Abstract
Unfolded protein response (UPR) is a cellular adaptive response which functions to reduce stress caused by misfolded proteins in the endoplasmic reticulum (ER). We and others have previously shown that infection with hepatitis C virus (HCV) or expression of the viral proteins can trigger the UPR. HCV is a single-stranded positive-sense RNA virus causing chronic diseases in humans. Its genome encodes two envelope proteins E1 and E2 that mature in the ER to form non-covalently bound native complex and disulphide-bonded aggregates. Apart from the ER targeting proteins, cytosolic forms have been documented. We have previously shown that the ER-targeting E1 and E2 are capable of eliciting the UPR whereas others have shown that the cytosolic-targeting E2 can bind to the ER stress kinase PERK to dampen the UPR. In this report, we further show that the other envelope protein E1, in its cytosolic form, can also bind PERK and dampen the UPR. Using GST-pulldown assay, we show that E1 binds to the cytoplasmic domain of PERK, suggesting interaction of E1 and PERK takes place in the cytoplasm. Using reporter gene assay and Western blotting, we show that cytosolic E1 can repress UPR-induced BiP and CHOP promoter activity and reduce UPR-induced CHOP expression level. Altogether these results suggest opposing functions of ER- and cytosolic forms of HCV envelope proteins depending on their subcellular localization.
Keywords: hepatitis C virus, envelope protein, unfolded protein response, PERK.
INTRODUCTION
The unfolded protein response (UPR) is a cellular homeostatic response to unfolded or malfolded proteins. The UPR is under the negative regulation of the immunoglobulin heavy-chain binding protein (BiP) and its induction is mediated by three proximal UPR sensors: the activating transcription factor 6 (ATF6), inositol-requiring enzyme 1 (IRE1) and RNA-activated protein kinase (PKR)-like ER-resident kinase (PERK) [1,2]. Activated ATF6 is a transcription factor which transactivates genes harboring an ER-stress element (ERSE) in their promoters [2]. IRE1 mediates productive translation of X-box binding protein 1 into an active transcription factor to transactivate UPR genes with an ERSE [3]. PERK is activated by oligomerization and autophosphorylation [1]. Upon activation, PERK phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α), resulting in global inhibition of protein synthesis but at the same time enhances translation of the transcription factor ATF4 which transactivates specific UPR genes via the ATF4 element in their promoters [4,5]. BiP and CCAAT/ enhancer-binding protein-homologous proteins (CHOP) are two downstream effectors of the UPR. Both contain the ERSE and the ATF element in their promoters, thus are potentially responsive to all three signaling pathways although one or two of the regulatory pathways may predominate depending on the cell types and stimuli [6,7].
Hepatitis C virus (HCV) is a single-stranded positive-sense RNA virus. Its genome encodes two envelope proteins, E1 and E2, that mature in the endoplasmic reticulum (ER) to form a non-covalently bound native complex and disulphide-bonded protein aggregates [8]. Apart from the ER-targeting proteins, cytosolic forms have been documented. The cytosolic E2 presumably arises via retrotranslocation from the ER or direct translocation from the ribosomes [9]. In vivo evidence suggests cytosolic existence of E1 from the detection of a deglycosylated-deamidated T-epitope, which indicates retrograde transport of E1 from the ER to the cytoplasm for proteasome degradation [10]. Intriguingly, the E2 protein can assume opposing functions depending on the subcellular localization. We have previously shown that the ER-targeting E2 is capable of eliciting the UPR [11,12]. On the other hand, cytosolic E2 can bind to the cytoplasmic kinase domain of PERK thus inhibiting its kinase activity [13]. Similarly, we have also shown that the ER-targeting E1 is capable of eliciting the UPR [11,12]. In this study, we therefore sought to investigate whether the cytosolic E1 could exert opposing function to that of ER-targeting E1 by binding PERK and dampening the UPR.
MATERIALS AND METHODS
Plasmids. The plasmids HisMax-E1, HisMax-E2 and HisMax-NS4B were generated using HCV genotype 1b as a template [14]. PCR fragments were subcloned into pcDNA4.1HisMax (Invitrogen) to allow T7 promoter-driven in vitro transcription and translation. The plasmids green fluorescent protein (gfp)-E1 and gfp-E2 were constructed by subcloning the E1 and E2 genes without signal peptide sequences into pEGFP-C1 (Clontech) as fusion proteins with N-terminally tagged GFP and have been previously described [11]. The BiP-luciferase reporter driven by –304bp to +7bp of the BiP promoter was obtained from Kazutoshi Mori [15]. The CHOP-luciferase reporter driven by 954bp of the CHOP promoter was obtained from Pierre Fafournoux [7].
Purification of proteins. The plasmids GST-PERK (WT) and GST-PERK (KR) were gifts of David Ron [4]. GST-fusion proteins were expressed in E. coli BL21 (RIL) and affinity purified using glutathione Sepharose 4B (Amersham).
GST-pulldown assay. Radiolabelled polypeptides generated using the TNT-coupled transcription and translation kit (Promega) and 35S[Met/Cys] (MP Biomedicals) were incubated with Sepharose-bound GST-fusion proteins in 0.1% Tween/PBS overnight at 4 oC. After resolving by SDS-PAGE, radiolabelled bands were visualized by a Typhoon 8600 phosphorimager (Amersham).
Cell culture. Huh-7 and HeLa cells were maintained in Dulbecco Modified Eagle Medium (Sigma) supplemented with 10% fetal calf serum (Sigma), 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM glutamate (Sigma). For Huh-7 cells, medium was supplemented with 1x non-essential amino acids (Sigma).
Transfection. Plasmid transfection was performed using Trans-IT LT1 (Mirus) for Huh-7 cells and PolyFect (Qiagen) for HeLa cells according to the manufacturers’ instructions.
Reporter assays. Luciferase and β-galactosidase activities were measured as previously described [11].
Western blotting. Protein lysates harvested in 1x SDS-PAGE sample buffer were resolved by SDS-PAGE, transferred to PVDF membrane and immunoblotted with primary antibodies (GADD153/CHOP (Santa Cruz), β-tubulin (Sigma)) followed by horseradish peroxidase-conjugated secondary antibodies. Immunocomplexes were detected with the ECL Plus Kit (Amersham).
Statistics. Statistical analyses were performed using ANOVA. A P value of less than 0.05 was considered statistically significant.
RESULTS
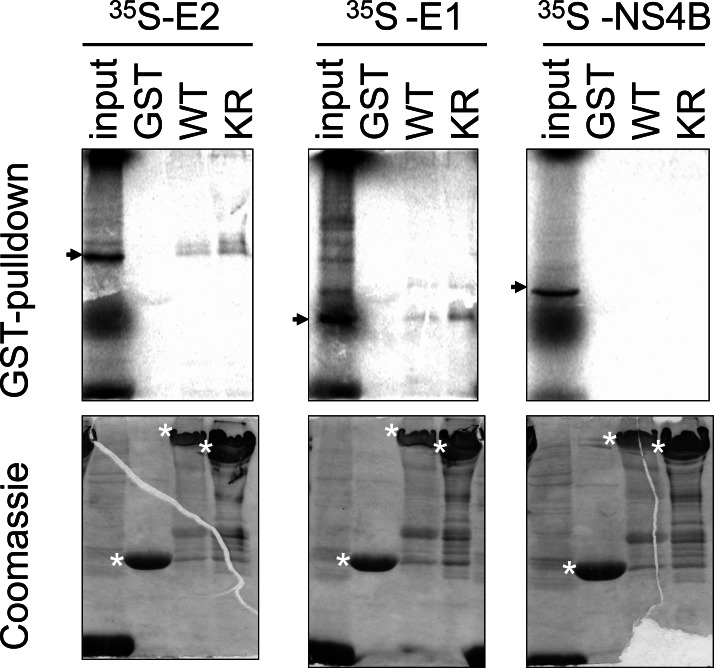
HCV E1 binds PERK. One of the envelope proteins of HCV, E2, was previously shown to bind the UPR sensor PERK [13], we therefore, investigated whether the other HCV envelope protein, E1, could also interact with PERK. Using a GST-pulldown assay employing a GST-PERK fusion protein and 35S radiolabelled in vitro translated polypeptides, binding was detected between the GST-PERK and E1 proteins (Fig. 1). Specificity of this assay was illustrated by the binding between GST-PERK and the positive control E2 and the absence of binding between GST-PERK and another HCV protein the non-structural 4B (NS4B). Binding was specific for the PERK domain, but not for the GST domain within the GST-PERK fusion protein as neither E1 nor E2 protein bound to the GST protein alone. PERK is a type I transmembrane protein with a lumenal regulatory domain and a cytoplasmic kinase domain [4]. As the GST-PERK fusion protein contains only the cytoplasmic kinase domain of PERK, the results indicate that E1 binds to the cytosolic domain of PERK. E1 and E2 also bound to PERK with a K-R mutation at amino acid residue 618 (Fig. 1). This mutation abolishes the ability of PERK to undergo autophosphorylation [4], therefore, autophosphorylation of PERK is not a pre-requisite for its interactions with either E1 or E2.
Fig. (1).
PERK binds E1 and E2. (upper panel) Phosphorimages of GST-pulldown reactions detecting binding of 35S-labelled in vitro translated E2, E1 and NS4B polypeptides with GST, GST fusion proteins with wild type PERK (WT) or K618R mutant PERK (KR). One-tenth of in vitro translated reactions were run as input controls. Arrows denote polypeptides of the correct sizes. (lower panel) Coomassie Blue staining of the same gels showing the corresponding amounts of proteins used in the pulldown reactions. Asterisks denote protein bands of the correct sizes.
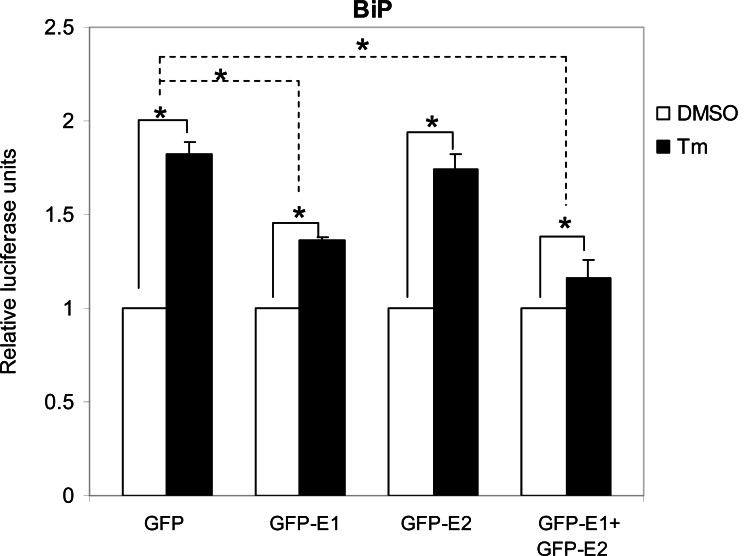
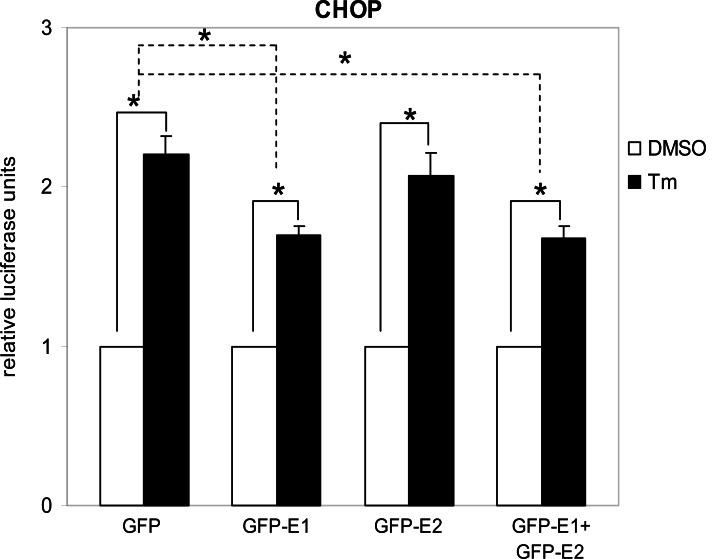
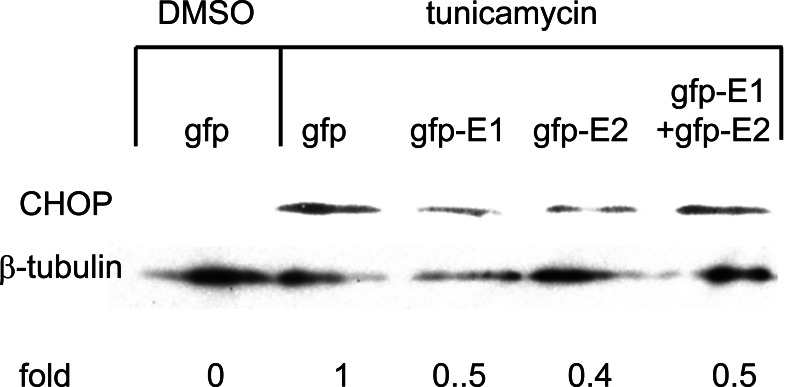
Cytosolic E1 represses UPR. We then sought to investigate the functional importance of this interaction. As PERK is a proximal sensor of the UPR, interaction between E1 and the cytoplasmic kinase domain of PERK may imply a role of the cytosolic-targeting E1 in the modulation of the UPR. During the maturation of E1, the protein is targeted to the ER by an N-terminal signal peptide [16]. Therefore, to target the E1 to the cytoplasm we removed the signal peptide from the E1 protein and fused the GFP to its N-terminus to create a fusion protein GFP-E1. Similarly, a cytosolic-targeting E2, GFP-E2, was generated in the same way. To study the effects of the cytosolic-targeting proteins on the UPR we co-transfected gfp-E1 or gfp-E2 with the respective reporter plasmids containing the BiP or CHOP promoter, into Huh-7 cells followed by treatment of cells with the UPR inducer, tunicamycin. Tunicamycin stimulation resulted in significant increases in the BiP and CHOP promoter activities in cells transfected with the gfp control plasmid, gfp-E1 and/or gfp-E2 plasmids (Figs. 2, 3). However, expression of GFP-E1 caused significant reduction in the levels of tunicamycin-induced BiP and CHOP activation compared with that of the GFP controls, suggesting an inhibitory effect of cytosolic E1 on the UPR. As E1 forms complex with E2, we then studied whether the effect of cytosolic E1 would be different in the presence of cytosolic E2. Co-expression of GFP-E1 and GFP-E2 in trans also resulted in significant reduction in the levels of tunicamycin-induced BiP and CHOP activation, suggesting that the ability of E1 to repress the UPR is not affected in the presence of its natural interacting partner, E2. Although GFP-E2 on its own did not cause significant reduction in the levels of tunicamycin-induced BiP and CHOP activation, these two promoter activities were consistently lower than that of the GFP controls. Importantly, when co-expressed with GFP-E1, GFP-E2 exhibited a synergistic inhibitory effect on tunicamycin-induced BiP activation, suggesting a role of the GFP-E2 in modulating the UPR (Fig. 2). Indeed, Western blotting revealed that expression of GFP-E1, GFP-E2 and co-expression of the two proteins could all reduce the levels of CHOP expression induced by tunicamycin treatment in HeLa cells (Fig. 4).
Fig. (2).
E1 represses UPR activation of BiP. Huh-7 cells were co-transfected with the BiP reporter gene plasmid and the respective plasmids as indicated together with the internal control plasmid encoding β-galactosidase for 32 h and then treated with 1 µg/ml of tunicamycin (Tm) or the dimethylsulphoxide (DMSO) solvent control for 16 h. The BiP promoter luciferase activity was normalized against β-galactosidase activity and expressed relative to the solvent controls, which are set as 1. The values obtained represent the mean ± SEM of three independent experiments performed in triplicate. *Significance of the difference (P<0.05).
Fig. (3).
E1 represses UPR activation of CHOP. Huh-7 cells were co-transfected with the CHOP reporter gene plasmid and the respective plasmids as indicated together with the internal control plasmid encoding β-galactosidase for 32 h and then treated with 1 µg/ml of tunicamycin (Tm) or the dimethylsulphoxide (DMSO) solvent control for 16 h. The CHOP promoter luciferase activity was normalized against β-galactosidase activity and expressed relative to the solvent controls, which are set as 1. The values obtained represent the mean ± SEM of three independent experiments performed in triplicate. *Significance of the difference (P<0.05).
Fig. (4).
Cytosolic E1 reduces CHOP activation. HeLa cells were transfected with plasmids encoding the envelope proteins as indicated with or without treatment with the solvent control DMSO or 2µg/ml tunicamycin (Tm) for 16hr. Protein lysates resolved on SDS-PAGE were immunoblotted with antibodies specific for CHOP and β-tubulin. The intensities of the bands were quantified with ImageQuant 5.0. The levels of CHOP were normalised against β-tubulin and expressed as fold change relative to the gfp control which was set as 1.
DISCUSSION
The UPR has emerged to play an important role in the life cycle of HCV as supported by in vitro and in vivo evidence [17,18]. Infection of Huh-7 cells with HCV results in the induction of the UPR [18]. Examination of HCV-infected liver samples for UPR markers confirms the existence of ER stress [17]. HCV encodes two envelope proteins, E1 and E2. The role of these envelope proteins in the regulation of the UPR is emerging. We have previously shown that targeting of the E1 and E2 proteins to the ER induces the UPR [11,12]. Apart from the ER, E1 and E2 are also targeted to the cytoplasm where they may assume different functions [9,10,12,13]. We and others have shown that cytosolic-targeting E2 can bind to PERK [13 and this study]. In this study, we further show that the E1 protein can also bind PERK and repress the UPR, suggesting that the envelope proteins can assume different and opposing functions depending on their subcellular localization. Moreover, the ability of E1 to repress the UPR is not affected in the presence of its natural interacting partner, E2. It is therefore tempting to speculate that the ability of the cytosolic envelope proteins to damp down the UPR may be important in regulating host responses to provide a favorable environment for virus replication and survival.
The UPR is a tripartite response originating from the PERK, ATF6 and IRE1 proximal sensors [1,2]. Normally, the PERK pathway is responsible for CHOP activation and the ATF6 and IRE1 pathways responsible for BiP activation but functional cross-talk exists as both BiP and CHOP promoters harbor similar responsive elements [6,7]. Using Perk+/+ and Perk-/- mouse embryonic fibroblasts, we and others have shown that CHOP induction by tunicamycin or the ER-targeting E1 and E2 proteins are highly dependent on PEKR whereas BiP induction is only partially dependent on PERK [5,11 and our data not shown]. Thus, inhibition of PERK should lead to a complete repression of CHOP activation and a partial repression of BiP activation. In this study, expression of cytosolic E1 resulted in a partial repression of BiP activation, as expected. However, expression of cytosolic E1 did not result in a complete repression of CHOP activation. This could be because of the use of a very potent UPR inducer, tunicamycin. Alternatively, CHOP activation in a different cell type, Huh-7, is dependent on pathways in addition to PERK signaling.
Cytosolic E1 and E2 synergistically inhibited BiP activation. As a synergistic inhibition of CHOP activation is not observed, it is unlikely that the synergistic inhibition of BiP activation by E1E2 is because the protein complex has a higher affinity for PERK. Therefore, the synergistic repression of BiP activation is probably not by means of PERK inhibition alone but via inhibition of the ATF6 and/or IRE1 signaling or a downstream effector of the PERK pathway, hinting at another novel function of the cytosolic envelope proteins.
ACKNOWLEDGEMENTS
We thank Makoto Hijikata, Kazutoshi Mori, Pierre Fafournoux, David Ron and Charlie Rice for plasmids and cell line. This work was in part supported by a Medical Research Council Career Establishment Grant G0000092 awarded to SWC.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Bertolotti A, Zhang YH, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–32. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 2.Shen JS, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 3.Calfon M, Zeng HQ, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 4.Harding HP, Zhan YH, Ro D. Protein translation and folding are coupled by an endoplasmic- reticulum-resident kinase. Nature. 1999;397:271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 5.Harding HP, Novoa I, Zhang YH, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 6.Luo SZ, Baumeister P, Yang SJ, Abcouwer SF, Lee AS. Induction of Grp78/BiP by translational block - Activation of the Grp78 promoter by ATF4 through an upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J Biol Chem. 2003;278:37375–85. doi: 10.1074/jbc.M303619200. [DOI] [PubMed] [Google Scholar]
- 7.Bruhat A, Jousse C, Carraro V, Reimold AM, Ferrara M, Fafournoux P. Amino acids control mammalian gene transcription.Activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol Cell Biol. 2000;20:7192–204. doi: 10.1128/mcb.20.19.7192-7204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubuisson J, Rice CM. Hepatitis C virus glycoprotein folding.Disulfide bond formation and association with calnexin. J Virol. 1996;70:778–86. doi: 10.1128/jvi.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavio N, Taylor DR, Lai MMC. Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR. J Virol. 2002;76:1265–72. doi: 10.1128/JVI.76.3.1265-1272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selby M, Erickson A, Dong C, et al. Hepatitis C virus envelope glycoprotein E1 originates in the endoplasmic reticulum and requires cytoplasmic processing for presentation by class I MHC molecules. J Immunol. 1999;162:669–76. [PubMed] [Google Scholar]
- 11.Chan SW, Egan PA. Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J. 2005;19:1510–2. doi: 10.1096/fj.04-3455fje. [DOI] [PubMed] [Google Scholar]
- 12.Chan SW, Egan PA. Effects of hepatitis C virus envelope glycoprotein unfolded protein response activation on translation and transcription. Arch Virol. 2009;154:1631–40. doi: 10.1007/s00705-009-0495-5. [DOI] [PubMed] [Google Scholar]
- 13.Pavio N, Romano PR, Graczyk TM, Feinstone SM, Taylor DR. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK. J Virol. 2003;77:3578–85. doi: 10.1128/JVI.77.6.3578-3585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marusawa H, Hijikata M, Chiba T, Shimotohno K. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-kappa B activation. J Virol. 1999;73:4713–20. doi: 10.1128/jvi.73.6.4713-4720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida H, Okada T, Haze K, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–67. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimotohno K, Tanji Y, Hirowatari Y, Komoda Y, Kato N, Hijikata M. Processing of the hepatitis C virus precursor protein. J Hepat. 1995;22:87–92. [PubMed] [Google Scholar]
- 17.Asselah T, Biechel I, Mansouri A, et al. In vivo hepatic endoplasmic reticulum stress in patients with chronic hepatitis C. J Pathol. 2010;221:264–74. doi: 10.1002/path.2703. [DOI] [PubMed] [Google Scholar]
- 18.Sekine-Osajima Y, Sakamoto N, Mishima K, et al. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology. 2008;371:71–85. doi: 10.1016/j.virol.2007.09.019. [DOI] [PubMed] [Google Scholar]