The 2009 H1N1 influenza pandemic showed the speed with which a novel respiratory virus can spread and the ability of a generally mild infection to induce severe morbidity and mortality in an unfortunate few. Recent in vitro studies show that the interferon-inducible transmembrane (IFITM) protein family members potently restrict the replication of multiple pathogenic viruses1-7. Both the magnitude and breadth of the IFITM proteins’ in vitro effects suggest they are critical for intrinsic resistance to such viruses, including influenza viruses. Using a knockout mouse model8, we now directly test this hypothesis and find that IFITM3 is essential for defending the host against influenza A virus in vivo. Mice lacking Ifitm3 display fulminant viral pneumonia when challenged with a normally low-pathogenicity influenza virus, mirroring the destruction inflicted by the highly pathogenic 1918 ‘Spanish’ influenza9, 10. Similar increased viral replication is seen in vitro, with protection rescued by the re-introduction of Ifitm3. To test the role of IFITM3 in human influenza virus infection, we assessed the IFITM3 alleles of individuals hospitalised with seasonal or pandemic influenza H1N1/09 viruses. We find that a statistically significant number of hospitalised subjects show enrichment for a minor IFITM3 allele (SNP rs12252-C) that alters a splice acceptor site, and functional assays show the minor CC genotype IFITM3 has reduced influenza virus restriction in vitro. Together these data reveal that the action of a single intrinsic immune effector, IFITM3, profoundly alters the course of influenza virus infection in mouse and man.
IFITM3 was identified in a functional genomic screen as mediating resistance to influenza A virus, dengue virus and West Nile virus infection in vitro1. However, the role of the Ifitm proteins in anti-viral immunity in vivo is unknown. Therefore, we infected mice that are homozygous for a disruptive insertion in exon 1 of the Ifitm3 gene that abolishes its expression8 (Ifitm3−/−), with a low-pathogenicity (LP) murine-adapted H3N2 influenza A virus (A/X-31). LP strains of influenza do not normally cause extensive viral replication throughout the lungs, or cause the cytokine dysregulation and death typically seen after infection with highly-pathogenic (HP) viral strains9, at the doses used (Fig. 1a). However, LP-infected Ifitm3−/− mice became moribund, losing >25% of their original body weight and exhibiting severe signs of clinical illness (rapid breathing, piloerection) 6 days after infection. In comparison, wild-type (WT) litter mates shed <20% of their original body weight, before fully recovering (Fig. 1a, b). There was little difference in virus replication in the lungs during the first 48 hours of infection. However, virus persisted and was not cleared as quickly in Ifitm3−/− mice, whose lungs contained 10-fold higher levels of replicating virus than the WT mice at 6 days post-infection (Fig. 1c). No viral RNA was detected in the heart, brain or spleen of infected WT or Ifitm3−/− mice over the course of infection, revealing that systemic viremia was not occurring. Full genome sequencing of virus removed from the lungs of WT and Ifitm3−/− mice showed no genetic variation. We demonstrated that Ifitm3 protein expression after influenza infection was absent in Ifitm3−/− mice but increased substantially in WT controls (Fig. 1b, Supp. Fig. S1). Infection of WT and Ifitm3−/− mice with a human isolate of pandemic influenza A H1N1 (pH1N1/09) resulted in the same severe pathogenicity phenotype in the Ifitm3−/− mice (Fig. 1a, b). Mouse embryonic fibroblast (MEF) lines generated from multiple matched littermates demonstrated that Ifitm3−/− cells are infected more readily in vitro, and lack much of the protective effects of interferon (IFN). Importantly, the stable restoration of Ifitm3 conferred WT levels of restriction against either X-31, or the more pathogenic Puerto Rico/8/34 (PR/8) influenza strain (Fig. 1d, Supp. Fig. S2). Further to IFITM3’s role in restriction of HP H5N1 avian influenza7, we also show that it limits infection by recent human influenza A isolates and influenza B virus (Supp. Fig. S3). Therefore, enhanced pathogenesis to diverse influenza viruses is attributable to loss of Ifitm3 expression and consequential changes in immune defence of the lungs.
Figure 1. Influenza A virus replicates to higher levels in Ifitm3−/− mice.
Change in body mass (a) and survival (b) of WT (•) and Ifitm3−/− (□) mice following intranasal inoculation with A/X-31 and pandemic H1N1/09 Eng/195 influenza (n>5). Absence of IFITM3 expression was verified in the Ifitm3−/− mice at all time points, but was seen to increase in WT mice (b). A/X-31 viral load in the lungs of mice (n>4) was calculated over the course of infection (c) by plaque assay. Ifitm3−/− murine embryonic fibroblasts (n=3 per condition) stably-expressing Ifitm3 (+), or the empty vector (−) were left untreated (blue), or incubated with IFN-α (red) or -γ (green), then challenged with either A/X-31 or PR/8 influenza. 12h after infection, the cells were assessed for either HA expression (PR/8), or NP expression (A/X-31) (d). Results show means ± s.d. Statistical significance was assessed by Student’s t-test (**: P < 0.01; ***: P < 0.001).
Examination of lung pathology showed fulminant viral pneumonia with substantial damage and severe inflammation in the infected Ifitm3−/− mice. Lung pathology was characterised by extensive oedema and red blood cell extravasation, as well as pneumonia, hemorrhagic pleural effusion and multiple, large lesions on all lung lobes (Fig. 2a, b, Supp. Fig. S4). We note this pathology is similar to that produced by infection of mice and primates with 1918 H1N1 virus9-11. Given the higher viral load in Ifitm3−/− mice and increased replication of influenza A virus in Ifitm3 deleted cells in vitro (Fig. 1d), we examined both viral nucleic acid and protein distribution in the lung. Influenza virus infection penetrated deeper into the lung tissue in Ifitm3−/− compared to WT mice whose infection was primarily restricted to the bronchioles, with minimal alveolar infection. Influenza virus was detected throughout the entire lung in Ifitm3−/− sections, spreading extensively in both bronchioles and alveoli (Fig. 2c). Histopathology showed marked infiltration of cells and debris into the bronchoalveolar space of Ifitm3−/− mice (Fig. 2b, Supp. Fig. S4b). The extent and mechanism of cell damage was investigated by TUNEL assay, showing widespread cellular apoptosis occurring 6 days post-infection in Ifitm3−/− mice, whereas apoptosis in WT lungs was very limited (Supp. Fig. S4c). Together, the Ifitm3−/− mouse pathology is consistent with infection by HP strains of influenza A virus, where widespread apoptosis occurs by day 6 post-infection, whilst lungs from LP infections were similar to those of WT mice, displaying minimal damage9, 12, 13.
Figure 2. Pathological examination of infected lungs.
WT mice showed few visible signs of external damage on lung lobes at day six post-infection, whilst Ifitm3−/− mice showed multiple large lesions (a: left, ventral view of intact lungs, right, all lobes displayed) resulting from severe oedema and hemorrhagic pleural effusion (b), as well as a markedly higher infiltration of cells and proteinaceous debris into the alveoli and bronchioles. Localisation of virus within the lungs on day six (c) indicated virus penetrated deeper and more extensively into the lung tissue in the Ifitm3−/− mice, as determined by immunohistochemistry for total influenza protein and detection of virus nucleic acid (virus: red, cell nuclei: blue, A: alveolus, B: bronchiole). Original magnifications were 5x (b) and 20x (c).
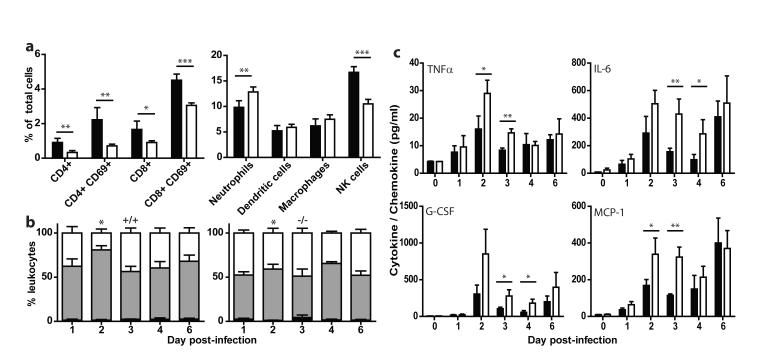
Analysis of cell populations resident in the lung tissue on day 6 post-infection showed that Ifitm3−/− mice displayed significantly reduced proportions of CD4+ (p=0.004) and CD8+ T-cells (p=0.02) and natural killer (NK) cells (p=0.0001) but an elevated proportion of neutrophils (p=0.007) (Fig. 3a). Despite the extensive cellular infiltration, (Supp. Fig. S4b, S5a) the absolute numbers of CD4+ T-lymphocytes in the lungs of the Ifitm3−/− mice were also lower and neutrophils increased compared to WT mice (Supp. Fig. S6). The peripheral blood of infected Ifitm3−/− mice showed leukopenia (Supp. Fig. S5c). Blood differential cell counts indicated marked depletion of lymphocytes on day 2 post-infection in the Ifitm3−/− mice (p=0.04) (Fig. 3b) reflecting changes observed previously in HP (but not LP) influenza infections in both humans and animal models9, 12, 14, 15.Heightened cytokine and chemokine levels are also hallmarks of severe influenza infection; having been observed in both human and animal models9, 16. We observed exaggerated pro-inflammatory responses in the lungs of Ifitm3−/− mice, with higher levels of TNFα, IL-6, G-CSF and MCP-1 showing the most marked changes (Fig. 3c, Supp. Fig. S7). This is indicative of the extent of viral spread within the lungs, as TNFα and IL-6 are released from cells upon infection17. Consistent with the immunopathology data above, these changes are comparable in level to those seen with non-H5N1 HP influenza infections9. Neutrophil chemotaxis, together with elevated proinflammatory cytokine secretion, has previously been reported as one of the primary causes of acute lung injury18.
Figure 3. Altered leukocyte and cytokine response to influenza A infection in Ifitm3−/− mice.
Cytometric analysis of proportional resident cell populations in the lungs of mice (a, +/+: black, −/−: white) showed evidence of lymphopenia in Ifitm3−/− mice six days post-infection. Systemic lymphopenia was confirmed through differential analysis of peripheral blood cell counts (b), which showed a significant depletion of lymphocytes on day two post-infection of Ifitm3−/− mice (monocytes: black, lymphocytes: grey, polymorphonuclear leukocytes: white). Levels of pro-inflammatory cytokines were also recorded as being elevated in Ifitm3−/− lungs over the course of infection (c, +/+: black, −/−: white). Results show means ± s.d., n=5. Statistical significance was assessed by Student’s t-test (*: P < 0.05, **: P < 0.01, ***: P < 0.001).
To further investigate the extensive pathogenesis observed with LP influenza A virus infection in the absence of Ifitm3, we infected both WT and Ifitm3−/− mice with a PR/8 influenza strain deficient for the multi-functional NS1 gene (delNS1)19, 20. NS1 is the primary influenza virus interferon antagonist, with multiple inhibitory effects on host immune pathways20, 21. We found that delNS1 virus was attenuated in both WT and Ifitm3−/− mice, whilst the isogenic PR/8 strain expressing NS1 exhibited typical high pathogenicity in all mice tested, lower doses of PR/8 influenza (whilst lethal in both genotypes of mice) caused accelerated weight loss in Ifitm3−/− compared to WT mice (Supp. Fig. S8). As delNS1 influenza A virus retains its pathogenicity in IFN-deficient mice19, this suggests that Ifitm3−/− mice can mount an adequate IFN-mediated anti-viral response without extensive morbidity. Therefore, unchecked lung viral replication and an enhanced inflammatory response accounts for the profoundly deleterious effects of viral infection in Ifitm3−/− mice.
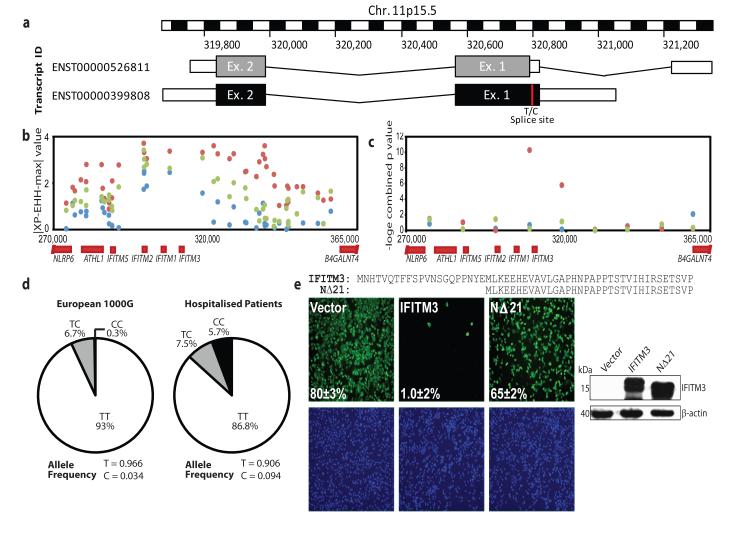
The human IFITM3 gene has two exons and is predicted to encode two splice variants that differ by the presence or absence of the first N-terminal 21 amino acids (Fig. 4a). Currently, 13 non-synonymous, 13 synonymous, one in-frame stop and one splice site acceptor-altering single nucleotide polymorphisms (SNPs) have been reported in the translated IFITM3 sequence (Supp. Table S1). Using tests sensitive to recent positive selection, we can find evidence for positive selection on the IFITM3 locus in human populations acting over the last tens of thousands of years in Africa (Fig. 4b, c). We therefore sequenced 1.8kb of the IFITM3 locus encompassing the exons, intron and untranslated regions from 53 individuals who required admission to hospital as a result of pandemic H1N1/09 or seasonal influenza virus infection in 2009-2010. Of these, 86.8% of patients carried majority alleles for all 28 SNPs in the coding sequence of the gene, but 13.2% possessed known variants. In particular, we discovered over-representation in cases of the synonymous SNP rs12252, wherein the majority T allele is substituted for a minority C allele, which alters the first splice acceptor site and may be associated with the IFITM3 splice variant (ENST00000526811), which encodes an IFITM3 protein lacking the first 21 amino acids due to the use of an alternative start codon.
Figure 4. Single nucleotide polymorphisms of the human IFITM3 gene.
Multiple single nucleotide polymorphisms have been identified within the coding region of the human IFITM3 gene. One such SNP, rs12252 (red) (a), encodes a splice acceptor site altering T/C substitution mutation, which results in a truncated protein with an N-terminal 21 amino acid deletion. Therefore two transcripts are predicted to be expressed from the IFITM3 gene. Positive selection analysis using a haplotype-based test (∣XP-EHH-max∣, b) where data points above 2.7 in the YRI (red), 3.9 in the CEU (blue) and 5.0 in the CHB+JPT (green) are in the top 1% of values and using a combination of three allele frequency spectrum-based test statistics (c), namely Tajima’s D, Fay and Wu’s H and Nielsen et al.’s CLR on 10 kb windows along chromosome 11 encompassing the IFITM3 locus. Evidence for positive selection is seen only in the YRI. Mutations recorded through sequencing of patients hospitalised with influenza virus during the H1N1/09 pandemic showed an overrepresentation of individuals with the C allele at SNP rs12252 (d), relative to matched Europeans. A549 cells transduced to express either full-length (IFITM3) or truncated (NΔ21) IFITM3 (e) (cell nuclei: blue, virus: green, 4× magnification), show a reduction in viral restriction when IFITM3’s N-terminal 21 amino acids are removed, relative to vector controls (Vector). Alignment of the N-termini of full length (IFITM3, top) and truncated IFITM3 (NΔ21, bottom). Values represent the mean of the percent infected cells ± s.d. (n=3).
The allele frequencies for SNP rs12252 vary in different human populations (Supp. Table S2). The ancestral (C) allele, reported in chimpanzees, is rare in sub-Saharan African and European populations (Derived Allele Frequency (DAF) 0.093 and 0.026-0.036 respectively), but more frequent in other populations (Supp. Table S2). SNP rs12252 is notable for its high level of differentiation between Europeans and East Asians, although the Fixation index (FST, a measure of population differentiation) does not reach statistical significance. The genotypes associated with rs12252 in Caucasians hospitalised following influenza infection differ significantly from ethnically matched Europeans in 1000 Genomes sequence data and from genotypes imputed against the June 2011 release of the 1000 Genomes phased haplotypes from the UK, Netherlands and Germany (WTCCC1: p=0.00006, Netherlands: p=0.00001, Germany: p=0.00007; Fisher’s exact test). Patients’ genotypes also depart from Hardy Weinberg equilibrium (p=0.003), showing an excess of C alleles in this population (Fig. 4d). Principal components analysis of over 100K autosomal SNPs showed no evidence of hidden population structure differences between WTCCC controls and a subset of the hospitalised individuals from this study (Supp. Fig S9a, b).
To test the functional significance of the IFITM3 rs12252 polymorphism in vitro, we confirmed the genotypes of HapMap lymphoblastoid cell lines (LCLs) homozygous for either the majority (TT) or minority (CC) variant IFITM3 alleles (Supp. Fig. S9c). We next challenged the LCLs with influenza A virus and found that the minority (CC) variant was more susceptible to infection, and this vulnerability correlated with lower levels of IFITM3 protein expression as compared to the majority (TT) variant cells (Supp. Fig. S10). Although we did not detect the IFITM3 splice variant protein (ENST00000526811) in the CC LCLs, we nonetheless investigated the possible significance of its presence by stably expressing the N-terminally truncated (NΔ21) and WT proteins to equivalent levels in human A549 lung carcinoma cell lines before infection with influenza A virus (A/WSN/1933 (WSN/33)). We found that cells expressing the NΔ21 protein failed to restrict viral replication when compared to WT IFITM3 (Fig. 4e); consistent with previous data showing that IFITM3’s N-terminal 21 amino acids are required for attenuation of vesicular stomatitis virus replication in vitro4. Similar results were obtained using other virulent viral strains (A/California/7/2009 [pH1N1], A/Uruguay/716/2007 [H3N2], and B/Brisbane/60/2008) (Supp. Fig. S3).
We show here that Ifitm3 expression acts as an essential barrier to influenza A virus infection in vivo and in vitro. The fulminant viral pneumonia that occurs in the absence of Ifitm3 arises because of uncontrolled virus replication in the lungs, resulting in profound morbidity. In effect, the host’s loss of a single immune effector, Ifitm3, transforms a mild infection into one with remarkable severity. Similarly, the enrichment of the rs12252 C-allele in those hospitalised with influenza infections, together with the decreased IFITM3 levels and the increased infection of the CC-allele cells in vitro, suggests that IFITM3 also plays a pivotal role in defence against human influenza virus infections. This innate resistance factor is all the more important during encounters with a novel pandemic virus, when the host’s acquired immune defences are less effective. Indeed, IFITM3-compromised individuals, and in turn populations with a higher percentage of such individuals, may be more vulnerable to the initial establishment and spread of a virus against which they lack adaptive immunity. In light of its ability to curtail the replication of a broad range of pathogenic viruses in vitro, these in vivo results suggest that IFITM3 may also shape the clinical course of additional viral infections in favour of the host, and may have done so over human evolutionary history.
Methods in Full
Mouse infection
Background-matched WT (>95% C57BL/6) and Ifitm3−/− mice8 8-10 weeks of age were maintained in accordance with UK Home Office regulations, UK Animals Scientific Procedures Act 1986 under the project licence PPL80/2099. This licence was reviewed by The Wellcome Trust Sanger Institute Ethical Review Committee. Groups of >5 isofluorane-anaesthetised mice of both genotype were intranasally inoculated with 104 PFU of A/X-31 influenza in 50μl of sterile PBS. In some experiments A/X-31 was substituted with 200 PFU of A/England/195/09 influenza, or 50-103 PFU of A/PR/8/34 (PR/8) or an otherwise isogenic virus with a deletion of the NS1 gene (delNS1)19, made as described23. Their weight was recorded daily and they were monitored for signs of illness. Mice exceeding 25% total weight loss were killed in accordance with UK Home Office guidelines. Littermate controls were used in all experiments.
Influenza virus quantification
Lungs from five mice per genotype were collected on days 1, 2, 3, 4 and 6 post-infection, weighed and homogenised in 5% weight / volume (w/v) of Leibovitz’s L-15 medium (Invitrogen) containing antibiotic-antimycotic (Invitrogen). Samples were quantified for viral load by plaque assay in 10-fold serial dilutions on Madin-Darby canine kidney (MDCK) cell monolayers overlaid with 1% Avicell medium24. Lungs were subjected to two freeze-thaw cycles before titration. Virus was also quantified by RT-qPCR, wherein RNA was first extracted from lung, heart, brain and spleen using the RNeasy Mini Plus Kit (Qiagen). Purified RNA was normalised by mass and quantified with SYBR Green (Qiagen) using the manufacturer’s instructions and 0.5μM primers for influenza matrix 1 protein (M1) Fw: 5′-TGAGTCTTCTAACCGAGGTC-3′, Rv: 5′GGTCTTGTCTTTAGCCATTCC-3′ (Sigma-Aldrich) and mouse β-actin (Actb) Fw: 5′CTAAGGCCAACCGTGAAAAG-3′, Rv: 5′-ACCAGAGGCATACAGGGACA-3′. qPCR was performed on a StepOnePlus machine (Applied Biosystems) and analysed with StepOne software v2.1 (Applied Biosystems).
Western blotting
Lungs were homogenised in 5% w/v of Tissue Protein Extraction Reagent (Thermo Scientific) containing “cOmplete Protease Inhibitor” (Roche). Total protein was quantified by BCA assay (Thermo Scientific) and was normalised before loading into wells. Proteins were visualised with the following indicated primary antibodies: Mouse Ifitm2 rabbit polyclonal was purchased from Santa Cruz Biotechnology (Cat# sc-66828); Anti-fragilis (Ifitm3) rabbit polyclonal antibody was from Abcam (Cat # ab15592). The IFITM3 and NΔ21 western blot using the A549 stable cell lines were probed with the anti-IFITM1 antibody from Prosci (Cat# 5807), which recognises a conserved portion of the IFITM1, 2 and 3 proteins which is still present even in the absence of the first twenty one N-terminal amino acids. The LCL blots (including the A549 cell line lysate controls) were probed with either an antibody which is specific for the N-terminus of IFITM3 (Rabbit anti-IFITM3 (N-term aa 8-38) (Abgent, #AP1153a)), or with anti-IFITM1 antibody from Prosci (Cat# 5807), as well as Rabbit anti-MX1 (Proteintech, #13750-1-AP) and mouse anti-GAPDH (clone GAPDH-71.1) (Sigma, #G8795). For the LCL immunoblots all antibodies were diluted in DPBS (Sigma) containing 0.1% Tween 20 (Sigma) and 5% non-far dried milk (Carnation) and incubated overnight at 4°C. All primary antibodies were consequently bound to the corresponding species-appropriate HRP-conjugated secondary antibodies (Dako). Actin antibody was purchased from either Abcam or Sigma, Mouse monoclonal, Cat# A5316.
Pathological examination
5-μm sections of paraffin-embedded tissue were stained with hematoxylin and eosin (Sigma-Aldrich) and were examined and scored twice, once by a pathologist under blinded conditions. The TUNEL assay for apoptosis was conducted using the TACS XL DAB In Situ Apoptosis Detection Kit (R&D Systems).
Immunofluorescent tissue staining: protein
Lung tissue was embedded in glycol methacrylate (GMA) to visualise the spread of viral protein, as described previously25. Briefly, 2-μm sections were blocked with 0.1% sodium azide and 30% hydrogen peroxide followed by a second block of RPMI 1640 (Invitrogen) containing 10% fetal calf serum (Sigma-Aldrich) and 1% bovine serum albumen (Invitrogen). Viral antigen was stained using M149 polyclonal antibody to influenza A, B (Takara) and visualised with a secondary goat anti-rabbit antibody conjugated to AP (Dako). Sections were counterstained with hematoxylin (Sigma-Aldrich). Murine Ifitm1 and Ifitm3 protein expression in lung sections from either uninfected mice, or those two days post-infection with A/X-31, were immunostained with either anti-IFITM1 antibody (Abcam, cat# ab106265) or anti-fragilis (anti-Ifitm3) rabbit polyclonal antisera (Abcam, cat# ab15592). Sections were also stained for DNA with Hoechst 33342 (Sigma).
Immunofluorescent staining: RNA
Viral RNA was visualised in 5-μm paraffin-embedded sections using the QuantiGene viewRNA kit (Affymetrix). Briefly, sections were rehydrated and incubated with Proteinase K. They were subsequently incubated with a viewRNA probe set designed against the negative stranded vRNA encoding the NP gene of A/X-31 (Affymetrix). The signal was amplified before incubation with labelled probes and visualised.
Flow cytometry
Single cell suspensions were generated by passing lungs twice through a 100μm filter before lysing red blood cells with RBC lysis buffer (eBioscience) and assessing for cell viability via Trypan blue exclusion. Cells were characterised by flow cytometry as follows: T-lymphocytes CD4+ or CD8+, T-lymphocytes (activated) CD4+CD69+ or CD8+CD69+, neutrophils CD11bhiCD11c−Ly6g+, dendritic cells CD11c+CD11bloLy6glo MHC class II high, macrophages CD11b+CD11c+F4/80hi, natural killer cells NKp46+CD4−CD8−. All antibodies (Supp. Table S3) were from BD Bioscience, except CD69 and F4/80, which were from AbD Serotec. Samples were run on a FACSAria II (BD Bioscience) and visualised using FlowJo 7.2.4. Data were analysed statistically and graphed using Prism 5.0 (GraphPad Software).
Peripheral leukocyte analysis
Mice (n=3 per genotype per day) were bled on days 0, 1, 2, 3, 4 and 6 by tail vein puncture. Leukocyte counts were determined by haemocytometer, whilst blood cell differential counts were calculated by counting from duplicate blood smears stained with Wright-Giemsa stain (Sigma-Aldrich). At least 100 leukocytes were counted per smear. All blood analyses were conducted in a blinded fashion. Data were analysed statistically and graphed using Prism 5.0 (GraphPad Software).
Cytokine/chemokine analysis
Lungs were collected and homogenised days 0, 1, 2, 3, 4 and 6 post-infection from four mice of each genotype. G-CSF, GM-CSF, IFNγ, IL-10, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-9, IP-10, KC-like, MCP-1, MIP-1α,. RANTES and TNFα were analysed using a mouse antibody bead kit (Millipore) according to the manufacturer’s instructions on a Luminex FlexMAP3D. Results were analysed and quality control checked using Masterplex QT 2010 and Masterplex Readerfit 2010 (MiraiBio). Data were analysed statistically and graphed using Prism 5.0 (GraphPad Software).
Murine embryonic fibroblast generation, transduction, and infectivity assays
Adult Ifitm3−/− mice8 were intercrossed and fibroblasts (MEFs) were derived from embryos at day 13.5 of gestation, as described previously1. MEFs were genotyped by PCR (Thermo-Start Taq DNA Polymerase, ABgene; Epsom, UK) on embryo tail genomic DNA using primers and the cycle profile described previously8 to detect the presence of the WT allele (450 bp band) and the targeted/knockout allele (650 bp band). MEFs were cultured in DMEM, containing 10% FBS, 1X MEM essential amino acids, 1X 2-Mercapto-ethanol (Gibco). MEFs were transduced with VSV-G pseudotyped retroviruses expressing either the empty vector control (pQXCIP, Clontech), or one expressing Ifitm3, as previous described1. After puromycin selection the respective cell lines were challenged with either A/X-31 virus (moi 0.3-0.4) or PR/8 (moi 0.4). For PR/8 infections, after 12h the media was removed and the cells were then fixed with 4% formalin and stained with purified anti-HA monoclonal antibody (Hybridoma HA36-4-5.2, Wistar Institute). For A/X-31 experiments, cells were processed comparably as above, but in addition were permeabilized, followed by immunostaining for NP expression (NP (clone H16-L10-4R5) mouse monoclonal (Millipore MAB8800)). Both sets of experiments were completed using an Alexa Fluor 488 goat anti-mouse secondary at 1:1,000 (A11001, Invitrogen). The cells were imaged on an automated Image Express Micro microscope (Molecular Devices), and images were analysed using the Metamorph Cell Scoring software program (Molecular Devices Inc.). Cytokines: Cells were incubated with cytokines for 24 h prior to viral infection. Murine interferon α (PBL Interferon Source, Cat # 12100-1) and IFN-γ (PBL Interferon Source, Cat # 12500-2) were used at 500-2500 U/ml., and 100-300 ng/ml respectively.
A549 transduction and infectivity assays
A549 cells (ATCC Cat#CCL-185) were grown in complete media (DMEM (Invitrogen Cat#11965) with 10% FBS (Invitrogen)). A549 stable cell lines were made by gamma-retroviral transduction using either the empty vector control virus (pQXCIP, Clontech), the full-length human IFITM3 cDNA, or a truncated human IFITM3 cDNA which is missing the first 21 amino acids (NΔ21). After puromycin selection, expression of the IFITM3 and NΔ21 proteins were confirmed by Western blotting using an 18% SDS-PAGE gel and an anti-IFITM3 antibody that was raised against the conserved intracellular loop (CIL) of IFITM3 (Proteintech). A549 cell lines were challenged with one of the following strains: A/WSN/33 (a kind gift of Dr. Peter Palese), A/California/7/2009, A/Uruguay/716/2007 and B/Brisbane/60/2008 (kind gift of Dr. Jan Malbry, CDC, Atlanta, GA, USA) for 12h, then fixed with 4% PFA and immunostained with anti-HA antibody (HA (Wistar collection) or anti-NP antibodies (Abcam), or Millipore clone H16-L10-4R5 anti-influenza A virus antibody). Percent infection was calculated from immunofluorescent images as described for the MEF experiments above. Alternatively, cells were transduced with lentiviral vectors to express green fluorescent protein (GFP) or IFITM3 and were stained with anti-NP antibody (Abcam) and analysed by flow cytometry following challenge with B/Bangladesh/3333/2007 virus (NIMR, England). For the immunofluorescence-based viral titering experiments, virus-containing supernatant was collected from the indicated A549 cell line cultures after 12h of infection with WSN/33 (Part One). Next this supernatant was used to infect MDCK cells (ATCC) in a well by well manner (Part Two). Both the A549 and MDCK cells were then processed to detect viral HA expression as described above.
LCL infectivity assays
LCL TT and LCL CC cells were grown in RPMI-1640 (Sigma-Aldrich) containing 10% FCS, 2mM L-glutamine, 1mM sodium pyruvate, 1× MEM non-essential amino acids solution, and 20mM HEPES (all from Invitrogen). For infectivity assays, LCL cells were either treated with recombinant human IFN-α2 (PBL Interferon Source, #11100) at 100 units/ml or DPBS (Sigma-Aldrich) for 16h. The LCL cells were then counted, resuspended at a concentration of 5 × 105 cells/ml, and plated on a 96-well round-bottom plate (200μl cell suspension/well). The cells were then challenged with WSN/33 influenza A virus (MOI 0.1). After 18h, the cells were washed twice with 250μl MACS buffer (DPBS containing 2% FCS and 2mM EDTA (Sigma-Aldrich)). The cells were fixed and permeabilised using the BD Cytofix/Cytoperm Fixation/Permeabilisation Kit (BD Biosciences), following the manufacturer’s instructions. Briefly, the cells were resuspended in 100μl of Cytofix/Cytoperm Fixation and Permeabilisation solution and incubated at 4°C for 20 minutes. The cells were then washed twice with 250μl 1× Perm/Wash buffer and resuspended in 50ml 1× Perm/Wash buffer containing a 2 μg/ml solution of a fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal antibody against influenza A virus NP (clone 431, Abcam, #ab20921). The cells were incubated in the diluted antibody solution for 1h at 4°C, washed twice with 250μl 1× Perm/Wash buffer, resuspended in 200μl MACS buffer, and analysed by flow cytometry using a BD FACS Calibur (BD Biosciences).
Ethics and sampling
We recruited patients with confirmed seasonal influenza A or B virus or pandemic influenza A pH1N1/09 infection who required hospitalisation in England and Scotland between November 2009 and February 2011. Patients with significant risk factors for severe disease, and patients whose daily activity was limited by co-morbid illness were excluded. 53 patients, 29 male and 24 female, average age 37 (range 2-62) were selected. 46 (88%) had no concurrent comorbidities. The remaining 6 had the following comorbid conditions: hypertension (3 patients), alcohol dependency and cerebrovascular disease (1 patient), bipolar disorder (1 patient) and kyphoscoliosis (1 patient). Four patients were pregnant. Where assessed, 36 patients had normal body mass (69%), one had a BMI <18.5 and 10 had a BMI between 25 and 39.9 and one a BMI>40. Seasonal influenza A H3N2, influenza B and pandemic influenza A pH1N1/09 were confirmed locally by viral PCR or serological tests according to regional protocols. Consistent with the prevalent influenza viruses circulating in the UK between 2009 and 201126 44 (85%) had pH1N1/09, 2 had pH1N1/09 and influenza B co-infection, 4 had influenza B and 2 had non-subtyped influenza A virus infection. Of the adults, 24 required admission to an intensive care unit (ICU) and 1 required admission to a high dependency unit (HDU). The remainder were managed on medical wards and survived their illnesses. The GenISIS study was approved by the Scotland ‘A’ Research Ethics Committee (09/MRE00/77) and the MOSAIC study was approved by the NHS National Research Ethics Service, Outer West London REC (09/H0709/52, 09/MRE00/67).
Consent was obtained directly from competent patients, and from relatives/friends/welfare attorneys of incapacitated patients. Anonymised 9ml EDTA blood samples were transported at ambient temperature. DNA was extracted using a Nucleon Kit (GenProbe) with the BACC3 protocol. DNA samples were re-suspended in 1 ml TE buffer pH 7.5 (10mM Tris-Cl pH 7.5, 1mM EDTA pH 8.0).
Sequencing and Genetics
Human IFITM3 sequences were amplified from DNA obtained from peripheral blood by nested PCR (GenBank accession numbers JQ610570 – 621). The first round utilised primers FW: 5′-TGAGGGTTATGGGAGACGGGGT-3′and Rv: 5′-TGCTCACGGCAGGAGGCC-3′, followed by an additional round using primers FW: 5′-GCTTTGGGGGAACGGTTGTG-3′and RV: 5′-TGCTCACGGCAGGAGGCCCGA-3′. The 1.8kb IFITM3 band was gel extracted and purified using the QiaQuick Gel Extraction Kit (Qiagen). IFITM3 was Sanger sequenced on an Applied Biosystems 3730×l DNA Analyzer (GATC Biotech) using a combination of eight sequencing primers (Supp. Table S4). Single nucleotide polymorphisms were identified by assembly to the human IFITM3 encoding reference sequence (Acc. No.: NC_000011.9) using Lasergene (DNAStar). Homozygotes were called based on high, single base peaks with high Phred quality scores, whilst heterozygotes were identified based on low, overlapping peaks of two bases with lower Phred quality scores relative to surrounding base calls (Supp. Fig. S9). We identified SNP rs12252 in our sequencing and compared the allele and genotype frequencies to allele and genotype frequencies from 1000 Genomes sequencing data from different populations (Supp. Table S3). In addition, we used the most recent release of phased 1000 Genomes data27 to impute the region surrounding SNP rs12252 to determine allele frequencies in the publicly available genotype dataset of WTCCC1 controls (n=2,938) and four previously published datasets genotyped from the Netherlands (n=8,892) and Germany (n=6,253)22. In the imputation, samples genotyped with Illumina 550k, 610k and 670k platforms were imputed against the June 2011 release of 1000 Genotypes phased haplotypes using the Impute software28, version 2.1.2. Only individuals with European ethnicities (CEU, FIN, GBR, IBS, TSI) were included from the 1000 Genomes reference panel. Recommended settings were used for imputing the region 200 kb in either direction from the variants of interest, along with 1Mb buffer size. The statistical significance of the allele frequencies was determined by Fisher’s exact test.
We assessed for population stratification by principal component analysis. Genotype data from the WTCCC1 1958 Birth Cohort dataset were obtained from the European Genotype Archive with permission, reformatted and merged with genotype data from the GenISIS study to match 113,819 SNPs present in both cohorts. Suspected strand mismatches were removed by identifying SNPs with more than 2 genotypes and using the LD method as implemented in Plink (v1.07)29, resulting in 105,362 matched SNPs. Quality control was applied in GenABEL version 1.6-9 to genotype data for these SNPs for the GenISIS cases and 1499 individuals from WTCCC. Thresholds for quality control (deviation from Hardy-Weinberg equilibrium (p<0.05), MAF<0.0005, call rate<98% in all samples) were applied iteratively to identify all markers and subjects passing all quality control criteria, followed by principal component analysis using GenABEL. We tested for positive selection using both a haplotype-based test (∣XP-EHH-max∣) and allele frequency spectrum-based test statistics, namely Tajima’s D, Fay and Wu’s H and Nielsen et al.’s CLR on 10 kb windows across the entire genome as described previously27, 30. The three statistics were combined and the combined p value was plotted corresponding to the 10 kb windows.
Supplementary Material
Acknowledgements
We would like to thank C. Brandt for maintaining mouse colony health and wellbeing and T. Hussell for provision of A/X-31 virus. We also thank D. Gurdasani for statistical analysis of genotype frequencies. We also thank M. Hu and I. Gallego Romero for calculating genome-wide selection statistics. This work was supported by the Wellcome Trust. The MOSAIC work was supported by Imperial’s Comprehensive Biomedical Research Centre (cBRC), the Wellcome Trust and Medical Research Council UK. The GenISIS work was supported by the Chief Scientist Office (Scotland). A.L.B. is the recipient of a Charles H. Hood Foundation Child Health Research Award, and is supported by grants from the Phillip T. and Susan M. Ragon Institute Foundation, the Bill and Melinda Gates Foundation’s Global Health Program and the National Institute of Allergy and Infectious Diseases (R01AI091786). J.K.B. is supported by a Wellcome Trust Clinical Lectureship (090385/Z/09/Z) through the Edinburgh Clinical Academic Track (ECAT).
Footnotes
Author information IFITM3 sequences are deposited in GenBank under the accession numbers JQ610570 – 621.
MOSAIC Core Investigators:
Benaroya Research Institute, USA: D. Chaussabel. Gartnavel General Hospital, Greater Glasgow, UK: W.E. Adamson, W.F. Carman. Health Protection Agency, UK: C. Thompson, M.C. Zambon. Imperial College London, UK: P. Aylin, D. Ashby, W.S. Barclay, S.J. Brett, W.O. Cookson, L.N. Drumright, J. Dunning, R.A. Elderfield, L. Garcia-Alvarez, B.G. Gazzard, M.J. Griffiths, M.S. Habibi, T.T. Hansel, J.A. Herberg, A.H. Holmes, T. Hussell, S.L. Johnston, O.M. Kon, M. Levin, M.F. Moffatt, S. Nadel, P.J. Openshaw, J.O. Warner. National Institute for Medical Research, UK: A. Hay, J. McCauley, A. O’Garra. Roche, Nutley, USA: J. Banchereau. University College London, UK: A. Hayward. University of Edinburgh, UK: K.J. Baillie, D.A. Hume, P. Simmonds University of Liverpool, UK: S.J. Aston, S.B. Gordon, P.S. McNamara; M.G. Semple; R.L. Smyth University of Nottingham, UK: J.S. Nguyen-Van-Tam University of Oxford, UK: L-P. Ho, A. J. McMichael Wellcome Trust Sanger Institute, UK: P. Kellam. We gratefully acknowledge the generous assistance of Alshafi, K.; Bailey, E.; Bermingham, A.; Berry, M.; Bloom, C.; Brannigan, E.; Bremang, S.; Clark, J.; Cox, M. C.; Cross, M.; Cumming, L. A.; Dyas, S.; England-Smith, J.; Enstone, J.; Ferreira, D.; Goddard, N.; Godlee, A.; Gormley, S.; Guiver, M.; Hassan-Ibrahim, M.O.; Hill, H.; Holloway, P.; Hoschler, K.; Houghton, G.; Hughes, F.; Israel, R.R.; Jepson, A.; Jones, K.D.; Kelleher, W.P.; Kidd, M.; Knox, K.; Lackenby, A.; Lloyd, G.; Longworth, H.; Minns, M.; Mookerjee, S.; Mt-Isa, S.; Muir, D.; Paras, A.; Pascual, V.; Rae, L.; Rodenhurst, S.; Rozakeas, F.; Scott, E.; Sergi, E.; Shah, N.; Sutton, V.; Vernazza, J.; Walker, A.W.; Wenden, C.; Wotherspoon, T.; Wright, A.D.; Wurie, F. and the clinical and laboratory staff of the Brighton & Sussex University Hospitals NHS Trust; Central Manchester University Hospitals NHS Foundation Trust; Chelsea and Westminster Hospital NHS Foundation Trust; Alder Hey Children’s Hospital and Liverpool School of Tropical Medicine; Imperial College Healthcare NHS Trust; Liverpool Women’s NHS Foundation Trust; Royal Liverpool and Broadgreen University Hospitals NHS Trust; Royal Brompton and Harefield NHS Foundation Trust; The Roslin Institute, Edinburgh; University Hospitals Coventry and Warwickshire NHS Trust. The MOSAIC consortium was supported by several Comprehensive Local Research Networks (CLRNs), the National Institute for Health Research (NIHR), UK and by Biomedical Research Centre (BRC) and Unit (BRU) funds. Finally, we thank all patients and their relatives for their generous agreement to inclusion in this study.
GenISIS Investigators (* Local lead investigator, ** Principal Investigator)
Critical Care Medicine, University of Edinburgh K. Everingham, H. Dawson, D. Hope, P. Ramsay, T. S. Walsh*; Generation Scotland, University of Edinburgh Molecular Medicine Centre A. Campbell, S. Kerr; Intensive Care National Audit & Research Centre, London D. Harrison, K. Rowan; Intensive Care Unit, Aberdeen Royal Infirmary J. Addison, N. Donald, S. Gait, D. Noble, J. Taylor, N. Webster*; Intensive Care Unit, Ayr Hospital I. Taylor*; Intensive Care Unit, Borders General Hospital Melrose, J. Aldridge*, R. Dornan, C. Richard; Intensive Care Unit, Crosshouse Hospital, Kilmarnock D. Gilmour, R. Simmons*, R. White*; Intensive Care Unit, Dumfries and Galloway Royal Infirmary C. Jardine, D. Williams*; Intensive Care Unit, Glasgow Royal Infirmary M. Booth*, T. Quasim; Intensive Care Unit, Hairmyres Hospital, Lanarkshire, V. Watson; Intensive Care Unit, Inverclyde Royal Hospital, Greenock P. Henry, F. Munro; Intensive Care Unit, Monklands Hospital, Airdrie L. Bell, J. Ruddy*; Intensive Care Unit, Ninewells Hospital, Dundee S. Cole*, J. Southward; Intensive Care Unit, Queen Margaret Hospital, Dunfermline P. Allcoat, S. Gray, M. McDougall*; Intensive Care Unit, Raigmore Hospital, Inverness J. Matheson, J. Whiteside*; Intensive Care Unit, Royal Alexandra Hospital, Paisley D. Alcorn, K. Rooney*, R. Sundaram; Intensive Care Unit, Southern General Hospital, Glasgow G. Imrie*; Intensive Care Unit, St John’s Hospital, Livingston J. Bruce, K. McGuigan, S. Moultrie*; Intensive Care Unit, Stirling Royal Infirmary C. Cairns*, J. Grant, M. Hughes; Intensive Care Unit, Stobhill Hospital, Glasgow C. Murdoch*; Intensive Care Unit, Victoria Hospital, Glasgow A. Davidson*; Intensive Care Unit, Western General Hospital, Edinburgh G. Harris, R. Paterson, C. Wallis*; Intensive Care Unit, Western Infirmary, Glasgow S. Binning*, M. Pollock; Wellcome Trust Clinical Research Facility, Edinburgh J. Antonelli, A. Duncan, J. Gibson, C. McCulloch, L. Murphy; Roslin Institute, University of Edinburgh C Haley, G. Faulkner, T. Freeman, D. A. Hume, J. K. Baillie**.
References
- 1.Brass AL, et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang D, et al. Identification of Five Interferon-Induced Cellular Proteins That Inhibit West Nile Virus and Dengue Virus Infections. J. Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yount JS, et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weidner JM, et al. Interferon-Induced Cell Membrane Proteins, IFITM3 and Tetherin, Inhibit Vesicular Stomatitis Virus Infection via Distinct Mechanisms. J. Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang IC, et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus. PLoS Pathog. 2011;7:13. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–U545. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feeley EM, et al. IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange UC, et al. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol. Cell. Biol. 2008;28:4688–4696. doi: 10.1128/MCB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belser JA, et al. Pathogenesis of Pandemic Influenza A (H1N1) and Triple-Reassortant Swine Influenza A (H1) Viruses in Mice. J. Virol. 2010;84:4194–4203. doi: 10.1128/JVI.02742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 11.Kobasa D, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 12.Tumpey TM, Lu XH, Morken T, Zaki SR, Katz JM. Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J. Virol. 2000;74:6105–6116. doi: 10.1128/jvi.74.13.6105-6116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobasa D, et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 14.Maines TR, et al. Pathogenesis of emerging avian influenza viruses in mammals and the host innate immune response. Immunol. Rev. 2008;225:68–84. doi: 10.1111/j.1600-065X.2008.00690.x. [DOI] [PubMed] [Google Scholar]
- 15.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:11. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuyama S, Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol. 2011;23:481–486. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julkunen I, et al. Inflammatory responses in influenza A virus infection. Vaccine. 2000;19:S32–S37. doi: 10.1016/s0264-410x(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 18.Yum HK, et al. Involvement of phosphoinositide 3-kinases in neutrophil activation and the development of acute lung injury. J. Immunol. 2001;167:6601–6608. doi: 10.4049/jimmunol.167.11.6601. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 20.Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 21.Billharz R, et al. The NS1 Protein of the 1918 Pandemic Influenza Virus Blocks Host Interferon and Lipid Metabolism Pathways. J. Virol. 2009;83:10557–10570. doi: 10.1128/JVI.00330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anttila V, et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nature Genet. 2011;42:869. doi: 10.1038/ng.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Wit E, et al. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 2004;103:155–161. doi: 10.1016/j.virusres.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational Analysis of cis-Acting RNA Signals in Segment 7 of Influenza A Virus. J. Virol. 2008;82:11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Britten KM, Howarth PH, Roche WR. IMMUNOHISTOCHEMISTRY ON RESIN SECTIONS - A COMPARISON OF RESIN EMBEDDING TECHNIQUES FOR SMALL MUCOSAL BIOPSIES. Biotech. Histochem. 1993;68:271–280. doi: 10.3109/10520299309105629. [DOI] [PubMed] [Google Scholar]
- 26.Ellis J, et al. Virological analysis of fatal influenza cases in the United Kingdom during the early wave of influenza in winter 2010/11. Eurosurveillance. 2011;16:2–7. [PubMed] [Google Scholar]
- 27.Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5:15. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81 doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang Z, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld J, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner M-M, Hunt T, Barnes IHA, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper D, Xue Y, Gallego Romero I, 1000 Genomes Project Consortium. Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012 doi: 10.1126/science.1215040. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.