Abstract
A novel γ-carboxyglutamate (Gla)-containing protein, named Gla-rich protein (GRP) after its high content in Gla residues or upper zone of growth plate and cartilage matrix associated protein after its preferential expression by cartilage chondrocyte, was recently identified in sturgeon, mice, and humans through independent studies. GRP is the most densely γ-carboxylated protein identified to date and its structure has been remarkably conserved throughout vertebrate evolution but is apparently absent from bird genomes. Several transcript and genomic variants affecting key protein features or regulatory elements were described and 2 paralogs were identified in the teleost fish genome. In the skeleton, most relevant levels of GRP gene expression were observed in cartilaginous tissues and associated with chondrocytes, suggesting a role in chondrogenesis. But GRP expression was also detected in bone cells, indicative of a more widespread role for the protein throughout skeletal formation. Although the molecular function of GRP is yet unknown, the high content of Gla residues and its accumulation at sites of pathological calcification in different human pathologies affecting skin or the vascular system and in breast cancer tumors suggest that GRP may function as a modulator of calcium availability. Because of its association with fibrillar collagens, GRP could also be involved in the organization and/or stabilization of cartilage matrix. Although transgenic mice did not reveal obvious phenotypic alterations in skeletal development or structure, zebrafish morphants lack craniofacial cartilage and exhibit limited calcification, suggesting a role for GRP during skeletal development, but additional functional data are required to understand its function.
Introduction
In the last decade, and following the acquisition of an almost complete draft of the human genome sequence, there was a renewed interest in identifying new genes/proteins associated with human pathologies. The concomitant development of more powerful and efficient sequencing techniques has also permitted the transcriptomic analysis of specific cell types and tissues at an affordable price, thus facilitating the identification of new molecular players involved in a given pathway and their relevance in health and disease. Although most work has been developed in mammals, fish have nevertheless been recognized as valuable resources to understand the basic mechanisms underlying vertebrate development as well as its genetics and pathological changes (1). The growing bulk of genetic data available have also been derived from many different organisms, thus allowing evolutionary studies to take place. As part of our goal to study the evolution of skeletal proteins, our laboratory and the Price laboratory have searched for γ-carboxyglutamate/Gla7-containing proteins in extracts of cartilage and bone tissues collected from the Adriatic sturgeon Acipenser naccarii, a fish that has retained many ancestral features (2). From this work, a novel Gla-containing protein was identified and named Gla-rich protein (GRP) due to its high content in Gla residues. Upon cloning the corresponding cDNA and gene, we found that it matched a human transcript not previously annotated with orthologs in most vertebrates (2). At about the same time, 2 independent studies were published reporting the identification, through either a human fetal cDNA library sequencing approach or a mouse embryonic microarray approach, of a new gene that appeared to be present during early development and was highly specific for distal chondrocytes. This gene was named UCMA for unique cartilage matrix associated protein (3) (later renamed as upper zone of growth plate and cartilage matrix associated protein) and claimed to be essential for cartilage development (4). However, neither study identified UCMA, alias GRP, as being γ-carboxylated and thus a new member of the large family of vitamin K-dependent proteins. This feature makes this protein even more interesting it has a primary structure and a genomic organization distinct from all other known Gla proteins, despite sharing a few unique features typical of the members of this family. In this review, we aim to summarize the available data on GRP, thus providing the reader with a comprehensive and updated summary of all relevant information available on this subject.
Gene and protein structures
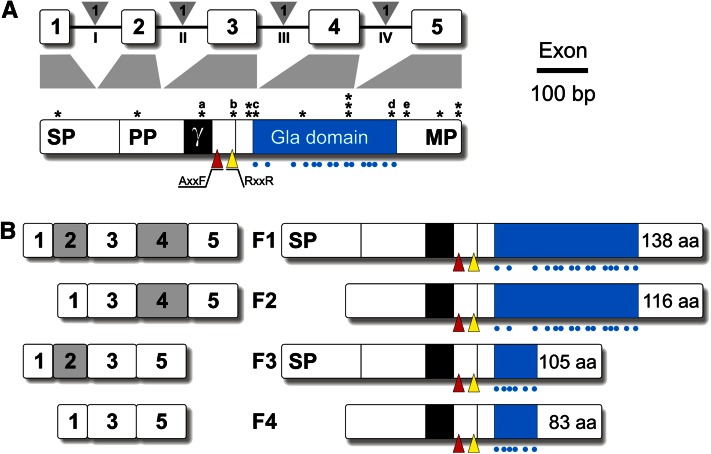
Vertebrate GRP genes are organized into 5 coding exons separated by phase-1 introns (Fig. 1) and encoding a pre-propeptide of ~135 amino acids. Following the removal of the transmembrane signal peptide, proGRP is cleaved by a furin-like protease (at the RXXR site) into a propeptide (38–39 amino acids) and a mature peptide (67–74 amino acids), which is characterized by a dense cluster of Gla residues. The presence of 16 and 15 Gla residues in sturgeon and human GRP, respectively, has made GRP the most densely γ-carboxylated protein identified to date (2). Interestingly, most of the Gla residues occur in exon 4, whose structure has been remarkably conserved throughout vertebrate evolution (2). Other notable features of GRP are: 1) a docking site for the γ-glutamyl carboxylase in the propeptide; 2) a AXXF motif (proteolytic cleavage site also present in the matrix Gla protein) in the propeptide; 3) several partially sulfated tyrosine residues (4); and 5) a coiled-coil oligomerization domain (3) in the mature peptide. No evidence for additional post-translational modifications of GRP, e.g., phosphorylation and glycosylation, were found. Information on the 3-dimensional structure of GRP and the conformation of the various protein domains is still unavailable.
Figure 1.
Schematic representation of vertebrate GRP gene and protein structures (A) and mouse transcript variants (B). In gene structure, exons (limited to coding sequence) are represented by white boxes/Arabic numbers and introns by black lines/Roman numbers; phase of intron insertion is indicated in gray triangles. In protein structure, Gla residues are indicated by blue dots and Gla domain is represented by a blue box; SNPs are indicated by asterisks and those affecting important protein domains by letters a, b, c, d, and e (Table 4); red and yellow triangles indicate AXXF and RXXR proteolytic cleavage sites, respectively; γ indicates the docking site for γ-glutamyl carboxylase. In transcript variants, mRNA structure (limited to coding sequence) of the 4 variants (F1–4) is indicated on the left side and peptide structure on the right side. Length of each peptide is indicated after the Gla domain. Gla, γ-carboxy glutamate; MP, mature peptide; PP, propeptide; SNP, single-nucleotide polymorphism; SP, signal peptide.
Molecular evolution
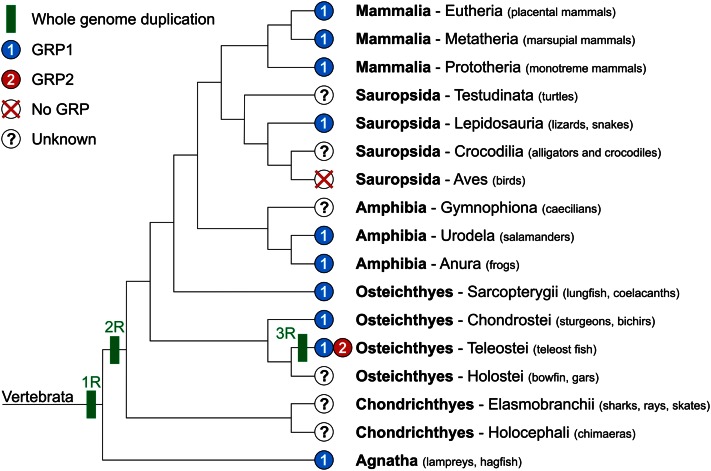
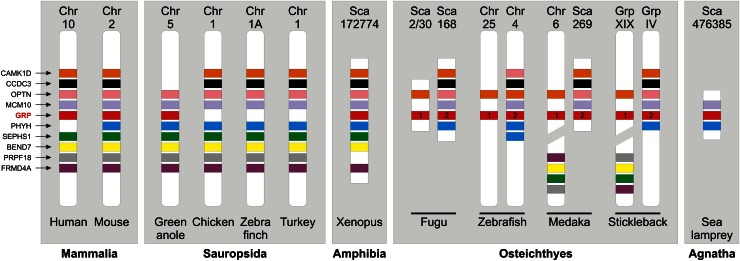
Absent in invertebrate genomes, the GRP gene was identified in most classes of vertebrates, including mammals, sauropsids, amphibians, bony fish, and jawless fish (Fig. 2). No GRP gene has been identified yet in chondricthyans, holosteans, caecilians, testudines, and archosaurians (birds and crocodilians). Although the lack of the GRP gene can be attributed to scarce genomic information in most of these groups, its absence in birds, in particular in chicken, zebra finch, and turkey, whose genomes are almost complete, is more puzzling. In-depth analysis of the genomic context around the GRP locus in several vertebrate genomes revealed that although almost all the genes flanking the GRP gene have been conserved in the same syntenic block (or 2 blocks in fish following the fish-specific whole genome duplication event) throughout vertebrate evolution, the GRP gene is absent from this block in bird genomes (Fig. 3). The occurrence of a GRP gene in the green anole genome indicates that it may have disappeared from the archosaurian genome after branching from lepidosaurians or later from the bird genome after branching from crocodilians; genomic information regarding crocodilian genomes is, however, not sufficient to clarify this issue. Although the biological importance of the absence of the GRP gene remains to be elucidated, we can already speculate that its function is either not required in archosaurians/birds or another gene(s) has acquired during evolution the capacity to fulfill the same function(s). Whereas only one GRP gene is present in tetrapods and cartilaginous and jawless fish, 2 paralogs (GRP1/UCMAa and GRP2/UCMAb) were identified in the teleost fish genome (2). This occurrence has been associated with a fish-specific whole genome duplication event that has reportedly occurred in the teleost fish lineage after divergence from tetrapods around 450 million years ago that likely affected most teleost fish (5). The high sequence homology between GRP1 and GRP2 and the rigorous conservation of protein features throughout evolution indicate that both fish GRP isoforms have most likely conserved a similar structure and function in modern teleost fish. As previously reported by Neacsu et al. (6), as well as our group (7), the genomic context of the fish GRP2 gene is more closely related to that of tetrapod GRP, although the fish GRP1 gene was proposed to be the vertebrate ortholog by sequence comparison. Finally, given the fact that they share several unique features and similar post-translational modifications, we can speculate that GRP may be evolutionary related to osteocalcin (OC), matrix Gla protein (MGP), and coagulation factors. Given their time of appearance, we propose that GRP would be ancestral to OC and MGP; this hypothesis will need to be further supported.
Figure 2.
Cladogram of vertebrate groups and occurrence of GRP forms. Sequence data used to construct this cladogram: Mammalia-Eutheria (Ailuropoda melanoleuca, Bos taurus, Callithrix jacchus, Canis familiaris, Cavia porcellus, Choloepus hoffmanni, Dipodomys ordii, Equus caballus, Erinaceus europaeus, Felis catus, Homo sapiens, Loxodonta africana, Macaca mulatta, Microcebus murinus, Mus musculus, Myotis lucifugus, Nomascus leucogenys, Ochotona princeps, Oryctolagus cuniculus, Otolemur garnettii, Pan troglodytes, Pongo pygmaeus, Procavia capensis, Pteropus vampyrus, Rattus norvegicus, Spermophilus tridecemlineatus, Sus scrofa, Tarsius syrichta, Tupaia belangeri, Tursiops truncatus); Mammalia-Metatheria (Macropus eugenii, Monodelphis domestica, Sarcophilus harrisii); Mammalia-Prototheria (Ornithorhynchus anatinus); Sauropsida-Reptilia (Anolis carolinensis); Amphibia-Urodela (Ambystoma mexicanum); Amphibia-Anura (Xenopus laevis, Xenopus tropicalis); Osteichthyes-Sarcopterygii (Latimeria chalumnae); Osteichthyes-Chondrostei (Acipenser naccarii); Osteichthyes-Teleostei (Cyprinus carpio, Danio rerio, Dicentrarchus labrax, Gasterosteus aculeatus, Ictalurus punctatus, Oncorhynchus mykiss, Oryzias latipes, Osmerus mordax, Salmo salar, Sparus aurata, Takifugu rubripes, Tetraodon nigroviridis); Agnatha (Petromyzon marinus).
Figure 3.
Schematic representation of the genomic region flanking vertebrate GRP genes using data from the Ensembl project (24). The physical localization of 9 genes present in the vicinity of mouse GRP gene is indicated (irrespectively of their orientation) for various mammalian, sauropsidian, amphibian, piscine, and agnathan species. BEND7, BEN domain-containing protein 7; CAMK1D, calcium/calmodulin-dependent protein kinase ID; CCDC3, coiled-coil domain-containing protein 3; FRMD4A, FERM domain-containing protein 4A; GRP, Gla-rich protein; MCM10, minichromosome maintenance complex component 10; OPTN, optineurin; PHYH, phytanoyl-CoA 2-hydroxylase; PRPF18, PRP18 pre-mRNA processing factor 18 homolog; SEPHS1, selenophosphate synthetase 1.
Expression patterns
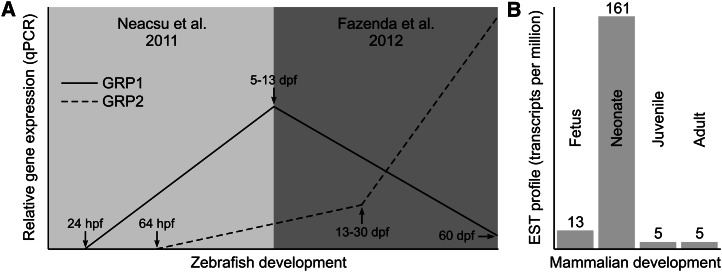
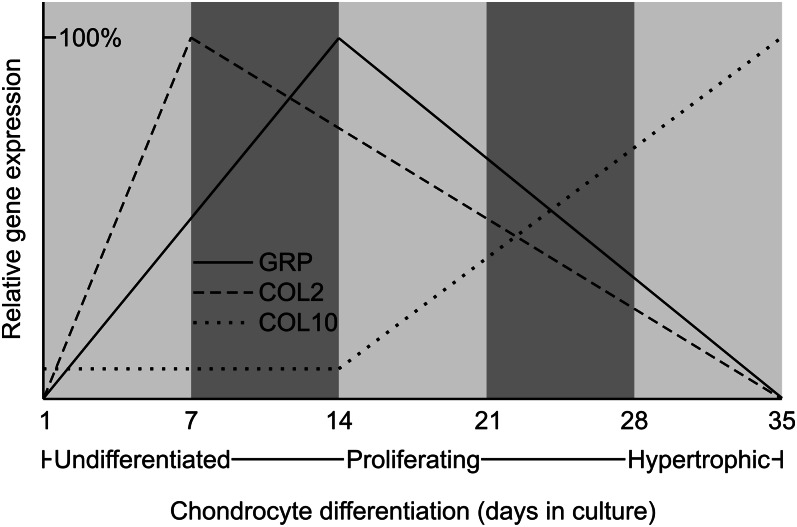
Levels and sites of GRP gene expression were determined in a variety of adult tissues and during the development of several vertebrate species. In sturgeon and zebrafish, the GRP transcript was detected in all tissues analyzed by qPCR, but the highest and more relevant levels were observed in cartilaginous tissues (Table 1). Both mature and immature chondrocytes of vertebra and mandibula, and also chordoblasts of the notochord, were shown by in situ hybridization (ISH) to be the primary sites of GRP gene expression in sturgeon (2). In zebrafish, GRP paralogs exhibited distinct patterns of expression, suggesting different regulatory pathways for each gene (6, 7). Indeed, their levels of expression appear to be somewhat inversely correlated, with GRP1 being expressed first and remaining high during early development, whereas expression of GRP2 appears more tardive but increases in late larval and juvenile stages and is also the most preponderant in adult tissues (Fig. 4A;Table 1). In mice, the GRP transcript was first detected by ISH in vertebra of E13.5 fetus, where the GRP gene was expressed in resting chondrocytes of the distal and peripheral zones of epiphysis and in vertebral cartilage (4), a pattern that was maintained up to neonate. Indeed, upon analysis of the large bulk of EST available from public databases, there appears to be a clear increase in GRP expression in neonate stages of development in mammals (Fig. 4B), thus suggesting that, at this time, GRP expression may be relevant. However, the available information on gene expression at these particular stages of development is sparse and therefore it is not possible to ascertain whether this prediction is correct. Interestingly, the location of GRP expression has been shown in one study to correspond, during mouse development, almost exclusively to areas of type II collagen (COL2) expression (8). In adult mice, the GRP transcript was observed in growth plate cartilage close to the cartilage-bone interface (9). These authors also showed that the processed mature GRP diffuses into the cartilage matrix and is retained there until chondrocytes reach hypertrophy, further emphasizing its possible involvement in chondrocyte differentiation/maturation. Accordingly, in rats, the GRP transcript was detected in immature, proliferating, mature, columnar, and hypertrophic chondrocytes of the ribs, tail, vertebra, and femur (2). Comparative analysis of expression sites for GRP and MGP (a vitamin K-dependent protein also expressed by chondrocytes) showed that, whereas the MGP transcript was restricted to immature, proliferating, and late hypertrophic chondrocytes, the GRP transcript was widely distributed in all chondrocyte types (2). Together, the available data provide evidence for a possible chondrocyte-associated function of GRP. This transcript was also detected in other skeletal cell types such as osteocytes and osteoblasts found in the trabecular bone of rat rib, tail, vertebra, and femur (2). Comparative analysis of expression sites for GRP and OC (a vitamin K-dependent protein expressed by osteoblasts) showed that, whereas the OC transcript was restricted to mature osteoblasts, the GRP transcript was strongly detected in both osteoblasts and osteocytes (2). The available data thus indicate that GRP expression is not restricted to chondrocytes but is also present in bone cells, suggesting a more widespread role for the protein throughout skeletal formation. Patterns of GRP gene expression were also analyzed in differentiating chondrocytes using several in vitro cell systems [e.g., mouse ATDC5 and MC615 cell lines (3, 4)] (Table 2). Although it was high or increasing at early stages of differentiation, it slowly decreased in proliferating and almost disappeared in hypertrophic chondrocytes (Fig. 5). It was also shown that GRP gene expression parallels the expression of the COL2 gene and is inversely correlated with that of type X collagen (COL10) (Fig. 5), a result that agrees with previously reported in vivo data (8). Therefore, based on the available results, it has been suggested that GRP may represent a suitable marker of chondrocyte differentiation/chondrogenesis (3, 10) and several authors have already used GRP in this way (11, 12). Furthermore, the GRP transcript was detected in other rat and human tissues such as skin and blood vessel walls and was also found in serum (13). These findings appeared to be in contrast with what was previously reported for mice, but a more recent report describing the activation of the lacZ reporter gene in heterozygous and homozygous GRP-null mice revealed that this protein was also expressed in other mouse tissues not previously identified, such as parietal calvaria, nasal cartilage, thyroid cartilage, and trachea (8, 9). Together, the available data confirm that GRP is not a cartilage-specific protein, although chondrocytes appear to be one of its major sites of expression. Further work is, however, required to relate its sites of expression with function.
Table 1.
Sites of GRP gene expression and protein accumulation in mammalian and bony fish tissues
| Tissue | Origin | Cell type expressing GRP | Detection method1 | Reference |
| Fetal growth plate cartilage | Human | Chondrocytes | cDNA sequence | (3) |
| Fetal limbs | Mouse | Not described | NB | (3) |
| Fetal skull and skeletal elements from endochondral calcification | Mouse | Not described | NB | (3) |
| Developing cartilage(E15.5–18.5) | Mouse | Distal chondrocytes, developing epiphysis | ISH, IHC | (3,4,11) |
| Epiphyseal cartilage(E18.5, P1, P4, P6, P10, P161) | Mouse | Resting zone chondrocytes | ISH, IHC, qPCR, Xgal | (4,9,11) |
| Cartilage of os occipitale, thyroid, and trachea (P4) | Mouse | Not described | Xgal | (9) |
| Mandible, branchial arches, vertebra | Sturgeon | Chordoblasts, chondrocytes | qPCR, ISH | (2) |
| Embryos (18–96 h postfertilization) | Zebrafish | GRP1 (UCMAa): notochord and cranio-facial cartilaginous structures GRP2 (UCMAb): trabecular cartilage, ceratohyal, and hypobranchial | ISH | (6) |
| Developing fish | Zebrafish | See Figure 4 | qPCR | (7) |
| Adults | Zebrafish | GRP2 > GRP1 in all tissues analyzed, expression higher in branchial arches, pectoral fin, and vertebra | qPCR | (7) |
| Vascular system | Rat, human | Vascular smooth muscle cells | qPCR, IHC, ISH | (2,13) |
| Skin and its appendages | Rat, human | Epidermis, dermis, fibroblasts, hair follicles, and sebaceous and sweat glands | qPCR, IHC, ISH | (2,13) |
| Rib | Rat | Chondrocytes, osteoblasts, and osteocytes | qPCR, IHC, ISH | (2) |
| Outer ear | Rat | Chondrocytes of elastic cartilage, skin, and its appendages | qPCR, IHC, ISH | (2,13) |
| Tail | Rat | Chondrocytes, osteocytes | qPCR, IHC, ISH | (2,13) |
| Nose | Rat | Chondrocytes of elastic cartilage, skin, and its appendages | qPCR, IHC, ISH | (13) |
IHC, immunohistochemistry; ISH, in situ hybridization; NB, Northern blot; qPCR, quantitative real-time PCR; Xgal, X-gal staining in GRP-deficient mice carrying a lacZ cassette under the control of GRP promoter.
Figure 4.
Relative expression of GRP gene(s) throughout zebrafish (A) and mouse (B) development. Expressed sequence tag (EST) profile in B is according to sequence data available in Unigene database (25). dpf, days postfertilization.
Table 2.
Levels of GRP gene expression in different in vitro cell systems
| System | Name (species) | Cell type | Expression level | Reference |
| Cell lines | ATDC5 (mouse) | Chondrocyte | ++ | (3,4) |
| MC615 (mouse) | Chondrocyte | ++++ | (4) | |
| C3H10T 1/2 (mouse) | Fibroblast | − | (4) | |
| C2C12 (mouse) | Myoblast | − | (4) | |
| MC3T3-E1 (mouse) | Preosteoblast | + | (4) | |
| NIH3T3 (mouse) | Fibroblast | + | (4) | |
| Primary cell cultures | RC (mouse) | Rib chondrocyte | ++++ | (4) |
| CC (mouse) | Calvaria osteoblast | ++ | (4) | |
| RZ (bovine) | Epiphyseal chondrocyte, resting zone | ++ | (4) | |
| HZ (bovine) | Epiphyseal chondrocyte, hypertrophic zone | − | (4) | |
| BAAn1F (sturgeon) | Vertebra-derived cells | +++ | (26) | |
| Van2H (sturgeon) | Branchial arches-derived cells | +++ | (26) | |
| E11.5 Msc (mouse) | Fetal limb buds undifferentiated cells | − | (10) | |
| E11.5 Msc (mouse) | Postchondrogenic differentiation cells | +++ | (10) | |
| E18.5 Chondr (mouse) | Freshly isolated chondrocytes | +++ | (10) |
Figure 5.
Expression of GRP gene throughout chondrocyte differentiation, from undifferentiated (day 1) toward hypertrophic cells (day 35). Highest levels of gene expression were set to 100% for each of the genes.
Regulation of grp expression
Data on the regulation of the GRP gene are scarce. First identified using an approach aiming at detecting chondrocyte-associated genes whose expression was suppressed during retinoic acid-induced dedifferentiation (4), the GRP gene was later also shown to be downregulated in chondrocytes exposed to bone morphogenetic protein 2 (BMP-2) and to transforming growth factor b1 (TGF-β1) (4, 10). Another study reported Indian hedgehog signaling-independent expression of GRP in developing mouse limbs (3). However, no data are available indicating a direct transcriptional regulation of the GRP gene by any of those potential regulators. Le Jeune et al. (14), who have identified GRP as an upregulated gene in transgenic chondrocytes expressing a dominant negative of ERG, a member of the E-twenty six (ETS) family of transcription factors, also identified a putative ETS binding site in mouse GRP gene promoter. They proposed that ERG, which is expressed during embryonic development of cartilage (15), could be directly involved in such regulation. However, no additional studies were published yet that confirm this hypothesis. Computational prediction of binding sites for transcription factors in the flanking region of the Japanese and green-spotted pufferfish GRP1 and GRP2 genes (16) revealed putative binding sites for nuclear factors essential for the patterning process involved in skeletal development, cartilage differentiation, and cartilage gene regulation. For 3 of them, i.e., E47, MEF2, and STAT1, putative binding sites were also predicted in human GRP gene promoter, making them good candidates for further works toward identifying transcriptional regulators of GRP gene expression.
Molecular function and physiological role
Although the molecular function of GRP is yet unknown, the high content of Gla residues responsible for the high-affinity binding of calcium ions and its accumulation at sites of pathological calcification suggest that GRP may function as a modulator of calcium availability. Its association with fibrillar collagens type II, V, IX, and XI, but not type I collagen, also suggests that GRP may be involved in the organization and/or stabilization of cartilage matrix (8) and may play a key role in chondrogenesis. Based on its negative action on the differentiation of preosteoblast MC3T3-E1 cell line and murine primary calvaria cells, it has been proposed that GRP may function as an inhibitor of osteogenesis (8). To verify these speculations, a GRP-deficient mouse was generated by homologous recombination (8, 9) and a transient GRP1 knockdown was performed in zebrafish by injecting morpholino antisense oligonucleotides in embryos (6). Whereas morphological and histological analysis of mouse heterozygous and homozygous F2 offspring did not reveal obvious phenotypic alterations of the cartilage structure and skeletal development (8, 9), zebrafish morphants showed a broad spectrum of phenotypes, including: 1) weak and transient phenotypes such as slight overall growth retardations, smaller head, reduced eye pigmentation, and irregular and undulating notochord; and 2) severe and definitive phenotypes such as growth arrest (head), incomplete eye pigmentation, compressed and folded body, no yolk sack extension, and no clearly defined somites. Morphants at 72-h postfertilization showed weak/transient phenotypes often lacking cartilages such as the lower jaw and gill arches and lost most of COL2 in the remaining cartilaginous structures. Morphants exhibiting severe/definitive phenotypes lacked craniofacial cartilage in almost all cases and their notochord was strongly spirally compressed. Calcification, evaluated through alizarin red staining in 7-d postfertilization morphants, occurred normally in weak/transient phenotypes but was largely reduced in severe/definitive phenotypes. These results should, however, be taken with care, because onset of calcification (∼6 d postfertilization) is almost coincident with loss of morpholino activity (∼5 d postinjection) (6). To evaluate the role of Gla residues in the function of GRP protein, zebrafish embryos were treated with sodium warfarin, an inhibitor of vitamin K-dependent carboxylation of Glu residues. The phenotype of warfarin-treated zebrafish provided additional data confirming the phenotypes encountered after treatment by morpholinos (6). Nevertheless, because warfarin affects carboxylation of all Gla proteins, care should be taken when claiming that the observed effects are solely due to lack of correct γ-carboxylation of GRP. Furthermore, it is not clear whether the phenotype observed is due also to a general retardation in growth development rather than to a specific defect in fish development.
Transcript and genomic variants of grp gene
In addition to the original GRP transcript, 3 variants characterized by the absence of exon 2 (coding for signal peptide) and/or exon 4 (coding for most of the Gla domain) have been identified in mice (10) and later in zebrafish (6, 7). In mice, F2 and F4 variants lacking exon 2 and therefore a functional signal peptide (Fig. 1) were not secreted and remained as intracellular aggregates (10). In zebrafish, whereas F2 and F3 variants were detected for both GRP paralogs (6, 7) the F4 variant was detected only for GRP1/UCMAa (6) or only for GRP2/UCMAb (7). The biological importance of GRP variants remains to be elucidated. The GRP-F1 isoform (complete protein) was shown to be mainly expressed during chondrogenesis, whereas the GRP-F3 isoform (lacking the Gla domain) was rapidly coexpressed during chondrocyte maturation (10). The resulting difference in number of Gla residues may contribute to regulate the function of GRP at sites of expression.
SNPs are the most common genetic variations in the human genome, accounting for ~90% of all known variations (17), and a total of 374 (290 validated) SNPs have been described so far for the GRP gene region. Among these, 22 are exonic SNPs (16 in the coding sequence and 6 in the untranslated regions) and 286 are intronic SNPs (data retrieved from NCBI dbSNP database and summarized in Tables 3 and 4). Various SNPs occurred in the 5′-flanking/promoter region of the GRP gene and some were shown to affect putative binding sites for transcription factors (Supplemental Table 1) and thus GRP gene transcription. Among those transcription factors, several are known to play critical roles in chondrogenesis, including the transcription factors of the SOX family. Similarly, several SNPs occurred in the 3′ untranslated region of the GRP transcript and some were shown to affect putative binding sites for microRNAs (Supplemental Table 2) and thus GRP post-transcriptional processing. Among those microRNAs, several are known to target genes involved in osteoblast differentiation (miR-206, miR-31, miR-210) or associated with osteosarcomas (miR-151–3p and miR-382).
Table 3.
SNPs occurring in the different genomic regions of the human GRP gene, according to data from dbSNP1
| GRP gene region | SNP occurrence (region size in bp) |
| 5′ near gene | 42 (1992) |
| 5′UTR | 1 (73) |
| Exon 1 (coding region) | 1 (131) |
| Intron I | 13 (400) |
| Exon 2 | 1 (66) |
| Intron II | 4 (101) |
| Exon 3 | 5 (96) |
| Intron III | 98 (3828) |
| Exon 4 | 4 (99) |
| Intron IV | 172 (7410) |
| Exon 5 (coding region) | 5 (434) |
| 3′UTR | 5 (339) |
| 3′ near gene | 23 (482) |
(27). SNP, single nucleotide polymorphism; UTR, untranslated region.
Table 4.
SNPs occurring in the coding sequence of human GRP gene, according to data from dbSNP1
| Exon | SNP reference | Flanking region | Amino acid substitution |
| 1 | rs187657704 | CAGGAC[A/G]GCC | Ala6 = |
| 2 | rs41291317 | ATCTGC[A/G]TGG | Met32Thr |
| 3 | rs183396474 | CGAGGC[A/C]TCT | Asp53Glu2 |
| rs140874143 | GCTTGC[C/T]GCG | Gly62Ser3 | |
| rs3829925 | CATCTC[G/T]GGA | Arg69 = | |
| rs150509047 | TCATCT[C/T]TGG | Arg69Gln | |
| rs142585346 | TTGACC[A/T]CAT | Glu71Val4 | |
| 4 | rs148425191 | TCTCTC[C/T]GCA | Arg86Gln |
| rs74123515 | CCTCCA[C/T]GAA | Val101Met | |
| rs80070283 | TCCTCC[A/G]CGA | Val101Ala | |
| rs184841299 | TTCCTC[C/G]ACG | Val101Ala | |
| 5 | rs139212237 | CACTGC[C/T]CCA | Glu117Gly4 |
| rs147328988 | ACTGGC[A/G]CCA | Arg120Cys5 | |
| rs2281796 | TAGAGA[C/T]AGG | Tyr131Cys | |
| rs11547943 | GGATCA[C/G]GTG | Thr138 = | |
| rs140771568 | GATCAG[C/G]TGT | Thr138Arg |
(27). Gla, γ-carboxy glutamate; SNP, single nucleotide polymorphism.
Amino acid substitution affecting GGCX docking site.
Amino acid substitution affecting furin-like cleavage site.
Amino acid substitution affecting Gla domain.
Amino acid substitution affecting conserved residue.
GRP-associated pathologies
The GRP protein was also shown to accumulate at sites of ectopic calcification in different pathologies affecting either the human skin, such as dermatomyositis with calcinosis and pseudoxanthoma elasticum, or the vascular system such as in chronic kidney disease (2, 13) (Table 5). GRP was also detected in healthy mammary gland tissue, but its accumulation is greatly enhanced in several types of breast cancer (18, 19). The presence of a large number of Gla residues in GRP is likely to promote its preferential accumulation at sites of ectopic calcifications, but the relationship between this localization and GRP function remains to be clarified. GRP has also been found in blood cells (e.g., neutrophils, lymphocytes, and plasmocytes) either circulating or infiltrated in human lesions (19). This finding could indicate the involvement of GRP in inflammatory processes that are known to play a role in human pathologies showing ectopic calcifications (20). Interestingly, a recent microarray study analyzing age-related differences in gene expression during development of post-traumatic osteoarthritis (OA) in mice identified GRP as one of the genes whose expression is increased in aging mice compared to younger mice with the same pathology (21). Other groups recently investigated GRP expression in OA-related tissues, but data are still preliminary and no conclusion can be drawn yet (14, 22). Nevertheless, it is possible that inflammation may be the common feature between abnormal expression of GRP and disease, because it is now accepted that inflammation plays a central role in the development of OA (23). As reported above, 2 GRP-related peptides originating from alternative splicing of exon 4 may exhibit different calcium binding capacities due to a very different number of Gla residues (10); thus, their differential expression could be relevant for the development of GRP-related pathologies, a hypothesis that remains to be analyzed.
Table 5.
Sites of human GRP gene expression and protein accumulation in pathological conditions
| Tissue | Origin | Cell type expressing GRP | Detection method1 | Reference |
| Dermatomyositis with calcinosis | Human | Human skin with calcifications | IHC | (13) |
| Pseudoxanthoma elasticum | Human | Human skin with calcifications | IHC | (13) |
| Scars | Human | Calcified scar tissue | IHC | (13) |
| Chronic kidney disease | Human | Arteries with calcifications | IHC | (13) |
| OA model with surgically induced destabilization of medial meniscus | Mouse | Increase in knee joint tissue of old (12 mo) vs. young (12 wk) animals | Microarray | (21) |
IHC, immunohistochemistry; OA, osteoarthritis.
Conclusions
Clearly, much remains to be discovered with regard to the Gla-rich protein. Discovered 4 years ago, we now know that it has an exceptional capacity in binding calcium ion through a dense network of Gla residues and the ability to bind collagen in the extracellular compartment. Thus, it has been proposed that GRP may function as a modulator of calcium availability in vertebrates and as a player in extracellular matrix organization. Future studies should aim at further understanding the mechanisms that regulate GRP gene expression and at clarifying the precise role of the protein in vertebrate physiology. In this aspect, this review has highlighted the importance of GRP for chondrocyte differentiation and chondrogenesis but has also raised the possibility that it may be involved in other mechanisms related to skeletal homeostasis. A great deal is yet to be learned about the tissue specificity of GRP transcripts and the biological relevance of spliced variants. Answering questions related to the molecular evolution of GRP will certainly deserve a greater attention, in particular in unveiling the absence of GRP in birds/archosaurians. Are physiological processes requiring GRP function absent in these species, or is there another gene capable of supplying a similar function? Also unclear is the role of GRP in the development of human diseases. Despite the fact that no obvious phenotypic alterations in null mice were reported, GRP expression and accumulation has been associated with ectopic calcification, cancers, and OA. Although the influence of GRP on the progression of these diseases will need to be further elucidated, it raises the possibility that GRP could be used as a molecular biomarker and/or target for disease prevention and therapy.
Acknowledgments
The authors thank Cindy Fazenda and Iris Silva for their help in collecting part of the data used in this review. All authors have read and approved the final manuscript.
Footnotes
Supported in part by the Centre of Marine Sciences. N.C. was supported by postdoctoral grant SFRH/BPD/48206/2008 from the Portuguese Science and Technology Foundation.
Author disclosures: M. L. Cancela, N. Conceição, and V. Laizé, no conflicts of interest.
Supplemental Tables 1 and 2 are available from the “Online Supporting Material” link in the online posting of the article and from the same link in the online table of contents at advances.nutrition.org.
All authors contributed equally to this review.
Abbreviations used: Gla, γ-carboxyglutamate; ISH, in situ hybridization; OA, osteoarthritis; SNP, single nucleotide polymorphism.
Literature Cited
- 1.Gong Z, Korzh V, Fish development and genetics: the zebrafish and medaka models. Singapore: World Scientific Publishing; 2004 [Google Scholar]
- 2.Viegas CSB, Simes DC, Laizé V, Williamson MK, Price PA, Cancela ML. Gla-rich protein (GRP), a new vitamin K-dependent protein identified from sturgeon cartilage and highly conserved in vertebrates. J Biol Chem. 2008;283:36655–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tagariello A, Luther J, Streiter M, Didt-Koziel L, Wuelling M, Surmann-Schmitt C, Stock M, Adam N, Vortkamp A, Winterpacht A. Ucma: a novel secreted factor represents a highly specific marker for distal chondrocytes. Matrix Biol. 2008;27:3–11 [DOI] [PubMed] [Google Scholar]
- 4.Surmann-Schmitt C, Dietz U, Kireva T, Adam N, Park J, Tagariello A, Önnerfjord P, Heinegard D, Schlötzer-Schrehardt U, Deutzmann R, et al. Ucma, a novel secreted cartilage-specific protein with implications in osteogenesis. J Biol Chem. 2008;283:7082–93 [DOI] [PubMed] [Google Scholar]
- 5.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. 2005;27:937–45 [DOI] [PubMed] [Google Scholar]
- 6.Neacsu CD, Grosch M, Tejada M, Winterpacht A, Paulsson M, Wagener R, Tagariello A. Ucmaa (Grp-2) is required for zebrafish skeletal development. Evidence for a functional role of its glutamate γ-carboxylation. Matrix Biol. 2011;30:369–78 [DOI] [PubMed] [Google Scholar]
- 7.Fazenda C, Silva IAL, Cancela ML, Conceição N. Molecular characterization of the two genes encoding Gla-rich protein (Grp) in zebrafish. J Appl Ichthyology. 2012 [Google Scholar]
- 8.Surmann-Schmitt C, Stock M, Eitzinger N, Adam N, Dietz U, von der Mark K. Role of Ucma, a novel cartilage specific protein in cartilage matrix and development; 2008 [cited 2011 Dec 4]. Available from: http://www.molmed.uni-erlangen.de/Ressourcen/REPORT2008.pdf.
- 9.Eitzinger N, Surmann-Schmitt C, Bösl M, Schett G, Engelke K, Hess A, von der Mark K, Stock M. Ucma is not necessary for normal development of the mouse skeleton. Bone. Epub 2011 Dec 2 [DOI] [PubMed] [Google Scholar]
- 10.Le Jeune M, Tomavo N, Tian TV, Flourens A, Marchand N, Camuzeaux B, Mallein-Gerin F, Duterque-Coquillaud M. Identification of four alternatively spliced transcripts of the Ucma/GRP gene, encoding a new Gla-containing protein. Exp Cell Res. 2010;316:203–15 [DOI] [PubMed] [Google Scholar]
- 11.Koyama E, Yasuda T, Minugh-Purvis N, Kinumatsu T, Yallowitz AR, Wellik DM, Pacifici M. Hox11 genes establish synovial joint organization and phylogenetic characteristics in developing mouse zeugopod skeletal elements. Development. 2010;137:3795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuelling M, Kaiser FJ, Buelens LA, Braunholz D, Shivdasani RA, Depping R, Vortkam A. Trps1, a regulator of chondrocyte proliferation and differentiation, interacts with the activator form of Gli3. Dev Biol. 2009;328:40–53 [DOI] [PubMed] [Google Scholar]
- 13.Viegas CSB, Cavaco S, Neves PL, Ferreira A, João A, Williamson MK, Price PA, Cancela ML, Simes DC. Gla-rich protein is a novel vitamin K-dependent protein present in serum that accumulates at sites of pathological calcifications. Am J Pathol. 2009;175:2288–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Jeune M, Duterque-Coquillaud M, Flajollet S, Tian T, Legeai-Mallet L, Tomavo N, Flourens A, Holder-Espinasse M, Galéra P, Mallein-Gerin F. The expression of GRP gene, encoding four new Gla-rich protein isoforms, is finely regulated in cartilage. Osteoporos Int. 2011;22 Suppl 1:S52 [Google Scholar]
- 15.Vlaeminck-Guillem V, Carrere S, Dewitte F, Stehelin D, Desbiens X, Duterque-Coquillaud M. The Ets family member Erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech Dev. 2000;91:331–5 [DOI] [PubMed] [Google Scholar]
- 16.Conceição N, Fazenda C, Cancela ML. Comparative promoter analysis and its application to the identification of candidate regulatory factors of Gla-rich protein (GRP). J Appl Ichthyology. 2012 [Google Scholar]
- 17.Collins FS, Brooks LD, Chakravarti AA. DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–31 [DOI] [PubMed] [Google Scholar]
- 18.Viegas CSB, Morera JL, Teixeira A, João A, Ferreira A, Neves PL, Smith E, Herfs M, Vermeer C, Simes DC. GRP is a new VKD protein associated with ectopic calcifications and potential application in clinical diagnostics. Molecular, Structural and Clinical Aspects of Vitamin K and Vitamin K-Dependent Proteins, FASEB Summer Conference. Carefree (AZ); 2011 [Google Scholar]
- 19.Viegas CSB. Identification and characterization of a new calcium binding protein from sturgeon, Gla rich protein or GRP. Contribution to unveil its function in vertebrates [dissertation]. Faro: University of Algarve; 2010 [Google Scholar]
- 20.Li X, Kramer MC, van der Loos CM, Koch KT, de Boer OJ, Henriques JP, Baan JJ, Vis MM, Piek JJ, Tijssen JG, et al. A pattern of disperse plaque microcalcifications identifies a subset of plaques with high inflammatory burden in patients with acute myocardial infarction. Atherosclerosis. 2011;218:83–9 [DOI] [PubMed] [Google Scholar]
- 21.Loeser RF, Olex A, McNulty MA, Carlson CS, Callahan M, Ferguson C, Chou J, Leng X, Fetrow JS. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. Epub 2011 Oct 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simes D, Cavaco S, Ramos A, Silva J, Morera JL, Teixeira A, Smith E, Herfs M, Vermeer C, Viegas C. Insights into the relation between Gla-rich protein (GRP) and osteoarthritis. Molecular, Structural and Clinical Aspects of Vitamin K and Vitamin K-Dependent Proteins, FASEB Summer Conference. Carefree (AZ); 2011 [Google Scholar]
- 23.Wang Q, Rozelle AL, Lepus CM, Scanzello CR, Song JJ, Larsen DM, Crish JF, Bebek G, Ritter SY, Lindstrom TM, et al. Identification of a central role for complement in osteoarthritis. Nat Med. 2011;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ensembl. release 65; December 2011 [cited 2011 Dec 4]. Available from: www.ensembl.org.
- 25. UniGene: a unified view of the transcriptome [cited 2011 Dec 4]. Available from: www.ncbi.nlm.nih.gov/unigene.
- 26.Viegas CSB, Conceição N, Fazenda C, Simes DC, Cancela ML. Expression of Gla-rich protein (GRP) in newly developed cartilage-derived cell cultures from sturgeon (Acipenser naccarii). J Appl Ichthyology. 2010;26:214–8 [Google Scholar]
- 27. dbSNP: short genetic variations. release 135; October 2011 [cited 2011 Dec 4]. Available from: www.ncbi.nlm.nih.gov/projects/SNP.