., 2010c), support the likelihood of an intrinsic deficiency in the regulation of complement activation under oxidative stress in glaucomatous eyes. It is highly motivating to determine whether such a dysregulation in the complement-mediated tissue cleaning process may lead to secondary injury to neurons by uncontrolled complement attack in glaucoma.
Another outcome of our proteomics studies was differential regulation of TLR signaling molecules in human glaucoma. Immunohistochemical analysis supported the up-regulated expression of TLRs on retinal glial cells, and the bioinformatics knowledge-based analysis of the high-throughput proteomics data linked many adaptor proteins and kinases detected in the retinal proteome to the TLR signaling leading to immune responses (Luo et al., 2010). In addition, our accompanying in vitro experiments provided evidence that exogenously applied heat shock proteins and H2O2-induced oxidative stress up-regulate glial TLR expression, cytokine secretion, and antigen presentation, as assessed by stimulated proliferation and cytokine production of co-cultured T cells. When we used specific treatments inhibiting MyD88 and TIRAP (adaptor proteins involved in the TLR signaling) for functional testing, we detected a significant decrease in stimulated antigen presentation by heat shock proteins and oxidants (Luo et al., 2010). Thus, components of the tissue stress may serve as intrinsic ligands of TLRs in glaucoma and by detecting the danger signals arisen from stressed or injured RGCs, glial cells can initiate an immune response through TLRs. Based on the proteomics data, ongoing studies using transgenic mouse models functionally test the importance of TLR signaling components in the regulation of immune homeostasis in glaucoma.
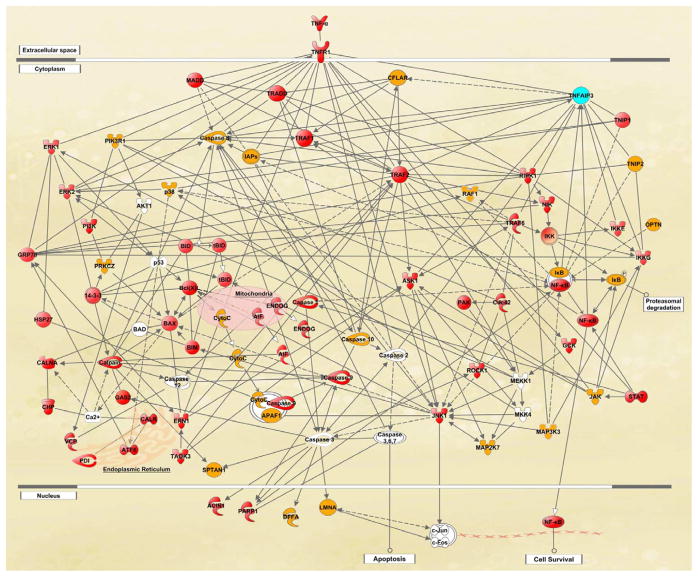
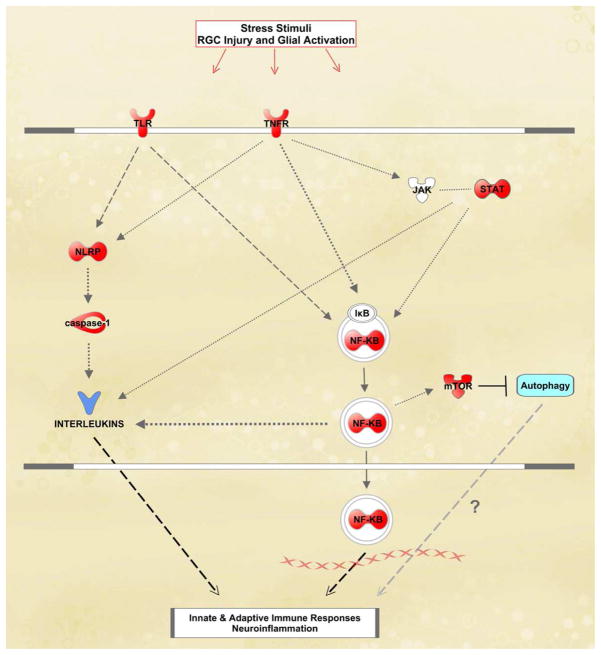
Our proteomics data collected from the analysis of glaucomatous human retina also highlighted a number of molecules linked to neurodegenerative and inflammatory pathways of TNF-α/TNFR1 signaling (Tezel, 2008a). Our high-throughput comparative data obtained by label-free quantitative 2D-LC-MS/MS analysis, along with the supportive findings of Western blot and immunohistochemical analyses, indicated a prominent up-regulation of apoptosis-related pathways and markers of inflammation in human glaucoma. The retinal proteins exhibiting increased expression in human glaucoma included TNF-α, TNFR1, and various downstream adaptor/interacting proteins and protein kinases known to regulate diverse consequences of TNF-α/TNFR1 signaling. Proteomics data supported co-activation of different apoptotic pathways in the glaucomatous human retina, including caspase- and calpain-mediated pathways, mitochondrial dysfunction, and endoplasmic reticulum stress, that may reinforce each other during the apoptotic process of glaucoma. Regarding the inflammation signaling, we detected NF-κB activation and various molecular components involved in JAK/STAT signaling and inflammasome formation in the glaucomatous human retina (Figure 4). By highlighting a number of signaling molecules and regulators involved in cell death and immune response pathways, the high-throughput proteomics data support that a complex cross-talk relationship between multiple signalling pathways determines diverse consequences of TNF-α/TNFR1 signaling in the glaucomatous human retina. Interestingly, the proteomics data from this study also led us to disclose a potential epigenetic modification, thereby expanding the informative scope of proteomics beyond conventional applications. The proteomics data indicating a prominent variability in the expression level of TNFAIP3 (a potent inhibitor of NF-κB activation and negative regulator of TNF-α signaling leading to apoptosis and inflammation) among glaucomatous donors motivated us to study the methylation pattern in the promoter region of TNFAIP3 using bisulfite sequence analysis. Findings of this analysis that correlated the differences in protein expression and promoter methylation encourage further research to determine whether an epigenetic modification alters an individual's response to TNF-α-mediated processes in glaucoma (Yang et al., 2011).
Figure 4.
Global proteomics analysis of the glaucomatous human retina. To determine alterations in retinal protein expression, retinal protein samples obtained from human donor eyes with or without glaucoma were analyzed by label-free quantitative 2D-LC-MS/MS analysis. The knowledge-based analysis of the generated high-throughput datasets established extended networks of diverse functional interactions between death-promoting and survival-promoting pathways and mediation of immune response. Up-regulated pathways included death receptor-mediated caspase cascade, mitochondrial dysfunction, endoplasmic reticulum stress, and calpains leading to apoptotic cell death; and NF-κB and JAK/STAT pathways, and inflammasome mediating inflammation. Proteins shown in red color exhibited significantly increased expression and shown in yellow color no significantly increased expression in glaucomatous samples relative to non-glaucomatous controls, while the protein shown in blue color exhibited prominent individual differences. Proteins shown in white color were not detectable by quantitative LC-MS/MS. This simplified network, generated using the Ingenuity Pathways Analysis, has recently been published (Yang et al., 2011).
Another proteomics study of the human retina focused on alterations in retinal protein expression in ocular hypertensive donors in comparison to glaucomatous donors and normotensive controls (Tezel et al., 2010b). The information gained from this study supported that a threshold of proteome alterations determines the initiation of neurodegenerative injury in glaucoma, which follows an initial period of the intrinsic defense response exhibiting individual differences. Findings of this study are unique to reveal ocular hypertension-related early molecular alterations in the human retina as opposed to alterations detected in eyes with clinically manifest glaucomatous damage and should be explored further.
A number of studies by other researchers have also used proteomics analysis techniques and provided valuable information illuminating different aspects of glaucoma. A proteomics study of the retina in rats with steroid-induced glaucoma used 2D-DIGE-based analysis coupled with MALDI-TOF/MS and revealed down-regulated expression of αA-crystalline (a molecular chaperone), superoxide dismutase 1 (an antioxidant enzyme), and triosephosphate isomerase 1 (a glycolytic enzyme), suggesting a potential vulnerability to oxidative stress (Miyara et al., 2008). Another study using a similar approach to DBA/2J mice detected differentially expressed 18 proteins, and based on in vitro data, correlated the down-regulated expression of integrin β7 to glutamate-induced death of RGCs (Kanamoto et al., 2009). The 2D-PAGE-based approach was also used to determine the effects of α2-adrenergic receptor agonist brimonidine tartrate on protein expression changes in cultured retinal pieces obtained from rats with or without episcleral vein cauterization-induced ocular hypertension. MALDI-TOF/MS analysis indicated treatment-related alterations in expression patterns of glial fibrillary acidic protein (GFAP), glucose-related protein 58, platelet-activating factor, and laminin-binding protein, thereby suggesting a role for α2-adrenergic receptor in retinal metabolism (Prokosch et al., 2010). Another study, also using the 2D-gel-based mass spectrometry approach, identified albumin as an abundant protein in the glaucomatous monkey retina and suggested that this antioxidant protein may provide an intrinsic protective mechanism against oxidative stress in experimental glaucoma (Carter-Dawson et al., 2010). 1D-gel-based MALDI-TOF/MS peptide mass fingerprinting was also used as a supplemental technique to demonstrate calpain-mediated cleavage of calcineurin in a rat model of experimental glaucoma (Huang et al., 2010b). In a more recent study, retinal proteome changes were studied in non-human primates by label-free quantitative LC-MS/MS analysis using an internal standard. By identifying over a hundred proteins, findings of this analysis indicated that early experimental glaucoma and optic nerve transection exhibit condition-specific alterations in protein expression. A prominent component of early retinal response to experimental glaucoma included up-regulation of proteins involved in the regulation of cytoarchitecture (Stowell et al., 2011).
A proteomics study of the optic nerves isolated from human donor eyes used 1D-gel-based LC-MS/MS analysis and detected 68 proteins, out of 250 proteins identified, only in glaucomatous samples, which included peptidyl arginine deiminase 2 (PAD2). This enzyme catalyzes a post-translational modification called deimination that refers to the conversion of protein bound arginine into citrulline. Subsequent immunoblotting verified the presence of PAD2 only in glaucomatous samples (Bhattacharya et al., 2006).
More recently, a proteomics study analyzed responses of cultured human optic nerve astrocytes to biomechanical strain. The identified pathways by LC-MS/MS iTRAQ included those previously implicated in the activation of astrocytes, such as TGFβ1 and TNF-α (Rogers et al., 2012).
Ongoing proteomics studies also analyze scleral protein expression and modifications to elucidate the corneoscleral connective tissue properties influencing the intraocular pressure-generated stress and its impacts on neuronal injury in experimental mouse glaucoma (Nguyen et al., 2012).
3.1.2. Proteomics findings in the trabecular meshwork, aqueous humor, and tear
Proteomics studies of glaucoma have also analyzed trabecular meshwork, aqueous humor, and tear samples to study intraocular pressure regulation mechanisms or topical treatment effects. Although these studies may not have direct implications for neurodegenerative mechanisms of glaucoma, they provide helpful information as reviewed below. Analysis of tear and aqueous humor is also advantageous for biomarker studies.
In a study of cultured human trabecular meshwork cells, 2D-gel-based analysis coupled with MALDI-TOF/MS determined responses of treatment with TGFβ1 and TGF β2. The treatment-related alterations in the expression of cytoskeleton proteins were suggested to be consistent with the potential effects of increased TGFβ expression in the glaucomatous trabecular meshwork (Zhao et al., 2004). Another study used LC-MS/MS iTRAQ to also analyze responses of cultured human trabecular meshwork cells to TGFβ2 and expanded the repertoire of proteins participating in the TGFβ2 signaling (Bollinger et al., 2011). In a follow-up study, the same group of researchers studied the responses of trabecular meshwork cell cultures derived from glaucomatous and normal donor tissues to dexamethasone treatment, and documented the similarities in proteome alterations induced by steroids and TGFβ2 (Bollinger et al., 2012).
In a different study, trabecular meshwork proteins obtained from glaucomatous donors and age-matched controls were analyzed by 2D-SDS-PAGE coupled with LC-MS/MS. This analysis demonstrated that cochlin, a protein associated with a deafness disorder, is present in the glaucomatous trabecular meshwork. Cochlin expression was also detected in the trabecular meshwork of DBA/2J mouse prior to intraocular pressure elevation and glaucoma development. Based on proteomics findings, in vitro cell aggregation, and immunohistochemical colocalization with mucopolysaccharide deposition, cochlin expression was suggested to contribute to intraocular pressure elevation in glaucoma (Bhattacharya et al., 2005). Using a similar 2D-gel-based approach for analysis of normal and glaucomatous human trabecular meshwork cells, another study indicated up-regulation of a calcium-dependent membrane-binding protein, copine1, and linked this protein to abnormal calcium signaling in the glaucomatous trabecular meshwork (Zhang et al., 2008).
A study of rats with steroid-induced ocular hypertension used 2D-DIGE and MALDI-TOF/MS to study the global protein expression profile in trabecular meshwork and detected down-regulation of type I collagen C-propeptides, thereby suggesting the impaired collagen turnover with steroid-treatment (Shinzato et al., 2007). A more recent study evaluated the effects of dexamethasone treatment on cytoskeleton changes in cultured human trabecular meshwork cells by comparative genomics and proteomics analysis. LC-MS/MS analysis of the cytoplasmic fraction of proteins identified 318 cytoskeleton-associated proteins. Based on spectral counting for label-free relative quantification, some of these proteins exhibited altered expression levels after dexamethasone treatment, with or without correlation to alterations in RNA expression levels analyzed by RT-PCR. It was suggested that the trabecular meshwork cytoskeleton is affected by dexamethasone treatment and the up-regulated cytoskeleton proteins may increase the cross-linked actin network formation (Clark et al., 2013).
Aqueous humor samples collected from patients with glaucoma and non-glaucomatous controls have also been analyzed using proteomics analysis techniques. In a study of aqueous humor samples collected from patients with primary open-angle glaucoma and control subjects, two different methods, protein chip array/SELDI-TOF and 2D-gel-based MALDI-TOF/MS, were used for analysis of protein expression. Differences detected in protein profiles between the glaucomatous and control samples included up-regulation of transthyretin that was previously shown to form amyloid deposits. Based on this observation, a similar mechanism to amyloid deposition was suggested as a potential mechanism causing the outflow obstruction and intraocular pressure elevation in glaucoma (Grus et al., 2008). A following study of aqueous humor samples through the 2D-gel-based analysis also indicated prominent differences in the aqueous humor protein composition between patients with glaucoma and non-glaucomatous controls (Duan et al., 2010). In addition, studies of aqueous humor samples using antibody microarrays supported dramatic variation of protein expression in glaucomatous samples in correlation to glaucoma-related neuronal damage (Izzotti et al., 2010), as well as anterior chamber endothelial damage (Sacca et al., 2012).
Gel-free LC-MS/MS analysis and label-free quantification has also been used to analyze aqueous humor samples from patients with primary congenital glaucoma. Many proteins exhibiting increased or decreased expression levels in congenital glaucoma compared to controls included the proteins linked to retinoic acid binding and transport, which have been implicated in the pathogenesis of Alzheimer's disease (Bouhenni et al., 2011). Direct LC-MS/MS analysis of aqueous humor samples collected from the buphthalmic rabbits also indicated differentially expressed proteins from a wide range of functional groups, including extracellular matrix modulation, regulation of apoptosis, oxidative stress, and protein transport (Edward and Bouhenni, 2011).
Analysis of aqueous humor samples to study intraocular pressure regulation in a rabbit model detected autotaxin, a major source for extracellular lysophosphatidic acid. Label-free quantitative LC-MS/MS analysis similarly detected autotaxin as an abundant protein in the glaucomatous human aqueous humor, and additional analysis indicated elevated lysophospholipase activity in these samples. Subsequent in vitro and in vivo experiments further explored the role of autotaxin in glaucoma. Based on intraocular pressure lowering response to topical treatment with a small molecule inhibitor of autotaxin in rabbits, and biochemical and structural alterations after siRNA suppression of autotaxin expression in cultured trabecular meshwork cells, the autotaxin/lysophosphatidic acid axis was suggested as a potential therapeutic target for lowering of intraocular pressure in glaucoma (Iyer et al., 2012).
In a study of pseudoexfoliation glaucoma, the pseudoexfoliative material isolated from the surgically removed anterior lens capsule was analyzed to identify protein constituents. LC-MS/MS analysis identified LOXL1, genetically associated with the risk of pseudoexfoliation syndrome, as well as other proteins in the pseudoexfoliative material (Sharma et al., 2009). More recently, tear samples were analyzed by LC-MS/MS using iTRAQ or label-free techniques for quantification. These studies have focused on profiling the inflammatory signaling molecules in tear samples collected from patients on anti-glaucoma medication and detected treatment-related alterations in the tear protein profile in patients with primary open-angle or pseudoexfoliative glaucoma (Wong et al., 2011; Pieragostino et al., 2012).
3.2. Targeted proteomics analysis of glaucoma
In addition to alterations in the expression level of numerous proteins in glaucoma, growing evidence supports post-translational modifications of proteins governing the molecular processes of glaucomatous neurodegeneration. For example, many retinal proteins undergo proteolytic cleavage during glaucomatous neurodegeneration as assessed by Western blot analysis and immunohistochemical labeling with cleavage site-specific antibodies (Tezel and Wax, 1999; Tezel and Wax, 2000; McKinnon et al., 2002; Huang et al., 2010b). As reviewed below, targeted proteomics studies characterize various other post-translational modifications in glaucoma, including protein phosphorylation and oxidation. Many other studies not included in this review have also analyzed protein phosphorylation using phosphorylation site-specific antibodies in Western blot analysis or immunohistochemical analysis of in vivo or in vitro models of glaucoma. It is worth noting that analysis of the proteome, not the transcriptome, is the efficient strategy to identify signaling molecules that function after post-translational modifications (Ideker et al., 2001; de Godoy et al., 2008). This is also supported by the inconsistency detected between mRNA and protein expression in stressed RGCs and glia (Tezel and Yang, 2005). Particularly the studies of protein phosphorylation highly promise to drive the glaucoma research toward improved understanding of specific signaling pathways.
3.2.1. Post-translational protein modifications
In a targeted proteomics study to identify phosphorylated retinal proteins in experimental glaucoma, we used a 2D-gel-based approach to detect phosphoproteins by Pro-Q Diamond staining of 2D-gels followed by Sypro Ruby staining for total protein profiling. For large-scale relative quantification, differential display analysis compared the intensity of Pro-Q Diamond-staining at matching spots on 2D-gel images from ocular hypertensive and control samples after spot normalization to the protein content measured by the intensity of Sypro Ruby staining. This analysis revealed over a hundred spots exhibiting a new or increased phosphorylation in ocular hypertensive samples. These protein spots were then identified by MALDI-TOF/MS and LC/MS/MS analyses. The proteins identified with significant matches included those involved in cell signaling and stress response (Yang et al., 2006). Different isoforms of 14-3-3 family of proteins were among the proteins exhibiting over two-fold increased phosphorylation in ocular hypertensive samples. 14-3-3 phosphorylation was also validated by an acidic shift of the 14-3-3 protein spot on 2D-gels and 2D-Western blots of ocular hypertensive samples relative to controls (Yang et al., 2008).
Our proteomics studies also assessed retinal protein oxidation by measuring the protein carbonyl content through the 2D-oxylot analysis coupled with LC-MS/MS. This analysis revealed increased protein oxidation in experimental glaucoma and identified retinal proteins targeted by carbonyls (Tezel et al., 2005). We detected approximately 60 protein spots, out of more than 400 spots, exhibiting a significant increase in protein carbonyl immunoreactivity in ocular hypertensive samples compared to controls. Among the oxidized retinal proteins detected on oxyblots, approximately 20 spots exhibited new protein carbonyls in ocular hypertensive samples. MALDI-TOF/MS and LC/MS/MS analysis of sister gel pieces corresponding to the immunoreactive spots identified target proteins of oxidation, which included glyceraldehyde-3-phosphate dehydrogenase, a glycolytic enzyme, HSP72, a stress protein, and glutamine synthetase, an excitotoxicity-related protein. Besides Western blots validating the proteomics data, retinal immunolabeling indicated increased anti-carbonyl reactivity in inner retinal cells, including RGCs, in ocular hypertensive rat eyes. Parallel to these observations in experimental glaucoma, we also detected increased retinal protein carbonyl immunolabeling in the glaucomatous human glaucoma, predominantly in the inner retinal layers. Protein carbonyls were co-localized with AGEs that exhibit enhanced accumulation in human glaucoma relative to age-matched controls (Tezel et al., 2007). In another study, Western blot analysis of human retinal protein samples obtained from 10 donors with glaucoma and 10 age-matched controls without glaucoma indicated a prominent increase in 4-hydroxy-2-nonenal (HNE) immunolabeling of glaucomatous samples compared to controls, which reflects protein modification induced by oxidative stress by-products (Tezel et al., 2010c). As reviewed in section 4.2, in a more recent study of serum samples, we analyzed protein oxidation by setting a search parameter to identify a mass shift corresponding to addition of the oxidation moiety to methionine (Tezel et al., 2012b).
Based on the evidence of an accelerated accumulation of AGEs in the glaucomatous human retina and optic nerve (Tezel et al., 2007), we also studied glycosylated retinal proteins in rats with experimentally induced intraocular pressure elevation or streptozosin-induced diabetes (Atmaca-Sonmez et al., 2006). The number and intensity of glycoprotein spots detected by the Pro-Q Emerald glycoprotein staining of 2D-gels (Steinberg et al., 2001; Wu et al., 2005) were greater in diabetic retinas relative to controls. In contrast to controls, increased glycoprotein staining was also detectable in ocular hypertensive retinas at over 20 spots shared with diabetic retinas. Cytoskeleton proteins, including α-actin and β-tubulin, as well as some glycolytic enzymes and signaling molecules, were identified at the glycoprotein spots by MALDI-TOF/MS and LC/MS/MS analysis
Many other researchers have also analyzed glaucoma-related post-translational modifications of proteins using immunoprecipitation and gel-based analysis by mass spectrometry. For example, in a study of the optic nerves isolated from human donor eyes, Western blot analysis and immunohistochemical analyses using specific antibodies indicated increased citrullination and decreased arginyl methylation in glaucoma. After anti-citrulline immunoprecipitation, 1D-gel-based LC-MS/MS analysis identified myelin basic protein as a major citrullinated protein in optic nerve samples from glaucomatous donors. This study also provided in vitro evidence of pressure-induced translational control of PAD2 expression in optic nerve astrocytes, suggesting a possible role for the enzyme in citrullination and structural disruption of myelination in the glaucomatous optic nerve (Bhattacharya et al., 2006).
Another study using Western blot analysis showed iso[4]LGE2-modified lipid oxidation products in astrocyte cultures derived from rat brain cortex and human optic nerve after exposure to elevated pressure, in vitro. In addition, after 1D-SDS-PAGE separation of the proteins co- immunoprecipitated with anti-iso[4]LGE2 antibody, the excised protein bands were analyzed by mass spectrometry. This analysis of cultured astrocyte proteins resulted in a list of 21 proteins as potential targets of oxidative modification. In addition, in vitro treatment with pyridoxamine, a pharmacological inhibitor of iso[4]LGE2 formation, protected astrocyte proteins against modification by pressure-induced iso[4]LGE2 (Govindarajan et al., 2009).
A study of cat eyes aimed to determine the proteome alterations induced by laser trabeculoplasty that is a commonly used treatment modality for glaucoma. Using 1D-SDS-PAGE, glycoprotein gel staining, and mass spectrometry, this study found protein expression changes and elevated glycosylation of three proteins, biglycan, keratocan, and prolargin, in the laser-treated trabecular meshwork (Amelinckx et al., 2009).
The more recent study of dexamethasone treatment effects on cultured human trabecular meshwork also analyzed treatment effects on the phosphorylation state of proteins. For this purpose, phosphopeptides were enriched using metal oxide affinity chromatography, and the following direct LC-MS/MS analysis provided a list of cytoskeleton proteins containing phosphorylated residues (Clark et al., 2013).
It is very likely that the reviewed examples of glaucoma-related post-translational modifications are just the tip of the iceberg. More systematical analysis should enhance our current understanding of protein modifications and their implications in pathogenic mechanisms and deliver new molecular targets for improved treatment of glaucoma.
3.2.2. Protein interactions
To explore the functional importance of 14-3-3 protein detected by phosphoproteomics analysis of the glaucomatous retina, we employed a targeted proteomics approach in an experimental rat model of glaucoma (Yang et al., 2008). For analysis of the interaction partners of 14-3-3, we eluted 14-3-3-containing protein complexes by two different approaches, co-immunoprecipitation using a 14-3-3 antibody and recombinant protein-based affinity pull-down using GST-tagged 14-3-3 protein. Eluted proteins were then subjected to gel-free 2D-LC-MS/MS analysis. Identified proteins included pro-apoptotic members of the Bcl-2 family, such as Bad, and many other proteins, such as calmodulin, zinc-finger proteins, ubiquitins, protein kinase C, and HSP70 (Yang et al., 2006). Western blot analysis of subcellular fractions and immunolabeling of retinal tissue sections with phosphorylation site-specific antibodies indicated that phosphorylated Bad normally remains sequestered in the cytoplasm by 14-3-3 scaffold. However, 14-3-3 phosphorylation and Bad de-phosphorylation in ocular hypertensive eyes lead to Bad translocation into mitochondria, where its binding to Bcl-2/Bcl-x(L) is known to initiate the mitochondrial apoptosis pathway. To further determine the association of 14-3-3 protein with cell death signaling in RGCs, we conducted a series of in vivo treatment experiments for functional testing in experimental rat glaucoma. Treatments inhibiting 14-3-3 phosphorylation (DJNKI) and Bad dephosphorylation (FK506) maintained the cytoplasmic 14-3-3/Bad interaction and resulted in decreased neuronal damage in ocular hypertensive rat eyes. These experimental findings support that 14-3-3 is involved in the control of subcellular localization and function of Bad in a phosphorylation-dependent manner (Figure 5). Since the Bcl-2 family proteins control the mitochondrial pathway of RGC death in glaucoma (Nickells et al., 2008; Nickells, 2010), the upstream 14-3-3 scaffolding may be a promising treatment target for neuroprotective interventions (Yang et al., 2008).
Figure 5.
Targeted proteomics analysis of experimental rat glaucoma. As an effort to explore specific protein interactions in an experimental rat model of glaucoma, 14-3-3-containing retinal protein complexes were eluted using co-immunoprecipitation and recombinant protein-based affinity pull-down for subsequent analysis by mass spectrometry. Based on the proteomics data and in vivo treatment experiments in rats, 14-3-3 proteins were found to control the subcellular localization of and function of Bad in a phosphorylation-dependent manner, and thereby constitute an important regulatory pathway of RGC death signaling during glaucomatous neurodegeneration. This flow diagram has recently been published (Yang et al., 2008).
We use bioinformatics tools to analyze the high-throughput datasets of identified proteins and their relative expression values in glaucomatous versus non-glaucomatous samples. This analysis defines functional patterns within the specific proteome composition and generates knowledge-based protein interactions networks relevant to glaucoma. Using the Ingenuity Pathways Analysis (Ingenuity® Systems, Mountain View, CA), recent studies revealed a number of canonical pathways most significantly associated with our high-throughput datasets and populated high-probability networks for complement activation (Tezel et al., 2010c), TLR signaling (Luo et al., 2010), and neurodegenerative and inflammatory components of TNF- a/TNFR signaling (Yang et al., 2011) in human glaucoma. These protein interactions networks generated based on the existing repository of knowledge can provide guidelines for proteins to pursue in future functional studies for gaining further information about the pathogenic importance of specific molecular pathways.
3.3. Cell-specific proteomics analysis of glaucomatous neurodegeneration
The complexity of retina and optic nerve tissues is a standing challenge of the glaucoma research aiming to identify pathogenic mechanisms of RGC death and axon loss. This is because the analysis of whole retina or optic nerve may only reflect the sum of opposing responses from many different cell types. To speed the scientific understanding of specific molecular responses during glaucomatous neurodegeneration, cell-specific analysis is essential. Undoubtedly, RGC axons, somas, and synapses are specific victims of neurodegeneration in glaucoma. In respect to diverse roles of glial cells, either neurosupportive or neurodestructive, analysis of glial responses constitutes another major focus of the glaucoma research to better understand neurodegenerative mechanisms (Tezel, 2009). The substantial need for dissection of the complexity of data obtained from whole tissue led us to isolate cell-specific samples to better understand alterations in protein expression during glaucomatous neurodegeneration. Our cell-specific proteomics studies that began in 2004 initially analyzed enriched samples of RGCs (Yang and Tezel, 2005; Tezel, 2008b; Yang et al., 2008; Tezel et al., 2010a; Tezel et al., 2012c), and more recently included isolated samples of astrocytes (Tezel et al., 2012c). Since human donor samples are not available in quantities sufficient for cell-specific sampling, or histological tissue sections are the preferable sampling strategy for human tissues which can be stored over years for multiple analyses, these studies mainly included animal models.
3.3.1. Proteomics analysis of RGC responses
For cell-specific proteome analysis, we isolate RGCs through an immunomagnetic cell separation technique originally established for RGC cultures (Tezel and Wax, 2000). This cell isolation process that can be completed within approximately one hour presents advantages over alternative techniques, such as laser-capture microdissection, that require much longer and complicated processes. Most importantly, the protein yields after immunomagnetic cell separation allow direct proteome analysis in pooled samples (Tezel et al., 2012c). RGCs isolated by this technique were verified in multiple studies by retrograde fluorescence labeling (Tezel and Wax, 2000), cell morphology and immunolabeling in culture (Tezel and Wax, 2000), and RT-PCR (Tezel et al., 2010a) or Western blot analysis of specific cell markers (Tezel, 2008b; Tezel et al., 2012c). Figure 6 shows most recent Western blots of enriched RGC samples with specific antibodies to NeuN (a neuronal marker) and Thy-1.1 (a marker for RGCs).
Figure 6.
Cell-specific proteomics analysis of glaucoma. A. Western blot analysis using antibodies to specific cell markers validated enriched samples of retinal astrocytes and RGCs through the immunomagnetic cell isolation process. Only neuronal markers (NeuN and Thy-1.1) were detectable in RGC samples, while astrocyte samples exhibited prominent immunolabeling for astrocyte markers, ASTRO1 and GFAP. Despite faint immunoreactivity of enriched astrocyte samples for GS (a marker for Müller cells) and CRALBP (expressed in Müller cells and retinal pigment epithelium), these samples were negative for RPE65, a marker for retinal pigment epithelium or neuronal markers. Presented data represent three independent sets of analyses with different samples. B. Bioinformatics analysis of the comparative proteomic data by the Ingenuity Pathways Analysis supported distinct responses of astrocytes and RGCs in ocular hypertensive rat eyes. Shown are functional groups of the up-regulated proteins in ocular hypertensive astrocytes and RGCs relative to their controls. While ocular hypertensive samples of RGCs predominantly exhibited stress response and cell death signaling, cellular activation and immune/inflammatory responses were most prominent in ocular hypertensive astrocytes. This figure has recently been published (Tezel et al., 2012c).
Gel-based or gel-free LC-MS/MS analysis of RGC-specific samples using 16O/18O labeling-based or label-free quantification techniques in multiple studies identified thousands of proteins with high confidence. In addition, quantitative mass spectral data indicating differential responses of ocular hypertensive RGCs and astrocytes supported the feasibility of cell-specific sampling and verified the sensitivity of cell-specific analysis to characterize distinct cellular responses in experimental glaucoma (Yang and Tezel, 2005; Tezel, 2008b; Yang et al., 2008; Tezel et al., 2010a; Tezel et al., 2012c). To provide overall information about our cell-specific high-throughput data, Figure 6 shows the functional groups of up-regulated proteins in ocular hypertensive RGCs and astrocytes. RGCs predominantly exhibited stress response and cell death signaling as opposed to cellular activation and immune/inflammatory responses of astrocytes in ocular hypertensive retinas.
A following report has validated the feasibility of global proteomics analysis of RGCs enriched by immunopanning. Among the 268 proteins quantified by LC-MS/MS iTRAQ, approximately 8% exhibited increased and 13% decreased expression after 8 weeks of induced ocular hypertension. Voltage-dependent anion channel protein 2, aldose reductase, and ubiquitin were among the proteins with significantly increased expression in glaucomatous samples, while the expression of prothymosin was decreased in these samples (Crabb et al., 2010).
By pursuing the outcome molecules in focus-in studies for functional testing, cell-specific proteomics should allow new molecular targets to be identified for cell-specific treatments to improve neuronal survival and function in glaucoma.
3.3.2. Proteomics analysis of glial responses
Glial cells, including astrocytes that are the most abundant glia in the retina and optic nerve, generate both neurosupportive and neurodestructive effects in glaucoma, and gial activation response and secondary neuroinflammatory processes are regarded as continuous components of glaucomatous neurodegeneration. It is commonly viewed that better characterization of glial responses, which can be possible by proteomics analysis, can help therapeutically manipulate neurodestructive consequences of glial responses and enhance glial homeostastic functions (Tezel, 2009; Tezel, 2011).
Using a technique similar for selection of RGCs, we isolated enriched samples of retinal astrocytes for cell-specific proteome analysis in experimental rat glaucoma. As shown in Figure 6, Western blot analysis using specific antibodies to astrocyte markers, such as GFAP and ASTRO1, verified enriched samples of astrocytes, while neuronal markers, including NeuN and Thy-1.1, were not detectable in these samples. Astrocyte samples were only faintly immunoreactive for GS (a marker for Müller cells) and CRALBP (expressed in Müller cells and retinal pigment epithelium) and negative for RPE65 (a marker for retinal pigment epithelium) (Tezel et al., 2012c). Isolation of enriched astrocyte samples allows direct analysis of the astrocyte proteome by avoiding any potential phenotypic changes in cell culture models.
The quantitative LC-MS/MS analysis of enriched astrocyte samples identified over two thousand proteins, up-regulated or down-regulated in ocular hypertensive samples, characterizing the distinct responses of astrocytes in experimental glaucoma (Tezel et al., 2012c). As summarized in Figure 6 the functional groups of up-regulated proteins in ocular hypertensive astrocytes included immune mediators/regulators, such as a number of downstream proteins linked to TNF-α/TNFR signaling, NF-κB activation, JAK/STAT signaling, TLR signaling, and inflammasome formation. These molecular components appear to be co-players of neuroinflammatory responses mediated by ocular hypertensive astrocytes (Figure 7). In addition, cell-specific proteomics data provided clues about up-regulation of autophagy signaling during glaucomatous neurodegeneration. Based on mass spectral data, Western blot analysis, and retinal immunolabeling, phosphorylation-mediated activation of mTOR, an upstream negative regulator of the autophagy signaling (Jung et al., 2010; Yang and Klionsky, 2010), was most prominently detectable in ocular hypertensive astrocytes. Although autophagic activation is a cell-protection mechanism, under specific conditions it may become an alternative cell-death mechanism (Gump and Thorburn, 2011) or induce autoimmunity (Deretic, 2009). Our findings therefore motivate further research to answer many arising questions as to whether NF-κB-dependent activation of mTOR controls the homeostatic balance between activator and inhibitor pathways of autophagy in astrocytes, or whether a potential dysregulation of autophagy compromises immune homeostasis in glaucoma (Tezel et al., 2012c).
Figure 7.
I nflammatory responses of astrocytes in ocular hypertensive rat retinas. In this simplified network generated by The Ingenuity Pathways Analysis, astrocyte proteins shown in red color exhibited significantly increased expression in ocular hypertensive samples relative to normotensive controls. Based on the high-throughput proteomics data validated by Western blot analysis and immunohistocemical labeling, TNF-α/TNFR signaling, NF-κB activation, JAK/STAT signaling, TLR signaling, and inflammasome appear to be co-players of inflammatory responses mediated by ocular hypertensive astrocytes. This figure has recently been published (Tezel et al., 2012c).
Impending functional testing in transgenic models is expected to translate the cell-specific proteomics data into improved understanding of glial molecular responses, including various immune mediators/regulators as potential treatment targets to provide immunomodulation in glaucoma.
3.4. Challenges in proteomics studies of glaucomatous neurodegeneration
Since neurodegeneration depends mostly on post-transcriptional processes, proteomics presents a powerful toolbox to elucidate molecular mechanisms of injury to RGCs and optic nerve axons in glaucoma. As reviewed in section 2, there are inherent challenges in mass spectrometry-based proteomics analysis techniques with regards to sensitivity (dynamic range), reproducibility, and comprehensiveness; however, many tools are available to assess the quality of the data, and technological advances promise to increasingly overcome current limitations.
Besides technical drawbacks, proteomics analysis suffers from the molecular complexity of neurodegenerative mechanisms in glaucoma. Evidently, pathogenic mechanisms of glaucomatous neurodegeneration are governed by spatially and temporally-regulated interplay of proteins with genes, other proteins, and environmental stress. Biological variability may be particularly important when human samples are analyzed due to patients’ heterogeneity. Studies of human tissues, although particularly important, rely on the availability of well-characterized patient material in eye banks and the ethical and legal regulations of the use of donor tissues. Data interpretation and determining the effects of confounding factors, such as other disease conditions or treatments, may also be challenging due to retrospective data collection. Despite excellent maintenance of tissue structure and no evidence for cellular autolysis, the possibility of perimortem tissue alterations may not fully be ruled out. To minimize such vulnerabilities, glaucomatous and non-glaucomatous samples should be matched for many parameters, such as donor age, cause of death, and postmortem period till tissue sampling. On the other hand, animal models are valuable research tools; however, the results from studies of experimental models cannot always be extrapolated to human glaucoma (Levin, 2004; McKinnon et al., 2009).
Concerning the high molecular complexity of retina and optic nerve tissues, the coverage of protein identification is particularly important in the field of glaucoma research. Although it is not without its shortcomings, 2D-LC-MS/MS approaches with continuously increasing sensitivity and speed have significantly aided protein identification coverage for downstream quantification studies (Domon and Aebersold, 2006; Kocher and Superti-Furga, 2007; Nilsson et al., 2010). As previously noted, measurement of multiple peptides from the same protein enhances the confidence of protein quantification. Consequently, the precision of quantification is higher for high-abundant proteins than low-abundant proteins, since they have more quantifiable peptides. Therefore, quantification software should determine the significance of a measured fold-change in the context of absolute protein abundance. More samples may be required for repeated analyses as well as for sample enrichment to increase the sensitivity to identify and quantify low-abundant proteins. Based on the accuracy of mass spectrometry-based quantification, a minimum of 1.3-fold to 2-fold expression change is meaningful for quantitative proteomics analysis as a cut-off for biological significance (Mann and Kelleher, 2008). The sample size should be adjusted according to the required statistical power of an experiment for detection of significant differences. Since small changes in protein expression and modification may be obscured by the background noise, the use of biological and technical replicates, and validation of mass spectral data using alternative analysis techniques of biochemistry and molecular biology are also critically important for data interpretation. Thus, experimental design for a proteomics study should consider the realistic balance between the desired power, the existing amount of samples, and the availability and cost of resources, including the type of equipment, software, and database searching tools.
Apart from the large number of proteins spanning a wide dynamic range in abundance in retina or optic nerve proteome, cellular complexity of retina and optic nerve tissues makes this difficulty even a larger challenge for glaucoma research. As opposed to analysis of the whole tissue that complicates data interpretation, cell-specific analysis is particularly important to reduce sample complexity, increase the sensitivity of detection, and improve the informative value of the proteomics data. However, cell-specific analysis requires additional isolation steps. Even the higher protein yields of immunomagnetic cell isolation through a shorter and simpler process, may require sample pooling to collect sufficient amount of protein sample (Tezel et al., 2012c). Collecting cell-specific protein samples may be problematic particularly in glaucomatous samples, since sufficient sampling of RGCs, but not glia, needs increasing number of ocular hypertensive eyes depending on the extent of neuronal injury during the experimental paradigm. Since RGC isolation is based on the cell surface expression of Thy1.1 (Tezel and Wax, 2000), decreased Thy1.1 expression prior to significant cell loss after intraocular pressure elevation in experimental models (Schlamp et al., 2001; Huang et al., 2006) may also amplify this difficulty.
It is also worth noting that the analysis of dynamic post-translational modifications, such as phosphorylation, requires fresh samples. Long-term sample storage may also cause difficulties for analysis of other modifications known to be more stable. Even carbonyl groups targeted for analysis of protein oxidation may undergo Schiff base formation with lysine residues on proteins, and major amounts of carbonylated proteins may be lost in stored samples (Madian and Regnier, 2010).
In summary, the sample type, proteomics analysis workflow, data analysis, and validation are critical steps determining the success of proteomics analysis. Despite several challenges, isotope labeling-based and label-free techniques of quantitative proteomics have been increasingly used in the field of glaucoma research. These analyses have provided with the lists of differentially expressed or modified proteins in glaucomatous samples and enhanced our knowledge beyond that achieved by earlier methods. By using latest generation mass spectrometers with increasing resolution, mass accuracy, dynamic range, and sequencing speed together with advances made to analyze post-translational modifications and protein interactions, valuable applications of proteomics in the field will grow even further. The information gained from proteomics analysis will continue to lay the foundation for more focused studies using loss-of-function analysis to delineate the molecular mechanisms of glaucomatous neurodegeneration and define molecular targets for new treatment strategies.
4. Proteomics toward clinical biomarker discovery in glaucoma
4.1. Overview of molecular biomarker discovery
Molecular biomarkers are in strong demand to help diagnose glaucoma early, assess risk profiles for disease progression, and monitor treatment responses of patients with glaucoma. Prediction of conversion from ocular hypertension to primary open-angle glaucoma or from pseudoexfoliation syndrome to pseudoexfoliative glaucoma is particularly important to halt disease progression. Besides glaucoma prediction, monitoring the treatment responses is a major need, particularly in clinical trials of imminent neuroprotective treatments. As discussed at the ARVO/Pfizer Ophthalmic Research Institute Conference, 2011 (Bhattacharya et al., 2013), development of clinically useful molecular biomarkers for glaucoma is an area of active investigation, and proteomics is at the center of these activities. Since the blood can be sampled easily, identification of biomarkers that can be measured in blood samples is a logical choice. Consequently, the proteins detectable in serum or plasma have formed the basis of commonly used tests to screen and monitor disease biomarkers in various fields. In addition to blood samples, analysis of more proximal fluids, such as tear or aqueous humor, may also be useful, or even more sensitive and specific, for biomarker applications in glaucoma.
Earlier studies comparing the measurements of individual proteins in normal and diseased blood samples using specific immunoassays (Sugiyama et al., 1995; Tezel et al., 1997; Leibovitch et al., 2005; Huang et al., 2010a) may help efforts to identify candidate biomarkers for glaucoma. However, most of the work that benefits for molecular biomarker discovery in glaucoma comprises the studies of autoantibodies and their target antigens. A number of techniques, including serological identification of antigens by recombinant expression cloning (SEREX), serological proteome analysis (SERPA), protein arrays, and open reading frame phage display, have been used to identify disease-associated antigens and their cognate autoantibodies in multiple diseases (Desmetz et al., 2009; Georgieva and Konthur, 2011; Kim et al., 2011). To harness the immune response appears to be a promising approach for molecular biomarker discovery in glaucoma, since a panel of antigenic proteins that elicit serum immunoreactivity at a high frequency among glaucoma patients can provide an effective tool for biomarker screening. Indeed, protein arrays containing a repertoire of proteins have been used to provide the molecular signature for immune responses in patients with glaucoma (Dervan et al., 2010; Boehm et al., 2012b). The potential usefulness of serum antibodies as biomarkers is supported by the unique antibody pattern among glaucoma patients, which exhibits specificity and sensitivity of approximately 93% (Boehm et al., 2012b), and the similarities in complex antibody profiles among different ethnic populations (Wax et al., 2001; Grus et al., 2006). An important challenge of serum biomarker detection is related to much lower abundance of most protein biomarkers than some disease-irrelevant serum proteins. Concerning this challenge, antibody response also holds the advantage of signal amplification for biomarker discovery in glaucoma. However, compared to protein arrays that contain limited number of proteins, which are pre-chosen, the mass spectrometry-based techniques offer a high-throughput analytical approach to glaucoma biomarker discovery.
The data obtained from proteomics, and also genomics, studies of human glaucoma or animal models can guide molecular biomarker discovery through the “targeted” approach. For example, LC-MS/MS analysis was used to measure serum levels of dimethylated isomeric derivatives of the amino acid L-arginine in patients with glaucoma and non-glaucomatous controls and detected elevated levels of dimethylarginines in glaucomatous samples (Javadiyan et al., 2012). However, availability of high resolution instruments, improved protein separation strategies, and simplified quantification algorithms increase the sensitivity and coverage of mass spectrometry to identify biomarker candidates through the "de novo" discovery approach (Liotta et al., 2003; Hanash et al., 2008; Filiou and Turck, 2011). The study of modified proteins as a source of potential glaucoma biomarkers can also be possible by proteomics analysis techniques (Liotta et al., 2003; Hanash et al., 2008; Filiou and Turck, 2011).
As reviewed in section 4.2, preliminary proteomics studies have resulted in the lists of potential biomarker candidates for glaucoma to be validated in larger patient populations. Figure 3 summarizes the studies aiming to identify molecular biomarkers of glaucoma. However, it should be recognized that the clinical validation phase after discovery of candidate molecules remains a major standing challenge common for the biomarker discovery through different approaches.
4.2. Serum biomarker candidates identified by antibody-based immunoproteomics or direct proteomics analysis
A traditional approach to identify autoantigens in glaucoma has involved separation of retina or optic nerve proteins by 1D- or 2D-SDS-PAGE, and after probing with the sera collected from patients with glaucoma, analysis of the immunoreactive bands or spots to identify proteins by mass spectrometry (Yang et al., 2001; Li et al., 2006; Joachim et al., 2008; Reichelt et al., 2008). A number of studies have also used protein arrays (Dervan et al., 2010; Boehm et al., 2012b). Another immunoproteomics approach has been the isolation of autoantibodies from the patient serum for either direct analysis of IgG elutes to identify co-eluted proteins by mass spectrometry, or affinity enrichment of antigens in targeted tissue extracts for following characterization. In a recent study, we employed an antibody-based immunoproteomics approach for high-throughput characterization of potential biomarker candidates for glaucoma. Serum samples were collected from 111 patients with primary open-angle glaucoma and an age-matched control group of 49 healthy subjects without glaucoma. We eluted serum IgG from five randomly selected samples from each group using Melon gel spin columns or protein G-Dynabeads. After verification of IgG elutes, LC-MS/MS analysis of IgG-containing serum fractions identified hundreds of proteins. As a narrowing down strategy to reduce the list of candidate autoantigens, we determined whether the mass spectral data obtained from the glaucomatous serum IgG elutes overlapped with our high-throughput proteomics data obtained from the glaucomatous human retina (Luo et al., 2010; Tezel et al., 2010a; Tezel et al., 2010c; Yang et al., 2011). Ninety-six proteins overlapped between the serum and retinal proteomics data. Table 1 lists fifty of these proteins which are also known to be present in the glaucomatous human retina. We selected ten of these proteins as candidate biomarkers for initial validation studies and measured their serum titers in all of our glaucomatous and control samples by specific ELISAs. Four of these candidates (AIF, CREB-binding protein, ephrin type-A receptor, and huntingtin) exhibited higher ELISA titers in the glaucomatous sera (Tezel et al., 2012b). Particularly respecting the individual variability detected in in serum titers of these proteins, once the clinical correlations of potential biomarker candidates are established, a set of biomarkers in an array format, rather than a single molecule, may potentially be more useful as a diagnostic and/or prognostic clinical tool in glaucoma.
Table 1.
Serum proteins co-eluted with IgG
| Protein Name | Accession Number |
|---|---|
| integrin alpha-9 precursor | gi|52485941 |
| disintegrin and metalloproteinase domain-containing protein 9 precursor * | gi|4501915 |
| neural cell adhesion molecule 2 precursor | gi|33519481 |
| SH3 and multiple ankyrin repeat domains protein 3 * | gi|122937241 |
| extended synaptotagmin-1 isoform 2 * | gi|14149680 |
| SLIT and NTRK-like protein 3 precursor | gi|40217820 |
| microtubule-actin cross-linking factor 1 isoform b | gi|33188443 |
| calmodulin-regulated spectrin-associated protein 2 | gi|44955929 |
| vesicle-associated membrane protein 7 isoform 3 * | gi|297747292 |
| synphilin-1 | gi|76563940 |
| huntingtin | gi|90903231 |
| cytoplasmic dynein 1 heavy chain 1 | gi|33350932 |
| dynein heavy chain 2, axonemal | gi|75677365 |
| dynein heavy chain 5, axonemal | gi|19115954 |
| dynein heavy chain 6, axonemal | gi|194353966 |
| dynein heavy chain 7, axonemal | gi|151301127 |
| dynein heavy chain 10, axonemal | gi|198442844 |
| ephrin type-A receptor 5 isoform a precursor | gi|221625401 |
| ephrin type-A receptor 8 isoform 1 precursor | gi|10140845 |
| ephrin type-A receptor 10 isofom 3 | gi|150456460 |
| V-type proton ATPase subunit B, brain isoform | gi|19913428 |
| Kv channel-interacting protein 2 isoform 2 | gi|27886670 |
| tax1-binding protein 1 isoform b * | gi|119943086 |
| serine/threonine-protein phosphatase 2A regulatory subunit A alpha isoform | gi|21361399 |
| adenylate cyclase type 8 | gi|4557257 |
| rho guanine nucleotide exchange factor 11 isoform 2 | gi|38026934 |
| activated CDC42 kinase 1 isoform 1 | gi|56549666 |
| pleckstrin homology domain-containing family A member 4 isoform 1 * | gi|238859651 |
| nuclear factor related to kappa-B-binding protein isoform 1 | gi|219802034 |
| paired box protein Pax-2 isoform a * | gi|34878699 |
| zinc finger protein 496 | gi|14249386 |
| zinc finger protein 561 * | gi|206725458 |
| zinc finger protein 611 isoform a | gi|239787080 |
| CREB-binding protein isoform b | gi|119943102 |
| protein argonaute-2 isoform 2 * | gi|257467482 |
| histone-lysine N-methyltransferase NSD3 isoform long * | gi|13699811 |
| E3 ubiquitin-protein ligase PDZRN3 * | gi|57529737 |
| E3 ubiquitin-protein ligase RBBP6 isoform 2 | gi|33620716 |
| 26S proteasome non-ATPase regulatory subunit 1 isoform 1 | gi|25777600 |
| apoptosis-inducing factor 1, mitochondrial isoform 4 * | gi|195927004 |
| baculoviral IAP repeat-containing protein 3 | gi|33946285 |
| baculoviral IAP repeat-containing protein 6 | gi|153792694 |
| protein S100-A8 | gi|21614544 |
| annexin A1 | gi|4502101 |
| NACHT, LRR and PYD domains-containing protein 2 isoform 1 * | gi|291463275 |
| complement C3 precursor | gi|115298678 |
| complement C5 preproprotein | gi|38016947 |
| complement component C8 alpha chain precursor | gi|4557389 |
| complement component C8 beta chain preproprotein | gi|4557391 |
| complement factor H-related protein 1 precursor * | gi|118442839 |
Potential biomarker candidates of glaucoma. Listed are fifty serum proteins detected by 2D-LC-MS/MS analysis of the glaucomatous serum IgG elutes, which are also known to be present in the glaucomatous human retina. All listed proteins were identified with high confidence (more than 99.0% probability assigned by the Protein Prophet algorithm) based on at least two identified peptides.
indicates methionine oxidation increased or detectable only in glaucomatous samples. This selected list of potential biomarker candidates awaits larger validation studies to establish their value as a clinical tool to determine diagnosis, prognosis, and/or treatment responses in glaucoma. This table has recently been published (Tezel et al., 2012b).
This study also determined oxidation of serum proteins by setting a search parameter to identify a mass shift corresponding to addition of the oxidation moiety to methionine (+16 Da) from the acquired mass spectra. When the number of methionine oxidation was normalized to the number of mass spectra corresponding to the modified protein, we found greater abundances in IgG elutes isolated from the glaucomatous sera compared to age-matched healthy controls. Methionine oxidation counts were two or more in 23% and 11% of proteins identified in glaucomatous and control samples, respectively, while for 62 proteins (27%) methionine oxidation was detectable only in glaucomatous samples (Tezel et al., 2012b). Table 1 also indicates the oxidation status of listed proteins.
The increase detected in serum protein oxidation in glaucomatous samples was also consistent with the findings of accompanying in vitro experiments. We found that the immunoreactivity of glaucomatous sera against oxidatively stressed retinal cell culture proteins was more than their immunoreactivity to control proteins (Tezel et al., 2012b). As also supported by previous studies, oxidative stress constitutes an important component of the immunostimulatory signals arising from glaucomatous tissues (Tezel, 2011), and oxidized epitopes of proteins may be particularly important in stimulation of immune responses to self-proteins in glaucoma. This observation encourages further research to determine whether modified serum proteins, as opposed to unmodified native proteins, are particularly useful as glaucoma-specific biomarkers.
Although many research groups follow the path of serum antibodies to serum biomarker discovery in glaucoma, there is only unpublished work that aimed to identify glaucoma-related biomarkers by direct proteomics analysis of serum samples. We analyzed serum samples collected from different groups of patients (n: 196) with primary open-angle glaucoma with normal intraocular pressure (normal-pressure glaucoma) or pseudoexfoliative glaucoma and control subjects without glaucoma (n: 92). Through the quantitative analysis of un-depleted serum samples by 2D-LC-MS/MS, we have been able to identify 325 proteins with high confidence (0.2% protein and 5.0% peptide false discovery rates). A total of 248 proteins were common to three sample groups, while 22 proteins were detected only in glaucomatous samples (Figure 8). Identified proteins included immune mediators and components of cell death signaling (Tezel et al., 2012a). These proteins may also serve as biomarker candidates and succeeding studies in larger patient populations should establish and validate their potential for future clinical applications as a diagnostic and/or prognostic tool in glaucoma.
Figure 8.
P otential biomarker candidates of glaucoma. Direct analysis of un-depleted serum samples collected from different groups of patients with primary open-angle glaucoma with normal intraocular pressure (POAG) or pseudoexfolative glaucoma (XFG) and control subjects without resulted in a list of 325 proteins, 248 of which were common to three sample groups. Listed are 22 proteins that were detected only in glaucomatous samples. This data has recently been presented (Tezel et al., 2012a).
In addition to plasma or serum analysis, analysis of isolated leukocytes has been suggested as another practical application in population screening of high-risk subjects for glaucoma (Golubnitschaja et al., 2010; Yeghiazaryan et al., 2010). This is based on the literature pointing to a potential predisposition of vasospastic individuals to glaucomatous optic nerve injury and the similarity in gene expression profiles of circulating leukocytes in glaucomatous patients and vasospastic individuals (Yeghiazaryan et al., 2009). Analysis of circulating leukocytes by 2D-PAGE/MALDI-TOF indicated a significant increase in protein expression of basic transcription factor activating protein-2beta in glaucomatous samples (Golubnitschaja et al., 2007).
4.3. Challenges in glaucoma biomarker discovery
Identification of biomarkers from blood samples possesses many difficulties. Owing to current limitations of the proteomics technology in dynamic range and threshold of detection, one difficulty is typically related to low serum concentrations of relevant proteins within complex mixtures of proteins. The high-abundant proteins, such as albumin, immunoglobulin, transferrin, fibrinogen, apolipoprotein, haptoglobin, constitute more than 95% of the total protein mass of human blood and mask the detection of low-abundant proteins. This seems to be particularly challenging for glaucoma biomarker discovery, since representation of ocular proteins in the large and complex pool of human blood may be even more limited. Blood samples may be depleted of highly abundant proteins by affinity chromatography to improve the analysis of low-abundant molecules in a high-throughput setting. However, although the affinity columns are thought to have little non-specific binding, depletion strategy may often result in co-depletion of low-abundant proteins of interest and cause their loss during the chromatography or downstream concentration step. In addition, potential biomarkers, particularly including the modified proteins, may often undergo degradation during sample collection, transportation, or storage (Hanash et al., 2008; Matt et al., 2008; Filiou and Turck, 2011).
Biomarker candidates identified by proteomics analysis have traditionally been evaluated using quantitative immunoassays, such as ELISA that is the most commonly used, reliable, and highly specific technique. Growing technological advances allow the use of antibodies coupled to a wide variety of new fluorescent molecules enabling sensitive detection of multiple molecules simultaneously. A variety of multiplexed immunoassays have been developed in recent years that provide quantitative information in a higher-throughput format and pg/mL sensitivity. These bioassays can be used as a powerful tool for validation of candidate biomarkers; however, the limiting factor for ELISA or multiplexed immunoassays is the availability of antibodies with the required affinity and specificity for biomarker validation (Hanash et al., 2008; Matt et al., 2008).
Multiple reaction monitoring (MRM) or selective reaction monitoring (SRM) has been introduced as an attractive alternative approach to detect and quantitate candidate biomarkers in a large number of samples, without requiring antibodies (Anderson and Hunter, 2006; Meng and Veenstra, 2011). This targeted mass spectrometry technique monitors fragments of specific peptides for quantification of the corresponding proteins, and also offers a feasible alternative for the analysis of post-translational modifications. In general, target peptides labeled with stable isotopes are used as internal standards allowing very accurate measurements. One of the advantages of this approach is the multiplexing feature that allows high-throughput and rapid characterization of multiple proteins simultaneously. This approach is receiving increased interest for the quantification of proteins and peptides in targeted validations studies; however, it still has limitations in sensitivity for detection of low-abundant biomarkers at the low ng/mL to pg/mL range unless complemented by enrichment strategies (Whiteaker et al., 2010; Picotti and Aebersold, 2012; Shi et al., 2012), and it is applicable only to previously well-characterized peptides. Emerging technical developments, such as the data-independent SWATH-MS technique that allows the generation of complete fragment ion data for all peptide precursors in a biological sample, provide a proteome-wide high-throughput quantification strategy with high reproducibility and sensitivity (Gillet et al., 2012). New developments in nanotechnology also offer smart nanoparticles for biomarker harvesting strategies which can achieve at least 100-fold amplification of low-abundant proteins and increase the detection limit of mass spectrometry, while protecting the biomarker candidates from degradation during subsequent sample handling (Tamburro et al., 2011).
Although proteomics is evolving quickly to increase both the throughput and confidence of the measurements for biomarker validation, the translation of potential biomarker candidates from bench to bedside remains highly challenging. For biomarker studies, the discovery phase normally takes place using specimens from a small, well-characterized cohort. When proceeding to the validation phase, well-designed studies of larger heterogeneous cohorts are needed for appropriate statistical power and blinding. Validation phase is critical to eliminate false positivity and calculate the sensitivity and specificity of candidate molecules for clinical prediction (Hanash et al., 2008; Matt et al., 2008; Filiou and Turck, 2011). Studies for validation of glaucoma-related biomarker candidates should similarly include large cohorts of patients with different types and stages of glaucoma under different treatments, patients with different demographics and comorbidities, and disease-free controls. Thus, molecular biomarker discovery requires the engagement of multidisciplinary teams and cooperative efforts of scientists, physicians, and funding organizations.
Because of the etiological complexity and the characteristic inter-patient heterogeneity of glaucoma, a single biomarker is unlikely to provide the sensitivity and specificity needed. Therefore, a panel of biomarkers in an array format may potentially be more useful. However, assembling and validating such a biomarker panel is even more challenging. Biomarkers derived from different platforms, such as direct serum analysis or antibody-based analysis, may also be integrated for complementary use of assays for protein or peptide biomarkers and autoantibodies. Such a complementary approach may ensure useful applications, since blood levels of biomarkers may be reduced as a result of antibody response, thereby contributing to a decrease in biomarker sensitivity. Alternatively, there may not be prominent autoantibody response because of a weak immune response in some patients.
5. Conclusions and future perspectives
Proteomics enables high-throughput quantitative analysis of protein expression, modifications, and interactions, and allows comparisons across multiple samples. The value of the information obtained from proteomics analysis continues to drive the use of proteomics techniques even with their shortcomings. The proteomics technology presenting a powerful hypothesis-generating engine has been used increasingly in the field of glaucoma research and provided with the lists of differentially expressed proteins in human glaucoma and animal models. The information gained from these studies has guided targeted studies to identify post-translational modifications, protein interactions, and the importance of identified proteome alterations in glaucomatous samples.
In order to translate the extensive information about glaucoma-related proteomics data into new treatment strategies, more work is needed for functional testing of the identified molecules. Translation of identified biomarker candidates from bench to bedside similarly requires further validation by collaboration of basic scientists and clinicians in well-designed population studies. Since the information obtained from proteomics analysis is growing, it will also be necessary to build glaucoma-related databases to handle and store the high amount of information and exchange the proteome datasets. Such a proteomics database, when integrated with the genomics data as well as data generated by other approaches of functional genomics, can yield an even more comprehensive database serving as a reference for glaucoma researchers.
There is no doubt that the proteomics technology holds great promise to move glaucoma research forward to pinpoint key molecular players of neurodegeneration, identify molecular targets for new treatments, and discover biomarkers for clinical testing. With growing capabilities for broad proteome coverage, excellent accuracy in quantification, and data analysis, proteomics will continue to advance our understanding of the molecular processes of glaucomatous neurodegeneration and provide insights into new treatment strategies and biomarker discovery for glaucoma.
Acknowledgments
Dr. Tezel’s work is supported by National Eye Institute, Bethesda, MD (R01 EY013813 and R01 EY017131) and Research to Prevent Blindness Inc., New York, NY providing an unrestricted grant to University of Louisville Department of Ophthalmology & Visual Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Briefings in functional genomics & proteomics. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Thelen JJ. A high-resolution two dimensional Gel- and Pro-Q DPS-based proteomics workflow for phosphoprotein identification and quantitative profiling. Methods Mol Biol. 2009;527:3–19. ix. doi: 10.1007/978-1-60327-834-8_1. [DOI] [PubMed] [Google Scholar]
- Almasieh M, Wilson AM, Morquette B, Cueva Vargas JL, Di Polo A. The molecular basis of retinal ganglion cell death in glaucoma. Prog Retin Eye Res. 2012;31:152–181. doi: 10.1016/j.preteyeres.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Amelinckx A, Castello M, Arrieta-Quintero E, Lee T, Salas N, Hernandez E, Lee RK, Bhattacharya SK, Parel JM. Laser trabeculoplasty induces changes in the trabecular meshwork glycoproteome: a pilot study. J Proteome Res. 2009;8:3727–3736. doi: 10.1021/pr900294g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Anderson NG, Matheson A, Anderson NL. Back to the future: the human protein index (HPI) and the agenda for post-proteomic biology. Proteomics. 2001;1:3–12. doi: 10.1002/1615-9861(200101)1:1<3::AID-PROT3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Armirotti A, Damonte G. Achievements and perspectives of top-down proteomics. Proteomics. 2010;10:3566–3576. doi: 10.1002/pmic.201000245. [DOI] [PubMed] [Google Scholar]
- Atmaca-Sonmez P, Yang X, Cai J, Eskan MA, Yolcu ES, Tezel G. Proteomic identification of glycated proteins shared in ocular hypertensive and diabetic rat retinas. Invest Ophthalmol Vis Sci. 2006;47:E-Abstract 197. Available at www.iovs.org. [Google Scholar]
- Azarkan M, Huet J, Baeyens-Volant D, Looze Y, Vandenbussche G. Affinity chromatography: a useful tool in proteomics studies. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2007;849:81–90. doi: 10.1016/j.jchromb.2006.10.056. [DOI] [PubMed] [Google Scholar]
- Banks CA, Kong SE, Washburn MP. Affinity purification of protein complexes for analysis by multidimensional protein identification technology. Protein Expr Purif. 2012;86:105–119. doi: 10.1016/j.pep.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Bauer A, Kuster B. Affinity purification-mass spectrometry. Powerful tools for the characterization of protein complexes. European journal of biochemistry / FEBS. 2003;270:570–578. doi: 10.1046/j.1432-1033.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annual review of biochemistry. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Rockwood EJ, Smith SD, et al. Proteomics reveal Cochlin deposits associated with glaucomatous trabecular meshwork. J Biol Chem. 2005;280:6080–6084. doi: 10.1074/jbc.M411233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SK, Crabb JS, Bonilha VL, Gu X, Takahara H, Crabb JW. Proteomics implicates peptidyl arginine deiminase 2 and optic nerve citrullination in glaucoma pathogenesis. Invest Ophthalmol Vis Sci. 2006;47:2508–2514. doi: 10.1167/iovs.05-1499. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Lee RK, Grus FH. Molecular Biomarkers in Glaucoma. Invest Ophthalmol Vis Sci. 2013;54:121–131. doi: 10.1167/iovs.12-11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn K, Cheng FY, Williamson JD, Goshe MB. Data-independent liquid chromatography/mass spectrometry (LC/MS(E)) detection and quantification of the secreted Apium graveolens pathogen defense protein mannitol dehydrogenase. Rapid communications in mass spectrometry: RCM. 2010;24:1009–1016. doi: 10.1002/rcm.4476. [DOI] [PubMed] [Google Scholar]
- Block H, Maertens B, Spriestersbach A, Brinker N, Kubicek J, Fabis R, Labahn J, Schafer F. Immobilized-metal affinity chromatography (IMAC): a review. Methods Enzymol. 2009;463:439–473. doi: 10.1016/S0076-6879(09)63027-5. [DOI] [PubMed] [Google Scholar]
- Bock JR, Gough DA. Whole-proteome interaction mining. Bioinformatics. 2003;19:125–134. doi: 10.1093/bioinformatics/19.1.125. [DOI] [PubMed] [Google Scholar]
- Boehm ME, Seidler J, Hahn B, Lehmann WD. Site-specific degree of phosphorylation in proteins measured by liquid chromatography-electrospray mass spectrometry. Proteomics. 2012a;12:2167–2178. doi: 10.1002/pmic.201100561. [DOI] [PubMed] [Google Scholar]
- Boehm N, Wolters D, Thiel U, Lossbrand U, Wiegel N, Pfeiffer N, Grus FH. New insights into autoantibody profiles from immune privileged sites in the eye: a glaucoma study. Brain Behav Immun. 2012b;26:96–102. doi: 10.1016/j.bbi.2011.07.241. [DOI] [PubMed] [Google Scholar]
- Bollinger KE, Crabb JS, Yuan X, Putliwala T, Clark AF, Crabb JW. Quantitative proteomics: TGFbeta(2) signaling in trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2011;52:8287–8294. doi: 10.1167/iovs.11-8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger KE, Crabb JS, Yuan X, Putliwala T, Clark AF, Crabb JW. Proteomic similarities in steroid responsiveness in normal and glaucomatous trabecular meshwork cells. Mol Vis. 2012;18:2001–2011. [PMC free article] [PubMed] [Google Scholar]
- Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem. 2002;74:4741–4749. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- Bouhenni RA, Al Shahwan S, Morales J, Wakim BT, Chomyk AM, Alkuraya FS, Edward DP. Identification of differentially expressed proteins in the aqueous humor of primary congenital glaucoma. Exp Eye Res. 2011;92:67–75. doi: 10.1016/j.exer.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Brun V, Masselon C, Garin J, Dupuis A. Isotope dilution strategies for absolute quantitative proteomics. Journal of proteomics. 2009;72:740–749. doi: 10.1016/j.jprot.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Butterfield DA. Proteomics: a new approach to investigate oxidative stress in Alzheimer's disease brain. Brain Res. 2004;1000:1–7. doi: 10.1016/j.brainres.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Dalle-Donne I. Redox proteomics. Antioxid Redox Signal. 2012;17:1487–1489. doi: 10.1089/ars.2012.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campostrini N, Areces LB, Rappsilber J, Pietrogrande MC, Dondi F, Pastorino F, Ponzoni M, Righetti PG. Spot overlapping in two-dimensional maps: a serious problem ignored for much too long. Proteomics. 2005;5:2385–2395. doi: 10.1002/pmic.200401253. [DOI] [PubMed] [Google Scholar]
- Canas B, Pineiro C, Calvo E, Lopez-Ferrer D, Gallardo JM. Trends in sample preparation for classical and second generation proteomics. J Chromatogr A. 2007;1153:235–258. doi: 10.1016/j.chroma.2007.01.045. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson L, Zhang Y, Harwerth RS, et al. Elevated albumin in retinas of monkeys with experimental glaucoma. Invest Ophthalmol Vis Sci. 2010;51:952–959. doi: 10.1167/iovs.09-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaga GS. Twenty-five years of immobilized metal ion affinity chromatography: past, present and future. Journal of biochemical and biophysical methods. 2001;49:313–334. doi: 10.1016/s0165-022x(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Chait BT. Chemistry. Mass spectrometry: bottom-up or top-down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J Proteome Res. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- Clark R, Nosie A, Walker T, Faralli JA, Filla MS, Barrett-Wilt G, Peters DM. Comparative genomic and proteomic analysis of cytoskeletal changes in dexamethasone-treated trabecular meshwork cells. Mol Cell Proteomics. 2013;12:194–206. doi: 10.1074/mcp.M112.019745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CH, Buckley JA, Fung ET. SELDI-TOF-MS proteomics of breast cancer. Clinical chemistry and laboratory medicine: CCLM / FESCC. 2005;43:1314–1320. doi: 10.1515/CCLM.2005.225. [DOI] [PubMed] [Google Scholar]
- Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Crabb JW, Yuan X, Dvoriantchikova G, Ivanov D, Crabb JS, Shestopalov VI. Preliminary quantitative proteomic characterization of glaucomatous rat retinal ganglion cells. Exp Eye Res. 2010;91:107–110. doi: 10.1016/j.exer.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, Tell G, Lungarella G, Colombo R, Rossi R, Milzani A. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass spectrometry reviews. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Deretic V. Multiple regulatory and effector roles of autophagy in immunity. Curr Opin Immunol. 2009;21:53–62. doi: 10.1016/j.coi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan EW, Chen H, Ho SL, et al. Protein macroarray profiling of serum autoantibodies in pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2010;51:2968–2975. doi: 10.1167/iovs.09-4898. [DOI] [PubMed] [Google Scholar]
- Desmetz C, Maudelonde T, Mange A, Solassol J. Identifying autoantibody signatures in cancer: a promising challenge. Expert Rev Proteomics. 2009;6:377–386. doi: 10.1586/epr.09.56. [DOI] [PubMed] [Google Scholar]
- Di Palma S, Hennrich ML, Heck AJ, Mohammed S. Recent advances in peptide separation by multidimensional liquid chromatography for proteome analysis. Journal of proteomics. 2012;75:3791–3813. doi: 10.1016/j.jprot.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- Duan X, Xue P, Wang N, Dong Z, Lu Q, Yang F. Proteomic analysis of aqueous humor from patients with primary open angle glaucoma. Mol Vis. 2010;16:2839–2846. [PMC free article] [PubMed] [Google Scholar]
- Edward DP, Bouhenni R. Anterior segment alterations and comparative aqueous humor proteomics in the buphthalmic rabbit (an American Ophthalmological Society thesis) Trans Am Ophthalmol Soc. 2011;109:66–114. [PMC free article] [PubMed] [Google Scholar]
- Fedorova M, Kuleva N, Hoffmann R. Identification of cysteine, methionine and tryptophan residues of actin oxidized in vivo during oxidative stress. J Proteome Res. 2010;9:1598–1609. doi: 10.1021/pr901099e. [DOI] [PubMed] [Google Scholar]
- Filiou MD, Turck CW. General overview: biomarkers in neuroscience research. Int Rev Neurobiol. 2011;101:1–17. doi: 10.1016/B978-0-12-387718-5.00001-8. [DOI] [PubMed] [Google Scholar]
- Filiou MD, Martins-de-Souza D, Guest PC, Bahn S, Turck CW. To label or not to label: applications of quantitative proteomics in neuroscience research. Proteomics. 2012;12:736–747. doi: 10.1002/pmic.201100350. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M, Tsangaris G, Oh JE, Maris A, Lubec G. Protein profile of the HeLa cell line. J Chromatogr A. 2004;1038:247–265. doi: 10.1016/j.chroma.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Georgieva Y, Konthur Z. Design and screening of M13 phage display cDNA libraries. Molecules. 2011;16:1667–1681. doi: 10.3390/molecules16021667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustarini D, Dalle-Donne I, Tsikas D, Rossi R. Oxidative stress and human diseases: Origin, link, measurement, mechanisms, and biomarkers. Critical reviews in clinical laboratory sciences. 2009;46:241–281. doi: 10.3109/10408360903142326. [DOI] [PubMed] [Google Scholar]
- Glish GL, Vachet RW. The basics of mass spectrometry in the twenty-first century. Nature reviews Drug discovery. 2003;2:140–150. doi: 10.1038/nrd1011. [DOI] [PubMed] [Google Scholar]
- Golubnitschaja O, Yeghiazaryan K, Wunderlich K, Schild HH, Flammer J. Disease proteomics reveals altered basic gene expression regulation in leukocytes of Normal-Tension and Primary Open-Angle glaucoma patients. Proteomics Clinical applications. 2007;1:1316–1323. doi: 10.1002/prca.200700150. [DOI] [PubMed] [Google Scholar]
- Golubnitschaja O, Yeghiazaryan K, Flammer J. Key molecular pathways affected by glaucoma pathology: is predictive diagnosis possible? The EPMA journal. 2010;1:237–244. doi: 10.1007/s13167-010-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- Govindarajan B, Junk A, Algeciras M, Salomon RG, Bhattacharya SK. Increased isolevuglandin-modified proteins in glaucomatous astrocytes. Mol Vis. 2009;15:1079–1091. [PMC free article] [PubMed] [Google Scholar]
- Grus FH, Joachim SC, Bruns K, Lackner KJ, Pfeiffer N, Wax MB. Serum autoantibodies to alpha-fodrin are present in glaucoma patients from Germany and the United States. Invest Ophthalmol Vis Sci. 2006;47:968–976. doi: 10.1167/iovs.05-0685. [DOI] [PubMed] [Google Scholar]
- Grus FH, Joachim SC, Sandmann S, Thiel U, Bruns K, Lackner KJ, Pfeiffer N. Transthyretin and complex protein pattern in aqueous humor of patients with primary open-angle glaucoma. Mol Vis. 2008;14:1437–1445. [PMC free article] [PubMed] [Google Scholar]
- Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends in cell biology. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- Harris LR, Churchward MA, Butt RH, Coorssen JR. Assessing detection methods for gel-based proteomic analyses. J Proteome Res. 2007;6:1418–1425. doi: 10.1021/pr0700246. [DOI] [PubMed] [Google Scholar]
- Heller M, Mattou H, Menzel C, Yao X. Trypsin catalyzed 16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J Am Soc Mass Spectrom. 2003;14:704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- Hengel SM, Murray E, Langdon S, Hayward L, O'Donoghue J, Panchaud A, Hupp T, Goodlett DR. Data-independent proteomic screen identifies novel tamoxifen agonist that mediates drug resistance. J Proteome Res. 2011;10:4567–4578. doi: 10.1021/pr2004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert B, Righetti PG. A turning point in proteome analysis: sample prefractionation via multicompartment electrolyzers with isoelectric membranes. Electrophoresis. 2000;21:3639–3648. doi: 10.1002/1522-2683(200011)21:17<3639::AID-ELPS3639>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Houde D, Kauppinen P, Mhatre R, Lyubarskaya Y. Determination of protein oxidation by mass spectrometry and method transfer to quality control. J Chromatogr A. 2006;1123:189–198. doi: 10.1016/j.chroma.2006.04.046. [DOI] [PubMed] [Google Scholar]
- Huang P, Qi Y, Xu YS, Liu J, Liao D, Zhang SS, Zhang C. Serum cytokine alteration is associated with optic neuropathy in human primary open angle glaucoma. J Glaucoma. 2010a;19:324–330. doi: 10.1097/IJG.0b013e3181b4cac7. [DOI] [PubMed] [Google Scholar]
- Huang W, Fileta J, Guo Y, Grosskreutz CL. Downregulation of Thy1 in retinal ganglion cells in experimental glaucoma. Curr Eye Res. 2006;31:265–271. doi: 10.1080/02713680500545671. [DOI] [PubMed] [Google Scholar]
- Huang W, Fileta J, Rawe I, Qu J, Grosskreutz CL. Calpain activation in experimental glaucoma. Invest Ophthalmol Vis Sci. 2010b;51:3049–3054. doi: 10.1167/iovs.09-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Ito T, Ota K, Kubota H, Yamaguchi Y, Chiba T, Sakuraba K, Yoshida M. Roles for the two-hybrid system in exploration of the yeast protein interactome. Mol Cell Proteomics. 2002;1:561–566. doi: 10.1074/mcp.r200005-mcp200. [DOI] [PubMed] [Google Scholar]
- Iyer P, Lalane R, 3rd, Morris C, Challa P, Vann R, Rao PV. Autotaxin-lysophosphatidic Acid axis is a novel molecular target for lowering intraocular pressure. PLoS One. 2012;7:e42627. doi: 10.1371/journal.pone.0042627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Longobardi M, Cartiglia C, Sacca SC. Proteome alterations in primary open angle glaucoma aqueous humor. J Proteome Res. 2010;9:4831–4838. doi: 10.1021/pr1005372. [DOI] [PubMed] [Google Scholar]
- Jaquinod M, Trauchessec M, Huillet C, et al. Mass spectrometry-based absolute protein quantification: PSAQ strategy makes use of "noncanonical" proteotypic peptides. Proteomics. 2012;12:1217–1221. doi: 10.1002/pmic.201100538. [DOI] [PubMed] [Google Scholar]
- Javadiyan S, Burdon KP, Whiting MJ, Abhary S, Straga T, Hewitt AW, Mills RA, Craig JE. Elevation of serum asymmetrical and symmetrical dimethylarginine in patients with advanced glaucoma. Invest Ophthalmol Vis Sci. 2012;53:1923–1927. doi: 10.1167/iovs.11-8420. [DOI] [PubMed] [Google Scholar]
- Jenkins RE, Pennington SR. Arrays for protein expression profiling: towards a viable alternative to two-dimensional gel electrophoresis? Proteomics. 2001;1:13–29. doi: 10.1002/1615-9861(200101)1:1<13::AID-PROT13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- Joachim SC, Reichelt J, Berneiser S, Pfeiffer N, Grus FH. Sera of glaucoma patients show autoantibodies against myelin basic protein and complex autoantibody profiles against human optic nerve antigens. Graefes Arch Clin Exp Ophthalmol. 2008;246:573–580. doi: 10.1007/s00417-007-0737-8. [DOI] [PubMed] [Google Scholar]
- Joos T, Bachmann J. Protein microarrays: potentials and limitations. Front Biosci. 2009;14:4376–4385. doi: 10.2741/3534. [DOI] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamoto T, Ue T, Yokoyama T, Souchelnytskyi N, Kiuchi Y. Proteomic study of DBA/2J mice retina: Down-regulation of Integrin beta7 correlated with retinal ganglion cell death. Proteomics. 2009;9:4962–4969. doi: 10.1002/pmic.200800978. [DOI] [PubMed] [Google Scholar]
- Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J Am Chem Soc. 1999;121:806–812. [Google Scholar]
- Kim Y, Caberoy NB, Alvarado G, Davis JL, Feuer WJ, Li W. Identification of Hnrph3 as an autoantigen for acute anterior uveitis. Clin Immunol. 2011;138:60–66. doi: 10.1016/j.clim.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nature methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- Krishna RG, Wold F. Post-translational modification of proteins. Advances in enzymology and related areas of molecular biology. 1993;67:265–298. doi: 10.1002/9780470123133.ch3. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy D, Levin Y, Harris LW, Umrania Y, Bahn S, Guest PC. Analysis of the human pituitary proteome by data independent label-free liquid chromatography tandem mass spectrometry. Proteomics. 2011;11:495–500. doi: 10.1002/pmic.201000496. [DOI] [PubMed] [Google Scholar]
- Lam TC, Chun RK, Li KK, To CH. Application of proteomic technology in eye research: a mini review. Clinical & experimental optometry : journal of the Australian Optometrical Association. 2008;91:23–33. doi: 10.1111/j.1444-0938.2007.00194.x. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Leibovitch I, Kurtz S, Kesler A, Feithliher N, Shemesh G, Sela BA. C-reactive protein levels in normal tension glaucoma. J Glaucoma. 2005;14:384–386. doi: 10.1097/01.ijg.0000176932.06606.6e. [DOI] [PubMed] [Google Scholar]
- Levin LA. Extrapolation of animal models of optic nerve injury to clinical trial design. J Glaucoma. 2004;13:1–5. doi: 10.1097/00061198-200402000-00001. [DOI] [PubMed] [Google Scholar]
- Li WH, Zhao J, Li HY, et al. Proteomics-based identification of autoantibodies in the sera of healthy Chinese individuals from Beijing. Proteomics. 2006;6:4781–4789. doi: 10.1002/pmic.200500909. [DOI] [PubMed] [Google Scholar]
- Libby RT, Gould DB, Anderson MG, John SW. Complex genetics of glaucoma susceptibility. Annu Rev Genomics Hum Genet. 2005;6:15–44. doi: 10.1146/annurev.genom.6.080604.162209. [DOI] [PubMed] [Google Scholar]
- Limb GA, Martin KR. Current prospects in optic nerve protection and regeneration: sixth ARVO/Pfizer Ophthalmics Research Institute conference. Invest Ophthalmol Vis Sci. 2011;52:5941–5954. doi: 10.1167/iovs.10-6894. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nature biotechnology. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Ferrari M, Petricoin E. Clinical proteomics: written in blood. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lopez-Ferrer D, Ramos-Fernandez A, Martinez-Bartolome S, Garcia-Ruiz P, Vazquez J. Quantitative proteomics using (16)O/(18)O labeling and linear ion trap mass spectrometry. Proteomics. 2006;6(Suppl 1):S4–S11. doi: 10.1002/pmic.200500375. [DOI] [PubMed] [Google Scholar]
- Lundgren DH, Hwang SI, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev Proteomics. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- Luo C, Yang X, Kain AD, Powell DW, Kuehn MH, Tezel G. Glaucomatous tissue stress and the regulation of immune response through glial toll-like receptor signaling. Invest Ophthalmol Vis Sci. 2010;51:5697–5707. doi: 10.1167/iovs.10-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal N, Heegaard S, Prause JU, Honore B, Vorum H. Ocular proteomics with emphasis on two-dimensional gel electrophoresis and mass spectrometry. Biological procedures online. 2009;12:56–88. doi: 10.1007/s12575-009-9019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nature biotechnology. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- Mann M, Kelleher NL. Precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci U S A. 2008;105:18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- Matt P, Fu Z, Fu Q, Van Eyk JE. Biomarker discovery: proteome fractionation and separation in biological samples. Physiol Genomics. 2008;33:12–17. doi: 10.1152/physiolgenomics.00282.2007. [DOI] [PubMed] [Google Scholar]
- Matthiesen R, Azevedo L, Amorim A, Carvalho AS. Discussion on common data analysis strategies used in MS-based proteomics. Proteomics. 2011;11:604–619. doi: 10.1002/pmic.201000404. [DOI] [PubMed] [Google Scholar]
- McKinnon SJ, Lehman DM, Kerrigan-Baumrind LA, et al. Caspase activation and amyloid precursor protein cleavage in rat ocular hypertension. Invest Ophthalmol Vis Sci. 2002;43:1077–1087. [PubMed] [Google Scholar]
- McKinnon SJ, Schlamp CL, Nickells RW. Mouse models of retinal ganglion cell death and glaucoma. Exp Eye Res. 2009;88:816–824. doi: 10.1016/j.exer.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Veenstra TD. Targeted mass spectrometry approaches for protein biomarker verification. Journal of proteomics. 2011;74:2650–2659. doi: 10.1016/j.jprot.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Minden JS, Dowd SR, Meyer HE, Stuhler K. Difference gel electrophoresis. Electrophoresis. 2009;30(Suppl 1):S156–161. doi: 10.1002/elps.200900098. [DOI] [PubMed] [Google Scholar]
- Mitchell P. A perspective on protein microarrays. Nature biotechnology. 2002;20:225–229. doi: 10.1038/nbt0302-225. [DOI] [PubMed] [Google Scholar]
- Miyagi M, Rao KC. Proteolytic 18O-labeling strategies for quantitative proteomics. Mass spectrometry reviews. 2007;26:121–136. doi: 10.1002/mas.20116. [DOI] [PubMed] [Google Scholar]
- Miyara N, Shinzato M, Yamashiro Y, Iwamatsu A, Kariya K, Sawaguchi S. Proteomic analysis of rat retina in a steroid-induced ocular hypertension model: potential vulnerability to oxidative stress. Japanese journal of ophthalmology. 2008;52:84–90. doi: 10.1007/s10384-007-0507-5. [DOI] [PubMed] [Google Scholar]
- Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- Nguyen C, Cone F, Hanes J, Kim A, Nguyen T, Tezel G, Pease ME, Steinhart M, Quigley HA. Studies of scleral microstructure, proteomic analysis, and biomechanical behavior suggest mechanisms of susceptibility to experimental glaucoma in mice. Invest Ophthalmol Vis Sci. 2012;53:E-Abstract 3187. Available at www.iovs.org. [Google Scholar]
- Nickells RW, Semaan SJ, Schlamp CL. Involvement of the Bcl2 gene family in the signaling and control of retinal ganglion cell death. Prog Brain Res. 2008;173:423–435. doi: 10.1016/S0079-6123(08)01129-1. [DOI] [PubMed] [Google Scholar]
- Nickells RW. Variations in the rheostat model of apoptosis: what studies of retinal ganglion cell death tell us about the functions of the Bcl2 family proteins. Exp Eye Res. 2010;91:2–8. doi: 10.1016/j.exer.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nature methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nature chemical biology. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Ong SE. Whole proteomes as internal standards in quantitative proteomics. Genome medicine. 2010;2:49. doi: 10.1186/gm170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud A, Scherl A, Shaffer SA, von Haller PD, Kulasekara HD, Miller SI, Goodlett DR. Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Anal Chem. 2009;81:6481–6488. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- Patton WF. A thousand points of light: the application of fluorescence detection technologies to two-dimensional gel electrophoresis and proteomics. Electrophoresis. 2000;21:1123–1144. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1123::AID-ELPS1123>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome research. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, Zivny J, Vulpe CD. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: translating benchside promise into bedside reality. Nature reviews Drug discovery. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nature methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- Pieragostino D, Bucci S, Agnifili L, et al. Differential protein expression in tears of patients with primary open angle and pseudoexfoliative glaucoma. Molecular bioSystems. 2012;8:1017–1028. doi: 10.1039/c1mb05357d. [DOI] [PubMed] [Google Scholar]
- Pieroni E, de la Fuente van Bentem S, Mancosu G, Capobianco E, Hirt H, de la Fuente A. Protein networking: insights into global functional organization of proteomes. Proteomics. 2008;8:799–816. doi: 10.1002/pmic.200700767. [DOI] [PubMed] [Google Scholar]
- Ping P. Identification of novel signaling complexes by functional proteomics. Circ Res. 2003;93:595–603. doi: 10.1161/01.RES.0000093221.98213.E0. [DOI] [PubMed] [Google Scholar]
- Powell DW, Merchant ML, Link AJ. Discovery of regulatory molecular events and biomarkers using 2D capillary chromatography and mass spectrometry. Expert Rev Proteomics. 2006;3:63–74. doi: 10.1586/14789450.3.1.63. [DOI] [PubMed] [Google Scholar]
- Prokosch V, Panagis L, Volk GF, Dermon C, Thanos S. Alpha2-adrenergic receptors and their core involvement in the process of axonal growth in retinal explants. Invest Ophthalmol Vis Sci. 2010;51:6688–6699. doi: 10.1167/iovs.09-4835. [DOI] [PubMed] [Google Scholar]
- Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3–10. [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Reichelt J, Joachim SC, Pfeiffer N, Grus FH. Analysis of autoantibodies against human retinal antigens in sera of patients with glaucoma and ocular hypertension. Curr Eye Res. 2008;33:253–261. doi: 10.1080/02713680701871157. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nature biotechnology. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rogers RS, Dharsee M, Ackloo S, Sivak JM, Flanagan JG. Proteomics analyses of human optic nerve head astrocytes following biomechanical strain. Mol Cell Proteomics. 2012;11:M111 012302. doi: 10.1074/mcp.M111.012302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sacca SC, Centofanti M, Izzotti A. New proteins as vascular biomarkers in primary open angle glaucomatous aqueous humor. Invest Ophthalmol Vis Sci. 2012;53:4242–4253. doi: 10.1167/iovs.11-8902. [DOI] [PubMed] [Google Scholar]
- Sakai J, Kojima S, Yanagi K, Kanaoka M. 18O-labeling quantitative proteomics using an ion trap mass spectrometer. Proteomics. 2005;5:16–23. doi: 10.1002/pmic.200300885. [DOI] [PubMed] [Google Scholar]
- Sardiu ME, Washburn MP. Building protein-protein interaction networks with proteomics and informatics tools. J Biol Chem. 2011;286:23645–23651. doi: 10.1074/jbc.R110.174052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlamp CL, Johnson EC, Li Y, Morrison JC, Nickells RW. Changes in Thy1 gene expression associated with damaged retinal ganglion cells. Mol Vis. 2001;7:192–201. [PubMed] [Google Scholar]
- Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Lenz C, Grote M, Luhrmann R, Urlaub H. Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal Chem. 2010;82:2784–2796. doi: 10.1021/ac902710k. [DOI] [PubMed] [Google Scholar]
- Schnolzer M, Jedrzejewski P, Lehmann WD. Protease-catalyzed incorporation of 18O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- Sharma S, Chataway T, Burdon KP, Jonavicius L, Klebe S, Hewitt AW, Mills RA, Craig JE. Identification of LOXL1 protein and Apolipoprotein E as components of surgically isolated pseudoexfoliation material by direct mass spectrometry. Exp Eye Res. 2009;89:479–485. doi: 10.1016/j.exer.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Shen Y, Smith RD. Advanced nanoscale separations and mass spectrometry for sensitive high-throughput proteomics. Expert Rev Proteomics. 2005;2:431–447. doi: 10.1586/14789450.2.3.431. [DOI] [PubMed] [Google Scholar]
- Shi T, Su D, Liu T, Tang K, Camp DG, 2nd, Qian WJ, Smith RD. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012;12:1074–1092. doi: 10.1002/pmic.201100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato M, Yamashiro Y, Miyara N, Iwamatsu A, Takeuchi K, Umikawa M, Bayarjargal M, Kariya K, Sawaguchi S. Proteomic analysis of the trabecular meshwork of rats in a steroid-induced ocular hypertension model: downregulation of type I collagen C-propeptides. Ophthalmic research. 2007;39:330–337. doi: 10.1159/000109989. [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid Redox Signal. 2003;5:577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- Steely HT, Jr, Clark AF. The use of proteomics in ophthalmic research. Pharmacogenomics. 2000;1:267–280. doi: 10.1517/14622416.1.3.267. [DOI] [PubMed] [Google Scholar]
- Steinberg TH, Pretty On Top K, Berggren KN, Kemper C, Jones L, Diwu Z, Haugland RP, Patton WF. Rapid and simple single nanogram detection of glycoproteins in polyacrylamide gels and on electroblots. Proteomics. 2001;1:841–855. doi: 10.1002/1615-9861(200107)1:7<841::AID-PROT841>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Steinberg TH, Agnew BJ, Gee KR, et al. Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics. 2003;3:1128–1144. doi: 10.1002/pmic.200300434. [DOI] [PubMed] [Google Scholar]
- Stowell C, Arbogast B, Cioffi G, Burgoyne C, Zhou A. Retinal proteomic changes following unilateral optic nerve transection and early experimental glaucoma in non-human primate eyes. Exp Eye Res. 2011;93:13–28. doi: 10.1016/j.exer.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007;6:1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Moriya S, Oku H, Azuma I. Association of endothelin-1 with normal tension glaucoma: clinical and fundamental studies. Surv Ophthalmol. 1995;39(Suppl 1):S49–56. doi: 10.1016/s0039-6257(05)80073-6. [DOI] [PubMed] [Google Scholar]
- Sultana R, Butterfield DA. Identification of the oxidative stress proteome in the brain. Free Radic Biol Med. 2011;50:487–494. doi: 10.1016/j.freeradbiomed.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburro D, Fredolini C, Espina V, et al. Multifunctional core-shell nanoparticles: discovery of previously invisible biomarkers. Journal of the American Chemical Society. 2011;133:19178–19188. doi: 10.1021/ja207515j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannu NS, Hemby SE. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nature protocols. 2006;1:1732–1742. doi: 10.1038/nprot.2006.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Kass MA, Kolker AE, Becker B, Wax MB. Plasma and aqueous humor endothelin levels in primary open-angle glaucoma. J Glaucoma. 1997;6:83–89. [PubMed] [Google Scholar]
- Tezel G, Wax MB. Inhibition of caspase activity in retinal cell apoptosis induced by various stimuli in vitro. Invest Ophthalmol Vis Sci. 1999;40:2660–2667. [PubMed] [Google Scholar]
- Tezel G, Wax MB. Increased production of tumor necrosis factor-alpha by glial cells exposed to simulated ischemia or elevated hydrostatic pressure induces apoptosis in cocultured retinal ganglion cells. J Neurosci. 2000;20:8693–8700. doi: 10.1523/JNEUROSCI.20-23-08693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Wax MB. Hypoxia-inducible factor 1alpha in the glaucomatous retina and optic nerve head. Arch Ophthalmol. 2004;122:1348–1356. doi: 10.1001/archopht.122.9.1348. [DOI] [PubMed] [Google Scholar]
- Tezel G, Yang X. Comparative gene array analysis of TNF-alpha-induced MAPK and NF-kappaB signaling pathways between retinal ganglion cells and glial cells. Exp Eye Res. 2005;81:207–217. doi: 10.1016/j.exer.2005.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Yang X, Cai J. Proteomic identification of oxidatively modified retinal proteins in a chronic pressure–induced rat model of glaucoma. Invest Ophthalmol Vis Sci. 2005;46:3177–3187. doi: 10.1167/iovs.05-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G. Oxidative stress in glaucomatous neurodegeneration: Mechanisms and consequences. Prog Retin Eye Res. 2006;25:490–513. doi: 10.1016/j.preteyeres.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Luo C, Yang X. Accelerated aging in glaucoma: immunohistochemical assessment of advanced glycation end products in the human retina and optic nerve head. Invest Ophthalmol Vis Sci. 2007;48:1201–1211. doi: 10.1167/iovs.06-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G. TNF-alpha signaling in glaucomatous neurodegeneration. Prog Brain Res. 2008a;173:409–421. doi: 10.1016/S0079-6123(08)01128-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G. Proteomics in Defining Pathogenic Processes Involved in Glaucomatous Neurodegeneration. In: Tombran-Tink J, Barnstable J, Shields MB, editors. Ophthalmology Research Series, Glaucoma: Mechanisms of the Glaucomas: Disease Processes and Therapeutic Modalities. Humana Press Inc; 2008b. pp. 425–441. [Google Scholar]
- Tezel G. The role of glia, mitochondria, and the immune system in glaucoma. Invest Ophthalmol Vis Sci. 2009;50:1001–1012. doi: 10.1167/iovs.08-2717. [DOI] [PubMed] [Google Scholar]
- Tezel G, Yang X, Luo C, Cai J, Kain AD, Powell DW, Kuehn MH, Pierce WM. Hemoglobin expression and regulation in glaucoma: insights into retinal ganglion cell oxygenation. Invest Ophthalmol Vis Sci. 2010a;51:907–919. doi: 10.1167/iovs.09-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Yang X, Luo C, Cai J, Pierce WM, Kuehn MH. Proteomic analysis of human retina: Ocular hypertension versus glaucoma. Invest Ophthalmol Vis Sci. 2010b;51:E-Abstract 2186. Available at www.iovs.org. [Google Scholar]
- Tezel G, Yang X, Luo C, Kain AD, Powell DW, Kuehn MH, Kaplan HJ. Oxidative stress and the regulation of complement activation in human glaucoma. Invest Ophthalmol Vis Sci. 2010c;51:5071–5082. doi: 10.1167/iovs.10-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G. The immune response in glaucoma: a perspective on the roles of oxidative stress. Exp Eye Res. 2011;93:178–186. doi: 10.1016/j.exer.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Merchant ML, Yang X, Luo C, Soltau JB, Liebmann JM, Ritch R, Klein J. Candidate serum biomarkers in patients with glaucoma. Invest Ophthalmol Vis Sci. 2012a;53:E-Abstract 3832. Available at www.iovs.org. [Google Scholar]
- Tezel G, Thornton IL, Tong MG, et al. Immunoproteomic analysis of potential serum biomarker candidates in human glaucoma. Invest Ophthalmol Vis Sci. 2012b;53:8222–8231. doi: 10.1167/iovs.12-10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Yang X, Luo C, Cai J, Powell DW. An astrocyte-specific proteomic approach to inflammatory responses in experimental rat glaucoma. Invest Ophthalmol Vis Sci. 2012c;53:4220–4233. doi: 10.1167/iovs.11-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G. Immune regulation toward immunomodulation for neuroprotection in glaucoma. Curr Opin Pharmacol. 2013;13:23–31. doi: 10.1016/j.coph.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck CW, Falick AM, Kowalak JA, Lane WS, Lilley KS, Phinney BS, Weintraub ST, Witkowska HE, Yates NA. The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 study: relative protein quantitation. Mol Cell Proteomics. 2007;6:1291–1298. doi: 10.1074/mcp.M700165-MCP200. [DOI] [PubMed] [Google Scholar]
- Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wax MB, Tezel G, Kawase K, Kitazawa Y. Serum autoantibodies to heat shock proteins in glaucoma patients from Japan and the United States. Ophthalmology. 2001;108:296–302. doi: 10.1016/s0161-6420(00)00525-x. [DOI] [PubMed] [Google Scholar]
- Wheelock AM, Buckpitt AR. Software-induced variance in two-dimensional gel electrophoresis image analysis. Electrophoresis. 2005;26:4508–4520. doi: 10.1002/elps.200500253. [DOI] [PubMed] [Google Scholar]
- Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics. 2010;9:184–196. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore AV, Libby RT, John SW. Glaucoma: thinking in new ways-a role for autonomous axonal self-destruction and other compartmentalised processes? Prog Retin Eye Res. 2005;24:639–662. doi: 10.1016/j.preteyeres.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004;76:6085–6096. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- Wong TT, Zhou L, Li J, Tong L, Zhao SZ, Li XR, Yu SJ, Koh SK, Beuerman RW. Proteomic profiling of inflammatory signaling molecules in the tears of patients on chronic glaucoma medication. Invest Ophthalmol Vis Sci. 2011;52:7385–7391. doi: 10.1167/iovs.10-6532. [DOI] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- Wu J, Lenchik NJ, Pabst MJ, Solomon SS, Shull J, Gerling IC. Functional characterization of two-dimensional gel-separated proteins using sequential staining. Electrophoresis. 2005;26:225–237. doi: 10.1002/elps.200406176. [DOI] [PubMed] [Google Scholar]
- Xie F, Smith RD, Shen Y. Advanced proteomic liquid chromatography. J Chromatogr A. 2012;1261:78–90. doi: 10.1016/j.chroma.2012.06.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tezel G, Patil RV, Romano C, Wax MB. Serum autoantibody against glutathione S-transferase in patients with glaucoma. Invest Ophthalmol Vis Sci. 2001;42:1273–1276. [PubMed] [Google Scholar]
- Yang X, Tezel G. Proteomic analysis of retinal ganglion cells: Toward retinal ganglion cell protein mapping. Invest Ophthalmol Vis Sci. 2005;46:E-Abstract 3771. Available at www.iovs.org. [Google Scholar]
- Yang X, Luo C, Cai J, Tezel G. Proteomic identification of phosphorylated proteins in a chronic pressure-induced rat model of glaucoma. Invest Ophthalmol Vis Sci. 2006;47:E-Abstract 198. doi: 10.1167/iovs.05-0208. Available at www.iovs.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Luo C, Cai J, Pierce WM, Tezel G. Phosphorylation-dependent interaction with 14-3-3 in the regulation of bad trafficking in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2008;49:2483–2494. doi: 10.1167/iovs.07-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Luo C, Cai J, Powell DW, Yu D, Kuehn MH, Tezel G. Neurodegenerative and inflammatory pathway components linked to TNF-alpha/TNFR1 signaling in the glaucomatous human retina. Invest Ophthalmol Vis Sci. 2011;52:8442–8454. doi: 10.1167/iovs.11-8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- Ye J, Zhang X, Young C, Zhao X, Hao Q, Cheng L, Jensen ON. Optimized IMAC-IMAC protocol for phosphopeptide recovery from complex biological samples. J Proteome Res. 2010;9:3561–3573. doi: 10.1021/pr100075x. [DOI] [PubMed] [Google Scholar]
- Yeghiazaryan K, Flammer J, Orgul S, Wunderlich K, Golubnitschaja O. Vasospastic individuals demonstrate significant similarity to glaucoma patients as revealed by gene expression profiling in circulating leukocytes. Mol Vis. 2009;15:2339–2348. [PMC free article] [PubMed] [Google Scholar]
- Yeghiazaryan K, Flammer J, Golubnitschaja O. Predictive molecular profiling in blood of healthy vasospastic individuals: clue to targeted prevention as personalised medicine to effective costs. The EPMA journal. 2010;1:263–272. doi: 10.1007/s13167-010-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fang A, Riley CP, Wang M, Regnier FE, Buck C. Multi-dimensional liquid chromatography in proteomics--a review. Analytica chimica acta. 2010;664:101–113. doi: 10.1016/j.aca.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao Q, Duan S, He Y, Sun X, Jiang R, Duan Y, Zhong X, Ge J. Upregulation of Copine1 in trabecular meshwork cells of POAG patients: a membrane proteomics approach. Mol Vis. 2008;14:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal Chem. 2009;81:6317–6326. doi: 10.1021/ac9004887. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ramsey KE, Stephan DA, Russell P. Gene and protein expression changes in human trabecular meshwork cells treated with transforming growth factor-beta. Invest Ophthalmol Vis Sci. 2004;45:4023–4034. doi: 10.1167/iovs.04-0535. [DOI] [PubMed] [Google Scholar]
- Zuo X, Speicher DW. Comprehensive analysis of complex proteomes using microscale solution isoelectrofocusing prior to narrow pH range two-dimensional electrophoresis. Proteomics. 2002;2:58–68. [PubMed] [Google Scholar]