Abstract
The nutritional essentiality of zinc for the growth of living organisms had been recognized long before zinc biochemistry began with the discovery of zinc in carbonic anhydrase in 1939. Painstaking analytical work then demonstrated the presence of zinc as a catalytic and structural cofactor in a few hundred enzymes. In the 1980s, the field again gained momentum with the new principle of “zinc finger” proteins, in which zinc has structural functions in domains that interact with other biomolecules. Advances in structural biology and a rapid increase in the availability of gene/protein databases now made it possible to predict zinc-binding sites from metal-binding motifs detected in sequences. This procedure resulted in the definition of zinc proteomes and the remarkable estimate that the human genome encodes ∼3000 zinc proteins. More recent developments focus on the regulatory functions of zinc(II) ions in intra- and intercellular information transfer and have tantalizing implications for yet additional functions of zinc in signal transduction and cellular control. At least three dozen proteins homeostatically control the vesicular storage and subcellular distribution of zinc and the concentrations of zinc(II) ions. Novel principles emerge from quantitative investigations on how strongly zinc interacts with proteins and how it is buffered to control the remarkably low cellular and subcellular concentrations of free zinc(II) ions. It is fair to conclude that the impact of zinc for health and disease will be at least as far-reaching as that of iron.
Introduction
The chronology of events in the research of zinc biology is revealing as it unfolds through different disciplines and demonstrates an ever-increasing variety of functions. Three phases of discovery can be discerned: 1) the recognition that zinc is essential for all forms of life, 2) the detection of zinc in enzymes and other proteins as a basis of its catalytic and structural functions, and 3) a role of zinc(II) ions in cellular regulation, which is the main subject of this article (Fig. 1). Knowledge about molecular functions of zinc emerged slowly as zinc is refractory to investigations with most spectroscopic techniques, and analysis in biological tissues was challenging before sufficiently sensitive methods such as flameless atomic absorption spectrophotometry became widely available. Also, many zinc proteins are of such low abundance that they escaped detection through direct analysis. So far, only bioinformatics and systems biology approaches have taught us the full extent to which zinc is involved in cellular biology. Yet, the role of zinc as a cofactor in proteins did not seem to explain all its biological functions. The third and most recent phase of discovery demonstrates that zinc(II) ions are regulatory ions akin to calcium(II) ions and are fundamentally important for cellular control. Again, new insights became possible only with the development of new tools, in this case the advent of chelating agents that fluoresce when binding zinc(II) ions and that allow measurement of zinc(II) ions in cells with a sensitivity exceeding that of most other analytical techniques. This article focuses on these latest developments that address the roles of zinc(II) ions in cellular regulation, which, so far, are unique among the essential transition metal ions. These roles increase the significance of zinc for cellular biology tremendously and suggest that the micronutrient zinc has much broader functions than any of the other transition metal ions, including iron.
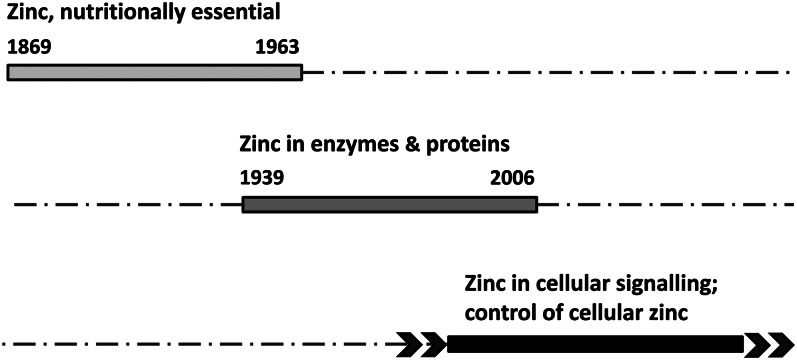
Figure 1.
The three phases of discoveries in zinc biology. Top: Discoveries that led to the recognition of zinc as an essential nutrient began with Jules Raulin’s seminal investigations of the dependence of Aspergillus niger on zinc for growth and culminated in the discovery of zinc deficiency in humans by Ananda Prasad. Middle: Discoveries of zinc as a cofactor of proteins also has a defined beginning, namely, David Keilin’s finding of zinc in carbonic anhydrase, and reached a certain end point by Ivano Bertini’s estimates of the number of zinc proteins in zinc proteomes. Bottom: Discoveries related to the role of zinc(II) ions in regulation do not have well-defined beginnings and are open in terms of future implications.
Current status of knowledge
Zinc as an essential transition metal ion
Jules Raulin, who trained under Louis Pasteur, reported in 1869 that zinc is essential for the growth of the fungus Aspergillus niger (1). His work is remarkable and decisive because he used chemically defined media to arrive at his conclusion. In the 1920s, it was discussed whether the presence zinc in human tissues is the result of environmental exposure that would affect humans adversely. Investigators at the Harvard School of Public Health in Boston measured zinc concentrations in human tissues and concluded that zinc is present endogenously and may serve biological functions (2, 3). It is instructive to read the conclusion of Drinker’s and Collier: “one is struck…by the...failure of investigators to recognize this normal universality of zinc...leads us to believe that zinc is indispensable for the normal functioning of the body” (3). Considerable time elapsed after Raulin’s original observation until zinc was shown to be essential for rats (1934) and for humans (1961) (4, 5). This early period of discovery was the subject of a Federation of American Societies for Experimental Biology symposium organized by Boyd O’Dell (6).
From a single zinc enzyme to #x223C;3000 human zinc proteins
The biochemistry of zinc began in 1939 when erythrocyte carbonic anhydrase was found to contain stoichiometric amounts of zinc and the zinc was shown to be essential for its enzymatic activity (7). Thus, knowledge of a role of zinc in proteins preceded Ananda Prasad’s seminal discovery of human zinc deficiency by more than twenty years (8). A second zinc enzyme, bovine pancreatic carboxypeptidase, was identified only fifteen years after the discovery of zinc carbonic anhydrase (9). Advances in analytical methods for isolating proteins and measuring metals in biological material, the latter largely pioneered by the late Bert L. Vallee and his colleagues (10), led to the discovery of hundreds of zinc proteins in all classes of enzymes, i.e., oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases (11).
A huge impact on the development of the field was the observation that Xenopus laevis transcription factor IIIA contains 9 repetitive sequences of C (cysteine) and H (histidine) residues that are involved in the binding of 9 zinc ions (12). This characteristic ligand signature of metal-coordinating amino acid side chains interspersed with noncoordinating amino acids (“spacers” a, b, c,) (CXaCXbHXcH, coordination type C2H2) forms relatively small protein domains that interact with nucleic acids like the fingers of a hand gripping a rod and hence became known generically as a “zinc finger.” In these sites, zinc has primarily a structural role. At about the same time, nucleotide sequence databases increased rapidly in number and size and led to the landmark event of the sequence of the human genome. These databases allowed searches for homologies with established signatures of metal-coordinating amino acids and made it possible to predict zinc-binding sites. Using only the signature of the classic C2H2 zinc finger motif, 5092 protein domains containing this motif were identified in the human genome (13). The actual number of zinc proteins with this motif is smaller, however, because many proteins contain several zinc fingers, with as many as 36 zinc fingers observed in one particular protein. Also, signatures and 3-dimensional structures of other structural zinc sites in protein domains were discovered, such as sites with 4 cysteine ligands (C4), with 3 cysteine ligands and 1 histidine ligand (C3H), with 1 aspartate (D) and 3 histidine ligands (DH3), and others (14). Mining of databases for these signatures further increased the number of zinc proteins. Thus, in addition to the hundreds of zinc enzymes, hundreds of zinc finger proteins were now known. Many of the zinc-binding domains do not interact with nucleic acids but rather with other proteins, and, in some cases, with lipids (15, 16). It became customary to assign zinc finger domains in newly discovered proteins based on sequence homology rather than performing a chemical analysis of zinc. Predictions were initially verified experimentally. A now classic example is the prediction of a zinc binding site in human leukotriene A4 (LTA4)2 hydrolase. LTA4 hydrolase has a signature HExxH…E (E, glutamate) that aligns with that of the catalytic zinc in the bacterial proteinase thermolysin (17). Metal analysis confirmed that LTA4 hydrolase is indeed a zinc enzyme (18). The analogy with thermolysin suggested that LTA4 hydrolase has peptidase activity, which was then also confirmed experimentally (19). The crystal structure of human LTA4 hydrolase proved the prediction about its zinc-binding site to be correct (20).
When the entire human genome was scanned for zinc-binding signatures, it was predicted that at least 3% of the genes encode zinc proteins (21). However, it was pointed out that the actual zinc proteome could be considerably larger because this approach does not account for all zinc-protein interactions (22). Indeed, a bioinformatics approach led to the prediction that at least 10% (∼2800 gene products) of the human genome encodes zinc proteins (23). This approach combines the information from databases for protein structures (24) and for protein domains (25) to scan gene databases, uses an automated procedure for analysis, and sets guidelines for manual curation of the information (26). Three data sets were obtained: metalloproteins with known signatures and unknown domains (2406 proteins), metalloproteins with known signatures and known domains (2506), and metalloproteins with unknown signatures and known domains (2407). The total number of zinc proteins is 3207; 2430 are identified by at least two methods and 1684 zinc proteins by all three methods (signatures, domains, annotations). As possible zinc proteins are counted: 397 hydrolases; 302 ligases; 167 transferases; 43 oxidoreductases; 24 lyases/isomerases; 957 transcription factors; 221 signaling proteins; 141 transport/storage proteins; 53 proteins with structural metal sites; 19 proteins involved in DNA repair, replication, and translation; 427 zinc finger proteins of unknown function; and 456 proteins of unknown function. The number of zinc proteins correlates linearly with the number of genes in a particular genome and is higher in eukarya (8.8%) than in bacteria and archaea (5–6%) (27). The increase in the zinc proteome in eukarya is due to the large number of additional zinc proteins involved in regulation, such as the zinc-binding domains of zinc finger proteins that extend considerably the conformational landscape of proteins and hence the possibilities for interactions, i.e., the interactome.
Predictions estimating the number of zinc proteins in the human zinc proteome demonstrated that the widespread use of zinc is in protein structure and function. However, they are based on known signatures and therefore are inherently limited (28–30). There are some examples of false positives, where zinc is not present in the protein because the site does not bind zinc or the site binds a different metal. Predictions do not take into account either where the protein is localized in the cell or the differences in metal utilization in some compartments such as mitochondria. Also, zinc binds between protein subunits, a function that it serves exceptionally well because it is redox inert and binds stronger than the other essential metal ions except copper(II). In these interface zinc sites, zinc has catalytic functions or, more frequently, structural functions in the quaternary (same subunits) or quinary (different subunits) structure of proteins (31). Because the ligands of interface zinc sites stem from different proteins and coordination may involve only one or two ligands from each of the protomers, signatures cannot be readily identified and used for homology searches, thus precluding estimates of the total number of such sites. Yet other proteins bind zinc transiently. This category includes proteins involved in the regulation of cellular zinc homeostasis and proteins that are regulated by zinc. Signatures of these sites are incomplete, raising questions as to the extent that additional signatures will increase the size of the zinc proteomes. Indeed, experimental work indicates that interactions of zinc with proteins are more frequent than predictions suggest (32). Extensive reviews on this second phase of discovery exist (33, 34). The third and ongoing phase of discovery focuses on the fact that free zinc(II) ions in addition to protein-bound zinc(II) ions are important for functions of zinc in biology.
New functions: Zinc ions as messengers
With so many proteins requiring zinc, the questions of how, when, and where zinc is made available for proteins to become zinc proteins came increasingly into focus. A comprehensive answer is lacking, however. It is also not known whether the allocation of zinc when zinc becomes limiting occurs with any type of hierarchy to conserve basic zinc-dependent functions while suspending others. Another critical question is which processes and molecules control systemic homeostasis of zinc. A hormone such as hepcidin controlling iron metabolism has not been identified for the control of zinc. With discoveries of proteins involved in cellular zinc homeostasis, it became clear how complex the control is. Dozens of transporters were identified, in contrast to iron, where there are only a few. However, the specificity of most of these transporters is not well established. They are called zinc transporters, but several of them also transport other metal ions. The transporters are localized on the plasma membrane and on intracellular membranes. This extensive compartmentalization is characteristic for cellular zinc metabolism.
Regulating zinc: control of cellular zinc homeostasis
Several dozen proteins regulate cellular zinc. They store and release zinc, bind any excess of zinc to avoid unspecific reactions, transport zinc through membranes and cellular compartments, or sense zinc(II) ion concentrations.
Transporting and sensing zinc.
Ten members of the zinc (Zn) transporter (ZnT) family (SLC30A) export zinc from the cytosol, whereas 14 members of the ZRT/IRT-like protein (Zip) family (SLC39A) import zinc into the cytosol. Together these transporters control cellular zinc and its traffic through the plasma membrane and between cytosol and various cellular compartments (35, 36). The transporters are regulated transcriptionally, translationally, and at the protein level through heterodimer formation, ubiquitination, phosphorylation, and proteolysis (35–37). Three-dimensional structures of Zip proteins are not available. Some of the members of the family may have additional zinc binding sites (38), which is the case for ZnT proteins. Escherichia coli Yiip is a homolog of the human ZnT proteins and a Zn2+/H+ antiporter residing in the inner membrane. Its 3-dimensional structure shows a dimeric protein with a transmembrane domain and a cytoplasmic domain and several zinc-binding sites in the monomer: a zinc site between the transmembrane helices involved in zinc transport, a zinc-binding site between the two domains with unknown function, and a binuclear zinc binding site in the cytoplasmic domain, which is thought to sense cytoplasmic zinc(II) ion concentrations and trigger a conformational change that allows transport of zinc through the membrane (39). Metal-response element–binding transcription factor-1 (MTF-1) is the only known eukaryotic zinc(II) ion sensor. It controls zinc-dependent gene expression of proteins at increased zinc concentrations. Sensing is thought to occur through a pair of its 6 zinc fingers with lower affinity for zinc than the other zinc fingers (40). In human MTF-1, the acidic activation domain also participates in metal sensing (41). MTF-1 is an essential protein in development as its genetic ablation in mice is embryonically lethal (42).
Storing and releasing zinc: proteins.
Humans have at least a dozen different metallothioneins (MTs), proteins with 60–68 amino acids, among which 20–21 are cysteines (43). The gene expression of most of them is under the control of MTF-1. Their expression increases the cellular zinc-binding capacity when the cellular zinc concentration increases. Characterization of the isolated protein demonstrated that zinc is bound exclusively to the sulfur donors of 20 cysteines in 2 zinc/thiolate clusters, 3 zinc(II) ions in the N-terminal cluster with 9 cysteine ligands, and 4 zinc(II) ions in the C-terminal cluster with 11 cysteine ligands (44, 45). Every one of the 7 zinc(II) ions binds to 4 sulfurs of cysteines. At least two properties of MTs are remarkable with regard to those of other zinc proteins. One property is the different affinities of binding sites for zinc in human MT-2 despite the fact that all 7 zinc(II) ions are in tetrathiolate coordination environments (46). With these characteristics, MTs are not simply trapping any zinc for storage but serve dynamic functions as both zinc acceptors and zinc donors. The other property is that MTs are redox proteins. Generally, zinc is redox inert in biology. However, in coordination environments with sulfur donors, sulfur is redox active, and redox reactions can modulate the stability of the complex (47). This chemistry makes it possible to mobilize zinc(II) ions from high-affinity sites. In this way, the redox environment affects the metal load of MT and the concentrations of cellular free zinc(II) ions. Oxidation of the sulfur donors leads to zinc release, whereas reduction of the oxidized sulfurs leads to zinc binding. An important consequence of these properties is that the structure of MT in vivo depends on both metal ion availability and redox state and is not necessarily the one derived from investigations in vitro with 7 zinc ions and all 20 cysteines reduced. It is important to recall that MTs isolated from natural sources are generally heterogeneous and that preparation of MT involves removing the metals, reducing any disulfides, and then reconstituting the protein with a complement of a given metal ion to obtain a homogeneous preparation. In vivo, the structure of MT is dynamic, and MT occurs with different metal loads, metal composition, and in different redox states. For example, the composition in eye tissue is Zn6Cu-MT, whereas it is Zn7-MT when the protein is induced with zinc (48). MT has a higher copper content when isolated from fetal or neonatal liver (49). The metal composition of isolated MT-3, which is a neuronal growth inhibitory factor, is Zn3–4Cu4-MT-3 (50). A crystal structure of MT was obtained from rat MT-2 induced with cadmium. This MT has an overall composition Cd5Zn2-MT with 2 zinc ions located at defined positions in the N-terminal domain (45). Thus, the metal composition of MT changes when the organism is exposed to metals in the environment, under physiological conditions, and in diseases that affect metal availability. MT is not fully saturated with zinc in the liver, as reflected by measurements of ∼10% of the apoprotein and by the capacity to bind additional zinc (51). These results are consistent with the observation that the affinity of the protein for one of the seven zinc ions is much weaker than the affinities for the others (46). Also, the cysteines of MT may not be fully reduced under physiological conditions. MT with some of its cysteines oxidized has been observed in a cell line that produces increased amounts of reactive oxygen species (52) and in the heart of mice that have been oxidatively stressed with doxorubicin (53).
Buffering zinc.
Zinc, like every other essential metal ion, is maintained at specific cellular concentrations to enable zinc-specific reactions and to avoid interference with the functions of other essential metal ions, such as preventing the coordination of the wrong metal ion to a metalloprotein. One component of the cellular homeostatic control is the buffering of zinc. The concept of metal buffering is similar to that of hydrogen ion (proton) buffering. Whereas the pKa of a ligand determines the pH for proton buffering, the pKd of a ligand determines the pMe for metal buffering. In analogy to the concept of pH, the concentration of free zinc(II) ions is expressed as the zinc potential, pZn = −log[Zn2+]i (54). These relationships explain why it is mandatory to know the affinity of proteins for zinc at physiological pH (pKd) to understand zinc buffering and the control of cellular zinc. Affinities of ligands for divalent metal ions follow the Irving-Williams series, according to which zinc(II) ions bind stronger than the other divalent metal ions of the first transition series, except for copper(II) ions. Affinities of cytosolic zinc proteins for zinc are in the range of picomoles per liter dissociation constants (55). As a consequence, most of the zinc is bound, and the free zinc(II) ion concentrations are commensurately low. Direct measurements from several laboratories with different methods provide estimates of free zinc(II) ion concentrations in the range of tens to hundreds of picomoles per liter (pZn = 10–11) (Table 1) (56–63) and confirm that the consideration based on binding equilibria is a valid approximation. Of note, estimates were already reported 40 years ago when the inhibition of the magnesium enzyme phosphoglucomutase by zinc was measured. The authors concluded that for the enzyme not to be zinc inhibited in muscle, the free zinc ion concentration must be <32 pmol/L (57). The typical total cellular zinc concentrations are a few hundred micromoles per liter, but at least six to seven orders of magnitude higher than those of free zinc(II) ions. Free zinc(II) ions, in contrast to protein-bound zinc(II) ions, are not devoid of any ligands, but the nature of the ligands is not known. Other terms, such as readily or easily exchangeable, mobile and kinetically labile zinc, are used in the literature for pools of zinc. Each term has a different meaning, and the terms should not be used interchangeably. As small as the free zinc(II) ion concentrations are, they are not negligible. In fact, they are critically important in cellular regulation.
Table 1.
Examples of measurements of free zinc(II) ion concentrations
| [Zn2+]i , pmol/L | Cell type/tissue | Method | Reference |
| <100; 32 | Rabbit muscle | Phosphoglucomutase inhibition | (56, 57) |
| 24 | Human erythrocytes | Determination of cellular zinc in a zinc-buffered system | (58) |
| 5–20 | Rat PC12, pheochromocytoma | Carbonic anhydrase sensor | (59) |
| 614 | Human HT29, colon cancer | FluoZin-3 | (60) |
| <1000 | Rat primary cortical neurons | ZnAF-2 | (61) |
| 400 | Rat INS-1, insulinoma | CALWY-FRET sensor | (62) |
| 80 | Human HeLa | ZapCY-FRET sensor | (63) |
The buffering causes a specific concentration of zinc(II) ions (pZn values) and is not to be confused with the buffering capacity, which describes the strength of buffering and the resistance to change and not the pZn at which buffering occurs. The physiological zinc-buffering capacity in an epithelial cell line is ∼15 μmol/L (60). It is not very high. In fact, if it were high, zinc(II) ions could not readily participate in biological regulation. In molecular terms, ∼30% of the buffering capacity is based on thiol-containing ligands (64). Oxidation of thiols under oxidative stress decreases the zinc-buffering capacity, making cells more vulnerable to the adverse effects of additional zinc(II) ions. Cellular zinc buffering is a dynamic process in which the pZn, the buffering capacity, or both may change. Increasing the zinc-buffering capacity by changing ligand concentrations allows the cell to handle more zinc while keeping the pZn value constant. Lowering the buffering capacity allows the cell to change its pZn under conditions such as zinc influx/efflux. According to the theory of buffering, both increases and decreases of the pZn value would then occur more readily. The same cell can have different pZn values when proliferating, differentiating, or undergoing apoptosis (60, 64). The significance of these dynamic processes is that global or local changes of zinc(II) ion concentrations provide a way to control cellular processes.
If affinities (pKd values), which control buffering, were the only factor to control cellular zinc(II) ion concentrations, zinc could move only in one direction of thermodynamic gradients, i.e., from lower to higher affinity sites. This is clearly not the case. Additional mechanisms are necessary to populate sites of somewhat lower affinity with zinc. One mechanism is the release of zinc(II) ions from a compartment in which zinc(II) ions are not in thermodynamic equilibrium with the cytosol.
Storing and releasing zinc: vesicles.
A characteristic feature of eukaryotic cells is the extensive compartmentalization of zinc. Cells restrict the availability of zinc(II) ions through compartmentalization and control zinc via dynamic coordination environments that modulate the kinetics of metal association and dissociation (65). The transfer of zinc between different cellular compartments requires a large number of transporters and contributes to buffering. Such movements of metal ions are a kinetic phenomenon. This additional component of buffering in biological systems has been referred to as muffling (66). By removing zinc from a particular compartment, such as the cytosol, cells can cope with much higher zinc loads than through buffering alone. Cellular zinc buffering is thus a combination of the cytosolic zinc-buffering capacity and the zinc-muffling capacity based on the activities of transporters that remove zinc(II) ions from the cytosol to an organelle or to the outside of the cell (67). Buffering and muffling are not just housekeeping functions. They are fundamentally important for allowing zinc(II) ion fluctuations to occur, and as such, they are critical for zinc(II) ions as regulatory ions. Sequestration of zinc(II) ions in a cellular compartment is one way to store zinc temporarily in the cell and then release it again on demand. Compartmentalization resolves a long-standing issue of whether cells store zinc. Zinc is mainly stored in vesicles rather than in a protein with a high zinc-binding capacity akin to ferritin in iron storage. The chemical state of zinc in vesicles is not known. Free zinc(II) concentrations in these vesicles are much higher (lower pZn values) than in the cytosol, indicating different buffering of zinc. Estimates are 1–100 μmol/L in insulin-storing granules (62). The determination of subcellular pZn values is an area of considerable current interest. Measurements indicate that some subcellular compartments have much lower free zinc(II) ion concentrations than cytosol. Estimates are 0.9 pmol/L for the endoplasmic reticulum (ER), 0.2 pmol/L for the Golgi (63), 0.2 pmol/L for the mitochondrial matrix (68), and 0.14 pmol/L for mitochondria (69).
Zinc in regulation: zinc(II) ions as signaling ions
Perhaps the most exciting area in contemporary zinc biology is the role of released zinc(II) ions in information transfer within cells and between cells (70). In pathways stimulus → increase of [Zn2+] → targets, zinc(II) ions are signals. The preference of zinc(II) ions for specific coordination environments in proteins endows these signals with a certain degree of specificity. The amplitudes of zinc(II) ion signals determine which proteins are targeted because any target must have a commensurate affinity for zinc. Under conditions of stimulation, cellular zinc(II) ion concentrations increase and reach concentrations of >2 nmol/L only under oxidative stress and in apoptosis. Unless the buffering capacity changes or is exhausted, they return to their resting concentrations within minutes (71). As for the estimates of the steady-state concentrations of free zinc(II) ions (Table 1), a consensus is building on the amplitudes of zinc(II) ion fluctuations. A variety of experimental paradigms, such as oxidative stress, exposure to glucose, electrical stimulation, and chemical displacement of zinc with the tighter binding mercury(II) ion, increase the cellular free zinc(II) ion concentrations to a similar degree (Table 2) (72–76). Although the control of zinc(II) ion fluctuations was discussed above, the following discussion focuses on how zinc(II) ion signals are generated, transmitted to, and terminated at targets.
Table 2.
Examples of measurements of fluctuations of free zinc(II) ion concentrations
| [Zn2+]i Fluctuation, nmol/L | Cell type/tissue | Condition | Reference |
| 0.1–0.7 | Cardiomyocytes | Electrical stimulation | (72) |
| 0.52–0.87 | Cardiomyocytes | Diabetes | (73) |
| 0.45–0.85 | Pancreatic islet β cells | Glucose exposure | (74) |
| 3–5 | Epithelial, colon mucosa | Oxidative stress | (75) |
| 0.78–1.25 | Epithelial, colon cancer cells (HT29) | Differentiation | (51, 64) |
| 0.60–0.78 | Proliferation | ||
| 0.78–2.51 | Apoptosis | ||
| 0.15–1.3 | Jurkat T-cells | Mercury (HgCl2) treatment | (76) |
Increasing the concentrations of free zinc(II) ions.
In addition to the release of zinc from proteins, two pathways of vesicular release of zinc(II) ions increase the concentrations of free zinc(II) ions. The first one is the release of zinc from vesicles into the extracellular space. The prime example is the release of zinc(II) ions from synaptic vesicles in a subset of glutamatergic neurons (77). One target of the released zinc(II) ions is the postsynaptic N-methyl-d-aspartate receptor, which zinc inhibits at low nanomoles per liter concentrations (78). Zinc(II) ions also modulate the activities of other neuroreceptors, and there is reuptake with functional effects in the presynaptic cell (79, 80). Which targets of zinc(II) ions are physiologically important depends on the effective concentrations of synaptic zinc. Zinc(II) ions also occur in other nonsynaptic vesicles and are released from cells by exocytosis from these vesicles that are loaded with zinc(II) ions by different zinc transporters, ZnT3 in neurons in the brain, ZnT8 in pancreatic β cells, or ZnT2 in epithelial cells of the mammary gland (81). Cellular release of zinc(II) ions has been observed in a number of cells in exocrine and endocrine glands. Somatotrophic cells in the pituitary gland, pancreatic acinar cells, β cells of the islets of Langerhans, Paneth cells in the crypts of Lieberkühn, cells of the tubuloacinar glands of the prostate, epithelial cells of the epididymal ducts, and osteoblasts all secrete zinc(II) ions (82). In pancreatic β cells, zinc(II) ions participate in the formation and structure of hexameric insulin, the storage form of insulin in the granules. Zinc(II) ions are secreted together with insulin from β cells. The purpose of co-secretion is not entirely clear but may relate to paracrine functions or to avoiding the formation of amyloid deposits from amyloidogenic proteins such as insulin and amylin. In mammary epithelial cells, exocytotic vesicles contain zinc(II) ions and supply the milk with zinc. Fluctuations of intracellular free zinc(II) ions occur in the mitotic cell cycle with two distinct peaks, one early in the G1 phase and another one early in the S phase (71). Mouse oocytes take up large amounts of zinc at the latest stage of maturation and then become growth arrested after the first meiotic division (83). On fertilization, the embryos resume progression through the cell cycle and, subsequent to calcium(II) ion oscillations, release intracellular zinc(II) ions into the extracellular space in a process that has been referred to as “zinc sparks.”
The second pathway involves intracellular release of zinc(II) ions from vesicles. Stimuli targeting the zinc transporter Zip7 control the release of zinc(II) ions from a store in the ER to the cytoplasm (84). The pathway involves growth factor stimulation and casein kinase 2–induced phosphorylation of Zip7, which opens the store for zinc release (85). Lysosomal release in IL-2–stimulated T cells is a required signal for proliferation (86). In macrophages, stimulation of the immunoglobulin E FcεRI receptor and extracellular signal-regulated kinase/inositol trisphosphate signaling triggers zinc release, which is preceded by Ca2+ release (87). This phenomenon has been referred to as a “zinc wave.” Zinc sparks occur within seconds, whereas zinc waves take minutes to develop. Thus, zinc(II) ion signals have different frequencies.
Targets of free zinc(II) ions: protein-bound zinc.
To transmit the zinc signal to targets, close proximity between the source of zinc(II) ions and their target(s) seems to be essential. Concentrations of picomoles per liter to nanomoles per liter cellular zinc(II) ions inhibit the activities of certain enzymes, such as protein tyrosine phosphatases (88). Protein tyrosine phosphatases are not recognized as zinc proteins, mainly because they are isolated with chelating agents to preserve their enzymatic activity. Kinetic investigation of isolated protein tyrosine phosphatases and buffered zinc(II) ion solutions demonstrate that the zinc inhibition constant of the cytosolic domain of receptor protein tyrosine phosphatase β is 27 pmol/L (89). Locally, the cellular zinc(II) ion concentration could be higher than the measured steady-state amounts indicate. Some protein tyrosine phosphatases are localized at the ER where zinc release occurs. The phosphorylation of Zip7, the zinc inhibition of protein tyrosine phosphatases, and fluctuations of zinc(II) ions indicate a role of zinc in controlling phosphorylation signaling. The match between free zinc(II) ion concentrations and the inhibition of target proteins is also evident from the measurement of 24 pmol/L free zinc(II) ions in erythrocytes and an IC50 of 80 pmol/L for zinc inhibition of erythrocyte Ca2+-ATPase (58, 90). The affinities of zinc proteins for zinc and the affinities of proteins that are targeted by signaling zinc(II) ions define the lower and upper ranges in which free zinc(II) ions regulate biological processes. Thirty years ago, before the molecules and mechanisms controlling cellular zinc were known, inhibitory control, vesicular sequestration of zinc, and complementarity of zinc and calcium in cellular regulation had been given as an answer to the question: “Zinc, what is its role in biology?” (91). The cellular roles of zinc(II) ions at exquisitely low concentrations, typically reserved for hormones, are most remarkable and constitute a novel principle for cellular signaling (Fig. 2).
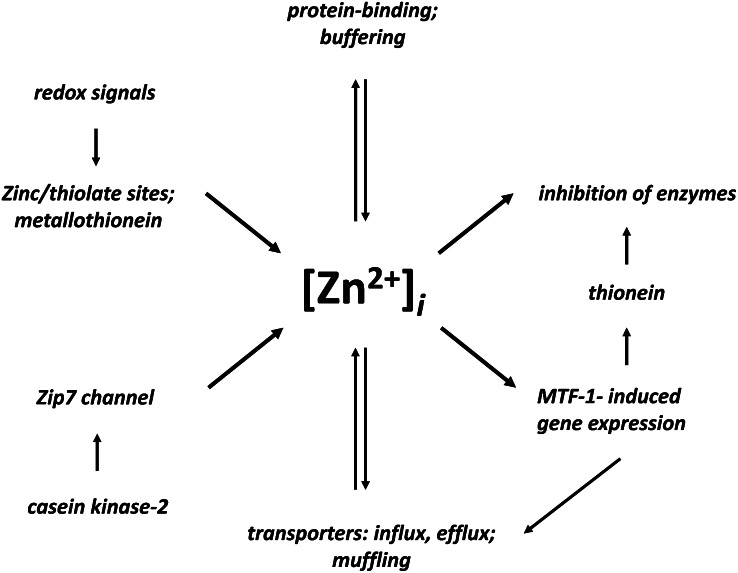
Figure 2.
Control and functions of free zinc(II) ion concentrations. Two pathways (left) increase the cytosolic free zinc(II) ion concentrations, [Zn2+]i, which serve as zinc signals (right) that inhibit enzymes and induce metal-response element–binding transcription factor 1 (MTF-1)–dependent gene transcription, which includes the synthesis of zinc transporters (ZnT1), and thionein that activates zinc-inhibited enzymes. Buffering and muffling control the free zinc(II) ion concentrations. Zip, ZRT/IRT-like protein.
Terminating zinc(II) ion signals.
The micromolar per liter concentrations of MTs in most cells provide a temporary reservoir of sufficiently tightly sequestered zinc and, compared with the picomoles per liter free zinc(II) ion concentrations, relatively large amounts of zinc that are available in a deliverable form on demand. This function of MT resembles those of metallochaperones, in particular because MTs have the capacity to transfer zinc via protein-protein interactions by an associative mechanism (92). MT has affinities for zinc in exactly the range of zinc(II) ion concentrations discussed here and therefore can participate in the distribution of zinc and the termination of zinc signals. Termination of zinc signals as inferred from experiments in vitro involves thionein, the apoprotein of MT. Thionein binds zinc and activates zinc-inhibited enzymes. In vivo, thionein is likely a molecule that is neither completely devoid of nor fully loaded with metal ions. The translocation of MTs in cells and their induction by many different cellular signal transduction pathways allow changes in the cellular zinc buffering and muffling capacities. Modulating the chelating capacity of MTs through changes of either their total amount or their redox state affords ways to control the regulatory functions of zinc(II) ions (93). Elevated zinc(II) ion concentrations induce MTF-1–controlled gene transcription of thioneins, the apoproteins of MTs, and the zinc transporter ZnT1, resulting in the binding of the surplus of zinc to MTs and the export of zinc from the cell. A host of other conditions also induce the apoprotein, indicating that forms of thioneins are produced for lowering the availability of cellular zinc(II) ions, and modulating zinc-dependent processes.
In summary, four observations define a range of picomoles per liter to low nanomoles per liter concentrations at which zinc(II) ions can serve regulatory functions: 1) the free zinc(II) ion concentrations and their fluctuations, 2) the affinities of MT for zinc, 3) the affinities of zinc proteins for zinc, and 4) the zinc affinities of proteins that are targets of zinc(II) ion signals (Fig. 3).
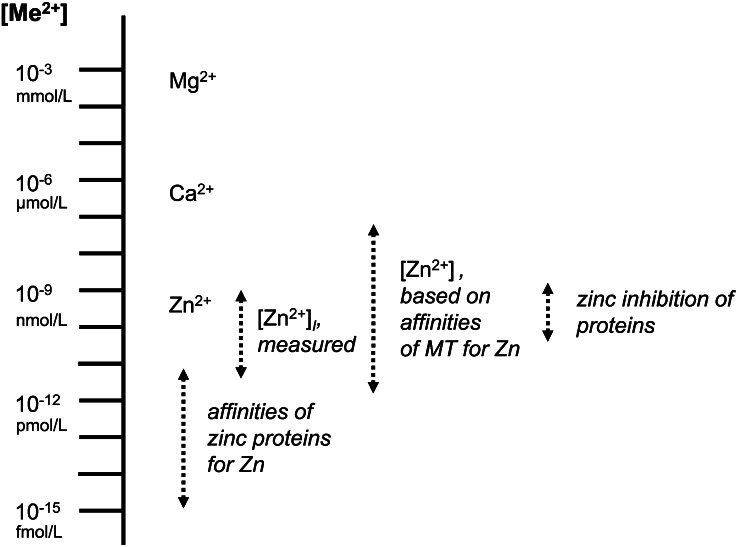
Figure 3.
Quantitative aspects underlying cellular zinc biochemistry. Zinc (Zn), calcium (Ca), and magnesium (Mg), the three major redox-inert metal ions involved in cellular regulation, cover many orders of magnitude in concentrations and affinities. Among the three, free zinc(II) ions are controlled at the lowest concentrations. Other essential divalent transition metal ions, with the exception of cupric ions, bind less tightly than zinc to proteins and need to be controlled at specific concentrations that are determined by affinities following the Irving-Williams series. Fluctuations of cytosolic free zinc(II) ion concentrations cover a range that corresponds to the affinities of metallothionein (MT)-2 for zinc and are bordered by zinc enzymes with the highest affinity for zinc and proteins, which zinc may regulate, with lower affinity for zinc.
Conclusions
The concentration of cellular zinc is rather high, almost as high as that of ATP. Therefore, at the cellular level, zinc can hardly be considered a trace element. Zinc is used as a cofactor in proteins much more frequently than most vitamins. The control of a fluctuating pool of zinc(II) ions at remarkably low concentrations and with the participation of many proteins provides a new perspective on the molecular functions of zinc in biology in general and for the impact of zinc on human health in particular (94). Roles of zinc(II) ions in biological phosphorylations and in redox signaling are already well documented and are part of the spectrum of actions of zinc in cellular proliferation, differentiation, and cell death. Zinc is not just required for the function of proteins; it participates in the control of cellular metabolism and paracrine and intracrine signaling (95). Not only the availability of zinc itself, but the limited cellular zinc buffering capacity and the many mutations that affect the functions of proteins involved in the cellular control of zinc have major implications for the balance between health and disease. The zinc-buffering system seems to be rather sensitive to environmental factors. Acute and long-term exposure to chemicals that interfere with zinc buffering should be a major concern for health, and so should be chelating agents that bind zinc and decrease its availability and that of other metal ions that compete with zinc, e.g., cadmium. The concentration range at which zinc(II) ions occur is critical for explaining the global functions of zinc. Zinc is generally considered to be an antioxidant. However, it is redox inert and thus can serve such a function only indirectly. The term proantioxidant is more appropriate (96). Whether zinc elicits antioxidant, anti-inflammatory, or antiapoptotic effects depends on its concentration. Outside the physiological or pharmacological range, under conditions of both zinc overload and zinc deficiency, zinc(II) ions have the opposite effect: they become pro-oxidants with proinflammatory and proapoptotic properties. This intricate balance at a relatively narrow and tightly controlled range of concentrations needs to be considered when the physiological significance of results from experiments performed with zinc are evaluated and in nutrition when supplementation with zinc is considered (97).
Acknowledgments
All authors have read and approved the final manuscript.
Footnotes
Abbreviations used: ER, endoplasmic reticulum; LTA4, leukotriene A4; MT, metallothionein; MTF-1, metal-response element–binding transcription factor 1; Zip, ZRT/IRT-like protein; Zn, zinc; ZnT, zinc transporter.
Literature Cited
- 1.Raulin J. Etudes chimiques sur la vegetation. Ann Sci Nat Bot Biol Veg. 1869;11:92–299 [Google Scholar]
- 2.Lutz RE. The normal occurrence of zinc in biologic material. J Ind Hyg. 1926;8:127 [Google Scholar]
- 3.Drinker KA, Collier ES. The significance of zinc in the living organism. J Ind Hyg. 1926;8:257 [Google Scholar]
- 4.Todd WR, Elvehjem CA, Hart EB. Zinc in the nutrition of the rat. Am J Physiol. 1934;107:146–56 [Google Scholar]
- 5.Prasad AS, Halsted JA, Nadimi M. Syndrome of iron deficiency anemia, hepatosplenomegaly, hypogonadism, dwarfism and geophagia. Am J Med. 1961;31:532–46 [DOI] [PubMed] [Google Scholar]
- 6.O’Dell BL. History and status of zinc in nutrition. Fed Proc. 1984;43:2821–2 [PubMed] [Google Scholar]
- 7.Keilin D, Mann T. Carbonic anhydrase. Nature. 1939;144:442–3 [Google Scholar]
- 8.Prasad AS. Discovery of human zinc deficiency: 50 years later. J Trace Elem Med Biol. 2012;26:66–9 [DOI] [PubMed] [Google Scholar]
- 9.Vallee BL, Neurath H. Carboxypeptidase, a zinc metalloprotein. J Am Chem Soc. 1954;76:5006–7 [Google Scholar]
- 10.Maret W, Bert L. Vallee 1919–2010. Angew Chem Int Ed. 2010;49:2–3 [Google Scholar]
- 11.Vallee BL, Galdes A. The metallobiochemistry of zinc enzymes. Adv Enzymol Relat Areas Mol Biol. 1984;56:283–430 [DOI] [PubMed] [Google Scholar]
- 12.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller A, MacCallum RM, Sternberg MJ. Structural characterization of the human proteome. Genome Res. 2002;12:1625–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auld DS. Structural zinc sites. In: Messerschmidt A, Bode W, Cygler M, editors. Handbook of metalloproteins. Vol. 3. Chichester, UK: Wiley; 2004, p. 403–15. [Google Scholar]
- 15.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46 [DOI] [PubMed] [Google Scholar]
- 16.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63–70 [DOI] [PubMed] [Google Scholar]
- 17.Vallee BL, Auld DS. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–59 [DOI] [PubMed] [Google Scholar]
- 18.Haeggström JZ, Wetterholm A, Shapiro R, Vallee BL, Samuelsson B. Leukotriene A4 hydrolase: a zinc metalloenzyme. Biochem Biophys Res Commun. 1990;172:965–70 [DOI] [PubMed] [Google Scholar]
- 19.Haeggström JZ, Wetterholm A, Vallee BL, Samuelsson B. Leukotriene A4 hydrolase: An epoxide hydrolase with peptidase activity. Biochem Biophys Res Commun. 1990;173:431–7 [DOI] [PubMed] [Google Scholar]
- 20.Thunnissen MM, Nordlund P, Haeggström JZ. Crystal structure of human leukotriene A(4) hydrolase, a bifunctional enzyme in inflammation. Nat Struct Biol. 2001;8:131–5 [DOI] [PubMed] [Google Scholar]
- 21.Clarke ND, Berg JM. Zinc fingers in Caenorhabditis elegans: Finding families and probing pathways. Science. 1998;282:2018–22 [DOI] [PubMed] [Google Scholar]
- 22.Maret W. Exploring the zinc proteome. J Anal At Spectrom. 2004;19:15–9 [Google Scholar]
- 23.Andreini C, Banci L, Bertini I, Rosato A. Counting the zinc-proteins encoded in the human genome. J Proteome Res. 2006;5:196–201 [DOI] [PubMed] [Google Scholar]
- 24.Protein Data Bank. http://www.rcsb.org, last accessed October 18, 2012.
- 25.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue 38):D211–22. [DOI] [PMC free article] [PubMed]
- 26.Bertini I, Cavallaro G. Bioinformatics in bioinorganic chemistry. Metallomics. 2010;2:39–51 [DOI] [PubMed] [Google Scholar]
- 27.Andreini C, Banci L, Bertini I, Rosato A. Zinc through the three domains of life. J Proteome Res. 2006;5:3173–8 [DOI] [PubMed] [Google Scholar]
- 28.Maret W. Zinc proteomics and the annotation of the human zinc proteome. Pure Appl Chem. 2008;80:2679–87 [Google Scholar]
- 29.Maret W. Metalloproteomics, metalloproteomes, and the annotation of metalloproteins. Metallomics. 2010;2:117–25 [DOI] [PubMed] [Google Scholar]
- 30.Maret W. Zinc and the zinc proteome. In: Banci L, guest editor. Metallomics and the cell. Sigel A, Sigel H, Sigel RKO, series editors. Metal ions in life sciences. Vol. 12. Dordrecht, the Netherlands: Springer Science + Business Media B.V; 2013 [Google Scholar]
- 31.Maret W. Protein Interface Zinc Sites, In: Messerschmidt A, Bode W, Cygler M, editors. Handbook of metalloproteins Vol. 3. Chichester, UK: Wiley; 2004 [Google Scholar]
- 32.Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL, II, Jenney FE, Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, et al. Microbial metalloproteomes are largely uncharacterized. Nature. 2010;466:779–82 [DOI] [PubMed] [Google Scholar]
- 33.Vallee BL, Falchuk K. The biochemical basis of zinc physiology. Physiol Rev. 1993;73:79–118 [DOI] [PubMed] [Google Scholar]
- 34.Andreini C, Bertini I, Cavallaro G. Minimal functional sites allow classification of zinc sites in proteins. PLoS ONE. 2011;6:e26325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukada T, Kambe T. Molecular and genetic features of zinc transporters in physiology and pathogenesis. Metallomics. 2011;3:662–74 [DOI] [PubMed] [Google Scholar]
- 36.Lichten LA, Cousins RJ. Mammalian zinc transporters: nutritional and physiologic regulation. Annu Rev Nutr. 2009;29:153–76 [DOI] [PubMed] [Google Scholar]
- 37.Gaither LA, Eide DJ. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14:251–70 [DOI] [PubMed] [Google Scholar]
- 38.Potocki S, Rowinska-Zyrek M, Valensin D, Krzywoszynska K, Wikowska D, Luczkowski M, Kozlowski H. Metal-binding ability of cysteine-rich peptide domain of Zip13 Zn2+ ions transporter. Inorg Chem. 2011;50:6135–45 [DOI] [PubMed] [Google Scholar]
- 39.Lu M, Chai J, Fu D. Structural basis for autoregulation of the zinc transporter Yiip. Nat Struct Mol Biol. 2009;16:1063–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laity JH, Andrews GK. Understanding the mechanism of zinc-sensing by metal-responsive element binding transcription factor-1 (MTF-1). Arch Biochem Biophys. 2007;463:201–10 [DOI] [PubMed] [Google Scholar]
- 41.Günther V, Lindert U, Schaffner W. doi: 10.1016/j.bbamcr.2012.01.005. The taste of heavy metals: gene regulation by MTF-1. Biochim Biophys Acta. 2012;1823:1416–25. [DOI] [PubMed] [Google Scholar]
- 42.Günes C, Heuchel R, Georgiev O, Müller KH, Lichtlen P, Blüthmann H, Marino S, Aguzzi A, Schaffner W. Embryonic lethality and liver degeneration in mice lacking the metal-response transcriptional activator MTF-1. EMBO J. 1988;17:2846–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Maret W. Human metallothionein metallomics. J Anal At Spectrom. 2008;23:1055–62 [Google Scholar]
- 44.Arseniev A, Schultze B, Wörgötter E, Braun W, Wagner G, Vasak M, Kägi JHR, Wüthrich K. Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance. J Mol Biol. 1988;201:637–57 [DOI] [PubMed] [Google Scholar]
- 45.Robbins AH, McRee DE, Williamson M, Collett SA, Xuong NH, Furey WF, Wang BC, Stout CD. Refined crystal structure of Cd,Zn metallothionein at 2.0 A resolution. J Mol Biol. 1991;221:1269–93 [PubMed] [Google Scholar]
- 46.Krężel A, Maret W. The nanomolar and picomolar Zn(II) binding properties of metallothionein. J Am Chem Soc. 2007;129:10911–21 [DOI] [PubMed] [Google Scholar]
- 47.Maret W, Vallee BL. Thiolate ligands in metallothionein confer redox activity on zinc clusters. Proc Natl Acad Sci U S A. 1998;95:3478–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarez L, Gonzales-Iglesias H, Garcia M, Gosh S, Sanz-Medel A, Coca-Prados M. The stoichiometric transition from Zn6Cu1-metallothionein to Zn7-metallothionein underlies the up-regulation of metallothionein (MT) expression. J Biol Chem. 2012;287:28456–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartmann HJ, Weser U. Copper-thionein from fetal bovine liver. Biochim Biophys Acta. 1977;491:211–22 [DOI] [PubMed] [Google Scholar]
- 50.Uchida Y, Takio K, Titani K, Ihara Y, Tomonaga M. The growth inhibitory factor that is deficient in the Alzheimer’s disease brain is a 68 amino acid metallothionein-like protein. Neuron. 1991;7:337–47 [DOI] [PubMed] [Google Scholar]
- 51.Krężel A, Maret W. Thionein/metallothionein control Zn(II) availability and the activity of enzymes. J Biol Inorg Chem. 2008;13:401–9 [DOI] [PubMed] [Google Scholar]
- 52.Krężel A, Maret W. Different redox states of metallothionein/thionein in biological tissues. Biochem J. 2007;402:551–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng W, Benz FW, Cai J, Pierce WM, Kang YJ. Metallothionein disulfides are present in metallothionein-overexpressing transgenic mouse heart and increase under conditions of oxidative stress. J Biol Chem. 2006;281:681–7 [DOI] [PubMed] [Google Scholar]
- 54.Maret W. Molecular aspects of human cellular zinc homeostasis: redox control of zinc potentials and zinc signals. Biometals. 2009;22:149–57 [DOI] [PubMed] [Google Scholar]
- 55.Maret W. Zinc and sulfur: a critical biological partnership. Biochemistry. 2004;43:3301–9 [DOI] [PubMed] [Google Scholar]
- 56.Peck EJ, Ray WJ. Metal complexes of phosphoglucomutase in vivo. J Biol Chem. 1971;246:1160–7 [PubMed] [Google Scholar]
- 57.Magneson GR, Puvathingal JM, Ray WJ. The concentration of free Mg2+ and free Zn2+ in equine blood plasma. J Biol Chem. 1987;262:11140–8 [PubMed] [Google Scholar]
- 58.Simons TJB. Intracellular free zinc and zinc buffering in human red blood cells. J Membr Biol. 1991;123:63–71 [DOI] [PubMed] [Google Scholar]
- 59.Bozym RA, Thompson RB, Stoddard AK, Fierke CA. Measuring picomolar intracellular exchangeable zinc in PC-12 cells using a ratiometric fluorescence biosensor. ACS Chem Biol. 2006;1:103–11 [DOI] [PubMed] [Google Scholar]
- 60.Krężel A, Maret W. Zinc buffering capacity of a eukaryotic cell at physiological pH. J Inorg Biol Chem. 2006;11:1049–62 [DOI] [PubMed] [Google Scholar]
- 61.Colvin RA, Bush AI, Volitakis I, Fontaine CP, Thomas D, Kikuchi K, Holmes WR. Insights into Zn2+ homeostasis in neurons from experimental and modeling studies. Am J Physiol Cell Physiol. 2008;294:C726–42 [DOI] [PubMed] [Google Scholar]
- 62.Vinkenborg JL, Nicholson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Methods. 2009;6:737–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin Y, Dittmer PJ, Park JG, Jansen KB, Palmer AE. Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc Natl Acad Sci U S A. 2011;108:7351–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krężel A, Hao Q, Maret W. The zinc/thiolate redox biochemistry of metallothionein and the control of zinc ion fluctuations in cell signaling. Arch Biochem Biophys. 2007;463:188–200 [DOI] [PubMed] [Google Scholar]
- 65.Maret W, Li Y. Coordination dynamics of zinc in proteins. Chem Rev. 2009;109:4682–707 [DOI] [PubMed] [Google Scholar]
- 66.Thomas RC, Coles JA, Deitmer JW. Homeostatic muffling. Nature. 1991;350:564. [DOI] [PubMed] [Google Scholar]
- 67.Colvin RA, Holmes WR, Fontaine CP, Maret W. Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis. Metallomics. 2010;2:306–17 [DOI] [PubMed] [Google Scholar]
- 68.McCranor BJ, Bozym RA, Vitolo MI, Fierke CA, Bambrick L, Polster BM, Fiskum G, Thompson RB. Quantitative imaging of mitochondrial and cytosolic free zinc levels in an in vitro model of ischemia/reperfusion. J Bioenerg Biomembr. 2012;44:253–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park JG, Qin Y, Galati DF, Palmer AE. New sensors for quantitative measurement of mitochondrial Zn(2+). ACS Chem Biol. 2012;7:1636–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haase H, Maret W. The regulatory and signaling functions of zinc ions in human cellular physiology. In: Zalups R, Koropatnick J, editors. Cellular and molecular biology of metals. Boca Raton, FL: Taylor and Francis; . 2009, p. 181–212. [Google Scholar]
- 71.Li Y, Maret W. Transient fluctuations of intracellular zinc ions in cell proliferation. Exp Cell Res. 2009;315:2463–70 [DOI] [PubMed] [Google Scholar]
- 72.Atar D, Backx PH, Appel MM, Gao WD, Marban E. Excitation-transcription coupling mediated by zinc influx through voltage-dependent calcium channels. J Biol Chem. 1995;270:2473–7 [DOI] [PubMed] [Google Scholar]
- 73.Ayaz M, Turan B. Selenium prevents diabetes-induced alterations in [Zn2+]i and metallothionein level of rat heart via restoration of cell redox cycle. Am J Physiol Heart Circ Physiol. 2006;290:H1071–80 [DOI] [PubMed] [Google Scholar]
- 74.Bellomo EA, Meur G, Rutter GA. Glucose regulates free cytosolic Zn2+ concentration, Slc39 (Zip), and metallothionein gene expression in primary pancreatic islet β-cells. J Biol Chem. 2011;286:25778–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cima RR, Dubach JM, Wieland A, Walsh BM, Soybel DI. Intracellular Ca2+ and Zn2+ signals during monochloramine-induced oxidative stress in isolated rat colon crypts. Am J Physiol Gastrointest Liver Physiol. 2004;290:G250–61 [DOI] [PubMed] [Google Scholar]
- 76.Haase H, Hebel S, Engelhardt G, Rink L. Flow cytometric measurement of labile zinc in peripheral blood mononuclear cells. Anal Biochem. 2006;352:222–30 [DOI] [PubMed] [Google Scholar]
- 77.Frederickson CJ, Koh J-Y, Bush AI. The neurobiology of zinc in health and disease. Nat Rev Neurosci. 2005;6:449–62 [DOI] [PubMed] [Google Scholar]
- 78.Paoletti P, Ascher P, Neyton J. High-affinity inhibition of NMDA NR1–NR2A receptors. J Neurosci. 1997;17:5711–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tóth K. Zinc in neurotransmission. Annu Rev Nutr. 2011;31:139–53 [DOI] [PubMed] [Google Scholar]
- 80.Sindreu C, Palmiter RD, Storm DR. Zinc transporter ZnT-3 regulates presynaptic Erk1/2 signaling and hippocampus-dependent memory. Proc Natl Acad Sci U S A. 2011;108:3366–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelleher SL, McCormick NH, Velasquez V, Lopez V. Zinc in specialized secretory tissues: roles in the pancreas, prostate, and mammary gland. Adv Nutr. 2011;2:101–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Danscher G, Stoltenberg M. Zinc-specific autometallographic in vivo selenium methods: tracing of zinc-enriched (ZEN) pathways, and pools of zinc ions in a multitude of other ZEN cells. J Histochem Cytochem. 2005;53:141–53 [DOI] [PubMed] [Google Scholar]
- 83.Kim AM, Bernhardt ML, Kong BY, Ahn RW, Vogt S, Woodruff TK, O’Halloran TV. Zinc sparks are triggered by fertilization and facilitate cell cycle resumption in mammalian eggs. ACS Chem Biol. 2011;6:716–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hogstrand C, Kille P, Nicholson RI, Taylor KM. Zinc transporters and cancer: a potential role for ZIP7 as a hub for tyrosine kinase activation. Trends Mol Med. 2009;15:101–11 [DOI] [PubMed] [Google Scholar]
- 85.Taylor KM, Hiscox S, Nicholson RI, Hogstrand C, Kille P. Protein kinase CK2 triggers cytosolic zinc signaling pathways by phosphorylation of zinc channel ZIP7. Sci Signal. 2012;5:ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaltenberg J, Plum JL, Ober-Blöbaum JL, Hönscheid A, Rink L, Haase H. Zinc signals promote IL-2-dependent proliferation of T-cells. Eur J Immunol. 2010;40:1496–503 [DOI] [PubMed] [Google Scholar]
- 87.Yamasaki S, Sakata-Sogawa K, Hasegawa A, Suzuki T, Kabu K, Sato E, Kurosaki T, Yamashita S, Tokunaga M, Nishida K, et al. Zinc is a novel intracellular second messenger. J Cell Biol. 2007;177:637–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maret W, Jacob C, Vallee BL, Fischer EH. Inhibitory sites in enzymes: zinc removal and reactivation by thionein. Proc Natl Acad Sci U S A. 1999;96:1936–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilson M, Hogstrand C, Maret W. Picomolar concentrations of free zinc(II) ions regulate receptor protein tyrosine phosphatase beta activity. J Biol Chem. 2012;287:9322–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hogstrand C, Verbost PM, Wendelaar Bonga SE. Inhibition of human Ca2+-ATPase by Zn2+. Toxicology. 1999;133:139–45 [DOI] [PubMed] [Google Scholar]
- 91.Williams RJP. Zinc: what is its role in biology? Endeavour. 1984;8:65–70 [DOI] [PubMed] [Google Scholar]
- 92.Maret W, Larsen KS, Vallee BL. Coordination dynamics of biological zinc clusters in metallothioneins and in the DNA-binding domain of the transcription factor Gal4. Proc Natl Acad Sci U S A. 1997;94:2233–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maret W. Redox biochemistry of mammalian metallothioneins. J Biol Inorg Chem. 2011;16:1079–86 [DOI] [PubMed] [Google Scholar]
- 94.Rink L. editor. Zinc in human health. Amsterdam, the Netherlands: IOS Press; 2011 [Google Scholar]
- 95.Maret W. Metals on the move: zinc ions in cellular regulation and in the coordination dynamics of zinc proteins. Biometals. 2011;24:411–8 [DOI] [PubMed] [Google Scholar]
- 96.Maret W. Metallothionein redox biology in the cytoprotective and cytotoxic functions of zinc. Exp Gerontol. 2008;43:363–9 [DOI] [PubMed] [Google Scholar]
- 97.Maret W, Sandstead HH. Zinc requirements and the risks and benefits of zinc supplementation. J Trace Elem Med Biol. 2006;20:3–18 [DOI] [PubMed] [Google Scholar]