Abstract
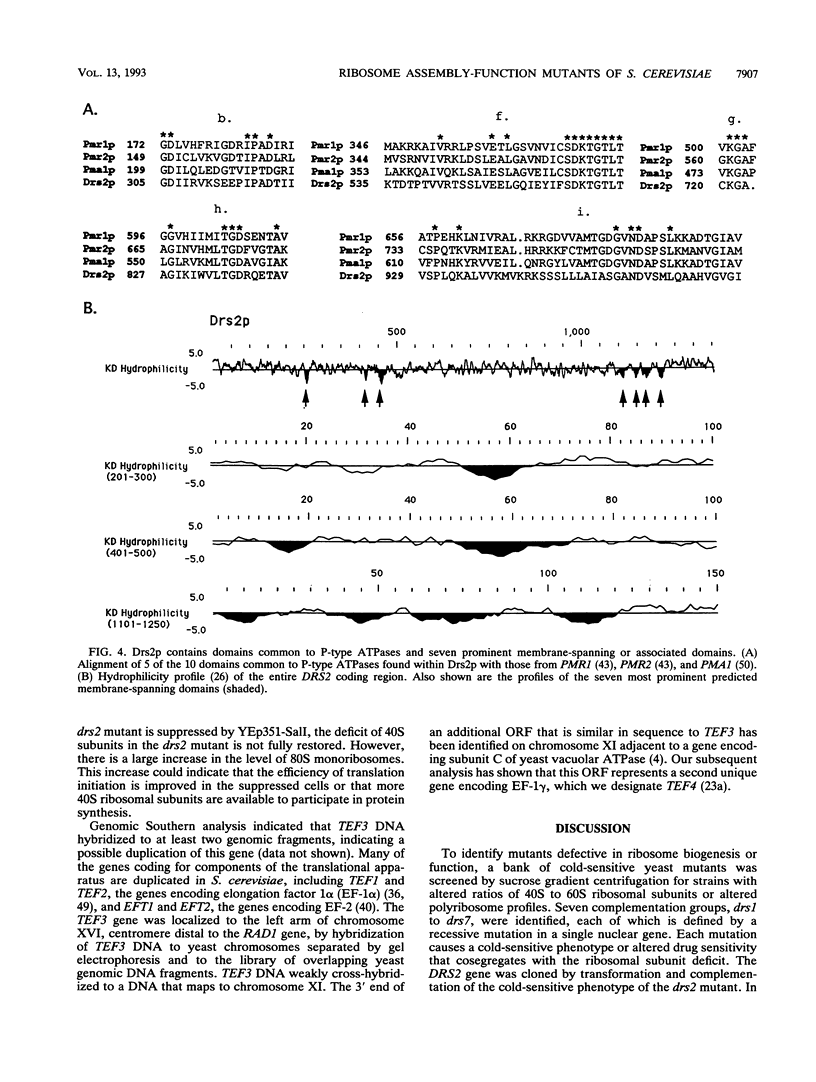
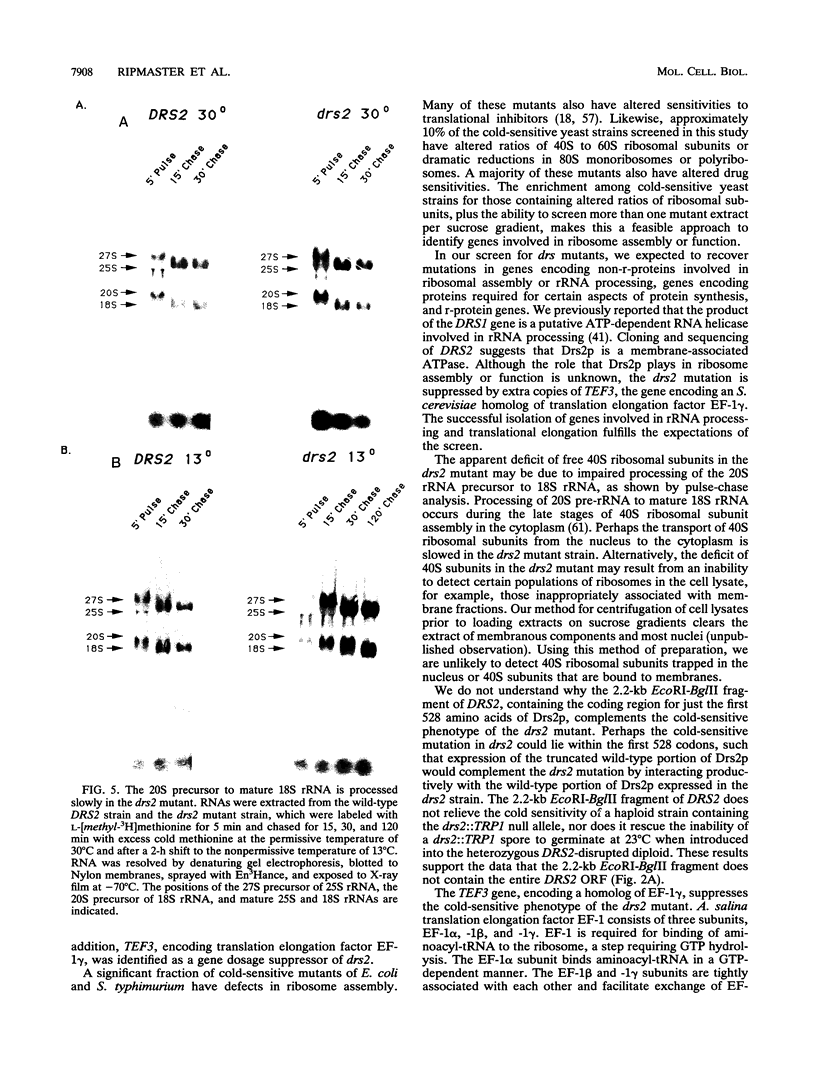
To identify Saccharomyces cerevisiae mutants defective in assembly or function of ribosomes, a collection of cold-sensitive strains generated by treatment with ethyl methanesulfonate was screened by sucrose gradient analysis for altered ratios of free 40S to 60S ribosomal subunits or qualitative changes in polyribosome profiles. Mutations defining seven complementation groups deficient in ribosomal subunits, drs1 to drs7, were identified. We have previously shown that DRS1 encodes a putative ATP-dependent RNA helicase necessary for assembly of 60S ribosomal subunits (T. L. Ripmaster, G. P. Vaughn, and J. L. Woolford, Jr., Proc. Natl. Acad. Sci. USA 89:11131-11135, 1992). Strains bearing the drs2 mutation process the 20S precursor of the mature 18S rRNA slowly and are deficient in 40S ribosomal subunits. Cloning and sequencing of the DRS2 gene revealed that it encodes a protein similar to membrane-spanning Ca2+ ATPases. The predicted amino acid sequence encoded by DRS2 contains seven transmembrane domains, a phosphate-binding loop found in ATP- or GTP-binding proteins, and a seven-amino-acid sequence detected in all classes of P-type ATPases. The cold-sensitive phenotype of drs2 is suppressed by extra copies of the TEF3 gene, which encodes a yeast homolog of eukaryotic translation elongation factor EF-1 gamma. Identification of gene products affecting ribosome assembly and function among the DNAs complementing the drs mutations validates the feasibility of this approach.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
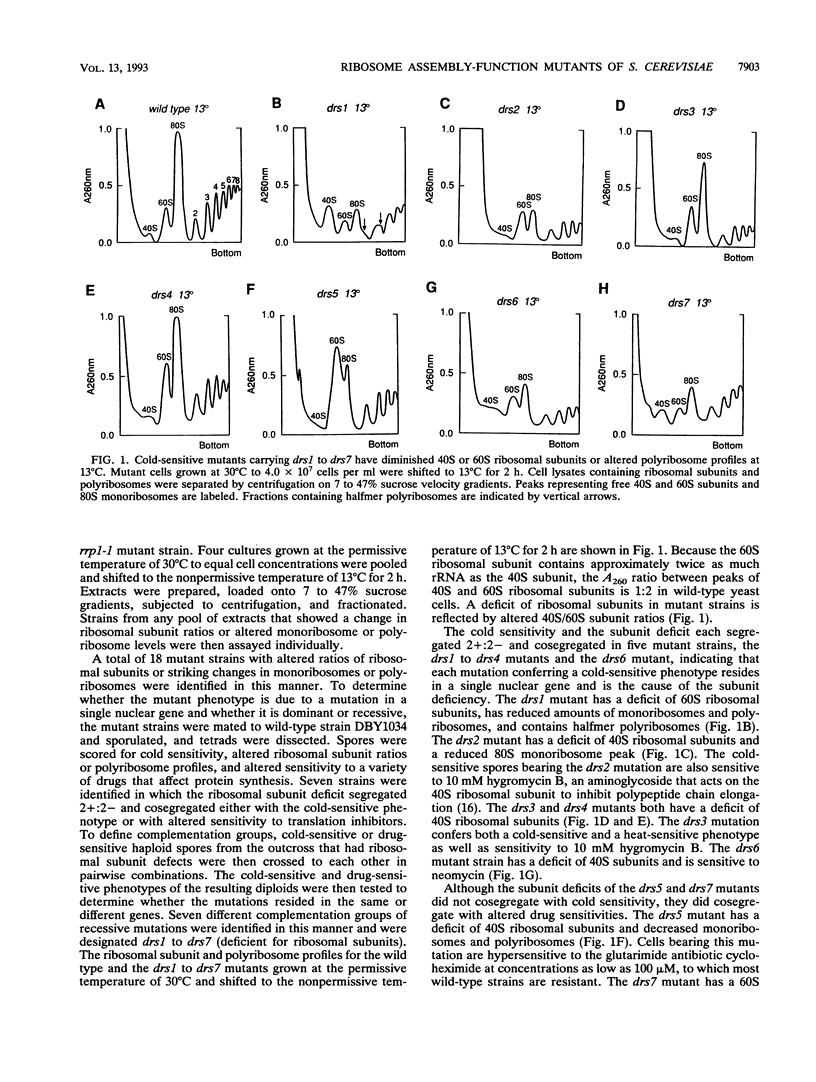
- Andrew C., Hopper A. K., Hall B. D. A yeast mutant defective in the processing of 27S r-RNA precursor. Mol Gen Genet. 1976 Feb 27;144(1):29–37. doi: 10.1007/BF00277300. [DOI] [PubMed] [Google Scholar]
- Baim S. B., Pietras D. F., Eustice D. C., Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985 Aug;5(8):1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss F. T., Ingrahm J. L. Mutation in Saccharomyces cerevisiae conferring streptomycin and cold sensitivity by affecting ribosome formation and function. J Bacteriol. 1974 May;118(2):319–328. doi: 10.1128/jb.118.2.319-328.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán C., Kopecky J., Pan Y. C., Nelson H., Nelson N. Cloning and mutational analysis of the gene encoding subunit C of yeast vacuolar H(+)-ATPase. J Biol Chem. 1992 Jan 15;267(2):774–779. [PubMed] [Google Scholar]
- Bender A., Pringle J. R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Mar;11(3):1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. J., Cannon M., Jiménez A. A trichodermin-resistant mutant of Saccharomyces cerevisiae with an abnormal distribution of native ribosomal subunits. Eur J Biochem. 1980;107(1):173–183. doi: 10.1111/j.1432-1033.1980.tb04638.x. [DOI] [PubMed] [Google Scholar]
- Cormier P., Osborne H. B., Morales J., Bassez T., Poulhe R., Mazabraud A., Mulner-Lorillon O., Bellé R. Molecular cloning of Xenopus elongation factor 1 gamma, major M-phase promoting factor substrate. Nucleic Acids Res. 1991 Dec 11;19(23):6644–6644. doi: 10.1093/nar/19.23.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto J. R., Tamm J., Parker R., Guthrie C. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987 Jul;1(5):445–455. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- Creutz C. E., Snyder S. L., Kambouris N. G. Calcium-dependent secretory vesicle-binding and lipid-binding proteins of Saccharomyces cerevisiae. Yeast. 1991 Apr;7(3):229–244. doi: 10.1002/yea.320070305. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra B., Skogerson L., Chakraburtty K. Protein synthesis in yeast. II. Purification and properties of the elongation factor 1 from Saccharomyces cerevisiae. J Biol Chem. 1981 Oct 10;256(19):10005–10011. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drain P., Schimmel P. Multiple new genes that determine activity for the first step of leucine biosynthesis in Saccharomyces cerevisiae. Genetics. 1988 May;119(1):13–20. doi: 10.1093/genetics/119.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J. P., Lehtonen H., Caizergues-Ferrer M., Amalric F., Tollervey D., Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992 Feb;11(2):673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Jiménez A., Vázquez D., Davies J. E., Schindler D. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim Biophys Acta. 1978 Dec 21;521(2):459–469. doi: 10.1016/0005-2787(78)90287-3. [DOI] [PubMed] [Google Scholar]
- Gritz L. R., Mitlin J. A., Cannon M., Littlewood B., Carter C. J., Davies J. E. Ribosome structure, maturation of ribosomal RNA and drug sensitivity in temperature-sensitive mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1982;188(3):384–391. doi: 10.1007/BF00330038. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Nashimoto H., Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci U S A. 1969 Jun;63(2):384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. M., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991 Dec;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. M., Möller W. Elongation factor 1 beta gamma from Artemia. Purification and properties of its subunits. Eur J Biochem. 1988 Jan 15;171(1-2):119–129. doi: 10.1111/j.1432-1033.1988.tb13766.x. [DOI] [PubMed] [Google Scholar]
- Kambouris N. G., Burke D. J., Creutz C. E. Cloning and genetic characterization of a calcium- and phospholipid-binding protein from Saccharomyces cerevisiae that is homologous to translation elongation factor-1 gamma. Yeast. 1993 Feb;9(2):151–163. doi: 10.1002/yea.320090206. [DOI] [PubMed] [Google Scholar]
- Kohno K., Uchida T., Ohkubo H., Nakanishi S., Nakanishi T., Fukui T., Ohtsuka E., Ikehara M., Okada Y. Amino acid sequence of mammalian elongation factor 2 deduced from the cDNA sequence: homology with GTP-binding proteins. Proc Natl Acad Sci U S A. 1986 Jul;83(14):4978–4982. doi: 10.1073/pnas.83.14.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumabe T., Sohma Y., Yamamoto T. Human cDNAs encoding elongation factor 1 gamma and the ribosomal protein L19. Nucleic Acids Res. 1992 May 25;20(10):2598–2598. doi: 10.1093/nar/20.10.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Larkin J. C., Woolford J. L., Jr Molecular cloning and analysis of the CRY1 gene: a yeast ribosomal protein gene. Nucleic Acids Res. 1983 Jan 25;11(2):403–420. doi: 10.1093/nar/11.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C., Zabetakis D., Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992 Sep;12(9):3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Archer R. H., Zengel J. M. A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Jan 25;20(2):295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maessen G. D., Amons R., Zeelen J. P., Möller W. Primary structure of elongation factor 1 gamma from Artemia. FEBS Lett. 1987 Oct 19;223(1):181–186. doi: 10.1016/0014-5793(87)80532-x. [DOI] [PubMed] [Google Scholar]
- Mizushima S., Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970 Jun 27;226(5252):1214–1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- Moritz M., Paulovich A. G., Tsay Y. F., Woolford J. L., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol. 1990 Dec;111(6 Pt 1):2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Pulaski B. A., Woolford J. L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991 Nov;11(11):5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic mapping in Saccharomyces. Genetics. 1966 Jan;53(1):165–173. doi: 10.1093/genetics/53.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S., Nagashima K., Tsunetsugu-Yokota Y., Fujimura K., Miyazaki M., Kaziro Y. Polypeptide chain elongation factor 1 alpha (EF-1 alpha) from yeast: nucleotide sequence of one of the two genes for EF-1 alpha from Saccharomyces cerevisiae. EMBO J. 1984 Aug;3(8):1825–1830. doi: 10.1002/j.1460-2075.1984.tb02053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H. G., Fried H. M. Effects of progressive depletion of TCM1 or CYH2 mRNA on Saccharomyces cerevisiae ribosomal protein accumulation. Mol Cell Biol. 1986 May;6(5):1535–1544. doi: 10.1128/mcb.6.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975 Aug 1;189(4200):347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Perentesis J. P., Phan L. D., Gleason W. B., LaPorte D. C., Livingston D. M., Bodley J. W. Saccharomyces cerevisiae elongation factor 2. Genetic cloning, characterization of expression, and G-domain modeling. J Biol Chem. 1992 Jan 15;267(2):1190–1197. [PubMed] [Google Scholar]
- Ripmaster T. L., Vaughn G. P., Woolford J. L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rudolph H. K., Antebi A., Fink G. R., Buckley C. M., Dorman T. E., LeVitre J., Davidow L. S., Mao J. I., Moir D. T. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989 Jul 14;58(1):133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Russell I. D., Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992 Nov;119(4):737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990 Mar 2;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Schirmaier F., Philippsen P. Identification of two genes coding for the translation elongation factor EF-1 alpha of S. cerevisiae. EMBO J. 1984 Dec 20;3(13):3311–3315. doi: 10.1002/j.1460-2075.1984.tb02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R., Kielland-Brandt M. C., Fink G. R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature. 1986 Feb 20;319(6055):689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- Shuai K., Warner J. R. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 1991 Sep 25;19(18):5059–5064. doi: 10.1093/nar/19.18.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Manney T. R. Genetic analysis of mutations affecting growth of Saccharomyces cerevisiae at low temperature. Genetics. 1974 Aug;77(4):651–659. doi: 10.1093/genetics/77.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989 May 19;244(4906):783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Surguchov A. P., Fominykch E. S., Smirnov V. N., Ter-Avanesyan M. D., Mironova L. N., Inge-Vechtomov S. G. Further characterization of recessive suppression in yeast. Isolation of the low-temperature sensitive mutant of Saccharomyces cerevisiae defective in the assembly of 60 S ribosomal subunit. Biochim Biophys Acta. 1981 Jun 26;654(1):149–155. doi: 10.1016/0005-2787(81)90148-9. [DOI] [PubMed] [Google Scholar]
- Tai P. C., Kessler D. P., Ingraham J. Cold-sensitive mutations in Salmonella typhimurium which affect ribosome synthesis. J Bacteriol. 1969 Mar;97(3):1298–1304. doi: 10.1128/jb.97.3.1298-1304.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991 Mar;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapman J., Retèl J., Planta R. J. Ribosomal precursor particles from yeast. Exp Cell Res. 1975 Jan;90(1):95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- Traub P., Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci U S A. 1968 Mar;59(3):777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem S. A., Warner J. R. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem. 1973 Feb 25;248(4):1412–1416. [PubMed] [Google Scholar]
- Walderhaug M. O., Post R. L., Saccomani G., Leonard R. T., Briskin D. P. Structural relatedness of three ion-transport adenosine triphosphatases around their active sites of phosphorylation. J Biol Chem. 1985 Mar 25;260(6):3852–3859. [PubMed] [Google Scholar]
- Warner J. R., Soeiro R. Nascent ribosomes from HeLa cells. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1984–1990. doi: 10.1073/pnas.58.5.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Kolb J. M., Dodd J., Yamagishi M., Mémet S., Buhler J. M., Nomura M. Conditional expression of RPA190, the gene encoding the largest subunit of yeast RNA polymerase I: effects of decreased rRNA synthesis on ribosomal protein synthesis. Mol Cell Biol. 1990 May;10(5):2049–2059. doi: 10.1128/mcb.10.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Jr The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]
- Yoon H. J., Donahue T. F. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol. 1992 Jan;12(1):248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]