Abstract
The genetics of sex determination remains mysterious in many organisms including some that are otherwise well-studied. Here we report the discovery and analysis of the mating-type locus of the model organism Dictyostelium discoideum. Three forms of a single genetic locus specifies this species’ three mating-types: two versions of the locus are entirely different in sequence, and the third resembles a composite of the other two. Single, unrelated genes are sufficient to determine two of the mating-types, while homologues of both these genes are required in the composite type. The key genes encode polypeptides that possess no recognisable similarity to established protein families. Sex determination in the social amoebae thus appears to use regulators unrelated to any currently known.
Most eukaryotes are sexual, but little is known in molecular detail about sex across most branches of the eukaryotic tree. One aspect, the genetic basis of sex determination, is well understood in several animal, fungal and plant lineages (1-5), but across the protozoan kingdoms we know little, and nothing in comparable detail. The social amoebae are members of the Amoebozoa, and have an unusual sexual cycle that leads to the formation of dormant, walled macrocysts (6) (Fig. 1 A and B). To produce a macrocyst, a pair of haploid amoebae of different sexes fuse (7) to form a diploid zygote, which then attracts surrounding haploid cells (8). These help to lay down external layers of cellulose around the developing mass of cells before being cannibalized by the zygote (9). After a period of dormancy the cyst germinates, releasing haploid progeny that arise most likely after meiosis and multiple mitoses (10). Population genetics of wild isolates indicate that mating and recombination are probably frequent in the wild (11).
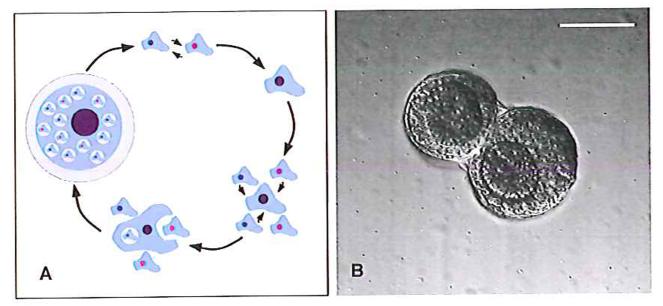
Figure 1. The sexual cycle of Dictyostelium discoideum.
A: Amoebae of different sexes (top) first fuse, and several hours later their two nuclei also fuse, resulting in a diploid zygote (top right). This then secretes cAMP to attract surrounding haploid cells (bottom right), which are ingested by the zygote (bottom); eventually a dormant macrocyst is formed, retaining part-digested haploids within vacuoles (left). Ultimately the macrocyst germinates, releasing tens or hundreds of progeny, all descending from the zygote. B: Early macrocysts (precysts) of strain AC4, formed in shaken suspension approximately 24 hours after removal of food bacteria. Here, two developing macrocysts are contained within the same outer wall. In each cyst inner walls envelop hundreds of haploid amoebae and the zygote, which is visible here as the darker cell mass at the centre of each cyst, and which slowly eats its way out to the limiting wall. Within the zygotes the structures of ingested amoebae are still clear. Scale bar = 50 μm.
The most-studied species of social amoeba, Dictyostelium discoideum, is notable for having three sexes (hereafter called mating-types I, II, and III; supporting online text, S1), as well as uncommon self-fertile homothallic strains (12-14). Each of the three sexes can pair with each of the other two, but not with itself, giving three possible classes of zygote: type-I/type-II, type-I/type-III, and type-II/type-III. Although several genes are known to be involved during the sexual cycle (15), the determinant of mating-type has proved elusive. Genetic analysis suggested that mating-type is stable, and determined by a single locus with two or more alleles (10, 14, 16). We argued that it might be possible to identify this postulated locus by searching for genes which are present in any member of one mating-type but absent (or highly diverged) in any member of another. For this purpose we performed comparative genomic hybridizations using DNA microarrays composed of probes for around 8500 of the 10500 predicted genes in the sequenced type-I D. discoideum genome (17).
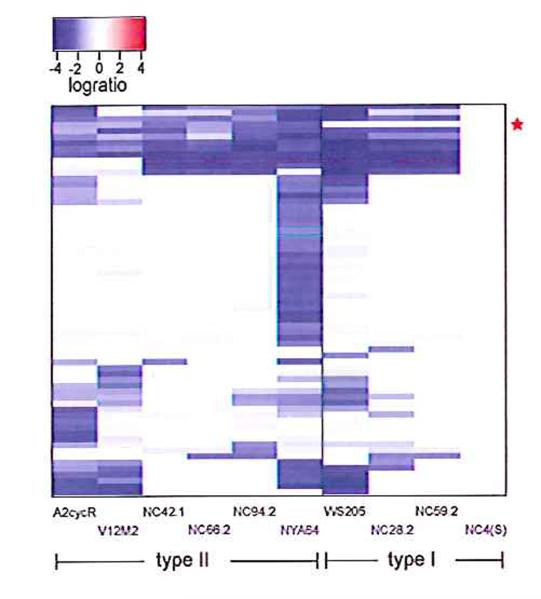
We analysed ten strains derived from independent wild isolates (table S1) using this microarray (18), and found a single candidate gene that follows the pattern expected of a sex-determining sequence: it is present in all type-I genomes but absent from all the type-II strains (Fig. 2, fig. S1, table S2). This open reading frame (ORF) is situated on chromosome 5 (see supporting online text S2), and is very short, encoding a 107 amino-acid polypeptide that contains no significant homology to previously studied proteins. No motifs or potential domains suggest a function, but it is relatively highly charged and so is most likely a soluble intracellular protein. We confirmed that the ORF is present in all the other type-I strains used in the microarray study (100% identical in amino acid sequence in all cases; table S3), and sequenced the entire locus from another type-I strain, WS205, to confirm that no other obvious coding sequences are present.
Figure 2. Identification of a candidate mating-type locus.
Our search assumed that the mating-type locus would differ substantially in sequence between mating-types, so using a microarray designed from type-I sequence we sought genes present in all examples of this mating-type and absent (or highly diverged) in all examples of type-II. Out of more than 8500 genes covered on the array only one, matA, (alongside the red star) behaved in this way when ten independent wild isolates were compared with our laboratory strain Ax2 in DNA to DNA comparisons. The heatmap shows only the set of genes giving a logratio below -2 in at least one of the wild strains compared to Ax2 (full data are presented in table S2). Each row in the plot represents a gene; each column a strain. Blocks are coloured according to log(2)ratio, from blue (negative – decreased copy number or sequence divergence in the test strain) through white (zero – no difference) to red (positive - increased copy number in the test strain). Several other sequences apart from matA are absent or diverged in different isolates, but none of these correlates with mating-type; it should be noted that NC4 is the ultimate parent of Ax2, accounting for the similarity between them.
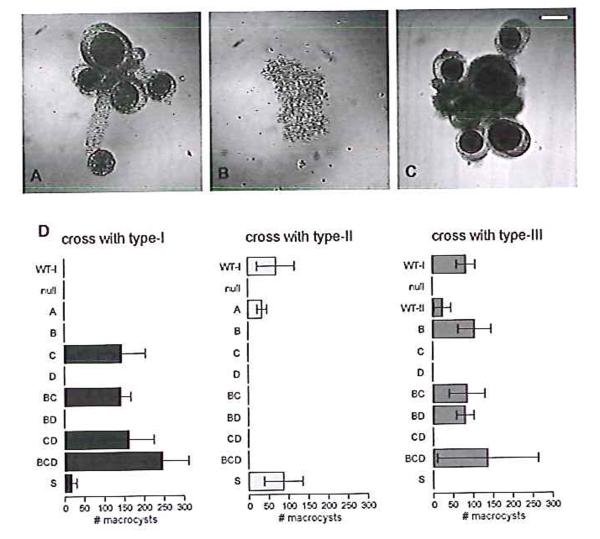
To test whether the gene is involved in the sexual cycle, we deleted it from our standard type-I strain (19). The resulting mutant, in contrast to its parent, is unable to form macrocysts when paired with a type-II strain (Fig. 3). When the coding sequence is reintroduced under the control of a constitutive promoter, mating competence is restored (Fig. 3D, table S4) showing that this gene, which we call matA, is necessary for correct type-I mating behaviour, and supporting the proposition that it is involved in sex determination.
Figure 3. Re-engineering the mating behaviour of a type-I strain.
The type-I strain Ax2, which can mate with type-II strain V12M2 (A), was first modified by the deletion of matA. The resultant strain is unable to mate with V12M2 (B) or any other strain. The introduction of matC with its own regulatory sequences into this null mutant gives a strain that is able to mate with its ultimate parent Ax2 (C). Structures were imaged seven days after mixing using differential interference contrast microscopy. D: Macrocysts formed in various crosses were counted eight days after mixture of strains. Eleven strains were crossed with a type-I strain, Ax2 (left), and with a type-II strain, V12M2 (middle): ‘WT-I’ is the parental Ax2 strain, ‘null’ is the matA deletion strain in this background. The next nine strains are the null plus one or more mat gene controlled by a constitutive promoter; each is designated by the gene’s letter. These strains, apart from the matA-expressing strain, and also the type-II strain V12M2 (‘WT-II’), were also crossed with the type-III strain WS2162 (right). The mean number of macrocysts plus and minus the standard error from three independent crosses are plotted.
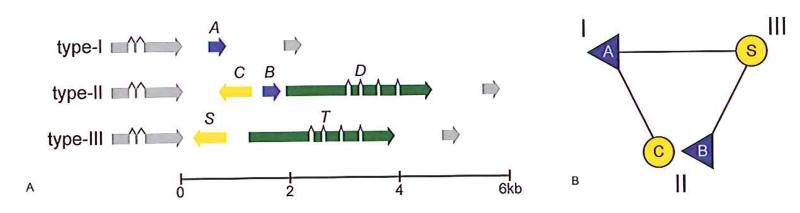
The two genes flanking matA in the reference type-I sequence are present in all strains, and do not vary in amino-acid sequence between mating-types (fig. S2). We could therefore amplify the entire type-II version of the locus by polymerase chain reaction from the NC66.2 isolate. The type-II version is larger and contains three genes (Fig. 4A), one of which, matB, is homologous to matA, but considerably diverged at approximately 60% identity in amino-acid sequence (fig. S3, and see supporting online text S1). A second small gene, matC, encodes a 208 amino-acid hydrophilic polypeptide that again has no similarity to known proteins (fig. S4). The third gene, matD, is larger, spliced, and encodes a 799 amino-acid preprotein that contains a predicted signal peptide and potential GPI-anchor attachment site (fig. S5). No homology is shared between these three genes. All of the other type-II strains tested contain the same three genes with >98% sequence identity (table S4), and the complete type-II locus from the Japanese type-II isolate NYA64 was sequenced, again confirming that no additional coding sequences are present.
Figure 4. The structure and logic of the D. discoideum mat locus.
A. Type-I strains are characterised by a single protein-coding gene, matA (coloured blue, marked above with ‘A’; dictyBase ID DDB_G0289165), which is homologous to matB (also coloured blue, marked ‘B’), one of the three genes present in the type-II version of the locus. The two other genes making up the type-II locus, matC (yellow, marked ‘C’) and matD (green, marked ‘D’), are homologous to the two genes that occupy the type-III version, matS and matT (coloured yellow and green according to homology, marked S and T; gene nomenclature is treated further in the supplementary discussion). The locus lies on chromosome 5 between the genes DDB_G0289171 and DDB_G0289163, which do not vary according to mating-type and are shown here coloured grey. B. Mating compatibility requires the presence of a matA-class gene (blue triangles) and a matS-class gene(yellow circles) in the two gametes. Type-II cells contain a gene of each class, allowing mating with both types -I and -III. The nature of the interactions between genes remains unknown, as does the molecular explanation of how the matB and matC pair are incompatible.
To prove that this locus is responsible for sex determination, we attempted to switch the mating-type of our type-I laboratory strain. The matA gene was first deleted and the resulting mutant then transformed with a construct bearing the entire type-II version of the locus. The resulting strain now behaves as type-II (see also supporting online text S3), forming macrocysts when paired with type-I cells but not with type-II (table S5). This reversal of mating orientation demonstrates that this locus is sufficient to specify mating-types -I and -II, and that we have identified the Dictyostelium mating-type locus.
Next we characterised the locus in the type-III strain WS2162. This version contains two genes homologous to matC and matD but divergent in sequence, and which we name matS and matT respectively (Fig. 4A, fig. S4, fig. S5). No sequence related to matA or matB is present. The same pattern is followed in another type-III isolate, WS112B (table S6). Each mating-type thus possesses a different version of the mating-type locus, and there is no overlap between the type-I and type-III versions. Strikingly, the type-II version resembles a composite of the other two, containing homologues of the genes from the type-I and type-III versions. Two homothallic isolates resemble type-III, containing genes related to matS and matT but no matA/matB homologue (table S6, see also supporting online text S4).
Since mating-type -I is determined by a single gene, we next asked whether any of the genes at the type-II and type-III versions of the locus could act as master regulators of sex determination, with others perhaps having subsidiary roles. To do this, we expressed individual genes, either alone or in combination, in the matA null strain, which is unable to mate with any strain. The type-II locus was analysed in most detail. Expressing matB alone from a constitutive promoter is sufficient to produce a strain to mate with a type-III strain, but not with types-I and -II (Fig. 3D, table S5). Thus the homologous matA and matB genes appear to have similar but not identical functions: the former specifies mating competence towards type-II and type-III while the latter is only effective towards type-III, and matB only partially accounts for the type-II phenotype.
Expressing matC alone in the same null background allows mating with type-I cells but not type-III nor type-II (Fig. 3D, table S5). We obtained the same result when we replaced the knocked-out version of the matA locus with a truncation of the NC66.2 mat locus containing only matC and its native regulatory sequences; again this strain, which now contains only matC in place at the mating locus on chromosome 5, mates efficiently with type-I cells but not at all with type-III nor type-II (Fig. 3C, table S5). When the knocked-out version is replaced with the genomic region bearing both matB and matC, under the control of their own promoters, the resulting strain can form macrocysts with both types-I and -III (table S5). Similarly, overexpression of both matB and matC in the same strain recapitulates normal type-II behaviour as adjudged by this assay (Fig. 3D, table S5). Importantly, these strains are not self-fertile (table S5). Therefore two unrelated mat genes together specify the type-II mating-type.
The final gene contained in the type-II version of the locus, matD, is not required for mating-type determination. When expressed alone in the mating null cells from a constitutive promoter it does not allow mating with any of the mating-types; and it does not affect the qualitative behaviour of matB or matC when expressed in combination with them, though there is some evidence of quantitative effects. Yields of macrocysts are higher when matD is present in some crosses (table S5); this is consistent with a possible role in promoting gamete fusion in these cases, an idea supported by distant homology to a known protein involved in this process (see below).
Turning to the type-III version, the presence of a matC homologue suggested that this gene, matS, might specify this third mating-type. As expected, expressing matS in the matA null background gives a strain which mates with type-I and type-II, but not with type-III (Fig. 3D, table S5). Furthermore, crosses between strains bearing just the master control genes give the expected results: null cells expressing just matS can mate with cells expressing just matB but not with cells expressing just matC (table S5). Finally, cells separately expressing matB and matC do not mate with each other (table S5). We have not analysed matT separately, but like matD it does not appear to be necessary for sex determination.
These results suggest a simple underlying picture: type-I and type-III mating behaviour can be specified by a single gene in each case: matA specifies type-I and matS specifies type-III. Type-II is a composite in which homologues of matA and matS (matB and matC respectively) allow it to mate with the other two mating-types but, for reasons that remain unclear, not with itself (Fig. 4B).
The molecular function of these genes remains to be addressed. No clear homologues are present in species outside of the dictyostelids, but Dictyostelium purpureum possesses two adjacent genes homologous to matS and matT (fig. S4, fig. S5). A clear matD/matT homologue is also evident in the more distantly related dictyostelid Acytostelium subglobusum (fig. S5). Adjacent to this gene is a small ORF very weakly similar to matC/matS, a possible matS orthologue in this species (fig. S6). MatD is distantly related to the Hap2-GCS1 family of gamete fusion proteins (20), though another Dictyostelium discoideum gene, hapA, encodes a protein much more closely related to the canonical Hap2 group, and is enriched in cells competent for mating (21).
The organisation of the Dictyostelium mating-type locus does not closely resemble previously studied sex-determining regions (supporting online text S5), although like them the mat locus must ultimately control a transcriptional cascade (21). Whether the key genes directly regulate transcription or through downstream targets remains to be determined. One further role could be in controlling mitochondrial inheritance, which is uniparental in related Amoebozoans (22, 23).
We have identified the sex-determining locus from Dictyostelium discoideum, a model organism and the best-studied member of the Amoebozoa. The master regulators of sex-determination at this locus encode homologues of two small, apparently soluble proteins, which are unrelated to previously studied proteins. The genetic logic of the system allows one to speculate that one sex, mating-type -II, may have arisen after a fusion of the versions of the locus of the other two sexes. Understanding the mating-type locus may help to overcome long-standing difficulties in making use of sexual genetics in Dictyostelium (24).
One sentence summary: Four genes contained in three versions of a single genetic locus determine the three sexes of the model organism Dictyostelium discoideum.
Supplementary Material
Acknowledgments
This work was supported by Wellcome Trust grant number 06724 and core funding from the MRC. We would like to thank the Dicty Stock Center (http://www.dictybase.org/StockCenter/StockCenter.html), David Francis, Hiromitsu Hagiwara, and Pauline Schaap for providing strains; Theresa Feltwell, Nefeli Nikolaidou-Katsaridou, and Kay Jagels for technical assistance; Maria Fookes, Karen Brooks, Catherine Pears, Nick Barry and the members and associate members of the Dictyostelium group at the LMB for valuable advice and discussions. A.I. is now at Fios Genomics, ETTC, King’s Buildings, Edinburgh, EH9 3JL, UK. We are grateful to the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) and co-workers, and to the Actyostelium Genome Consortium for making the Dictyostelium purpureum and Acytostelium subglobosum genome data available before publication. Microarray data are available under the accession E-TABM-394 and the array design under A-SGRP-3 at http://www.ebi.ac.uk/arrayexpress/ and DNA sequences have been deposited in EMBL-Bank under the accessions FN543120 – FN543124, FN994780, FR666792, and FR666793.
Footnotes
Supporting Online Material www.sciencemag.org Materials and Methods Supporting online text S1, S2, S3, S4, S5 Tables S1, S2, S3, S4, S5, S6 Figs. S1, S2, S3, S4, S5, S6
References
- 1.Marín I, Baker BS. Science. 1998;281:1990–1994. doi: 10.1126/science.281.5385.1990. [DOI] [PubMed] [Google Scholar]
- 2.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, et al. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 3.Lee SC, Ni M, Li W, Shertz C, Heitman J. Microbiol. Mol. Biol. Rev. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanurdzic M, Banks JA. Plant Cell. 2004;16:S61–71. doi: 10.1105/tpc.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, et al. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 6.Blaskovics JC, Raper KB. Biol. Bull. 1957;113:58–88. [Google Scholar]
- 7.Saga Y, Okada H, Yanagisawa K. J. Cell Sci. 1983;60:157–168. doi: 10.1242/jcs.60.1.157. [DOI] [PubMed] [Google Scholar]
- 8.O’Day D. Can. J. Microbiol. 1979;25:1416–1426. doi: 10.1139/m79-221. [DOI] [PubMed] [Google Scholar]
- 9.Filosa MF, Dengler RE. Dev. Biol. 1972;29:1–16. doi: 10.1016/0012-1606(72)90038-3. [DOI] [PubMed] [Google Scholar]
- 10.Wallace MA, Raper KB. J. Gen. Microbiol. 1979;113:327–337. doi: 10.1099/00221287-113-2-327. [DOI] [PubMed] [Google Scholar]
- 11.Flowers JM, Li SI, Stathos A, Saxer G, Ostrowski EA, et al. PLoS Genet. 2010;6:e1001013. doi: 10.1371/journal.pgen.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurst LD. Proc. R. Soc. London Ser. B. 1996;263:415–422. [Google Scholar]
- 13.Erdos GW, Raper KB, Vogen LK. Proc. Natl. Acad. Sci. U.S.A. 1973;70:1828–1830. doi: 10.1073/pnas.70.6.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robson GE, Williams KL. Curr. Genet. 1980;1:229–232. doi: 10.1007/BF00390948. [DOI] [PubMed] [Google Scholar]
- 15.Urushihara H, Muramoto T. Eur. J. Cell Biol. 2006;85:961–968. doi: 10.1016/j.ejcb.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Robson GE, Williams KL. Genetics. 1979;93:861–875. doi: 10.1093/genetics/93.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichinger L, Pachebat JA, Glockner G, Rajandream M, Sucgang R, et al. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloomfield G, Tanaka Y, Skelton J, Ivens A, Kay RR. Genome Biol. 2008;9:R75. doi: 10.1186/gb-2008-9-4-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supporting material on Science Online.
- 20.Wong JL, Johnson MA. Trends Cell Biol. 2010;20:134–141. doi: 10.1016/j.tcb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Muramoto T, Suzuki K, Shimizu H, Kohara Y, Kohriki E, et al. Mech. Dev. 2003;120:965–975. doi: 10.1016/s0925-4773(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 22.Kawano S, Anderson RW, Nanba T, Kuroiwa T. J. Gen. Microbiol. 1987;133:3175–3182. doi: 10.1099/00221287-133-11-3175. [DOI] [PubMed] [Google Scholar]
- 23.Mirfakhrai M, Tanaka Y, Yanagisawa K. Genetics. 1990;124:607–613. doi: 10.1093/genetics/124.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raper KB. The Dictyostelids. Princeton Univ. Press; Princeton: 1984. chap. 9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.