Abstract
Although cadmium (Cd) is a widespread environmental contaminant and human carcinogen, our studies indicate an organic Cd complex to be a potent inhibitor of proteasomal chymotrypsin-like (CT-like) activity, further capable of inducing apoptosis in a cancer cell-specific manner. It has been reported that the ligands indole-3-butyric acid (L1) and indole-3-propionic acid (L2) have cancer-fighting effects when tested in a rat carcinoma model. In addition, 3, 5-diaminobenzoic acid o-vanillin Schiff bases (L3) have high antimicrobial activity and a large number of Schiff base complexes have been reported to have proteasome-inhibitory activity. We therefore hypothesized that synthetic forms of Cd in combination with L1, L2 and L3 may have proteasome-inhibitory and apoptosis-inducing activities, which would be cancer cell-specific. To test this hypothesis, we have synthesized three novel Cd-containing complexes: [Cd2(C12H12O2N)4(H2O)2]·2H2O (Cd1), [Cd2(C11H10O2N)4(H2O)2]·2H2O (Cd2) and [Cd(C7H4N2O2)(C8H6O2)2]·2H2O (Cd3), by using these three ligands. We sought out to characterize and assess the proteasome-inhibitory and anti-proliferative properties of these three Cd complexes in human breast cancer cells. Cd1, Cd2 and Cd3 were found to effectively inhibit the chymotrypsin-like activity of purified 20S proteasome with IC50 values of 2.6, 3.0 and 3.3 μM, respectively. Moreover, inhibition of cancer cell proliferation also correlated with this effect. As a result of proteasomal shutdown, the accumulation of ubiquitinated proteins and the proteasome target IκB-α protein as well as induction of apoptosis were observed. To account for the cancer specificity of this effect, immortalized, non-tumorigenic breast MCF10A cells were used under the same experimental conditions. Our results indicate that MCF10A cells are much less sensitive to the Cd1, Cd2 and Cd3 complexes when compared to MDA MB 231 breast cancer cells. Therefore, our study suggests that these Cd organic complexes are capable of inhibiting tumor cellular proteasome activity and consequently induce cancer cell-specific apoptotic death.
Keywords: Cadmium, Proteasome inhibitor, Apoptosis, Indole-3-butyric acid, Indole-3-propionic acid, 3 5-diaminobenzoic acid o-vanillin Schiff bases
1. Introduction
The ubiquitin proteasome system (UP-S) regulates a number of key cellular processes, including cell proliferation and death through degradation of specific proteins involved [1,2]. Selective degradation of proteins is indeed a slippery slope, critical for protein homeostasis in normal cells, but dysregulated in cancer cells. The UP-S has therefore been extensively studied as a novel molecular target for the development of novel drugs in an attempt to restore protein homeostasis as the ultimate therapeutic strategy. The 20S proteasome, the main component of the UP-S, is a high molecular weight protease complex with a proteolytic core containing subunits including β1, β2 and β5, which are responsible for its caspase-like, trypsin-like and chymotrypsin-like (CT-like) activities, respectively [3]. It is well established that inhibition of the β5 proteasomal subunit, and therefore its CT-like activity, is primarily associated with apoptosis induction in tumor cells [4–6]. Furthermore, this shutdown also leads to the accumulation of several target proteins (i.e., IκB-α), followed by an induction of programmed cell death, or apoptosis [7–9].
Metal-based anti-cancer drugs were developed many years ago. Our laboratory has studied a number of the metal-based drugs, including organic copper-, zinc-, and gold-based complexes, all of which are capable of inhibiting the tumor cell proteasome and thus proliferation, thereby inducing cancer cell death [10–14]. We have also reported that mixtures of disulfiram (DSF) and cadmium (Cd) can selectively inhibit proteasome activity in human breast cancer cells, over their “normal”, immortalized, non-tumorigenic counterparts, and ultimately result in apoptosis [15]. Cd is an environmental hazard whose effects from a long term exposure are substantially controversial. Several incidents have been reported about general population of Cd poisoning due to continual consumption of contaminated food and water [16]. It has recently been proposed that Cd ingestion may increase one's risk of developing breast cancer [17]. This however, is an area of ongoing investigation and Cd's actual carcinogenic effects, in regard to breast cancer incidence, remain to be determined.
Nevertheless, the literature does point to cases where Cd has been shown to affect cell proliferation, differentiation, as well as apoptosis. Studies have shown that Cd has an effect on p38/MAPK isoforms [18] and plays an important role in the promotion of breast cancer cell growth by potentiating the interaction between ERα and c-Jun [19]. Other reports link Cd exposure to genomic instability, aberrant gene expression, and inhibition of DNA damage repair and apoptosis through complex and multifactorial mechanisms [20,21]. Conversely though, Cd has also been shown to induce p53-dependent apoptosis [22] and down-regulation of the x-linked inhibitor of apoptosis protein (XIAP) in human prostate cancer cells [23]. Interestingly, synthetically made meclofenamic acid-Cd complexes have anti-proliferative activity in the breast cancer cell line MCF7, bladder cancer cell line T24, and the non-small cell lung carcinoma line A549 [24]. So the question becomes: is Cd just a causal factor in these studies, or does it really possess the potential to inhibit cancer cell proliferation? The answer and involved mechanism(s) remain unknown. It is likely that Cd may exert a paradoxical effect in breast cancer [25] perhaps dependent on the form it exists in: free Cd, protein-bound Cd, and Cd complexed with novel ligands such as those described in this study, namely, indole-3-butyric acid (L1) or indole-3-propionic acid (L2), may exert either favorable or unfavorable effects in a breast cancer system. We propose that Cd, at least when it is complexed to the above ligands, exerts a very favorable anti-tumor effect in breast cancer cells. Many years ago, L1 and L2 were shown to possess cancer-preventive effects in a rat carcinoma model [26]. L2 also potently and via iron-inducing mechanisms, caused oxidative damage to cell membranes and, potentially, prevented carcinogenesis [27]. We have previously reported that the L-glutamine Schiff base copper complex [11], taurine Schiff base copper complex [12] and quinoline-2-carboxaldehyde Schiff base copper complexes [28] could act as potent proteasome inhibitors and induced apoptosis. Similarly, 3, 5-diaminobenzoic acid o-vanillin Schiff base (L3) has also been used in the synthesis of such compounds [29].
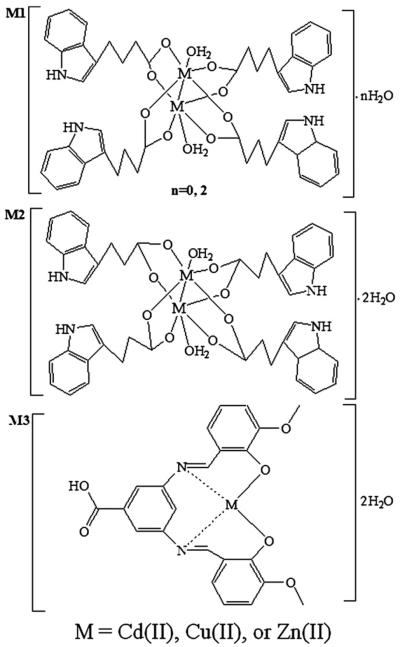
We hypothesized that synthetic forms of Cd with L1, L2 and L3 may have cancer-specific proteasome-inhibitory and apoptosis-inducing activities. To test this hypothesis, we have synthesized three novel Cd-containing complexes: [Cd2(C12H12O2N)4(H2O)2]·2H2O (Cd1), [Cd2(C11H10O2N)4(H2O)2]·2H2O (Cd2) and [Cd(C7H4N2O2)(C8H6O2)2]· 2H2O (Cd3) (Fig. 1) by using indole-3-butyric acid (L1), indole-3-propionic acid (L2) and 3, 5-diaminobenzoic acid o-vanillin Schiff base (L3), respectively, as ligands. We report here that these Cd complexes are potent inhibitors of the proteasome and inducers of apoptosis, effects which appear to be specific to tumor cells. We have first characterized and assessed these newly synthesized Cd complexes. We then compared the ability of the similar metal complexes containing copper (Cu), zinc (Zn) or Cd to inhibit breast cancer cell proliferation using the estrogen receptor (ER)-positive MCF7 and ER-negative MDA MB 231 breast cancer cell lines. Of the compounds tested, the Cd-containing versions appear to be the most potent inhibitors of cellular proteasome CT-like activity and effective inducers of apoptosis in breast cancer cells, but not in non-tumorigenic breast epithelial MCF10A cells. Additionally, these newly synthesized Cd compounds are superior in potency and cancer selectivity to the DSF-Cd mixture.
Fig. 1.
Chemical structures of M1, M2 and M3 (M=Cd(II), Cu(II), or Zn(II)).
2. Experimental
2.1. Materials
Indole-3-butyric acid, indole-3-propionic acid, 3, 5-diaminobenzoic acid and cadmium acetate were all purchased from Aladdin (Los Angeles, CA). The chemical agents, DMSO and 3-[4, 5-dimethyltiazol-2-yl]-2.5-diphenyl-tetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO). All compounds were made as 50 mM stocks in DMSO and stored at 4 °C. DMEM/F12 (1:1), RPMI-1640 and penicillin/streptomycin were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Aleken Biologicals (Nash, TX, USA). The fluorogenic peptide substrate Suc-LLVY-AMC (for the CT-activity assay) was purchased from Calbiochem (San Diego, CA). Rabbit polyclonal antibody against human poly (ADP-ribose) polymerase (PARP) (H-250) was purchased from BD Bioscience Pharmingen (San Diego, CA). Mouse monoclonal antibodies against ubiquitin (P4D1) and IκB-α (H-4), goat polyclonal antibody against β-actin (C-11) and all secondary antibodies were purchased from Santa Cruz (Santa Cruz, CA).
2.2. Metal complex syntheses
Cd1, Cd2, Cu1, Cu2, Zn1, Zn2: Synthesis of these complexes followed the previously described procedure [30]. The ligand C12H13O2N or C11H11O2N (1 mM) was dissolved in 5 ml of H2O. M (CH3COO)2·2H2O (0.5 mM) dissolved in 10 ml of anhydrous ethanol was added drop wise to the above solution with stirring and reacted for 3 h to give a milky precipitate, which was filtered off, to produce the final complex.
Cd1: Yield, 73%; Anal. Calc. for Cd1 (%, [Cd2(C12H12O2N)4(H2O)2]·2H2O, FW=1105.80 g·mol−1); C, 52.13; H, 5.10; N, 5.07. Found (%): C, 51.75; H, 5.21; N, 5.19. UV: λmax (nm): 238, 285. IR data (KBr, cm−1): 3393.68, υ (–NH–); 1570.95, υas (COO−); 1457.26, υs (COO−); 475.99, υ (Cd–O). 1H NMR (DMSO, 600 MHz, s, singlet; d, doublet; t, triplet): δ (ppm) 10.718 (1H, s, –NH–); 7.506 (1H, d, C(9)H); 7.321 (1H, d, C(12)H); 7.077 (1H, d, C(5)H); 7.039 (1H, t, C(10)H); 6.941 (1H, t, C(11)H); 3.331 (2H, s, C(2)H); 2.693 (2H, t, C(3)H); 2.191 (2H, t, C(4)H). 13C NMR (DMSO): δ (ppm) 180.15 (–COO), 136.16, 127.14, 122.00, 120.63, 118.28, 117.90, 114.41 and 111.15 (indole ring carbons), 34.40, 26.91 and 24.41 (–CH2). TG analysis: lost 3.70% (calculated 3.25%, 2H2O) in the first step at 25–150 °C; residue 23.65% (calculated 23.22%, CdO). Molar conductivity: Λm (S·cm2·mol−1): 9.41.
Cd2: Yield, 78%; Anal. Calc. for Cd2 (%, [Cd2(C11H10O2N)4(H2O)2]·2H2O, FW=1049.69 g·mol−1); C, 50.34; H, 4.61; N, 5.34. Found (%): C, 50.79; H, 5.01; N, 5.62. UV: λmax (nm): 236, 284. IR data (KBr, cm−1): 3390.36, υ (–NH–); 1548.05, υas (COO−); 1414.59, υs (COO−); 498.15, υ (Cd–O). 1H NMR (DMSO, 600 MHz, s, singlet; d, doublet; t, triplet): δ (ppm) 10.726 (1H, s, –NH–); 7.498 (1H, d, C(8)H); 7.325 (1H, d, C(11)H); 7.116 (1H, d, C(4)H); 7.050 (1H, t, C(9)H); 6.964 (1H, t, C(10)H); 3.338 (2H, s, C(2)H); 2.934 (2H, t, C(3)H). 13C NMR (DMSO): δ (ppm) 179.79 (–COO), 136.10, 127.01, 121.95, 120.67, 118.12, 117.98, 114.37 and 111.14 (indole ring carbons), 35.40, 26.91 and 21.71 (–CH2). TG analysis: lost 3.56% (calculated 3.43%, 2H2O) in the first step at 25–165 °C; residue 25.08% (calculated 24.46%, CdO). Molar conductivity: Λm (S·cm2·mol−1): 9.39.
Cu1: Yield, 68%; Anal. Calc. for Cu1 (%, [Cu2(C12H12O2N)4(H2O)2], FW=972.04 g·mol−1); C, 59.31; H, 5.39; N, 5.76. Found (%): C, 58.98; H, 5.65; N, 5.24. UV: λmax (nm): 237, 286. IR data (KBr, cm−1): 3394.81, υ (–NH–); 1588.13, υas (COO−); 1419.04, υs (COO−); 472.16, υ (Cu–O). 1H NMR (DMSO, 600 MHz): δ (ppm) 10.973 (1H, s, –NH–); 7.719 (1H, d, C(9)H); 7.423 (1H, d, C(12)H); 7.091 (1H, d, C(5)H); 7.052 (1H, t, C(10)H); 7.010 (1H, t, C(11)H); 3.357 (2H, s, C(2)H); 2.833 (2H, t, C(3)H); 2.21 (2H, t, C(4)H). TG analysis: lost 82.88% (calculated 83.63%) in only one step at 25–800 °C; residue 17.12% (calculated 16.37%, CuO). Molar conductivity: Λm (S·cm2·mol−1): 11.00.
Cu2: Yield, 62%; Anal. Calc. for Cu2 (%, [Cu2(C11H10O2N)4(H2O)2]·2H2O, FW=951.96 g·mol−1); C, 55.51; H, 5.08; N, 5.88. Found (%): C, 55.11; H, 5.22; N, 5.97. UV: λmax (nm): 234, 285. IR data (KBr, cm−1): 3408.16, υ (–NH–); 1591.13, υas (COO−); 1464.37, υs (COO−); 501.32, υ (Cu–O). 1H NMR (DMSO, 600 MHz): δ (ppm) 10.189 (1H, s, –NH–); 7.858 (1H, d, C(8)H); 7.627 (1H, d, C(11)H); 7.321 (1H, d, C(4)H); 7.139 (1H, t, C(9)H); 6.885 (1H, t, C(10)H); 3.797 (2H, s, C(2)H); 3.312 (2H, t, C(3)H). TG analysis: lost 3.84% (calculated 3.78%, 2H2O) in the first step at 25–125 °C; residue 17.33% (calculated 16.71%, CuO). Molar conductivity: Λm (S·cm2·mol−1): 6.59.
Zn1: Yield, 79%; Anal. Calc. for Zn1 (%, [Zn2(C12H12O2N)4(H2O)2]·2H2O, FW=1011.80 g·mol−1); C, 56.98; H, 5.58; N, 5.54. Found (%): C, 56.71; H, 5.66; N, 5.35. UV: λmax (nm): 234, 286. IR data (KBr, cm−1): 3389.91, υ (–NH–); 1539.23, υas (COO−); 1414.64, υs (COO−); 475.65, υ (Zn–O). 1H NMR (DMSO, 600 MHz): δ (ppm) 10.577 (1H, s, –NH–); 7.630 (1H, d, C(9)H); 7.415 (1H, d, C(12)H); 7.001 (1H, d, C(5)H); 6.887 (1H, t, C(10)H); 6.823 (1H, t, C(11)H); 3.509 (2H, s, C(2)H); 2.944 (2H, t, C(3)H); 2.368 (2H, t, C(4)H). TG analysis: lost 3.65% (calculated 3.27%, 2H2O) in the first step at 25–165°C; residue 17.42% (calculated 16.09%, ZnO). Molar conductivity: Λm (S·cm2·mol−1): 7.89.
Zn2: Yield, 69%; Anal. Calc. for Zn2 (%, [Zn2(C11H10O2N)4(H2O)2]·2H2O, FW=955.69 g·mol−1); C, 55.30; H, 5.06; N, 5.86. Found (%): C, 55.61; H, 5.43; N, 5.52. UV: λmax (nm): 236, 287. IR data (KBr, cm−1): 3399.26, υ (–NH–); 1579.18, υas (COO−); 1439.09, υs (COO−); 481.65, υ (Zn–O). 1H NMR (DMSO, 600 MHz): δ (ppm) 11.145 (1H, s, –NH–); 7.956 (1H, d, C(8)H); 7.743 (1H, d, C(11)H); 7.544 (1H, d, C(4)H); 7.273 (1H, t, C(9)H); 7.041 (1H, t, C(10)H); 3.626 (2H, s, C(2)H); 3.135 (2H, t, C(3)H). TG analysis: lost 3.59% (calculated 3.77%, 2H2O) in the first step at 25–155 °C; residue 17.47% (calculated 17.04%, ZnO). Molar conductivity: Λm (S·cm2·mol−1): 8.97.
Cd3, Cu3 and Zn3: Synthesis of these complexes followed the previously described procedure [29] and these compounds have been characterized by the following elemental analysis, IR, 1H NMR and TG analysis.
Cd3: Yield, 56%; Anal. Calc. for Cd3 (%, [Cd(C7H4N2O2) (C8H6O2)2]· 2H2O, FW=564.82 g·mol−1); C, 48.91; H, 3.57; N, 4.96. Found (%): C, 48.60; H, 3.74; N, 4.87. IR data (KBr, cm−1): 1635.46, υ (COOH); 493.27, υ (Cd–O); 415.04, υ (Cd–N). 1H NMR (DMSO, 600 MHz): δ (ppm) 12.772 (1H, s, COOH); 9.347 (2H, s, CH=N); 6.392–7.855 (6H, m, Ar–H); 3.911 (6H, s, OCH3). TG analysis: lost 3.28% (calculated 3.19%, 2H2O) in first step at 25–175 °C; residue 23.11% (calculated 22.73%, CdO).
Cu3: Yield, 60%; Anal. Calc. for Cu3 (%, [Cu(C7H4N2O2) (C8H6O2)2]·2H2O, FW=515.96 g·mol−1); C, 53.54; H, 3.91; N, 5.43. Found (%): C, 53.13; H, 3.99; N, 5.35. IR data (KBr, cm−1): 1597.58, υ (COOH); 475.36, υ (Cu–O); 448.19, υ (Cu–N). 1H NMR (DMSO, 600 MHz): δ (ppm) 11.933 (1H, s, COOH); 9.289 (2H, s, CH=N); 6.676–7.883 (6H, m, Ar–H); 3.435 (6H, s, OCH3). TG analysis: lost 6.82% (calculated 6.99%, 2H2O) in first step at 25–155 °C; residue 15.55% (calculated 15.42%, CuO).
Zn3: Yield, 58%; Anal. Calc. for Zn3 (%, [Zn(C7H4N2O2) (C8H6O2)2]·2H2O, FW=517.82 g·mol−1); C, 53.35; H, 3.89; N, 5.41. Found (%): C, 52.95; H, 4.01; N, 5.48. IR data (KBr, cm−1): 1615.52, υ (COOH); 483.04, υ (Zn–O); 431.86, υ (Zn–N). 1H NMR (DMSO, 600 MHz): δ (ppm) 12.016 (1H, s, COOH); 9.376 (2H, s, CH=N); 6.319–7.721 (6H, m, Ar–H); 3.893 (6H, s, OCH3). TG analysis: lost 7.30% (calculated 6.95%, 2H2O) in first step at 25–140 °C; residue 16.04% (calculated 15.72%, ZnO).
2.3. Elemental analysis and NMR spectrum
Elemental analysis (C, H and N) was performed using a 2400 Perkin-Elmer analyzer. Infrared spectrum was recorded as KBr pellets on the Nicolet 170SX spectrophotometer in the 4000–400 cm−1 region. 1H NMR spectrum was recorded on an AVANCE III (600 MHz) spectrometer and the 13C NMR spectra on an AV-600 Bruker (600 MHz) spectrometer.
2.4. Cell culture and whole cell extract preparation
MCF7 and MDA MB 231 cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and cultured in RPMI-1640 or DMEM/F-12 (1:1) respectively, supplemented with: 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 μg/ml streptomycin from Life Technology (Carlsbad, CA). MCF10A cells (immortalized, but non-tumorigenic) were generously provided by Dr. Fred Miller (Karmanos Cancer Institute, Detroit, MI, USA) and grown in 1:1 DMEM/F12 supplemented with: 5% (v/v) horse serum, 0.029 M sodium bicarbonate, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer solution, 100 U/ml of penicillin, 5 mg insulin, 10 μg of epidermal growth factor, and 250 μg hydrocortisone. All cell lines were maintained at 37 °C in an atmosphere containing 5% CO2. The cells were harvested, lysed and supernatants were collected as whole cell extracts used for Western blot analysis as previously described [31].
2.5. Cell proliferation assay
The effect of each compound on cell proliferation was determined by 1-(4,5-dimethylthiazol-2-yl)-3,5-diphenylformazan, thiazolyl (MTT) assay. MCF7 and MDA MB 231 cells were seeded in triplicate in a 96-well plate and incubated at 37 °C until 70–80% confluent, then treated with the indicated concentration of each compound for 24 h. Media was then removed and MTT solution (1 mg/ml) was added followed by a 2 h incubation period. MTT was then removed and 100 μl DMSO added to dissolve the metabolized MTT product, followed by measuring the absorbance values on a Victor 3 multi-label plate reader (PerkinElmer Wellesley, MA).
2.6. Nucleophilic susceptibility analysis
The CAChe work system (v 6.1) (Fujitsu America Inc Beaverton, OR) was used for performing the Extended Huckel molecular orbital calculations. The chemical structures were constructed and molecules were subjected to geometry optimization. Nucleophilic susceptibility was treated and the electron density surface was colored by nucleophilic susceptibility. The highest susceptibility towards nuceophilic attack was signified by a `bull's-eye' with a white center.
2.7. In vitro proteasomal activity assay
Purified human 20S proteasome (35 ng) was incubated in 100 μl of assay buffer (20 mM Tris–HCl, pH 7.5) and 20 μM of chymotrypsin-like substrate Suc-LLVY-AMC, using different concentrations of Cd1, Cd2 and Cd3 or DMSO as a vehicle control for 2 h at 37 °C. Following incubation, proteasome CT-like activity was measured using the Wallac Victor 3 Multi-label Counter with an excitation filter of 365 nm and emission filter of 460 nm.
2.8. Proteasomal CT-like assay using breast cancer cells
MCF7 and MDA MB 231 cells were treated as indicated, lysed and protein concentrations measured by the Bio-Rad Protein Assay (Bio-Rad Hercules, CA). Whole cell extracts (10 μg) were incubated for 2 h at 37 °C in 100 μl of assay buffer (20 mM Tris–HCl, PH 7.5) with 20 μmol/l fluorogenic peptide substrate Suc-LLVY-AMC. Proteasomal CT-like activity was measured using the Wallac Victor 3 Multi-label Counter with an excitation filter of 365 nm and emission filter of 460 nm.
2.9. Western blot assay
Proteins (40 μg) from whole cell extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane, followed by visualization with an enhanced chemiluminescence reagent (Denville Scientific Metuchen, NJ), as previously described [32].
2.10. Cellular morphology analysis
Cellular morphology changes were observed using a Zeiss (Thornwood, NY) Axiovert 25 microscope with phase contrast, as previously described [10].
3. Results
3.1. Synthesis and characterization of Cd complexes
We previously reported that the DSF-Cd mixture could selectively inhibit proliferation and induce apoptosis in human breast cancer cells, but not in their non-tumorigenic counterparts [15]. However, the effect of synthetic Cd complexes on tumor cells has not been studied, and the comparison of Cd to other metals in similar complexes has not been done. We therefore synthesized three Cd-based compounds: Cd1, Cd2 and Cd3 (Fig. 1) as well as several similar Cu and Zn complexes (Cu1, Zn1, Cu2, Zn2, Cu3, and Zn3) for a comparison. The contents of carbon, hydrogen and nitrogen in each complex were measured by an elemental analysis instrument, which confirmed the composition of these complexes.
After analysis of the infrared spectra of the synthesized dinuclear metal containing complexes (Cd1, Cd2, Cu1, Cu2, Zn1 and Zn2), the strong –NH– absorption peaks did indeed appear in the range of 3389.91–3408.16 cm−1 without any obvious shift compared to the ligands (data not shown). This observation suggests that the nitrogen within the indole ring did not form a coordination bond with the metal ions. In comparison, the infrared spectra of the ligands within the metal complexes produced two new peaks at 1539.23–1591.13 cm−1 and 1414.59–1464.37 cm,−1 which could be attributed to υas (COO−) and υas (COO−), respectively. Furthermore, the magnitude of νas (COO−)– νs) (COO−) was less than 200 cm−1 for these complexes, indicating that the oxygen in the –COO− group is coordinated to the metal ion in a bidentate fashion [33]. A new peak also appeared at 472.16–501.32 cm−1 in these complexes, which could be due to the vibration of M–O. According to the Cd3, Cu3 and Zn3 IR data, the 3563.94 cm−1 band present in the spectra of the metals complexes to L3 is attributed to the ν (OH) vibrations. In these complexes, this band disappears confirming that oxygen coordinates to the metal ions. The COOH group remains free as shown by infrared bands at 1635.46 cm−1. New bands in the 493.27 cm−1 and 415.04 cm−1 could be attributed to ν (M–O) and ν (M–N) modes, respectively.
The specifics of the 1H NMR and 13C NMR spectra are described in the Experimental section. From the 1H NMR spectra analysis we conclude that the complexes do consist of coordination between the ligands L1, L2 and the M (II) ion, and that the small shifts seen in the signals are present due to the electronic redistribution of the ligand and its interaction with M (II). Additionally, the hydrogen atom of -NH- was found to still be present, while the hydrogen atom of –COOH was in fact displaced by a metal ion. Our observations from the 1H NMR spectra are further supported by 13C NMR data. Compared with the free ligands, L1 and L2, the 13C NMR signals have values corresponding to a downfield shift upon coordination with Cd (II) (1H NMR and 13C NMR data of ligands L1 and L2 are not shown). According to the Cd3, Cu3 and Zn3 1H NMR data, broad bands in the 11.933–12.772 ppm range are assigned to the COOH groups. The singlet due to azomethine (–CH=N–) is in the 9.289–9.376 ppm range. Aromatic ring protons are shown in 6.319–7.883 ppm ranges. The very strong singlet in the 3.435–3.911 ppm range are attributed to –OCH3. The hydrogen atom of phenolic hydroxyl disappears and displaced by a metal ion. Consequently, the newly synthesized complexes were indeed formed by coordination with the metal ion, a conclusion further supported by IR output data.
The UV-Vis absorption spectra obtained of Cd1, Cd2, Cu1, Cu2, Zn1 and Zn2 complexes, which were dissolved in DMSO, was recorded in the 200–500 nm range. The λmax values, 234–238 nm and 284–287 nm can be attributed to the π–π* and n–π* transition of ligands, respectively. Furthermore, the absorption bands tend to shift towards longer wavelengths, which can also be ascribed to the metal-to-ligand charge transfer transitions taking place.
TG-analysis of these complexes was recorded in the range of 25–800 °C. These complexes were decomposed primarily in two steps except Cu1. In the first step, 3.28%–7.30% of these metal complexes (calculated as 3.25%–6.99%) were lost, respectively, which was attributed to the two molecules of crystal water. The residue rates of the metal complexes were 15.55–25.08%, respectively, consistent with the calculated values (15.42%–24.46%).
All of the compounds are soluble in DMF and DMSO and stable in air. The molar conductivities (Λm) of these complexes in DMSO were 6.59–11.00 S·cm2·mol−1, respectively, which is less than 35 S·cm2·mol−1 [34]. Hence, these complexes were considered to be nonelectrolytes and are quite stable in DMSO.
3.2. Inhibition of purified 20S proteasome activity by Cd1, Cd2 and Cd3
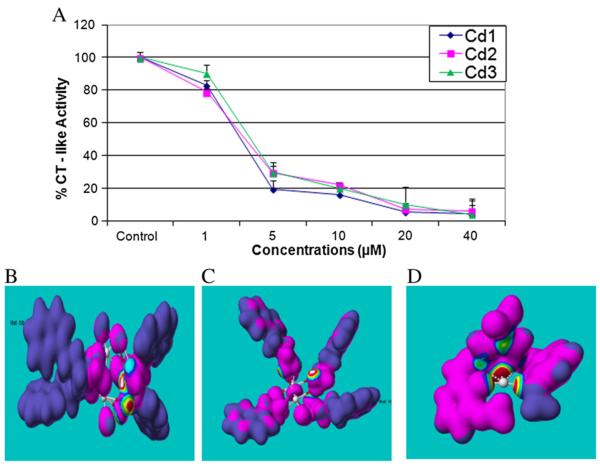
After chemical analysis of the three Cd complexes, we then investigated whether they were capable of inhibiting proteasome CT-like activity. A proteasome activity assay (using a fluorogenic substrate specific for the CT-like subunit) was performed using purified human 20S proteasome in the presence of various concentrations of Cd1, Cd2 or Cd3. The results indicate that all three complexes are capable of inhibiting proteasomal CT-like activity with IC50 values of 2.6, 3.0, and 3.3 μM, respectively (Fig. 2A). These data suggest that these Cd complexes could target the 20S proteasomal catalytic β5 subunit.
Fig. 2.
Purified 20S proteasome inhibition by Cd1, Cd2 and Cd3, supported by nucleophilic susceptibity analysis. A: Purified human proteasome (35 ng) was incubated with DMSO or various concentrations of Cd1, Cd2 and Cd3 for 2 h, followed by proteasomal chymotrypsin-like activity assay. Points, mean+/−SE of three independent experiments; bars, SD. B, C, D: The nucleophilic susceptibility of Cd1, Cd2 and Cd3 was analyzed by using CAChe software. Molecular orbital energy analysis is shown by drawing an electron density isosurface and the highest susceptibility towards nuceophilic attack was signified by white center.
3.3. Cd1, Cd2 and Cd3 bind to and inhibit the proteasome β5 subunit in an in silico model
In order to further explain how and why the Cd complexes act as proteasomal CT activity inhibitors, computational electron density analysis was performed in order to speculate whether they could actually bind to and inhibit the β5 subunit-mediated CT-like activity of the proteasome in an in silico model. The Cd complex structures were refined by performing an optimized geometry calculation in MOPAC, using PM5 parameters in the CAChe software. First, the chemical structures were constructed and the geometry molecules optimized. Second, the nucleophilic susceptibility was determined, and finally the electron density surface colored accordingly. Indeed, the results depict Cd1, Cd2 and Cd3 as all highly susceptible to nucleophilic attack, forming a `bull's-eye' with either a white center (Cd1, Cd2) (Fig. 2B-C) or red center (Cd3) (Fig. 2D). These observations further suggest that nucleophilic susceptibility of these Cd complexes is associated with their potency and ability to inhibit 20S proteasome activity. However, the detailed mechanism of inhibition needs to be further studied.
3.4. Cd compounds inhibit cell proliferation and proteasome activity in ER-positive MCF7 and ER-negative MDA MB 231 breast cancer cells
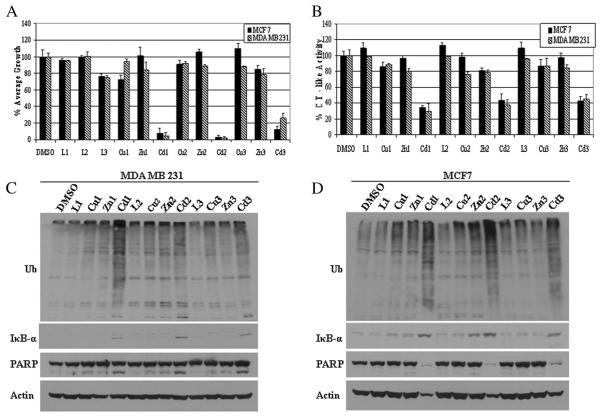
For this portion of our study, we asked the question of whether or not our Cd complexes were more valuable as proteasome inhibitors, and thus inhibitors of cell proliferation, than other metal complexes. We began by first testing their effects on cell proliferation using the ER-positive human breast cancer MCF7 and ER-negative MDA MB 231 cells and treating them with 40 μM of each compound for 24 h, after which a MTT cell proliferation assay was performed. From this experiment we found that Cd1 and Cd2 were most potent, causing more than 90% growth inhibition in both cell lines, while Cd3 was responsibly potent for an 88% (MCF7) or 74% (MDA MB 231) growth decline (Fig. 3A). However, all the ligands L1, L2, L3 as well as copper and zinc complexes had little or no growth inhibition on the breast cancer cells tested.
Fig. 3.
Comparison of cell proliferation and proteasome inhibition in MCF-7 and MDA-MB-231 cells treated with different compounds. MCF-7 and MDA-MB-231 cells were treated with each indicated compound at 40 μM for 24 h, followed by MTT assay (A), the chymotrysin-like activity assay (B), and Western blot analysis of polyubiquitinated proteins, PARP, IκB-α (C, D). Molecular weight of intact PARP is 116 kDa and the cleaved PARP fragment is 85 kDa. Molecular weight of IκB-α is 37 kDa. β-actin was used as a loading control. Columns, mean+/−SE of three independent experiments; bars, SD.
Next, we measured the ability of these metal complexes to inhibit proteasome activity and induce apoptotic cell death. MCF7 and MDA MB 231 cells were again treated using 40 μM of each compound for 24 h and the effect on proteasomal CT-like activity, accumulated ubiquitinated proteins and the level of the proteasome target protein IκB-α were assessed. The CT-like activity assay produced results which signify the Cd complexes as potent inhibitors, with a 55%–71% drop in CT-like activity in both breast cancer cell lines (Fig. 3B). In contrast, all the ligands failed to inhibit proteasome activity and the inhibitory effects of the Cu and Zn complexes were not significant (Fig. 3B). Consistently, the accumulation of ubiquitinated proteins and the proteasome target protein IκB-α (37 kDa) was also observed in the cells treated with Cd1, Cd2 or Cd3, but not others (Fig. 3C & D). A vast amount of literature exists linking tumor cell apoptosis as a result of proteasome inhibition [5,8,35], which therefore supported our interest in determining if this too was a result of the Cd complexes in question. In the same experiment, we noticed the appearance of the cleaved fragment (85 kDa), specific to the cell death-specific protein Poly (ADP-ribose) polymerase (PARP), in response to treatment of MDA MB 231 cells with the Cd complexes (Fig. 3C). Interestingly, no PARP cleavage was observed in the MCF7 cells, however the 116 kDa full-length PARP decreased, and even disappeared (Fig. 3D). We also noticed low levels of PARP cleavage produced in MDA MB 231 cells and no changes in full length PARP levels in MCF-7 cells, after treatment with Cu or Zn complexes (Fig. 3C). Our results suggest that Cd1, Cd2 and Cd3 are more potent in their ability to inhibit the proteasome and induce tumor cell apoptosis than those other compounds tested (L1, L2, L3, Cu1, Cu2, Cu3, Zn1, Zn2 and Zn3). Although the effect of Cd3 is slightly less robust than that of Cd1 and Cd2, these Cd complexes have very similar effect on the two breast cancer cell lines tested, ER-positive MCF7 and ER-negative MDA MB 231, suggesting an ER-independent mechanism of action.
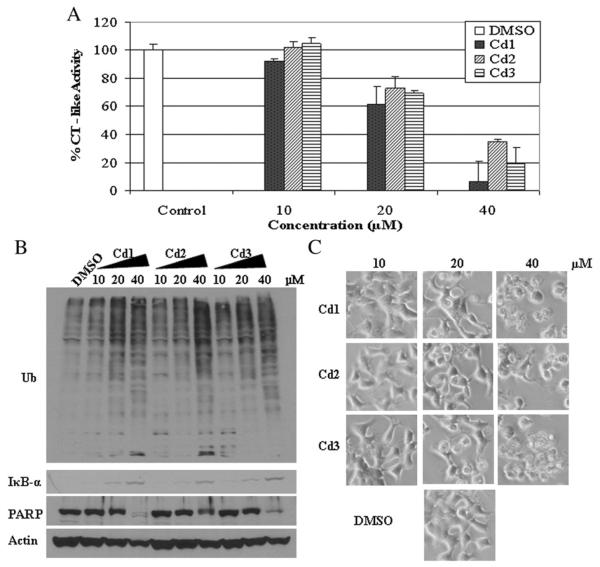
3.5. Concentration-dependent effect of Cd1, Cd2 and Cd3 on proteasome inhibition and apoptosis induction in ER-negative MDA MB 231 cells
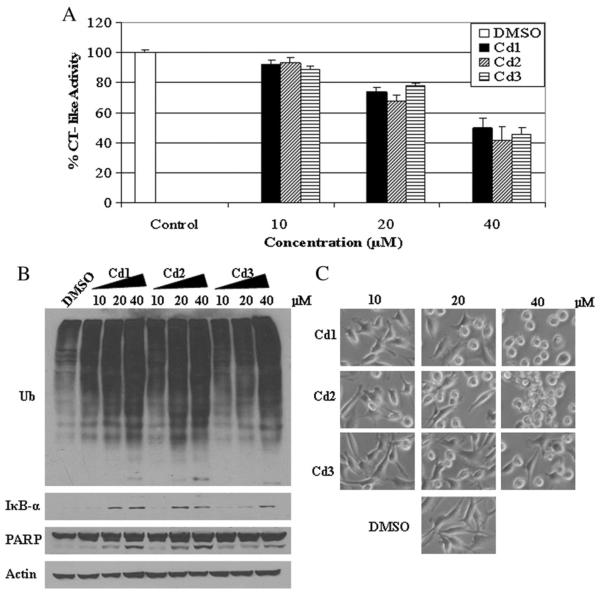
Because Cd1, Cd2 and Cd3 were all able to inhibit CT-like activity of the proteasome, we next sought out to determine if this effect is concentration-dependent. MDA MB 231 cells were treated with the Cd complexes at concentrations of 10, 20 and 40 μM for 24 h. Cells treated with DMSO were used as a vehicle control. The results show that all compounds at 10 μM generate about 10% inhibition of proteasome CT-like activity, and on average 55% inhibition at 40 μM (Fig. 4A). Consistently, the accumulation of ubiquitinated proteins and IκB-α was also observed in MDA MB 231 cells treated with Cd1, Cd2 and Cd3 in a concentration-dependent manner (Fig. 4B). In the same experiment and at the 40 μM concentration, we detected cellular morphological changes (shrunken and rounded up cell appearance) (Fig. 4C) as well as PARP cleavage (Fig. 4B), indicative of cellular apoptosis. The PARP cleavage fragment p85 appeared at 20 μM and 40 μM of Cd1 and Cd2 and at 40 μM of Cd3 (Fig. 4B). Our results demonstrate that Cd1, Cd2 and Cd3 all possess proteasome inhibition capability and induce apoptosis in a concentration-dependent manner in the ER-negative MDA MB 231 human breast cancer cells.
Fig. 4.
Concentration effects of Cd1, Cd2 and Cd3 on breast negative cancer MDA-MB-231 cells. MDA-MB-231 cells were treated with DMSO (as control) or different concentrations of Cd1, Cd2 and Cd3 for 24 h, followed by the chymotrysin-like activity assay (A), Western blot analysis of polyubiquitinated proteins, PARP and IκB-α (B), and cellular morphologic changes (C). Columns, mean+/−SE of three independent experiments; bars, SD.
3.6. Concentration-dependent effect of Cd1, Cd2 and Cd3 on proteasome inhibition and apoptosis induction in ER-positive MCF7 cells
To investigate whether these complexes have a similar concentration-dependent effect in ER-positive MCF7 breast cancer cells we treated MCF-7 cells with Cd1, Cd2 or Cd3 using the same experimental conditions as above. The results indicate that at 10 μM, only Cd1 was able to inhibit proteasomal CT-like activity by about 10% (Fig. 5A). However, Cd1, Cd2 and Cd3 at 40 μM were very potent, with degrees of inhibition being 93% (Cd1), 65% (Cd2) and 80% (Cd3) (Fig. 5A), respectively. Consistently, the accumulation of ubiquitinated proteins and IκB-α was also observed in MCF7 cells treated with Cd1, Cd2 and Cd3 in a concentration-dependent manner (Fig. 5B).
Fig. 5.
Concentration effects of Cd1, Cd2 and Cd3 on breast cancer MCF-7 cells. MCF-7 cells were treated with DMSO (as control) or different concentrations of Cd1, Cd2 and Cd3 for 24 h, followed by the chymotrysin-like activity assay (A), Western blot analysis of polyubiquitinated proteins, PARP and IκB-α (B), and cellular morphologic changes (C). Columns, mean+/−SE of three independent experiments; bars, SD.
When assessing PARP cleavage in characterizing the apoptosis-inducing ability of these compounds in MCF7 cells, we observed a reduction in the p116 full length PARP which disappeared at the 40 μM concentration of Cd1, Cd2 and Cd3 (Fig. 5B). Consistently, morphological changes, indicative of cellular apoptosis, were observed at the 20 μM and 40 μM concentrations (Fig. 5C). Our results demonstrate that the Cd complexes possess the ability to inhibit the proteasome and induce apoptosis in a concentration-dependent manner in ER-positive MCF7 cells.
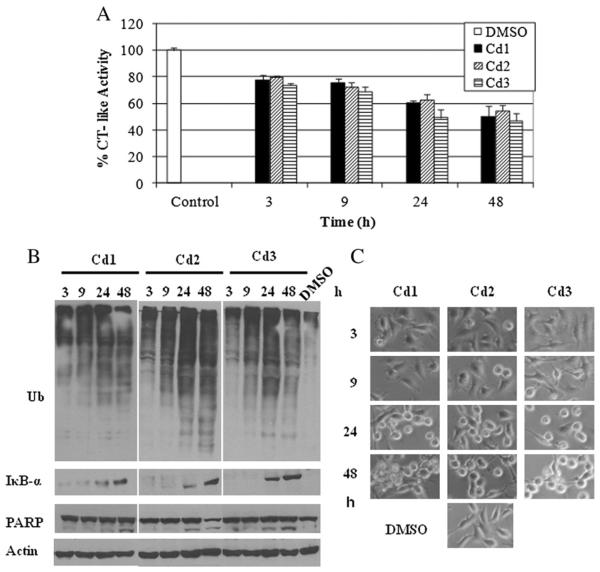
3.7. Cd1, Cd2 and Cd3 sequentially induce time-dependent proteasome inhibition and apoptosis in MDA-MB-231 cells
To ascertain the relationship between proteasome inhibition and apoptosis induction, we performed a kinetic experiment. MDA MB 231 cells were treated with 20 μM of Cd1, Cd2 and Cd3 for 3–48 h (DMSO treated cells as a vehicle control), followed by measurement of proteasomal inhibition and cell death (Fig. 6). We found that Cd1, Cd2 and Cd3 were able to inhibit 22%, 20% and 26% of proteasomal CT-like activity after 3 h of treatment, respectively (Fig. 6A). Up to the 48 h time-point, ~50% CT-like inhibition by Cd1, ~45% by Cd2 and ~53% by Cd3 was observed (Fig. 6A). Furthermore, Western blot analysis showed that the accumulation of ubiquitinated proteins appeared as early as 3 h of treatment and enhanced gradually as the time went on, peaking at 24 h (Fig. 6B). Also, increased levels of IκB-α were detected at 24 and 48 h of treatment with all three Cd complexes (Fig. 6B). In the same kinetic experiment, the cleaved PARP fragment p85 appeared 24 h after treatment (Fig. 6B). Furthermore, apoptotic morphological changes were detected after 24 h of treatment with each complex, also increasing gradually as time progressed (Fig. 6C). Our results support the notion that Cd1, Cd2 and Cd3 induce proteasome inhibition, followed by apoptosis induction in breast tumor cells.
Fig. 6.
Kinetic effect of proteasome inhibition and apoptosis induction by Cd1, Cd2 and Cd3 in MDA-MB-231 cells. MDA-MB-231 cells were exposed to 20 μM of Cd1, Cd2 and Cd3 for the indicated times, followed by the proteasomal chymotrypsin-like activity assay (A), Western blot assay (ubiquitin, PARP, IκB-α and β-actin, B), and the apoptosis morphologic changes (C.) Columns, mean+/−SE of three independent experiments; bars, SD.
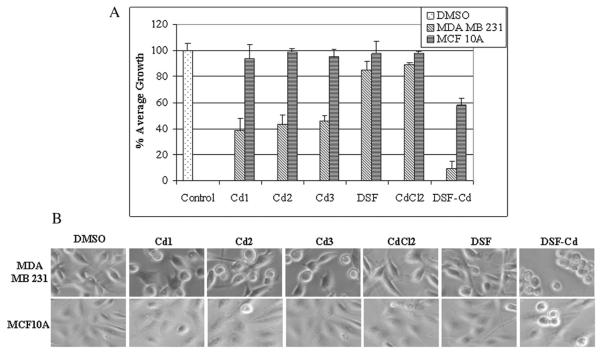
3.8. Cd1, Cd2 and Cd3 exhibit a less toxic effect in immortalized, non-tumorigenic breast MCF10A cells when compared to MDA MB 231 breast cancer cells
It is an important criterion for novel anti-cancer drugs to have the ability to induce apoptosis in tumor, but not normal cells. To investigate whether Cd1, Cd2 and Cd3 are nontoxic in “normal” or non-tumorigenic cells, highly metastatic MDA MB 231 breast cancer cells and the immortalized, but non-tumorigenic breast MCF10A cells were treated with 20 μM of Cd1, Cd2, Cd3 for 24 h, with DSF, CdCl2, DSF-Cd and DMSO as a comparison, followed by MTT assay and cellular morphological analysis. Based on the MTT results using MDA MB 231 cells, Cd1, Cd2 and Cd3 all appear to have a similar growth-inhibitory potency, causing 38%, 43% and 46% growth inhibition, respectively (Fig. 7A). Meanwhile, DSF and CdCl2 alone caused only slight growth inhibition, however, the combined DSF-Cd mixture was the most potent (~90% inhibition). In this regard, it is important to note that though DSF-Cd mixture was most robust against MDA MB 231 cell proliferation, the Cd complexes are much less toxic to the non-tumorigenic breast MCF10A cells than the DSF-Cd (5% growth inhibition versus 42%) (Fig. 7A), making our novel Cd complexes more favorable for further pre-clinical studies. Furthermore, consistent with MTT assay results, visual indications of apoptosis were almost nonexistent in MCF10A cells treated with the Cd complexes, as opposed to the shrunken and rounded up features seen in the MDA MB 231 cells under the same conditions (Fig. 7B). Most cell death in MDA MB 231 cells and some cell round up in MCF10A cells were observed after DSF-Cd mixture treatment (Fig. 7B). Taken together, our study demonstrates that Cd1, Cd2 and Cd3 are certainly potent in breast cancer cells and less toxic than the DSF-Cd mixture to immortalized, non-tumorigenic MCF10A cells.
Fig. 7.
Differential effects of Cd complexes on cancer and non-tumorigenic breast cells. Breast cancer MDA-MB-231 cells and non-cancer MCF-10A cells treated with 20 μM of Cd1, Cd2, Cd3, CdCl2, DSF and DSF-Cd mixture, or DMSO for 24 h, followed by MTT assay (A) and morphological changes evaluation (B). Columns, mean+/−SE of three independent experiments; bars, SD.
4. Discussion
Although Cd has been recognized as a human carcinogen and a relationship between Cd and breast, lung and prostate cancer occurrence may exist [36–38], a strong demonstration of Cd as such a factor in human cancer remains unseen. Furthermore, studies have shown that Cd can actually delay the onset of tumors [25] and that Cd containing compounds can inhibit tumor cell proliferation and induce apoptosis [24]. We previously reported that the complex formed by DSF and Cd (DSF–Cd) in solution could selectively inhibit proteasome activity and induce apoptosis in human cancer cells [15]. However, the shortcomings of that study included our inability to determine the nature of its coordination and chemical structure in solution and therefore posed a limit to our quantitative evaluation of this compound. Therefore, in order to further study the potential anti-tumor effect of Cd-containing complexes and to investigate the mechanism by which these complexes can inhibit tumor cell proliferation, in the current study we have synthesized three novel Cd-containing complexes Cd1, Cd2 and Cd3 using indole-3-butyric acid, indole-3-propionic acid and 3, 5-diaminobenzoic acid o-vanillin Schiff base as ligands, and have shown that they are tumor-specific proteasome inhibitors and apoptosis inducers.
First, we investigated whether these compounds were capable of proteasome inhibition using the purified 20S proteasome in an in vitro assay. The results proved that Cd1, Cd2 and Cd3 do inhibit CT-like activity of the purified 20S proteasome with IC50 values of 2.6, 3.0 and 3.3 μM (Fig. 2A), respectively. It is well established that the CT-like activity of the 20S proteasome, primarily associated with the β5 subunit [3] depends on the presence of the N-terminal threonine (Thr 1) residue that is responsible for catalyzing the cleavage of peptides by nucleophilic attack [39]. Our electron density analysis indicates that our newly designed Cd complexes are highly susceptible to nucleophilic attack and therefore are very likely to inhibit proteasomal CT-like function. However, the computational electron density analysis only suggests an association between nucleophilic susceptibility of the Cd complexes and their potency, and furthermore, ability to inhibit 20S proteasome activity (Fig. 2B). The detailed mechanism of inhibition needs to be further studied.
We expanded on this knowledge and have herein compared the proteasome-inhibitory potential of various metal containing complexes. We found that copper and zinc complexes with the same ligands have little activity, compared to Cd1, Cd2 and Cd3. The involved molecular basis is currently unknown to us. We found that Cd-coordinating compounds were most potent in their ability to inhibit breast cancer cell proliferation using the ER-positive MCF7 and ER-negative MDA MB 231 cell lines. This inhibition was strongly associated with shutdown (>90% at 40 μM) of CT-like activity of the proteasome (Figs. 3B, 4A and 5A), accumulation of ubiquitinated proteins, and aggregation of a prime proteasome target protein, IκB-α (Figs. 3C, 4B, and 5B). Correlating positively with these results was the observation that our Cd complexes also induced the cleavage of, or decrease in, full length PARP (Figs. 4B and 5B), indicating apoptosis occurrence, which was also supplemented nicely with phenotypic morphologic changes (Figs. 4C and 5C). Accumulation of ubiquitinated proteins occurred as early as 3 h, followed by PARP cleavage and cellular morphologic changes occurring 24 h post-treatment (Fig. 6B & C). Collectively, these findings indicate that Cd1, Cd2 and Cd3 inhibit tumor cell proteasome activity and induce apoptosis, an effect that coincides with the current literature.
As stated earlier, the growth inhibitory effects of the DSF–Cd complex in cancer cells have previously been reported and are herein also shown in Fig. 7, where its effect on MCF10A cells were seen. In addition to the inhibition of MDA MB 231 breast cancer cell growth, immortalized, non-tumorigenic MCF10A cells are affected by this compound under the tested experimental condition, an undesirable effect in the analysis of novel pre-clinical drugs. However, in the case of our newly synthesized Cd complexes, Fig. 7 also clearly shows that these immortalized breast cells remain unharmed and are insensitive to the cytotoxic effects of these agents. Using the same experimental conditions, we noted that the Cd complexes are potent cell proliferation inhibitors, specific to the breast cancer MDA MB 231 cells used (Fig. 7A). Also noted was the fact that these Cd complexes are indeed less toxic than DSF–Cd in MCF10A. The use of Cd in the synthesis of novel anti-tumor agents may therefore be a useful strategy after all. While more pre-clinical studies, including analysis in animal models, remain to be done, the cancer cell-specific effects seen in this study and reported by others imply a bright future for Cd in the search, design, and development of novel therapeutics for this disease. This study indicates that the nontoxic organic compounds indole-3-propionic acid, indole-3-butyric acid and 3, 5-diaminobenzoic acid o-vanillin Schiff base bind with Cd and that these Cd complexes are potent proteasome inhibitors and apoptosis inducers with potential as novel anti-cancer treatment modalities. Therefore our Cd complexes will undergo further biological analysis and pre-clinical testing.
Acknowledgements
The authors thank Ms. Sara Schmitt for critical reading of the manuscript. This research was supported by the National Science Foundation of China to Caifeng Bi (No.21071134) and to Yuhua Fan (No. 20971115), the National Cancer Institute to Q. P. Dou (1R01CA20009, 3R01CA120009-04S1 and 5R01CA127258-05), and the Special Foundation for Young Teachers of Ocean University of China to Xia Zhang (No. 201113025), and scholarship from the Chinese Scholarship Council to Zhen Zhang.
5. Abbreviations
- IC50
Half maximal (50%) inhibitory concentration of a substance
- DSF
Disulfiram
- XIAP
X-linked inhibitor of apoptosis protein
- CT
Chymotrypsin
- MTT
3-[4, 5-dimethyltiazol-2-yl]-2.5-diphenyl-tetrazolium bromide
- FBS
Fetal bovine serum
- PARP
Polyclonal antibody against human poly (ADP-ribose) polymerase
- HEPES
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
References
- [1].Hochstrasser M. Curr. Opin. Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- [2].Murray RZ, Norbury C. Anticancer Drugs. 2000;11:407–417. doi: 10.1097/00001813-200007000-00001. [DOI] [PubMed] [Google Scholar]
- [3].Clechanover Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- [4].An B, Goldfarb RH, Siman R, Dou QP. Cell Death Differ. 1998;5:1062–1075. doi: 10.1038/sj.cdd.4400436. [DOI] [PubMed] [Google Scholar]
- [5].Lopes UG, Erhardt P, Yao R, Cooper GM. J. Biol. Chem. 1997;272:12893–12896. doi: 10.1074/jbc.272.20.12893. [DOI] [PubMed] [Google Scholar]
- [6].Dou QP, Li B. Drug Resist. Updat. 1999;2:215–223. doi: 10.1054/drup.1999.0095. [DOI] [PubMed] [Google Scholar]
- [7].Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- [8].Yang HJ, Chen D, Cui QC, Yuan X, Dou QP. Cancer Res. 2006;66:4758–4765. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- [9].Freeza M, Schmitt S, Dou QP. Curr. Top. Med. Chem. 2011;11:2888–2905. doi: 10.2174/156802611798281311. [DOI] [PubMed] [Google Scholar]
- [10].Kenyon GD, Chen D, Shirley O, Cui QC, Fred RM, Dou QP. Breast Cancer Res. 2005;7:897–908. doi: 10.1186/bcr1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xiao Y, Bi CF, Fan YH, Cui QC, Zhang X, Dou QP. Int. J. Oncol. 2008;33:1073–1079. [PubMed] [Google Scholar]
- [12].Zhang X, Bi CF, Fan YH, Cui QC, Chen D, Xiao Y, Dou QP. Int. J. Mol. Med. 2008;22:677–682. [PMC free article] [PubMed] [Google Scholar]
- [13].Milacic V, Chen D, Giovagnini L, Diez A, Fregona D, Dou QP. Toxicol. Appl. Pharmacol. 2008;231:24–33. doi: 10.1016/j.taap.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Milacic V, Chen D, Ronconi L, Kristin R, Piwowar L, Fregona D, Dou QP. Cancer Res. 2006;66:10478–10486. doi: 10.1158/0008-5472.CAN-06-3017. [DOI] [PubMed] [Google Scholar]
- [15].Li LH, Yang HJ, Chen D, Cui QC, Dou QP. Toxicol. Appl. Pharmacol. 2008;229:206–214. doi: 10.1016/j.taap.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nogawa K, et al. Biometals. 2004;17:581–587. doi: 10.1023/b:biom.0000045742.81440.9c. [DOI] [PubMed] [Google Scholar]
- [17].Siewit CL, et al. Mol. Endocrinol. 2010;24:981–992. doi: 10.1210/me.2009-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Casano C, Agnello M, Sirchia R, Luparello C. Biometals. 2010;23:83–92. doi: 10.1007/s10534-009-9268-6. [DOI] [PubMed] [Google Scholar]
- [19].Christina L, Siewit BG, Esera V, Rachel P, Maggie C. Mol. Endocrinol. 2010;24:981–992. doi: 10.1210/me.2009-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Joseph P. Toxicol. Appl. Pharmacol. 2009;238:272–279. doi: 10.1016/j.taap.2009.01.011. [DOI] [PubMed] [Google Scholar]
- [21].Filipic M. Mutat. Res. 2011;733:69–77. doi: 10.1016/j.mrfmmm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- [22].Aimola P, Carmignani M, Volpe AR, Benedetto A, Claudio L, Waalkes MP, Bokhoven A, Tokar EJ, Claudio PP. PLoS One. 2012;7:e33647. doi: 10.1371/journal.pone.0033647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Golovine K, Makhov P, Uzzo RG, Kutikov A, Kaplan DJ, Fox E, Kolenko VM. Mol. Cancer. 2010;9:183. doi: 10.1186/1476-4598-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kovala DD, Staninska M, Garcia-Santos I, Castineiras A, Demertzis MA, Inorg J. Biochem. 2011;105:1187–1195. doi: 10.1016/j.jinorgbio.2011.05.025. [DOI] [PubMed] [Google Scholar]
- [25].Pacini S, Punzi T, Morucci G, Gulisano M, Ruggiero M. J. Environ. Pathol. Toxicol. Oncol. 2009;28:85–88. doi: 10.1615/jenvironpatholtoxicoloncol.v28.i1.90. [DOI] [PubMed] [Google Scholar]
- [26].Kline BE, Wasley WL, Rusch HP. Cancer Res. 1942;2:645–648. [Google Scholar]
- [27].Maøgorzata K, Russel JR, Joaquin JG, Javier C, Susanne B, Carmen O, Andrzej L. J. Cell. Biochem. 2001;81:507–513. [Google Scholar]
- [28].Adsule S, Barve V, Chen D, Ahmed F, Dou QP, Padhye S, Sarkar FH. J. Med. Chem. 2006;49:7242–7246. doi: 10.1021/jm060712l. [DOI] [PubMed] [Google Scholar]
- [29].Tümer M, Akgün E, Toroğlu S, Kayraldiz A, Dönbak L. J. Coord. Chem. 2008;61:2935–2949. [Google Scholar]
- [30].Fan YH, Dong LL, et al. Russ. J. Coord. Chem. 2012;38:1–7. [Google Scholar]
- [31].Daniel KG, Gupta P, Harbach RH, Guida WC, Dou QP. Biochem. Pharmacol. 2004;67:1139–1151. doi: 10.1016/j.bcp.2003.10.031. [DOI] [PubMed] [Google Scholar]
- [32].Chen D, Daniel KG, Chen MS, Kuhn DJ, Landis-Piwowar KR, Dou QP. Biochem. Pharmacol. 2005;69:1421–1432. doi: 10.1016/j.bcp.2005.02.022. [DOI] [PubMed] [Google Scholar]
- [33].Kazuo N, Huang DR, Wang QR. Infrared and Raman spectra of inorganic and coordination compounds. Chemical Industry Press; Beijing: 1988. [Google Scholar]
- [34].Gong QJ, Jin WJ, Dong C. Appl. Chem. 2000;17:227–229. [Google Scholar]
- [35].Drexler HC. Proc. Natl. Acad. Sci. 1997;94:855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Romanowicz-Makowska H, Forma E, Bryś M, Krajewska WM, Smolarz B. Pol. J. Pathol. 2011;62:257–261. [PubMed] [Google Scholar]
- [37].Park RM, Stayner LT, Petersen MR, Finley-Couch M, Hornung R, Rice C. Occup. Environ. Med. 2012;69:303–309. doi: 10.1136/oemed-2011-100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Neslund-Dudas C, Mitra B, Kandegedara A, Chen D, Schmitt S, Shen M, Cui QC, Rybicki BA, Dou QP. Biol. Trace Elem. Res. 2012 Mar 16; doi: 10.1007/s12011-012-9391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]