Abstract
Vitamin D deficiency is becoming more apparent in many populations. Genetic factors may play a role in the maintenance of vitamin D levels. The objective of this study was to perform a genome-wide analysis (GWAS) of vitamin D levels, including replication of prior GWAS results. We measured 25-hydroxyvitamin D (25(OH)D) levels in serum collected at the time of enrollment and at year 4 in 572 Caucasian children with asthma, who were part of a multi-center clinical trial, the Childhood Asthma Management Program. Replication was performed in a second cohort of 592 asthmatics from Costa Rica and a third cohort of 516 Puerto Rican asthmatics. In addition, we attempted replication of three SNPs that were previously identified in a large GWAS of Caucasian individuals. The setting included data from a clinical trial of childhood asthmatics and two cohorts of asthmatics recruited for genetic studies of asthma. The main outcome measure was circulating 25(OH)D levels. The 25(OH)D levels at the two time-points were only modestly correlated with each other (intraclass correlation coefficient = 0.33) in the CAMP population. We identified SNPs that were nominally associated with 25(OH)D levels at two time-points in CAMP, and replicated four SNPs in the Costa Rican cohort: rs11002969, rs163221, rs1678849, and rs4864976. However, these SNPs were not significantly associated with 25(OH)D levels in a third population of Puerto Rican asthmatics. We were able to replicate the SNP with the strongest effect, previously reported in a large GWAS: rs2282679 (GC), and we were able to replicate another SNP, rs10741657 (CYP2R1), to a lesser degree. We were able to replicate two of three prior significant findings in a GWAS of 25(OH)D levels. Other SNPs may be additionally associated with 25(OH)D levels in certain populations.
Introduction
Vitamin D is both a nutrient and a hormone. Environmental and behavioral determinants of vitamin D status vary widely, including exposure to the sun and time spent outdoors (Sahota et al. 2008; van der Mei et al. 2006), latitude, season, skin coverage (Webb 2006), skin color, age, diet, and supplement use (Sahota et al. 2008). As a nutrient, vitamin D is contained only in a few foods, including oily fish and fish liver oil, egg yolk, and offal (Lamberg-Allardt 2006). As such, excluding supplementation, food is not the primary way that we receive the necessary quantities of vitamin D. Sun exposure is one of the primary mechanisms by which we receive large amounts of vitamin D. Specifically, 7-dehydrocholesterol (7-DHC) is distributed in the skin and after exposure to sunlight, 7-DHC is converted to previtamin D3, which is then transformed to vitamin D3 by a thermally induced isomerization. Vitamin D3 then undergoes hydroxylation in the liver to 25-hydroxyvitamin D3 (henceforth, 25(OH)D) and then in the kidney to its biologically active form 1,25-dihydroxyvitamin D3 (1,25[OH]2D). Vitamin supplementation is another primary way that our bodies absorb vitamin D.
Serum 25(OH)D is the major circulating metabolite of vitamin D and is the standard measure of vitamin D status. Evaluations of most relations between vitamin D and health, and various disorders lead to the conclusion that a sufficient circulating vitamin D level for bone health is at least 20 ng/ml, but for other organ systems, is at least 30–40 ng/ml. Levels of 25(OH)D between 20 and 30 ng/ml are considered relatively insufficient and there are suggestions that levels even higher than 40 ng/ml may be necessary (Hollis et al. 2007; Taback and Simons 2007) for optimal immune functioning and overall health.
Vitamin D deficiency has been documented in many populations worldwide, and has been reported in individuals of all ages (Holick 2006; Nesby-O’Dell et al. 2002). Low vitamin D levels have been associated with a range of disorders. The association of low vitamin D and bone diseases such as rickets (Heaney 2003) and osteoporosis (Heaney 2004) is well known. While there are no controlled intervention studies, several lines of evidence have shown that vitamin D reduces the risk of colorectal cancer (Giovannucci 2005, 2006; Gorham et al. 2005). Other cancers that may be vitamin D-responsive include breast, lung, ovarian, and prostate cancer (Grant 2006). Other disorders in which the role of vitamin D is being actively investigated are the autoimmune disorders such as multiple sclerosis (MS), type 1 diabetes mellitus (1999b; Hypponen et al. 2001), and rheumatoid arthritis (Merlino et al. 2004). Our group is actively studying the effect of vitamin D on asthma pathogenesis and control (Brehm et al. 2009, 2010; Litonjua and Weiss 2007).
Despite the relatively large amount that it known about the epidemiology of vitamin D, little is known about how genetics influence vitamin D levels. Family and twin studies suggest that a substantial portion of the variability in vitamin D can be attributed to genetics, with heritability estimates of 53 % (Hunter et al. 2001; Shea et al. 2009). Research identifying genetic variants that could influence vitamin D levels has been minimal until recently. Specific candidate genes have been studied in relation to vitamin D using relatively small and underpowered studies (Engelman et al. 2008; Ramos-Lopez et al. 2007). Recently, three genome-wide association studies (GWAS) of vitamin D were published (Ahn et al. 2010; Engelman et al. 2010; Wang et al. 2010), with the largest study using 15 cohorts of 33,996 individuals (Wang et al. 2010). That study identified several genetic variants for vitamin D including rs2282679 located in GC on chromosome 4p12 (overall p = 1.9 × 10−109); rs12785878 located near DHCR7 on chromosome 11q12 (p = 2.1 × 10−27), and rs10741657 located near CYP2R1 on 11p15 (p = 3.3 × 10−20). In this manuscript, we sought to further investigate genetic determinants of vitamin D levels, and to replicate the previous GWAS hits.
Methods
Initial study population
The Childhood Asthma Management Program (CAMP) was a multicenter clinical trial of the effects of anti-inflammatory medications in children with mild to moderate asthma. Detailed descriptions of subject recruitment and study protocol have been published elsewhere (1999a). All participants had asthma defined by symptoms greater than two times per week, the use of an inhaled bronchodilator at least twice weekly or the use of daily medication for asthma, and airway responsiveness to ≤12.5 mg/ml of methacholine. Children were 5–12 years of age at randomization, and were randomly assigned to one of three treatment arms (inhaled budesonide, nedocromil, or placebo). Children with severe asthma or other clinically significant conditions were excluded. In CAMP 1,041 asthmatic children were followed up for 4–6 years. Of these, 968 children and 1,518 of their parents contributed DNA samples. Our analysis was restricted to the non-Hispanic white children. Two final samples were generated: (1) A total of 422 of white (non-Hispanic) CAMP subjects and their parents were genotyped on Illumina’s Human-Hap550 Genotyping BeadChip. (2) An additional 150 CAMP probands who did not have parental DNA were genotyped with the Illumina Infinium HD Human610-Quad BeadChip. From these data, two separate datasets were generated: (1) 422 parent–offspring trios genotyped on Illumina’s Human-Hap550 Genotyping BeadChip, and (2) a merged HumanHap550/Human610-Quad data set composed of 572 CAMP probands. The CAMP was approved by the Institutional Review Boards of Brigham and Women’s Hospital and the other participating centers.
Phenotype
Serum 25-hydroxyvitamin D3 (25(OH)D)
Serum levels of 25(OH)D are considered as the best circulating biomarker of vitamin D metabolic status and reflect contributions from all sources of vitamin D (i.e., diet and sun exposure) (Hollis 2005; Hollis and Wagner 2005). Two measures of vitamin D were available on 1,024 subjects (98 % of enrolled subjects) using a radioimmunoassay method in Dr. Bruce Hollis’ laboratory at the Medical University of South Carolina (Hollis et al. 1993; Hollis and Napoli 1985). This analysis was done on the 572 with both vitamin D values and GWAS data. Serum 25(OH)D was measured from stored samples drawn shortly prior to enrollment into the trial and at the end of the 4-year trial. Vitamin D levels have been shown to be relatively stable when specimens have been properly stored (Agborsangaya et al. 2010).
Genotyping and quality control
Genome-wide SNP genotyping was performed by Illumina, Inc. (San Diego, CA) on the HumanHap550v3 BeadChip for CAMP subjects and their parents. Genotype reproducibility was assessed by analyzing four subjects that were repeated once on each of the 14 genotyping plates; all replicates had at least 99.8 % concordance. The data were cleaned in several steps. 6,257 markers were removed due to low Illumina clustering scores (see supplementary file for additional information). An additional 1,329 markers were removed because their flanking sequences did not map to a unique position on the hg17 reference genome sequence. We used PLINK (Purcell et al. 2007) to further QC the remaining markers. All markers had greater than 90 % genotyping completion rate, while the average completion rate for each marker was over 99 %. 3,790 markers were removed because they were monomorphic in our sample. 2,445 markers were removed due to five or more parent–child genotype inconsistencies. No filtering was done based on Hardy–Weinberg equilibrium due to ascertainment of the cohort through affected probands. From 561,466 markers present on the BeadChip, 547,645 markers (97.54 %) passed quality control metrics. The 422 CAMP probands and their parents were successfully genotyped. The average genotyping completion rate for each subject was 99.75 %. SNPs with minor allele frequencies (MAF) <1 % were excluded. An additional 150 CAMP probands were available who had genotyping performed with the Illumina Infinium HD Human610-Quad BeadChip.
For replication studies of significant findings from prior GWAS studies, two additional SNPs were genotyped on the Sequenom platform, since they were not on the Illumina chip. SNPs rs10741657 (CYP2R1) and rs12785878 (DHCR7) were genotyped at the Children’s Hospital Boston Sequenom SNP Genotyping Facility. The genotyping completion rate for both these 2 SNPs was 99.2 % and there were no discordant genotypes.
Statistical methods
Since we had measured circulating vitamin D levels in the CAMP population at two time-points 4 years apart, we investigated the correlation between these two measures. We assessed the correlation between the two measures by calculating the Spearman correlation coefficient and by assessing the intra-class correlation (ICC) coefficient. We used mixed-effects regression models to estimate the ICC between the two values, adjusting for season of blood collection, using SAS version 9.1 (SAS Institute, Cary, NC).
We identified the most robust genetic associations for vitamin D by performing four genetic analyses: (1) a family-based association analysis of baseline vitamin D in 422 parent–offspring trios, (2) a family-based association analysis of vitamin D at year 4 in 422 parent–offspring trios, (3) a population-based analysis of baseline vitamin D in 572 probands, and (4) a population-based analysis of vitamin D at year 4 in 551 probands. Family-based association testing (FBAT) is a generalization of the transmission disequilibrium test (TDT), which allows for valid testing of association with any phenotype, sampling structure, and pattern of missing marker allele information. The FBATs have several attractive properties, including being inherently robust to population stratification. The genome-wide population-based analyses used linear regression analysis in PLINK (Purcell et al. 2007) to evaluate the association between SNPs and vitamin D. We used an additive model and adjusted for age, sex, season, and latitude. To correct for population stratification, we used Eigenstrat to generate the main eigenvectors that describe the underlying population substructure (Price et al. 2006), which were also included as covariates in the population-based analysis. The results from these analyses were then merged together and we identified SNPs with nominally significant p values and consistent direction of effect in all the four analyses. SNPs meeting these characteristics were then ranked by the average population-based p value and the top 50 The SNPs were identified and selected to be replicated in the Costa Rican population. We focused on population-based analyses as these analyses contained additional probands, making these analyses substantially more powerful. SNPs that did not have a consistent direction of effect or nominally significant results in one of the analyses were excluded. HWE was checked for these 50 SNPs, and none of them were out of equilibrium.
Primary replication population
Replication genotyping was performed in 592 Hispanic asthmatic children participating in a study of the genetics of asthma in Costa Rica. Detailed information about this sample can be found elsewhere (Hunninghake et al. 2007). In brief, screening questionnaires were sent to the parents of 13,125 children aged 6–14 years enrolled in 113 schools in the Central Valley of Costa Rica from February 2001 to December 2006. Children were eligible for inclusion in the study of parent–child trios if they had physician-diagnosed asthma, at least two respiratory symptoms (wheezing, cough, or dyspnea)or a history of asthma attacks in the previous year, and high probability of having at least six great-grandparents born in the Central Valley of Costa Rica (as determined by the study genealogist on the basis of the paternal and maternal last names of each of the child’s parents) (Escamilla et al. 1996). Blood samples were obtained on all probands on entry into the study. Measurement of 25(OH)D levels was performed as in the CAMP study, and the residuals were calculated adjusting for age and gender. Replicate genotyping was performed using the Illumina BeadStation 500G system (Oliphant et al. 2002). The genotyping completion rate was >99.8 % with no discordance among replicate genotypes. Written parental consent was obtained for the participating children, for whom written assent was also obtained. The study was approved by the Institutional Review Boards of the Hospital Nacional de Niños (San José, Costa Rica), and Brigham and Women’s Hospital (Boston, MA).
For replication studies of significant findings from prior GWAS studies, three additional SNPs were genotyped. SNPs rs2282679 (GC), rs10741657 (CYP2R1), and rs12785878 (DHCR7) were genotyped using the Taqman assay in our laboratory. Genotyping completion rates for these SNPs were 99.4, 99.6, and 99.2 %, respectively. No discordant genotypes were noted in duplicate genotyping.
Second replication population
Replication studies were conducted in a study of asthma genetics and epidemiology in Puerto Rican children in Hartford (CT) and San Juan (Puerto Rico). At both the sites, the main recruitment tool was a screening questionnaire given to parents of children aged 6–14 years. All participants had to have four Puerto Rican grandparents. Children with asthma (cases) were selected on the basis of physician-diagnosed asthma and wheeze in the prior year. Controls had no physician-diagnosed asthma or wheeze in the prior year.
From September 2003 to July 2008, the children were recruited from public schools enrolling Puerto Rican children in Hartford. Flyers with a study description were distributed to all parents of children in grades K to 8. Of the 640 children whose parents completed a screening questionnaire, 585 (91.4 %) were eligible; 449 (76.8 %) of these children agreed to participate. From March 2009 to June 2010, children were chosen from households selected by a multistage probability design in San Juan, using a scheme similar to that of a prior study (Bird et al. 2006). In an effort to reach a sample size of 700 children, a random sample of 783 of 1,111 eligible households was contacted; parents of 106 of these 783 households refused to participate or could not be reached, leaving 677 participants. The study was approved by the Institutional Review Boards of Connecticut Children’s Medical Center (Hartford, CT), the University of Puerto Rico (San Juan, PR), Brigham and Women’s Hospital (Boston, MA), and the University of Pittsburgh (Pittsburgh, PA).
Of the 1,126 participants at both study sites, 999 had blood samples and sufficient DNA for genotyping. Genome-wide genotyping was completed in these 999 children using the Illumina HumanOmni2.5-Quad v1.0 DNA Analysis BeadChip. After excluding subjects with >5 % missing genotypes, and SNPs with: missingness >2 %, extreme deviations from Hardy–Weinberg equilibrium, and minor allele frequency <1 %, there were 943 subjects and ~ 1.9 million SNPs available for analysis. Of these 943 children, 516 had asthma and were included in this analysis. Plasma 25(OH)D was measured with a liquid chromatography–mass spectrophotometry assay (Holick 2005). The association analysis of vitamin D was adjusted for study site (Hartford, CT vs. San Juan, PR), age, sex, season of blood draw, and GEM clusters (to adjust for population stratification) (Lee et al. 2010).
Results
The characteristics of children included in the three asthma cohorts are presented in Table 1. After data cleaning, there were 572 probands in CAMP and 592 parent–offspring trios in the Costa Rican population. Despite the disparity in climate and latitude, Table 1 illustrates the striking similarities between the CAMP and Costa Rican cohorts, particularly with regard to the vitamin D levels. The 25(OH)D levels were slightly lower in the Puerto Rico cohort compared with the other two cohorts. Other characteristics of the Puerto Rican children were similar to CAMP and Costa Rica. In the CAMP population, serum vitamin D levels were lower at the end of the 4-year trial than at the start. While the values were correlated, this was modest at best (Pearson correlation coefficient = 0.39, p < 0.001, Fig. 1), and the ICC coefficient adjusted for season of collection of the samples was only 0.33. The correlation did not change when the analysis was limited to the children who had their two measurements taken during the same season, 4 years apart (Pearson correlation coefficient = 0.39).
Table 1.
Main characteristics of study participants
| CAMP | Costa Rica asthma cohort | Puerto Rico asthma cohort | |
|---|---|---|---|
| N | 572 | 592 | 516 |
| Male gender | 341 (59.6 %) | 351 (59.3 %) | 285 (55.2 %) |
| Age at enrollment, years [mean (SD)] | 8.9 (2.1) | 9.0 (1.8) | 10.0 (2.7) |
| Circulating vitamin D at baseline, ng/ml [mean (SD)] | 40.46 (15.75) range: 10.1–110.4 | 37.44 (11.79) range: 12.5–98.1 | 28.9 (8.7) range: 8–62 |
| Circulating vitamin D at year 4a, ng/ml [mean (SD)] | 32.50 (13.50) range: 6.4–90.9 |
SD standard deviation
Only 551 individuals at the second time point
Fig. 1.
Scatter plot of 25(OH)D values at baseline and at year 4 in children with asthma. Trendline corresponds to Pearson correlation coefficient = 0.39, p < 0.001
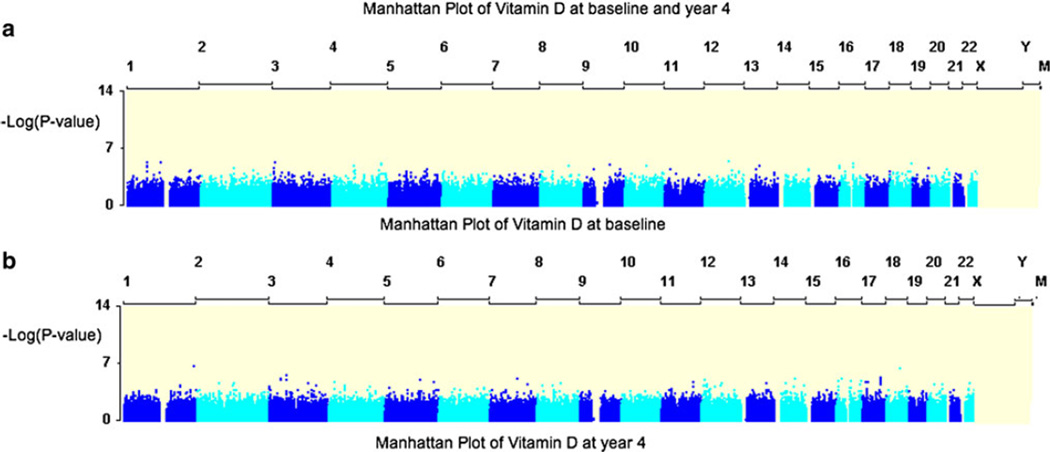
Supplementary Table S1 shows details of the genotyping process in CAMP, which indicates that there were 512,296 autosomal SNPs meeting our QC criteria for the family-based analyses. The “manhattan plots” and QQ plots at baseline and year 4 are shown in Figs. 2, 3, which indicate no substantial deviation from what is expected by chance (genetic inflation factor = 1.015 and 1.006 for baseline and year 4, respectively). None of the SNPs achieved genome-wide significance after a Bonferroni correction for multiple comparisons; however, the QQ plots indicate that the top hits are greater than what is expected by chance. The average population-based p value was calculated and the top 50 SNPs with low p values and nominally significant family-based p values were identified and replicated in the Costa Rican cohort (Table 2). Four of these SNPs had nominally significant associations in the Costa Rican sample, as depicted in Table 3. Although none of these SNPs meet genome-wide significance, they have consistent nominal associations in both studies. However, none of these SNPs were associated with 25(OH)D levels in a Puerto Rican cohort, despite having the same direction of effect as in the other two cohorts (Table 3, all p values >0.1). Even when the evidences from the three cohorts were combined, Liptak p values did not reach genome-wide significance.
Fig. 2.
a, b Manhattan plot of the population based analyses at baseline and year 4. The y-axis has the –log(p value) while the x-axis has the 22 chromosomes. In this figure, while none of the findings reached genome-wide significance, there were several strong associations
Fig. 3.
a, b QQ plots for the population-based analyses at baseline and year 4. In both cases, this line does not differ significantly from what is expected by chance. The top p values do seem slightly higher than what is expected by chance, which would indicate that there are true positive findings among the SNPs more strongly associated with vitamin D
Table 2.
Results for the top 50 SNPs from the 4 statistical analyses
| SNP | Chromosome | BP | Related gene(s) |
p value population- based baseline |
p value population- based year 4 |
Average population- based p value |
p value family-based baseline |
p value family- based year 4 |
|---|---|---|---|---|---|---|---|---|
| rs26673 | 5 | 116,107,966 | 3.3 × 10−4 | 3.6 × 10−4 | 3.4E–04 | 4.5E–03 | 4.5E–03 | |
| rs26674 | 5 | 116,107,928 | 3.1 × 10−4 | 5.6 × 10−4 | 4.4E–04 | 3.4E–03 | 3.4E–03 | |
| rs5753362 | 22 | 29,594,281 | OSBP2 | 4.2 × 10−4 | 6.6 × 10−4 | 5.4E–04 | 1.6E–03 | 6.7E–03 |
| rs739894 | 22 | 29,588,108 | OSBP2 | 6.5 × 10−4 | 1.3 × 10−3 | 9.6E–04 | 9.4E–04 | 3.2E–03 |
| rs6837397 | 4 | 55,828,386 | 5.1 × 10−4 | 1.4 × 10−3 | 9.8E–04 | 1.9E–02 | 6.4E–03 | |
| rs7231689 | 18 | 42,532,983 | ST8SIA5 | 1.9 × 10−4 | 3.5 × 10−4 | 1.1E–03 | 8.6E–03 | 2.4E–06 |
| rs1119208 | 5 | 76,524,369 | 1.8 × 10−4 | 4.3 × 10−4 | 1.1E–03 | 3.2E–02 | 4.4E–02 | |
| rs4920935 | 5 | 116,134,125 | 2.6 × 10−4 | 2.6 × 10−3 | 1.4E–03 | 6.1E–03 | 3.4E–02 | |
| rs7337310 | 13 | 41,198,323 | KIAA0564 | 1.8 × 10−3 | 1.3 × 10−3 | 1.5E–03 | 5.3E–03 | 4.2E–02 |
| rs680617 | 5 | 116,007,082 | 3.2 × 10−3 | 8.7 × 10−6 | 1.6E–03 | 1.1E–02 | 1.2E–03 | |
| rs1458825 | 4 | 55,804,268 | 1.1 × 10−3 | 2.3 × 10−3 | 1.7E–03 | 1.4E–02 | 1.8E–02 | |
| rs6554253 | 4 | 55,812,933 | 1.1 × 10−3 | 2.3 × 10−3 | 1.7E–03 | 1.3E–02 | 1.6E–02 | |
| rs4301169 | 4 | 55,826,496 | 1.2 × 10−3 | 2.5 × 10−3 | 1.8E–03 | 2.5E–02 | 7.6E–03 | |
| rs4864972 | 4 | 55,837,040 | 3.6 × 10−3 | 1.8 × 10−3 | 1.9E–03 | 6.8E–03 | 1.8E–03 | |
| rs340286 | 3 | 160,065,698 | 4.5 × 10−3 | 5.7 × 10−5 | 2.3E–03 | 1.7E–03 | 1.1E–03 | |
| rs1948429 | 5 | 116,022,674 | 4.3 × 10−3 | 5.0 × 10−4 | 2.4E–03 | 2.3E–02 | 1.1E–02 | |
| rs1111721 | 16 | 67,255,962 | CDH3 | 2.7 × 10−4 | 4.8E–03 | 2.5E–03 | 2.6E–02 | 1.3E–02 |
| rs12897195 | 14 | 61,029,474 | PRKCH | 1.8 × 10−4 | 5.0E–03 | 2.6E–03 | 3.4E–04 | 1.8E–04 |
| rs1888557 | 13 | 41,297,445 | KIAA0564 | 2.7 × 10−3 | 2.5E–03 | 2.6E–03 | 2.1E–03 | 3.9E–02 |
| rs13026792 | 2 | 156,324,389 | 3.7 × 10−4 | 4.9E–03 | 2.7E–03 | 3.2E–02 | 1.5E–02 | |
| rs12136973 | 1 | 173,406,942 | KIAA0040 | 2.3 × 10−3 | 3.1E–03 | 2.7E–03 | 1.5E–03 | 1.4E–02 |
| rs1585289 | 4 | 55,797,233 | 5.5 × 10−3 | 9.3E–05 | 2.8E–03 | 7.8E–03 | 2.1E–02 | |
| rs11002969 | 10 | 80,850,968 | C10orf56 | 1.9 × 10−3 | 3.9E–03 | 2.9E–03 | 3.1E–02 | 3.9E–02 |
| rs1467118 | 4 | 55,865,036 | 5.9 × 10−3 | 4.8E–05 | 3.0E–03 | 1.2E–02 | 1.4E–03 | |
| rs7843995 | 8 | 2,301,430 | 3.6 × 10−3 | 2.5E–03 | 3.1E–03 | 3.2E–02 | 4.0E–03 | |
| rs6847603 | 4 | 55,812,539 | 6.2 × 10−3 | 1.0E–04 | 3.2E–03 | 6.6E–03 | 1.1E–02 | |
| rs163221 | 18 | 22,763,991 | C18orf16 | 4.9 × 10−4 | 6.0E–03 | 3.2E–03 | 1.4E–02 | 6.0E–03 |
| rs1398765 | 3 | 132,964,104 | CPNE4 | 6.7 × 10−4 | 2.8E–05 | 3.4E–03 | 1.2E–02 | 4.5E–02 |
| rs1319922 | 2 | 126,291,398 | 6.8 × 10−3 | 3.0E–05 | 3.4E–03 | 6.1E–03 | 1.5E–02 | |
| rs4077725 | 4 | 55,851,899 | 2.3 × 10−3 | 5.8E–03 | 4.0E–03 | 9.8E–03 | 1.5E–02 | |
| rs1389164 | 5 | 116,027,419 | 8.0 × 10−3 | 3.3E–04 | 4.2E–03 | −3.1E–02 | −1.6E–02 | |
| rs3793825 | 10 | 71,357,498 | COL13A1 | 1.6 × 10−3 | 7.0E–03 | 4.3E–03 | 1.7E–03 | 3.0E–02 |
| rs1739644 | 20 | 36,460,639 | 1.4 × 10−4 | 7.6E–03 | 4.5E–03 | 2.2E–04 | 3.1E–03 | |
| rs4237510 | 10 | 120,983,651 | GRK5 | 6.4 × 10−3 | 2.5E–03 | 4.5E–03 | −1.4E–02 | −7.7E–05 |
| rs4864977 | 4 | 55,855,356 | 1.7 × 10−3 | 7.7E–03 | 4.7E–03 | 8.9E–03 | 1.8E–02 | |
| rs1678849 | 19 | 5,787,964 | FUT6 | 8.2 × 10−3 | 1.3E–03 | 4.7E–03 | 3.9E–02 | 8.6E–03 |
| rs205651 | 2 | 74,709,631 | LOC130951 | 9.5 × 10−3 | 2.3E–05 | 4.7E–03 | −1.6E–02 | −2.4E–04 |
| rs4864976 | 4 | 55,848,489 | 3.6 × 10−3 | 6.3E–03 | 5.0E–03 | 1.7E–02 | 1.9E–02 | |
| rs17831158 | 8 | 57,630,069 | 6.8 × 10−3 | 3.6E–03 | 5.2E–03 | 1.8E–04 | 1.8E–03 | |
| rs1406381 | 11 | 61,825,261 | SCGB1D4 | 2.7 × 10−3 | 7.6E–03 | 5.2E–03 | −3.1E–03 | −2.2E–02 |
| rs2025323 | 9 | 110,162,093 | 7.9 × 10−3 | 2.6E–03 | 5.2E–03 | 1.7E–02 | 3.4E–02 | |
| rs12785918 | 11 | 100,218,437 | 8.6 × 10−3 | 2.2E–03 | 5.4E–03 | −3.3E–02 | −3.3E–02 | |
| rs1780634 | 20 | 36,463,239 | 1.7 × 10−3 | 9.3E–03 | 5.5E–03 | 1.2E–04 | 8.9E–04 | |
| rs388383 | 5 | 111,731,534 | EPB41L4A | 4.9 × 10−4 | 1.1E–02 | 5.5E–03 | 1.2E–02 | 4.6E–02 |
| rs11013514 | 10 | 23,799,607 | 1.2 × 10−4 | 1.1E–02 | 5.6E–03 | −7.4E–04 | −9.1E–03 | |
| rs13103626 | 4 | 99,536,274 | RAP1GDS1 | 9.5 × 10−3 | 2.2E–03 | 5.8E–03 | 1.5E–02 | 6.6E–03 |
| rs232319 | 18 | 22,754,149 | C18orf16 | 1.4 × 10−3 | 1.2E–02 | 5.9E–03 | 3.3E–02 | 6.3E–03 |
| rs2470911 | 15 | 42,834,426 | RNF36 | 1.1 × 10−2 | 6.6E–04 | 5.9E–03 | −1.1E–02 | −8.6E–04 |
| rs137906 | 22 | 48,827,920 | 8.2 × 10−3 | 3.9E–03 | 6.1E–03 | 3.9E–03 | 1.9E–03 | |
| rs26670 | 5 | 116,116,302 | 1.2 × 10−3 | 1.1 × 10−2 | 6.2E–03 | −1.6E–02 | −4.3E–03 |
SNP single nucleotide polymorphism, BP base pair distance
Table 3.
SNPs that replicate in the Costa Rican cohort, with attempted replication in the Puerto Rico cohort
| SNP | Chr | BP | Gene(s) | CAMP |
Costa Rica asthma cohort |
Puerto Rico asthma cohort |
Combined (Liptak) p value |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β for pop-based |
Average pop-based p value |
MAF | β estimate | p value | MAF | β estimate | p value | MAF | |||||
| rs 11002969 | 10 | 80,850,968 | C10orf56/ ZCCHC24 |
−2.137 | 2.9E–03 | 0.46 | −0.013 | 4.4E–02 | 0.44 | −0.0941 | 0.8428 | 0.34 | 1.6E–02 |
| rs163221 | 18 | 22,763,991 | C18orf16, CHST9 | −2.011 | 3.2E–03 | 0.46 | −0.015 | 3.8E–02 | 0.43 | −0.7550 | 0.1031 | 0.43 | 3.8E–04 |
| rs1678849 | 19 | 5,787,964 | FUT6 | 2.459 | 4.7E–03 | 0.29 | 0.019 | 3.6E–02 | 0.15 | 0.0649 | 0.906 | 0.26 | 2.7E–02 |
| rs4864976 | 4 | 55,848,489 | – | 2.864 | 5.0E–03 | 0.15 | 0.019 | 4.6E–02 | 0.13 | 0.8248 | 0.1179 | 0.24 | 7.4E–04 |
β estimates are obtained from linear regression models in PLINK
SNP single nucleotide polymorphism, CHR chromosome, BP base pair distance, MAF minor allele frequency
Three SNPs were previously reported to be associated with vitamin D levels in a recent GWAS study (Wang et al. 2010), published after we had completed our analyses. None of these SNPs were selected for replication in our analyses. However, after publication of that study, we evaluated the associations for these three SNPs using both the baseline and year 4 vitamin D measurements in the CAMP population. Significant associations were found with baseline and 4-year 25(OH)D levels in rs2282679 and rs10741657 (Table 4). Our population-based analyses found an extremely strong association with rs2282679 as was observed previously. In fact, rs2282679 was among the top 25 associated SNPs for all of the population-based analyses and was the 11th best p value at the baseline vitamin D measurement (see online supplement, Supplementary Table S2). In Costa Rica, all three SNPs were associated with 25(OH)D levels, with the same direction of effects as that in CAMP. Taken together, the results in our initial and primary replication cohorts are consistent with the findings that this SNP in GC had the strongest association with 25(OH)D levels. Finally, in the Puerto Rico cohort, only one of the three SNPs were genotyped, and rs2282679 was significantly associated with 25(OH) levels (p value = 6.3 × 10−09), and the association was in the same direction as in the previous cohorts. The combined p value (Liptak) of the one-sided p values for this SNP from all three cohorts was 1.243E–14.
Table 4.
Replication of the genome-wide significant SNPs found in Wang et al
| SNP | CHR | Gene | Wang et al. p value |
Direction of effect in Wang et al. |
CAMP | Costa Rica asthma cohort |
Puerto Rico asthma cohort |
Combined (Liptak) p values |
Direction of effect in CAMP, Costa Rica, and Puerto Rican cohorts |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S3 pop- based p value* |
F48 pop- based p value* |
MAF | p value* | MAF | p valuea | MAF | |||||||
| rs2282679 | 4p12 | GC | 1.9E–109 | Minor allele undertransmitted | 6.0E–06 | 3.8E–03 | 0.29 | 9.3E–04 | 0.22 | 3.2E–09 | 0.21 | 2.06E–14 | Minor allele is undertransmitted |
| rs10741657a | 11q12 | CYP2R1 | 2.1E–27 | Minor allele overtransmitted | 3.7E–03 | 1.2E–04 | 0.43 | 0.0119 | 0.32 | – | 2.47E–4 | Minor allele is undertransmitted | |
| rs12785878a | 11p15 | DHCR7 | 3.3E–20 | Minor allele undertransmitted | 9.5E–01 | 9.7E–01 | 0.26 | 2.4E–05 | 0.47 | – | 0.037 | Minor allele is overtransmitted | |
SNP single nucleotide polymorphism, CHR chromosome, BP base pair distance, MAF minor allele frequency
One-sided p values
rs10741657 and rs12785878 were not genotyped in the Puerto Rico asthma cohort
Discussion
Vitamin D insufficiency has become a common problem for many individuals and is now linked to various diseases. This therefore has led to substantial interest in identifying determinants of vitamin D. As many of environmental determinants of vitamin D are known, identifying genetic determinants of vitamin D is likely to help in our overall understanding of the biologic processes which may increase or decrease vitamin D levels. In order to further elucidate the genetic determinants of vitamin D, we performed a genome-wide association study and replicated the top 50 findings in an independent population, then pursued replication of the top 4 SNPs in a third population.
We identified four SNPs associated with serum vitamin D in two independent cohorts of asthmatic children. These variants were in/near four genes that were identified in the CAMP population and had consistent nominal associations in the Costa Rican Cohort: (1) rs11002969 is located in an intronic region of ZCCHC24 on chromosome 10q22.3 and has previously been associated with right ventricular cardiomyopathy; (2) rs163221 is located in an intronic region of C18orf16/CHST9 (carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9); (3) rs1678849 is located in an intronic region of FUT6; and (4) rs4864976 is located on chromosome 4q12 and is not proximal to any relevant candidate genes. None of these SNPs or the nearby genes has a known clear involvement in biologic pathways related to vitamin D. We then attempted replication of these four SNPs in an additional population. However, these SNPs were not associated in a third cohort of asthmatic Puerto Rican children. One likely explanation is that the findings in the first two cohorts were due to chance alone. In addition, differences in genetic architecture between the Puerto Rico cohort and the previous cohorts is also possible, since Puerto Ricans have been reported to have a much higher proportion of African ancestry (18 %) (Tang et al. 2007) than Costa Ricans of Central Valley ancestry (3 %) (Celedon et al. 2002).
Three GWAS studies of vitamin D levels were recently published after we had completed our analyses. One study was conducted in Hispanic Americans and did not find any associations that were significant at a genome-wide level (Engelman et al. 2010). In the second study performed in five adult Caucasian cohorts, three SNPs from GC—rs2282679, rs7041, and rs1155563—were noted to be significantly associated with circulating vitamin D levels (Ahn et al. 2010). Additional analyses by the authors showed that the strongest signal came from rs2282679. Finally, a well-powered GWAS on insufficient vitamin D levels was published (Wang et al. 2010) with the strongest association being with rs2282679, similar to the previous study. We, therefore, examined the top SNPs from the report of Wang and colleagues, and examined these in our cohort. There was replication of the top findings from the recent vitamin D GWAS by Wang et al., with some notable discrepancies. SNP rs12785878 was not associated in CAMP, whereas it was associated in Costa Rica. The top association finding from the previous article, rs228269, had a very strong association in CAMP at baseline and year 4,and in Costa Rica in the correct direction. Finally, this SNP was also strongly associated with 25(OH)D levels in Puerto Rico. Therefore, our findings further substantiate all three of the initial associations to some degree, with the most consistent replication being for rs2282679, which was the strongest finding in the initial paper. We have confirmed this association, and the combined evidence from these three asthma cohorts was significant at a genome-wide level.
In the two recent studies of twins, vitamin D was noted to have high heritability (Karohl et al. 2010; Orton et al. 2008). However, we are not aware of any studies that have looked at the correlation between vitamin D levels over time in the same individuals. Since vitamin D levels are known to vary over time, a strength of our analysis is that we have measurements at two time-points in our initial population. As seen from the correlation analysis, the levels at the end of the 4-year time point were lower than the baseline, and the correlation between the two values were very modest, at best. The correlation coefficient value that we obtained, was similar to the correlation between dizygotic twins that have previously been reported (Karohl et al. 2010; Orton et al. 2008). Restricting the analysis to measurements obtained during the same season, 4 years apart, did not change the correlation. Thus, assessing the two measurements separately provides some robustness to the results obtained from this initial cohort, on which the choice of SNPs to be genotyped in the second population was based. In addition, since participants in these two populations arose from eight centers across North America and from Costa Rica, it provided for a reasonably wide range of latitudes. Despite this fact, vitamin D levels from CAMP participants were similar to levels from participants from Costa Rica, reinforcing the notion that behaviors are likely more important that simple latitude in determining the amount of sun exposure. With respect to the Puerto Rican cohort, an additional potential determinant of vitamin D levels is the higher African ancestry of the population.
Despite the given strengths of this study, there are certain limitations that must be discussed. First, the sample size of our initial and replication populations are underpowered given that we are testing over 500,000 SNPs. Secondly, in the analysis, we required that all genetic associations were nominally significant in the four analyses including both family-based and population-based analyses at baseline and year 4. Our intention of doing this was to identify genetic associations that are robust, and therefore limit the number of false positive findings. Although this approach is effective in limiting false positive associations, it is likely that we missed true genetic associations. This is likely what we observed with the SNP, rs2282679, previously reported by Wang et al. In our analysis, this SNP had a very strong association in both the baseline and year 4 population-based analyses; however, it did not reach nominal significance in one of the family-based association analyses and therefore was not selected to be replicated in the top 50 SNPs. A better strategy would be to use a more liberal threshold for the family-based analysis, as these analyses are underpowered. Third, these findings are investigating genetic associations with vitamin D in general; however, the probands used in this analysis were ascertained for asthma. This can complicate the interpretation of some of the association, as there is evidence implicating vitamin D in the development of asthma. Significant associations between polymorphisms in the VDR gene with asthma have been reported in several genetic association studies (Poon et al. 2004; Raby et al. 2004), but has not been consistently replicated (Vollmert et al. 2004; Wjst 2005). Since research suggests that genetic variants for vitamin D may also influence asthma affection status, the association results that we have observed must be interpreted with caution, as these associations may be specific to asthmatic individuals. In particular, children with asthma may have different behaviors with regard to sun exposure than a sample from the general population.
In conclusion, we have performed a GWAS analysis on vitamin D in three independent samples of asthmatic individuals. We identified four SNPs that had consistent genetic associations in the first two cohorts, but not the third cohort. In addition, we performed analyses to verify the top three SNPs that were identified in the previously reported GWAS of vitamin D, with replication of the findings in the top SNP (rs2282679) from that analysis; there was weak evidence for replication of the second SNP (rs10741657). Therefore, we have confirmed the association of the GC SNP from prior GWAS studies. Other SNPs may be associated with 25(OH)D levels in certain populations.
Supplementary Material
Acknowledgments
The authors acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data. All work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women’s Hospital under appropriate CAMP policies and human subject’s protections. This work was funded by R21 HL089842 from the National Heart, Lung and Blood Institute, National Institutes of Health. The CAMP Genetics Ancillary Study is supported by U01 HL075419, U01 HL65899, P01 HL083069, R01 HL086601, and T32 HL07427 from the National Heart, Lung and Blood Institute, National Institutes of Health.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-012-1185-z) contains supplementary material, which is available to authorized users.
Contributor Information
Jessica Lasky-Su, Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital, 181 Longwood Avenue, Boston, MA 02115, USA; Harvard Medical School, Boston, MA, USA.
Nancy Lange, Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital, 181 Longwood Avenue, Boston, MA 02115, USA; Harvard Medical School, Boston, MA, USA; Division of Pulmonary and Critical Care Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA.
John M. Brehm, Division of Pediatric Pulmonary Medicine, Allergy and Immunology, Children’s Hospital of Pittsburgh of UPMC, Pittsburgh, PA, USA
Amy Damask, Novartis Institutes for Biomedical Research, Cambridge, MA, USA.
Manuel Soto-Quiros, Division of Pediatric Pulmonology, Hospital Nacional de Niños, San José, Costa Rica.
Lydiana Avila, Division of Pediatric Pulmonology, Hospital Nacional de Niños, San José, Costa Rica.
Juan C. Celedón, Division of Pediatric Pulmonary Medicine, Allergy and Immunology, Children’s Hospital of Pittsburgh of UPMC, Pittsburgh, PA, USA
Glorisa Canino, Behavioral Sciences Institute, University of Puerto Rico, San Juan, PR, USA.
Michelle M. Cloutier, University of Connecticut Health Center, Farmington, CT, USA
Bruce W. Hollis, Darby Children’s Research Institute, Medical University of South Carolina, Charleston, SC, USA
Scott T. Weiss, Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital, 181 Longwood Avenue, Boston, MA 02115, USA Harvard Medical School, Boston, MA, USA; Department of Medicine, Center for Genomic Medicine, Brigham and Women’s Hospital, Boston, MA, USA.
Augusto A. Litonjua, Email: augusto.litonjua@channing.harvard.edu, Channing Laboratory, Department of Medicine, Brigham and Women’s Hospital, 181 Longwood Avenue, Boston, MA 02115, USA; Harvard Medical School, Boston, MA, USA; Division of Pulmonary and Critical Care Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA.
References
- The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials. 1999a;20:91–120. [PubMed] [Google Scholar]
- Vitamin D supplement in early childhood and risk for Type I (insulin-dependent) diabetes mellitus. The EURODIAB Sub-study 2 Study Group. Diabetologia. 42:51–54. doi: 10.1007/s001250051112. [DOI] [PubMed] [Google Scholar]
- Agborsangaya C, Toriola AT, Grankvist K, Surcel HM, Holl K, Parkkila S, Tuohimaa P, Lukanova A, Lehtinen M. The effects of storage time and sampling season on the stability of serum 25-hydroxy vitamin D and androstenedione. Nutr Cancer. 2010;62:51–57. doi: 10.1080/01635580903191460. [DOI] [PubMed] [Google Scholar]
- Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs KB, Li Q, Weinstein SJ, Purdue M, Virtamo J, Horst R, Wheeler W, Chanock S, Hunter DJ, Hayes RB, Kraft P, Albanes D. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19:2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird HR, Canino GJ, Davies M, Duarte CS, Febo V, Ramirez R, Hoven C, Wicks J, Musa G, Loeber R. A study of disruptive behavior disorders in Puerto Rican youth: I. Background, design, and survey methods. J Am Acad Child Adolesc Psychiatry. 2006;45:1032–1041. doi: 10.1097/01.chi.0000227878.58027.3d. [DOI] [PubMed] [Google Scholar]
- Brehm JM, Celedon JC, Soto-Quiros ME, Avila L, Hunninghake GM, Forno E, Laskey D, Sylvia JS, Hollis BW, Weiss ST, Litonjua AA. Serum vitamin D levels and markers of severity of childhood asthma in Costa Rica. Am J Respir Crit Care Med. 2009;179:765–771. doi: 10.1164/rccm.200808-1361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm JM, Schuemann BS, Fuhlbrigge AL, Hollis BW, Strunk RC, Zeiger RS, Weiss ST, Litonjua AA. Serum vitamin D levels and severe asthma exacerbations in the childhood asthma management program study. J Allergy Clin Immun. 2010;126(1):52–58. doi: 10.1016/j.jaci.2010.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon JC, Soto-Quiros ME, Palmer LJ, Senter J, Mosley J, Silverman EK, Weiss ST. Lack of association between a polymorphism in the interleukin-13 gene and total serum immunoglobulin E level among nuclear families in Costa Rica. Clin Exp Allergy. 2002;32:387–390. doi: 10.1046/j.1365-2222.2002.01348.x. [DOI] [PubMed] [Google Scholar]
- Engelman CD, Fingerlin TE, Langefeld CD, Hicks PJ, Rich SS, Wagenknecht LE, Bowden DW, Norris JM. Genetic and environmental determinants of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D levels in Hispanic and African Americans. J Clin Endocrinol Metab. 2008;93:3381–3388. doi: 10.1210/jc.2007-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman CD, Meyers KJ, Ziegler JT, Taylor KD, Palmer ND, Haffner SM, Fingerlin TE, Wagenknecht LE, Rotter JI, Bowden DW, Langefeld CD, Norris JM. Genome-wide association study of vitamin D concentrations in Hispanic Americans: the IRAS family study. J Steroid Biochem Mol Biol. 2010;122:186–192. doi: 10.1016/j.jsbmb.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla MA, Spesny M, Reus VI, Gallegos A, Meza L, Molina J, Sandkuijl LA, Fournier E, Leon PE, Smith LB, Freimer NB. Use of linkage disequilibrium approaches to map genes for bipolar disorder in the Costa Rican population. Am J Med Genet. 1996;67:244–253. doi: 10.1002/(SICI)1096-8628(19960531)67:3<244::AID-AJMG2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Giovannucci E. The epidemiology of vitamin D and cancer incidence and mortality: a review (United States) Cancer Causes Control. 2005;16:83–95. doi: 10.1007/s10552-004-1661-4. [DOI] [PubMed] [Google Scholar]
- Giovannucci E. The epidemiology of vitamin D and colorectal cancer: recent findings. Curr Opin Gastroenterol. 2006;22:24–29. doi: 10.1097/01.mog.0000196150.36701.c2. [DOI] [PubMed] [Google Scholar]
- Gorham ED, Garland CF, Garland FC, Grant WB, Mohr SB, Lipkin M, Newmark HL, Giovannucci E, Wei M, Holick MF. Vitamin D and prevention of colorectal cancer. J Steroid Biochem Mol Biol. 2005;97:179–194. doi: 10.1016/j.jsbmb.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Grant WB. Epidemiology of disease risks in relation to vitamin D insufficiency. Prog Biophys Mol Biol. 2006;92:65–79. doi: 10.1016/j.pbiomolbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Heaney RP. Long-latency deficiency disease: insights from calcium and vitamin D. Am J Clin Nutr. 2003;78:912–919. doi: 10.1093/ajcn/78.5.912. [DOI] [PubMed] [Google Scholar]
- Heaney RP. Functional indices of vitamin D status and ramifications of vitamin D deficiency. Am J Clin Nutr. 2004;80:1706S–1709S. doi: 10.1093/ajcn/80.6.1706S. [DOI] [PubMed] [Google Scholar]
- Holick MF. 25-OH-vitamin D assays. J Clin Endocrinol Metab. 2005;90:3128–3129. doi: 10.1210/jc.2005-0162. [DOI] [PubMed] [Google Scholar]
- Holick MF. High prevalence of vitamin D inadequacy and implications for health. Mayo Clin Proc. 2006;81:353–373. doi: 10.4065/81.3.353. [DOI] [PubMed] [Google Scholar]
- Hollis BW. Circulating 25-hydroxyvitamin D levels indicative of vitamin D sufficiency: implications for establishing a new effective dietary intake recommendation for vitamin D. J Nutr. 2005;135:317–322. doi: 10.1093/jn/135.2.317. [DOI] [PubMed] [Google Scholar]
- Hollis BW, Napoli JL. Improved radioimmunoassay for vitamin D and its use in assessing vitamin D status. Clin Chem. 1985;31:1815–1819. [PubMed] [Google Scholar]
- Hollis BW, Wagner CL. Normal serum vitamin D levels. N Engl J Med. 2005;352:515–516. doi: 10.1056/NEJM200502033520521. [DOI] [PubMed] [Google Scholar]
- Hollis BW, Kamerud JQ, Selvaag SR, Lorenz JD, Napoli JL. Determination of vitamin D status by radioimmunoassay with an 125I–labeled tracer. Clin Chem. 1993;39:529–533. [PubMed] [Google Scholar]
- Hollis BW, Wagner CL, Drezner MK, Binkley NC. Circulating vitamin D(3) and 25-hydroxyvitamin D in humans: an important tool to define adequate nutritional vitamin D status. J Steroid Biochem Mol Biol. 2007;103(3–5):631–634. doi: 10.1016/j.jsbmb.2006.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunninghake GM, Soto-Quiros ME, Avila L, Su J, Murphy A, Demeo DL, Ly NP, Liang C, Sylvia JS, Klanderman BJ, Lange C, Raby BA, Silverman EK, Celedon JC. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol. 2007;120:84–90. doi: 10.1016/j.jaci.2007.04.032. [DOI] [PubMed] [Google Scholar]
- Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res. 2001;16:371–378. doi: 10.1359/jbmr.2001.16.2.371. [DOI] [PubMed] [Google Scholar]
- Hypponen E, Laara E, Reunanen A, Jarvelin MR, Virtanen SM. Intake of vitamin D and risk of type 1 diabetes: a birth-cohort study. Lancet. 2001;358:1500–1503. doi: 10.1016/S0140-6736(01)06580-1. [DOI] [PubMed] [Google Scholar]
- Karohl C, Su S, Kumari M, Tangpricha V, Veledar E, Vaccarino V, Raggi P. Heritability and seasonal variability of vitamin D concentrations in male twins. Am J Clin Nutr. 2010;92:1393–1398. doi: 10.3945/ajcn.2010.30176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberg-Allardt C. Vitamin D in foods and as supplements. Prog Biophys Mol Biol. 2006;92:33–38. doi: 10.1016/j.pbiomolbio.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Lee AB, Luca D, Klei L, Devlin B, Roeder K. Discovering genetic ancestry using spectral graph theory. Genet Epidemiol. 2010;34:51–59. doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litonjua AA, Weiss ST. Is vitamin D deficiency to blame for the asthma epidemic? J Allergy Clin Immunol. 2007;120:1031–1035. doi: 10.1016/j.jaci.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Merlino LA, Curtis J, Mikuls TR, Cerhan JR, Criswell LA, Saag KG. Vitamin D intake is inversely associated with rheumatoid arthritis: results from the Iowa Women’s Health Study. Arthritis Rheum. 2004;50:72–77. doi: 10.1002/art.11434. [DOI] [PubMed] [Google Scholar]
- Nesby-O’Dell S, Scanlon KS, Cogswell ME, Gillespie C, Hollis BW, Looker AC, Allen C, Doughertly C, Gunter EW, Bowman BA. Hypovitaminosis D prevalence and determinants among African American and white women of reproductive age: third National Health and Nutrition Examination Survey, 1988–1994. Am J Clin Nutr. 2002;76:187–192. doi: 10.1093/ajcn/76.1.187. [DOI] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl. 2002;56–8:60–61. [PubMed] [Google Scholar]
- Orton SM, Morris AP, Herrera BM, Ramagopalan SV, Lincoln MR, Chao MJ, Vieth R, Sadovnick AD, Ebers GC. Evidence for genetic regulation of vitamin D status in twins with multiple sclerosis. Am J Clin Nutr. 2008;88:441–447. doi: 10.1093/ajcn/88.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon AH, Laprise C, Lemire M, Montpetit A, Sinnett D, Schurr E, Hudson TJ. Association of vitamin D receptor genetic variants with susceptibility to asthma and atopy. Am J Respir Crit Care Med. 2004;170:967–973. doi: 10.1164/rccm.200403-412OC. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raby BA, Lazarus R, Silverman EK, Lake S, Lange C, Wjst M, Weiss ST. Association of vitamin D receptor gene polymorphisms with childhood and adult asthma. Am J Respir Crit Care Med. 2004;170:1057–1065. doi: 10.1164/rccm.200404-447OC. [DOI] [PubMed] [Google Scholar]
- Ramos-Lopez E, Bruck P, Jansen T, Herwig J, Badenhoop K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab Res Rev. 2007;23:631–636. doi: 10.1002/dmrr.719. [DOI] [PubMed] [Google Scholar]
- Sahota H, Barnett H, Lesosky M, Raboud JM, Vieth R, Knight JA. Association of vitamin D related information from a telephone interview with 25-hydroxyvitamin D. Cancer Epidemiol Biomarkers Prev. 2008;17:232–238. doi: 10.1158/1055-9965.EPI-07-0632. [DOI] [PubMed] [Google Scholar]
- Shea MK, Benjamin EJ, Dupuis J, Massaro JM, Jacques PF, D’Agostino RBSr, Ordovas JM, O’Donnell CJ, Dawson-Hughes B, Vasan RS, Booth SL. Genetic and non-genetic correlates of vitamins K and D. Eur J Clin Nutr. 2009;63:458–464. doi: 10.1038/sj.ejcn.1602959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taback SP, Simons FE. Anaphylaxis and vitamin D: a role for the sunshine hormone? J Allergy Clin Immunol. 2007;120:128–130. doi: 10.1016/j.jaci.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Tang H, Choudhry S, Mei R, Morgan M, Rodriguez-Cintron W, Burchard EG, Risch NJ. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am J Hum Genet. 2007;81:626–633. doi: 10.1086/520769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Mei IA, Blizzard L, Ponsonby AL, Dwyer T. Validity and reliability of adult recall of past sun exposure in a case-control study of multiple sclerosis. Cancer Epidemiol Biomarkers Prev. 2006;15:1538–1544. doi: 10.1158/1055-9965.EPI-05-0969. [DOI] [PubMed] [Google Scholar]
- Vollmert C, Illig T, Altmuller J, Klugbauer S, Loesgen S, Dumitrescu L, Wjst M. Single nucleotide polymorphism screening and association analysis-exclusion of integrin beta 7 and vitamin D receptor (chromosome 12q) as candidate genes for asthma. Clin Exp Allergy. 2004;34:1841–1850. doi: 10.1111/j.1365-2222.2004.02047.x. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, Cooper JD, O’Reilly PF, Houston DK, Glazer NL, Vandenput L, Peacock M, Shi J, Rivadeneira F, McCarthy MI, Anneli P, de Boer IH, Mangino M, Kato B, Smyth DJ, Booth SL, Jacques PF, Burke GL, Goodarzi M, Cheung CL, Wolf M, Rice K, Goltzman D, Hidiroglou N, Ladouceur M, Wareham NJ, Hocking LJ, Hart D, Arden NK, Cooper C, Malik S, Fraser WD, Hartikainen AL, Zhai G, Macdonald HM, Forouhi NG, Loos RJ, Reid DM, Hakim A, Dennison E, Liu Y, Power C, Stevens HE, Jaana L, Vasan RS, Soranzo N, Bojunga J, Psaty BM, Lorentzon M, Foroud T, Harris TB, Hofman A, Jansson JO, Cauley JA, Uitterlinden AG, Gibson Q, Jarvelin MR, Karasik D, Siscovick DS, Econs MJ, Kritchevsky SB, Florez JC, Todd JA, Dupuis J, Hypponen E, Spector TD. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AR. Who, what, where and when-influences on cutaneous vitamin D synthesis. Prog Biophys Mol Biol. 2006;92:17–25. doi: 10.1016/j.pbiomolbio.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Wjst M. Variants in the vitamin D receptor gene and asthma. BMC Genet. 2005;6:2. doi: 10.1186/1471-2156-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.