Abstract
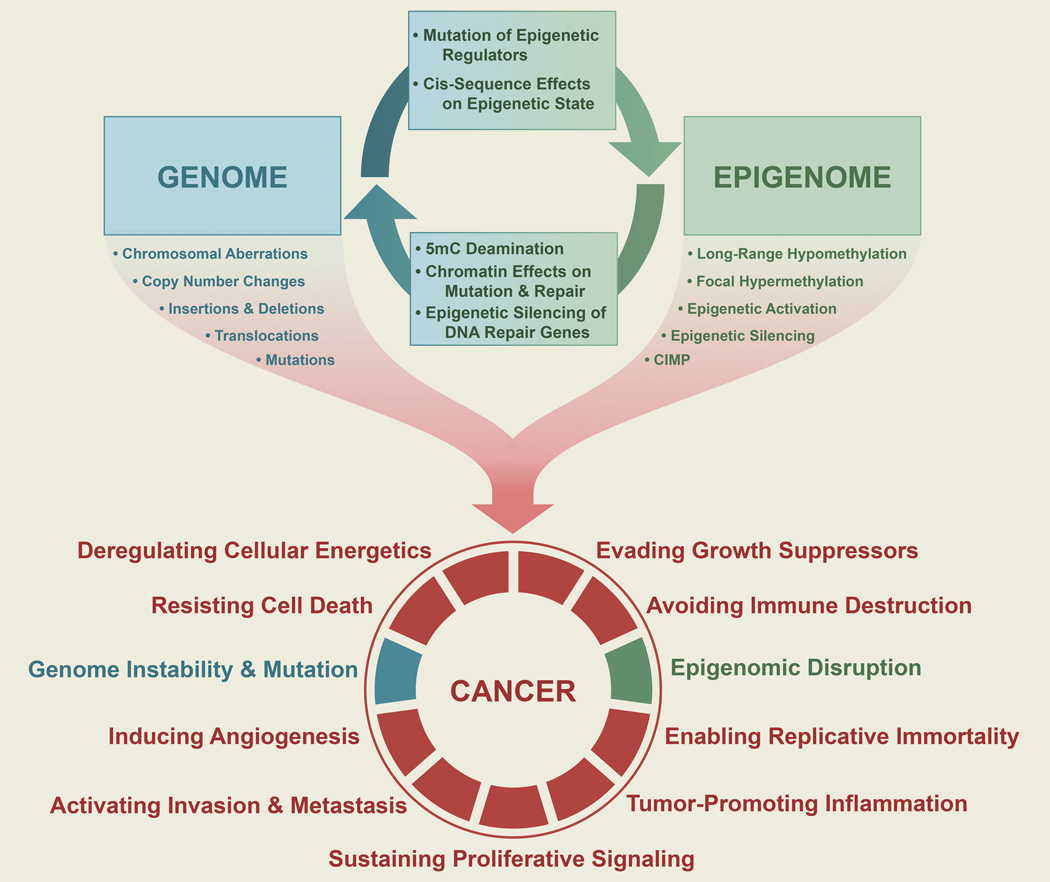
Cancer arises as a consequence of cumulative disruptions to cellular growth control, with Darwinian selection for those heritable changes which provide the greatest clonal advantage. These traits can be acquired and stably maintained by either genetic or epigenetic means. Here we explore the ways in which alterations in the genome and epigenome influence each other and cooperate to promote oncogenic transformation. Disruption of epigenomic control is pervasive in malignancy, and can be classified as an enabling characteristic of cancer cells, akin to genome instability and mutation.
Introduction
Cancer develops through successive disruptions to the controls of cellular proliferation, immortality, angiogenesis, cell death, invasion and metastasis. This evolutionary process requires new malignant traits to be stably encoded so that oncogenic events can accumulate in clonal lineages. Genetic mechanisms of mutation, copy number alteration, insertion, deletion and recombination are particularly well suited as vehicles of persistent phenotypic change. For this reason, cancer has long been viewed as a disease based principally on genetics. Nevertheless, genetic events occur at low frequency, and are thus not a particularly efficient means for malignant transformation. Some cancer cells overcome this bottleneck by acquiring DNA repair defects, thus boosting the mutation rate. Mechanisms of epigenetic control offer an alternative path to acquiring stable oncogenic traits. Epigenetic states are flexible yet persist through multiple cell divisions, and exert powerful effects on cellular phenotype. Although cancer cells have long been known to undergo epigenetic changes, genome-scale genomic and epigenomic analyses have only recently revealed the widespread occurrence of mutations in epigenetic regulators and the breadth of alterations to the epigenome in cancer cells (You and Jones, 2012). It is now clear that genetic and epigenetic mechanisms influence each other, and work cooperatively to enable the acquisition of the hallmarks of cancer (Hanahan and Weinberg, 2011).
Shaping the Epigenome
Epigenetic mechanisms allow genetically identical cells to achieve diverse stable phenotypes by controlling the transcriptional availability of various parts of the genome through differential chromatin marking and packaging. These embellishments include direct DNA modifications, primarily CpG cytosine-5 methylation (Jones, 2012), but also hydroxylation, formylation and carboxylation (Ito et al., 2011), as well as nucleosome occupancy and positioning (Gaffney et al., 2012; Valouev et al., 2011), histone variants, and dozens of different histone modifications (Tan et al., 2011b), interacting proteins (Ram et al., 2011), and non-coding RNAs (Fabbri and Calin, 2010; Lee, 2012). These epigenetic marks do not act in isolation, but form a network of mutually reinforcing or counteracting signals. Genome-scale projects charting the human epigenome are rapidly extending our understanding of epigenetic marks and how they interact (Adams et al., 2012; Encode-Project-Consortium et al., 2012; Ernst et al., 2011).
A key facet of epigenetics is that these marks can be stably maintained, yet adapt to changing developmental or environmental needs. This delicate task is accomplished by initiators, such as long non-coding RNAs, writers, which establish the epigenetic marks, readers, which interpret the epigenetic marks, erasers, which remove the epigenetic marks, remodelers, which can reposition nucleosomes, and insulators, which form boundaries between epigenetic domains. Epigenetic writers are directed to their target locations by sequence context, existing chromatin marks and bound proteins, non-coding RNAs, and/or nuclear architecture. Those marks are then recognized by reader proteins to convey information for various cellular functions. The establishment, maintenance, and change of epigenetic marks are intricately regulated, with crosstalk among the marks and writers to help guide changes to the epigenetic landscape.
DNA Methylation
De novo methylation of DNA is catalyzed by the enzymes DNMT3A and DNMT3B, and is then maintained by the major DNA methyltransferase DNMT1, with participation from DNMT3A and DNMT3B (Jones and Liang, 2009). DNA methylation patterns are guided in part by primary DNA sequence context (Cedar and Bergman, 2012; Lienert et al., 2011) and influenced by germline variation (Gertz et al., 2011; Kerkel et al., 2008). Much of the mammalian genome consists of vast oceans of DNA sequence containing sparsely distributed, but heavily methylated CpG dinucleotides, punctuated by short regions with unmethylated CpGs occurring at higher density, forming distinct islands in the genome (Bird et al., 1985). These CpG islands (CGIs) are protected from DNA methylation in part by GC strand asymmetry and accompanying R-Loop formation (Ginno et al., 2012) and possibly also by active demethylation mediated by the TET family members (Williams et al., 2012). The unmethylated state of CpG islands in the germline, along with biased gene conversion, helps to preserve CpG islands, despite ongoing attrition of methylated CpG dinucleotides by cytosine deamination throughout most of the genome (Cohen et al., 2011). Transition zones between CpG islands and CpG oceans are called CpG shores, and display more tissue-specific variation in DNA methylation (Irizarry et al., 2009). CpG islands span the transcription start sites of about half of the genes in the human genome, largely representing genes that are either actively expressed or poised for transcription (Figure 1).
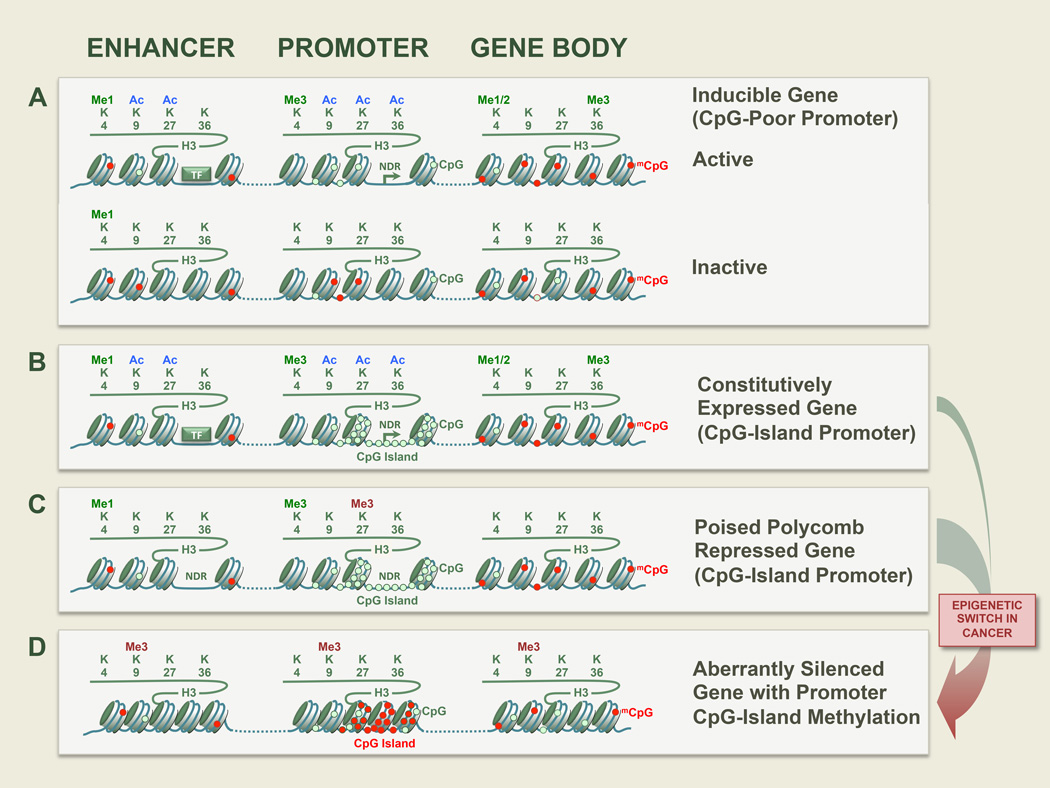
Figure 1. Representative Epigenetic States.
Examples of representative epigenetic states are shown for several typical categories of genes and in different cellular contexts. A. CpG-poor promoters are often tissue-specific and/or reside in inducible genes which can be readily turned on or off. Transcription factor (TF) binding at regulatory elements and the promoter initiates nuclear depleted regions (NDR). B. Many genes with CpG island promoters are constitutively expressed housekeeping genes. C. Some genes with CpG island promoters, such as transcription factor master regulators of differentiation and development are repressed by the Polycomb complexes in stem cells and kept in a bivalent state with both active and repressive marks. D. Polycomb targets in stem cells are predisposed to cancer-specific promoter hypermethylation.
Methylated DNA is recognized by methyl-CpG binding domains (MBD) or C2H2 zinc fingers. The MBD-containing DNA methylation readers include MBD1, MBD2, MBD4 and MeCP2, whereas Kaiso (ZBTB33), ZBTB4 and ZBTB38 proteins use zinc fingers to bind methylated DNA. MBDs and Kaiso are believed to participate in DNA methylation mediated transcriptional repression of tumor suppressor genes with promoter DNA methylation.
Histone Modifications
Post-translational modifications of histones are coordinated by counteracting histone-methyltransferases (HMTs) and demethylases (e.g. KDMs), histone acetyltransferases (HATs) and deacetylases (HDACs), and writers and erasers of phosphorylation, as well as many other modifications (Chi et al., 2010; Tan et al., 2011b). These histone modifiers generally act in complexes, such as the repressive Polycomb (PcG) and activating Trithorax (TrxG) group complexes, which counterbalance each other in the regulation of genes important for development, but which have also been implicated in cancer (Mills, 2010). Polycomb repressive complexes (PRCs) are guided to their targets in part by intrinsic signals in the genome sequence (Ku et al., 2008; Tanay et al., 2007). The histone H3K27me3 mark deposited by PRC2 provides docking sites for PRC1, whose enzymatic core unit RING1B monoubiquitinylates histone H2A at lysine 119 (H2AK119ub1) thereby blocking RNA polymerase II elongation. The Trithorax group complex, containing MLL, which lays down the H3K4 methylation mark, counteracts Polycomb function. The transcription factors encoding master regulators of differentiation and development are targeted by PRC2 in embryonic stem cells and held in a bivalent chromatin state poised for transcription, with both the activating H3K4me3 and the repressive H3K27me3 (Bernstein et al., 2006) (Figure 1). During differention the Trithorax demethylase, KDM6A/UTX removes the repressive H3K27me3 mark, allowing transcription elongation to proceed for genes required in that particular lineage, while genes not required in that cell type may acquire H3K9me3, which is recognized by readers like HP1 to reinforce a repressive state. Other histone marks have various readers with binding motifs including bromodomain, PHD domain, chromodomain, and tudor domain (Musselman et al., 2012) (Figure 2). Trithorax and Polycomb complexes recruit HATs and HDACs, respectively, to counteract each other, and the establishment of histone acetylation can block Polycomb binding (Mills, 2010).
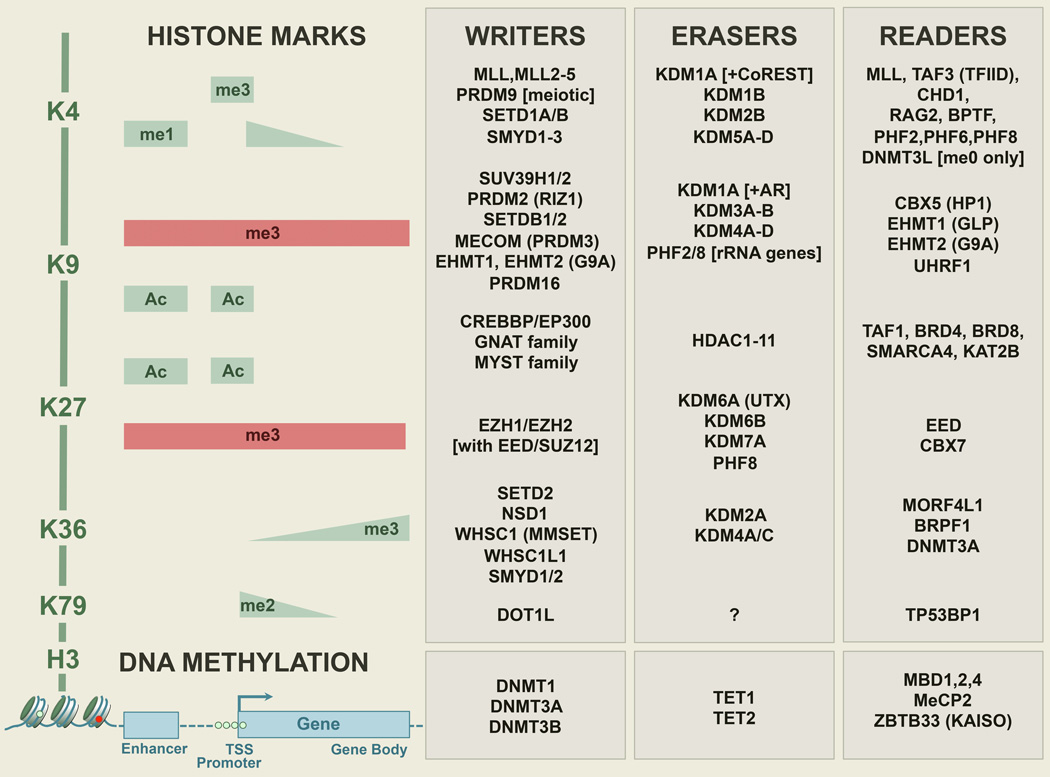
Figure 2. Histone H3 Lysine Writers, Erasers and Readers.
Although many other important histone modifications also occur, only major histone H3 lysine modifications (Ac: Acetylation; me1: monomethylation; me3: trimethylation) with well-defined functions are shown above a representative gene. The distribution of the marks are shown as colored bars and wedges to indicate approximate abundance. Repressive marks are shown in red, and active marks in blue. Epigenetic regulators are listed to the right of each mark. Acetylation across different lysines share writers and erasers, while methylation usually has dedicated enzymes. Readers (which can also be writers and erasers themselves) recognize different chromatin states and propagate the signal in various ways, including self reinforcement or cross talk, transcriptional activation or repression, or DNA repair. Crosstalk can also occur between histone modification and DNA methylation, since DNMT3A, DNMT3L, UHRF1 all contain reader domains for chromatin states.
Histone Variants
Histone variants provide an additional layer of regulation. The main histone genes have multiple copies in the genome and are expressed during S-phase. Single-copy variants are also expressed at other phases of the cell cycle and have distinct functions and/or locations. H2A has the largest number of variants, including H2A.Z, MacroH2A, H2A–Bbd, H2AvD, and H2A.X (Kamakaka and Biggins, 2005). The H3 variants include H3.3 and centromeric H3 (CenH3, or CENP-A), as well as a mammalian testis-specific histone H3 variant called H3.4. Nucleosomes containing H3.3 and H2A.Z are located at dynamic regions requiring nucleosome mobility and exchange, such as at actively expressed gene promoters (Jin et al., 2009). Wide presence of H2A.Z in embryonic stem cells (Zhu et al., 2013) suggests prevalent chromatin exchange, consistent with the emerging idea that the genome of ESC is generally kept highly accessible. During differentiation H2A.Z quickly redistributes. The mechanisms of recruitment have not been fully delineated, but various chromatin remodeler complexes and/or chaperones have been shown to be involved. For example, SRCAP is involved in H2A.Z loading into promoter/TSS, while H3.3 is loaded to telomeric/pericentric regions by the ATRX/DAXX complex and promoter/TSS by HIRA (Boyarchuk et al., 2011).
Nucleosome Positioning and Remodeling
The positioning of nucleosomes displays a weak 10-bp periodicity associated with minor sequence composition fluctuations in phase with the DNA helical repeat. Some nucleosomes are more consistently positioned in phased arrays anchored by sequence-specific binding of proteins such as CTCF or adjacent to nucleosome-free regions at transcription start sites (Gaffney et al., 2012; Valouev et al., 2011). CpG islands have been associated with transcription-independent nucleosome-depletion at mammalian promoters (Fenouil et al., 2012). ATP-dependent chromosome remodeling complexes are responsible for sliding of the nucleosomes, as well as insertion and ejection of histone octamers, processes important for transcriptional repression and activation, and other important cellular functions such as DNA replication and repair. The remodeling complexes can be divided into four families: SWI/SNF, CHD (chromodomain and helicase-like domain), ISWI, and INO80 (including SWR1, or SRCAP in mammals).
Insulators
The CCCTC-binding factor CTCF (and its paralogue CTCFL/BORIS, expressed in the germline) are the only insulator proteins that have been identified so far in vertebrates. CTCF has a strong binding motif and there is extensive overlap of the occupied CTCF-binding sites among different cell types (Kim et al., 2007). CTCF binds to enhancer blocking elements to prevent enhancer interactions with unintended promoters (‘enhancer blocking insulator’), and also demarcates active and repressive chromatin domains (‘barrier insulator’).
Nuclear Architecture
The genome can be compartmentalized based on nuclear architecture and associated genomic features into mostly heterochromatic late-replicating regions attached to the nuclear lamina at the nuclear periphery, and more gene-rich early-replicating regions closer to the nuclear interior (Encode-Project-Consortium et al., 2012; Meuleman et al., 2012). Lamina-associated sequences (LASs) enriched for a GAGA motif are bound by transcriptional repressors, and appear to contribute to the establishment of lamina associated domains (LADs) in the mammalian genome (Zullo et al., 2012).
Maintaining the Epigenetic State
The persistence of epigenetic traits in a growing tumor requires that the epigenome be faithfully copied during cell division. The chromatin structure is dismantled for passage of the replication fork (RF). Newly synthesized DNA and histone octamers are then assembled at the RF by Chromatin assembly factor I (CAF1), tethered to the RF by PCNA. Similarly, the dedicated maintenance DNA methyltransferase DNMT1, the euchromatic H3K9 methyltransferase G9a, among other epigenetic maintainers, are loaded to RFs and copy the epigenetic marks. The Trithorax and Polycomb complexes are recruited prior to replication and distributed evenly to the mother and daughter strands at the RF, and restore the correct marks on the daughter molecules during G1 (Petruk et al., 2012). The histone marks are self-reinforcing and self-propagating, as PcG, SUV39H1/2, SETDB1 and TrxG all bind to the marks that they are responsible for catalyzing, via an intrinsic reader domain or by interacting with a reader protein, thus helping to maintain the epigenetic state. Nucleosomes containing methylated DNA also stabilize DNMT3A/3B, which is a self-reinforcing mechanism for DNA methylation maintenance (Sharma et al., 2011).
Disruption of Epigenetic Control in Cancer
Most studies of cancer epigenetics have focused on DNA methylation, as the epigenetic mark that most easily survives various forms of sample processing, including DNA extraction, and even formalin fixation and paraffin embedding (Laird, 2010). However, other epigenetic marks also undergo broad changes, including long non-coding RNAs and miRNAs (Baer et al., 2013; Baylin and Jones, 2011; Dawson and Kouzarides, 2012; Sandoval and Esteller, 2012), and loss of K16 acetylation and K20 trimethylation at histone H4 (Fraga et al., 2005; Hon et al., 2012; Kondo et al., 2008; Seligson et al., 2005; Yamazaki et al., 2013). Loss of 5-methylcytosine in cancer cells was discussed more than three decades ago (Ehrlich and Wang, 1981), with global DNA hypomethylation reported in cancer cell lines (Diala and Hoffman, 1982; Ehrlich et al., 1982) and reduced levels of DNA methylation found at selected genes in primary human tumors compared to normal tissues (Feinberg and Vogelstein, 1983). The widespread loss of DNA methylation contrasted starkly with the subsequent finding of hypermethylation of CpG islands in cancer (Baylin et al., 1986), including of promoter CpG islands of tumor-suppressor genes (Jones and Baylin, 2002). These seemingly contradictory findings have been widely reported for many types of cancer (Baylin and Jones, 2011).
The causal relevance of epigenetic changes in cancer was initially questioned but this concern has now largely been laid to rest. First, many known tumor-suppressor genes have been shown to be silenced by promoter CpG island hypermethylation (Jones and Baylin, 2002). Importantly, the finding that these silencing events are mutually exclusive with structural or mutational inactivation of the same gene, such as the case for BRCA1 in ovarian cancer (TCGA, 2011) and for CDKN2A in squamous cell lung cancer (TCGA, 2012a), reinforces the concept that epigenetic silencing can serve as an alternative mechanism in Knudson's two-hit hypothesis (Jones and Laird, 1999). Second, mouse models of cancer have been shown to require epigenetic writers and readers for tumor development (Laird et al., 1995; Prokhortchouk et al., 2006; Sansom et al., 2003). Third, some DNA methylation changes appear to be essential for cancer cell survival, suggesting an acquired addiction to epigenetic alterations (De Carvalho et al., 2012). Finally, a plethora of significantly mutated epigenetic regulators have now been reported for many types of human cancer, as discussed further below.
Long-Range Coordinated Disruptions and Nuclear Architecture
The genome of undifferentiated embryonic stem cells is uniformly heavily methylated across CpG oceans, punctuated by unmethylated CpG islands. As stem cells differentiate and proliferate, the late-replicating lamin-associated domains (LADs) undergo progressive loss of DNA methylation within CpG oceans, and the LADs become recognizable as long partially methylated domains (PMDs), which become even more strikingly demarcated as hypomethylated domains in cancer cells (Berman et al., 2012; Hansen et al., 2011; Hon et al., 2012; Lister et al., 2009). This loss of DNA methylation is associated with an increase of repressive chromatin with large organized chromatin-lysine-(K) modification regions (LOCKs) (Hansen et al., 2011; Hon et al., 2012; Lister et al., 2009). CpG island hypermethylation is enriched in the hypomethylated domains, suggesting that these two events may be mechanistically linked, but confined to distinct areas of the genome near the nuclear periphery (Berman et al., 2012). These long regions of DNA hypomethylation and repressive chromatin are consistent with prior reports of coordinated epigenetic silencing events located across megabase distances, a phenomenon termed Long-Range Epigenetic Silencing (LRES) (Clark, 2007; Coolen et al., 2010).
It is noteworthy that the euchromatic part of the genome associated with the interior of the nucleus is generally much more epigenetically stable during cell differentiation, aging and malignant transformation. However, loss of the DNA methyltransferase Dnmt3a can promote tumor progression with uniform hypomethylation across the genome, and moderate deregulation of genes in euchromatic regions (Raddatz et al., 2012).
Disruption of Differentiation and Development
Differences between cell types are guided by the expression of tissue-specific transcription factors and consolidation of associated epigenetic states. Therefore, the epigenome of a cancer cell is determined in part by the cell of origin for that cancer and includes passenger hypermethylation events at genes not required in that particular lineage (Sproul et al., 2012). Epithelial to mesenchymal transition (EMT) of cancer cells is partly under reversible epigenetic control (Craene and Berx, 2012). For example, primary breast tumors display heterogeneous and unstable silencing of the CDH1 (E-cadherin) gene, which facilitates the plasticity required during extravasation, metastasis and establishment of a solid tumor at the metastatic site (Graff et al., 2000).
It has long been debated whether cancer cells arise by dedifferentiation or instead originate from stem cells or early progenitors by a differentiation block. Polycomb repressors mark genes in stem cells encoding master regulators of differentiation and development, poised to either be turned on to coordinate differentiation of a lineage, or to be fully repressed if it is not needed in that particular lineage (Bernstein et al., 2006). These genes occupied by Polycomb repressors in stem cells are particularly prone to acquiring CpG island hypermethylation during cell proliferation, aging and particularly malignant transformation (Ohm et al., 2007; Schlesinger et al., 2007; Teschendorff et al., 2010; Widschwendter et al., 2007) (Figure 1). Although the genes affected by this process are primarily those not required or expressed in that particular cell lineage, cancer cells do also show evidence of silencing of genes essential for differentiation of their cell of origin (Berman et al., 2012; Easwaran et al., 2012; Gal-Yam et al., 2008; Mohn et al., 2008; Teschendorff et al., 2010). This predisposition of Polycomb target genes to aberrant permanent epigenetic silencing is consistent with a model in which stem cells slowly acquire irreversible silencing of poised master regulators required for successful differentiation. As a consequence, some stem cells lose their ability to properly differentiate while retaining their self-renewal capabilities, and become attractive candidates for malignant transformation by subsequent genetic and epigenetic events. One provocative implication of this model is that the first steps of oncogenesis may in some cases be an epigenetic defect affecting the differentiation capabilities of stem cells, as opposed to a gatekeeper mutation.
Hematopoietic cell lineages and their corresponding malignancies also offer insights into the role of epigenetics in differentiation and transformation. For example, the DNMT3A gene is commonly mutated in human cases of Acute Myeloid Leukemia (AML) (Ley et al., 2010; Yan et al., 2011), whereas loss of Dnmt3a in mice progressively impairs hematopoietic stem cell differentiation (Challen et al., 2012), suggesting that epigenetic perturbation can lead to differentiation block and subsequent malignant transformation.
CpG Island Methylator Phenotypes
Aberrant DNA methylation of promoter CpG islands in cancer was initially viewed as a spontaneous or stochastic event with selection for functionally relevant silencing events. However, the discovery of cases of colorectal cancer with an exceptionally high frequency of CpG island hypermethylation suggested a coordinated event, possibly attributable to an epigenetic control defect. This phenomenon was referred to as a "CpG Island Methylator Phenotype" (CIMP) (Toyota et al., 1999), analogous to the mutator phenotypes observed in mismatch repair deficient cancers. Although the existence of CIMP subsets of cancer was initially disputed (Yamashita et al., 2003), more recent genome-scale analyses have unambiguously documented distinct epigenetic subtypes for some types of cancer, such as colorectal cancer (Hinoue et al., 2012; TCGA, 2012b) and glioblastoma (Noushmehr et al., 2010), and not for others, such as serous ovarian cancer (TCGA, 2011). The most distinct examples of CIMP show exceptionally strong associations with other molecular or pathological features of the tumors, lending further validity to the biological relevance to this classification. For example, colorectal CIMP is very tightly associated with the V600E mutation of the BRAF oncogene (Weisenberger et al., 2006), while glioma CIMP (G-CIMP) is exceptionally tightly associated with mutation of the IDH1 gene (Noushmehr et al., 2010). In the case of G-CIMP, IDH1 mutation appears to be a causal contributor to the phenotype (Turcan et al., 2012), whereas BRAF mutation does not appear to be directly implicated in colorectal CIMP (Hinoue et al., 2009). The affected gene subsets,differ between colorectal CIMP and glioblastoma G-CIMP, and their predisposition to aberrant methylation appears to be distinct from the susceptibility of stem cell polycomb targets in lamin-attachment domains (Hinoue et al., 2012), which is generally not restricted to cancer subtypes. Despite a clear rationale for the association of IDH1 mutation with G-CIMP, the mechanistic basis for the coordinated hypermethylation events in most cases of CIMP is unknown, and will remain an active area of investigation.
Epigenetic Influences on Genomic Integrity
Mutation rates vary strikingly across the genome, with strong local influences of base composition on single nucleotide variation (SNV), and regional effects of sequence composition, chromatin structure, replication timing, transcription and nuclear architecture, among others on both SNVs and structural alterations (Hodgkinson and Eyre-Walker, 2011). Despite widespread misuse of the term in the literature, it should be recognized that mutation rates of a tumor cannot be inferred directly from observed mutation numbers or frequencies in a tumor without consideration of the number of cell divisions that have occurred since a shared reference genome, although comparisons across the genome obviate the need for Luria-Delbrück fluctuation modeling and analysis. Epigenetic mechanisms can influence both the rates at which lesions arise and the rates at which they are repaired. For example, the epigenetic mark 5-methylcytosine undergoes spontaneous deamination at higher rates than unmethylated cytosines (Wang et al., 1982), while epigenetic silencing of the MLH1 mismatch repair gene increases mutation frequencies by several orders of magnitude, providing an adaptive advantage to mismatch repair deficient cancer cells.
Unmethylated and methylated cytosine residues both undergo spontaneous hydrolytic deamination but yield uracil and thymine respectively. Uracil is not a normal constituent base in DNA, and is repaired much more efficiently than thymine in a mismatch with guanine. As a consequence, the rate of C-to-T mutations in the context of CpG dinucleotides, most of which contain methylated cytosines, is about ten-fold higher than any other SNV in the human genome (Hodgkinson and Eyre-Walker, 2011). This effect is particularly pronounced in highly proliferative tissues because deamination of 5-methylcytosine in the parent strand just prior to DNA replication results in a full T:A base substitution that is not recognizable as a lesion for repair. Approximately a quarter of all TP53 mutations in human cancer are thus attributable to this epigenetic mark (Olivier et al., 2010).
Regional Effects of Chromatin Organization
Chromatin regulators play a role in maintaining genomic integrity (Papamichos-Chronakis and Peterson, 2012) and regional chromatin structure has a major impact on mutation frequencies. Megabase regions of repressive chromatin, represented by the H3K9me3 mark are positively correlated with single nucleotide variations in cancer (Schuster-Bockler and Lehner, 2012), while open chromatin associated with DNAse I hypersensitive sites (DHS) have a lower inferred mutation rate, but this is partly due to evolutionary constraints on this compartment (Hodgkinson and Eyre-Walker, 2011). Transcription-coupled repair may also play a role in suppressing observed mutation frequencies in gene-rich euchromatic regions.
Other types of mutation and structural change also appear to be associated with chromatin states. For example, retrotransposition occurs more frequently in hypomethylated regions (Lee et al., 2012). Genes resistant to cancer-associated hypermethylation are more likely to have SINE and LINE retrotransposons near their transcription start sites than methylation-prone genes (Estecio et al., 2010). Severe hypomethylation appears to be associated with genomic instability. Mouse models of DNA methyltransferase deficiency display chromosomal instability (Eden et al., 2003), and germline mutations of the DNMT3B gene cause ICF syndrome, characterized by centromeric instability (Okano et al., 1999). Indeed, areas of hypomethylation in the human germline showed higher frequencies of structural mutability (Li et al., 2012). DNA breakpoints associated with somatic copy-number alterations are also enriched in hypomethylated domains (De and Michor, 2011).
Epigenetic Influences on DNA Repair
Depletion of DNA methyltransferases causes increased microsatellite instability (Guo et al., 2004; Kim et al., 2004), destabilization of repeats (Dion et al., 2008), and dramatically increased telomere length, telomeric recombination, and alternative telomere lengthening (Gonzalo et al., 2006). These effects of DNA methyltransferase depletion appear to be mediated in part by a drop in DNA repair proteins as part of DNA damage response (Loughery et al., 2011). The Dnmt1 protein has also been shown to be recruited to areas of irradiation-induced DNA damage, possibly to facilitate repair of epigenetic information following DNA repair (Mortusewicz et al., 2005). It is increasingly appreciated that chromatin can serve as a cellular sensor for DNA damage and other genomic events (Johnson and Dent, 2013).
Epigenetic silencing of DNA repair genes such as MLH1, MGMT, BRCA1, WRN, FANCF, and CHFR can boost mutation rates and promote genomic instability in cancer cells (Toyota and Suzuki, 2010). Familial cases of tumors with microsatellite instability (MSI) in Lynch syndrome result from germline mutations in mismatch repair genes, primarily MSH2 and MLH1. However, most MSI-high tumors arise from an epigenetic defect in sporadic cases of cancer. Approximately 15% of sporadic cases of colorectal cancer display MSI as a consequence of epigenetic silencing of the MLH1 mismatch repair gene by promoter CpG island hypermethylation (Herman et al., 1998) in the context of CIMP (Toyota et al., 1999; Weisenberger et al., 2006). MSI caused by epigenetic silencing of MLH1 has also been reported in other types of cancer, including about a quarter of sporadic endometrial cancers, (Simpkins et al., 1999). Germline variants of MLH1 and MSH2 can predispose to extensive somatic epigenetic silencing of these genes, and thereby increase cancer risk (Hitchins et al., 2011; Ligtenberg et al., 2009). Such familial cases of systemic epigenetic abnormalities can masquerade as germline transmission of epigenetic defects. True transgenerational epigenetic inheritance is evident in genomic imprinting, and in mouse models, but has been difficult to demonstrate directly in human populations, although there is indirect evidence for its existence (Daxinger and Whitelaw, 2012).
The O6-Methylguanine DNA methyltransferase (MGMT) enzyme repairs O6-alkylated guanine residues in genomic DNA. O6-methylguanine pairs with thymine, and would lead to a G to A transition during DNA replication if left unrepaired. MGMT promoter methylation in colorectal cancer is associated with G-to-A mutations in KRAS (Esteller et al., 2000b) and in TP53 (Esteller et al., 2001). Alkylating agents such as Temozolomide are current standard of care for malignant glioblastoma (GBM), but are counteracted by MGMT-mediated repair of the alkylation damage. Epigenetic silencing of MGMT by promoter CpG island hypermethylation inactivates this repair pathway and renders the tumor more sensitive to the Temozolomide treatment (Esteller et al., 2000a; Hegi et al., 2005).
A Genetic Basis for Epigenetic Disruption in Cancer
The discovery of mutations in SMARCB1/SNF5 driving malignant rhabdoid tumours first introduced genetic disruption of epigenetic control as a mechanism of oncogenesis (Versteege et al., 1998),. Mutations in epigenetic regulators continued to emerge from subsequent cancer studies, and have surged in recent large-scale sequencing efforts (Figure 3). Epigenetic control genes are mutated in about half of hepatocellular carcinomas (Fujimoto et al., 2012) and bladder cancer (Gui et al., 2011) and represent six of the twelve most significantly mutated genes in medulloblastoma (Pugh et al., 2012). It is conceivable that disruption of epigenetic control by mutation of a key regulator has the capacity to cause widespread changes to the transcriptome, multiplying the effect of the single genetic alteration. It should be recognized that some of the mutations reported for epigenetic regulators may be passenger events, particularly in tumors with high background mutation rates. Therefore, we have emphasized hotspot mutations and genes recurrently mutated at significant frequencies. We focus here on somatic mutations, but germline variation have also been shown to play a role in cancer. For example, germline mutations in BAP1 have been found to be linked to a tumor predisposition syndrome characterized by melanocytic tumors, mesothelioma, and uveal melanoma (Testa et al., 2011; Wiesner et al., 2011), and rare germline allelic forms of PRDM9 have been found to be associated with childhood leukemia (Hussin et al., 2013).
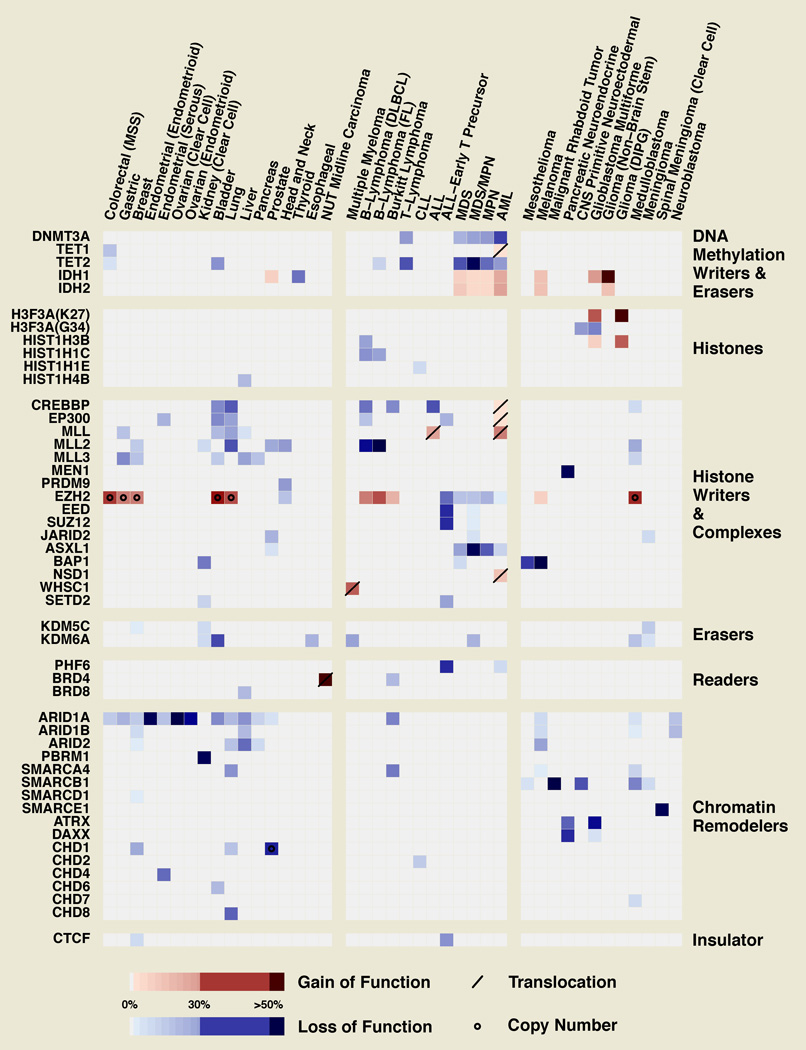
Figure 3. Genetic Alterations in Epigenetic Regulators.
Mutations and other genetic alterations reported for selected epigenetic regulators are shown for various types of human cancer in a heatmap. Malignancies are grouped by epithelial, hematological and other cancers. Mutations, represented by colored cells, are deemed loss of function (blue) unless evidence for gain of function (either hypermorphic or neomorphic, red) has been shown. Other genetic alterations are plotted with different symbols, with a slash indicating translocation events and a dot indicating copy number alterations. Translocations that generate oncogenic fusion proteins are represented in red as well. The mutation frequencies, represented by the darkness of the shade, are based on recent whole genome-exome studies, with adjustments made where whole-genome/exome studies are not available or have a small sample size. Different subtypes of lung cancers are combined without adjusting for subtype prevalence and certain mutations may only represent one subtype. Cells showing no entry may represent false negatives in our curation or in the literature, and cancer types highly covered with whole-genome/exome studies (e.g. breast cancer) might have fewer false negatives than those that are not. MSS/MSI – microsatellite stable/instable (MSI CRCs are excluded due to the high background mutation rate); DLBCL - Diffuse large B-cell lymphoma; FL – follicular lymphoma.
DNA Methylation Writers and Erasers
The DNA methyltransferase DNMT3A is recurrently mutated in acute myeloid leukemia (AML) and other myeloid malignancies (Ley et al., 2010; Yan et al., 2011), as well as T-cell lymphoma (Couronne et al., 2012). The mutations often occur at a R882 hotspot, but nevertheless likely reflect loss of function of DNMT3A. Mutations in the DNA methylation eraser TET2 have also been identified in the same cancer types (Abdel-Wahab et al., 2009; Langemeijer et al., 2009; Quivoron et al., 2011), and bone marrow from patients with TET2 mutations show reduced levels of 5hmC (Ko et al., 2010). The isocitrate dehydrogenases IDH1 and IDH2 are also recurrently mutated in AML. IDH1 enzymes with the R132 hotspot mutation and IDH2 enzymes containing R140 or R172 mutations have lost the ability to produce alpha-ketoglutarate (α-KG), but instead convert α-KG to an aberrant metabolite 2-hydroxyglutarate (2-HG), a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases, such as the TETs and JmjC-domain containing histone demethylases (Lu et al., 2012; Xu et al., 2011). IDH1/2 mutations are mutually exclusive with TET2 mutations in AML, consistent with the inhibitory effect of 2-HG on TETs as a mediator of the effects of IDH1/2 mutations (Figueroa et al., 2010; Weissmann et al., 2012). The same hotspot mutation for IDH1, and less often IDH2 is also found in gliomas and glioblastomas. Both glioblastomas with IDH1 mutations (Noushmehr et al., 2010) and cases of AML with mutation of IDH1 or IDH2 (Figueroa et al., 2010) display CpG island methylator phenotypes.
Histone Gene Mutations
Mutations in histone variants H3.3 (H3F3A) or sometimes H3.1 (HIST1H3B) have been found in pediatric (Schwartzentruber et al., 2012; Wu et al., 2012) and adult brain tumors (Sturm et al., 2012) with K27M and G34R or G34V mutation hotspots. Tumors with G34 mutations display extensive DNA hypomethylation, particularly in subtelomeric regions (Sturm et al., 2012), perhaps contributing to alternative lengthening of telomeres (ALT) (Schwartzentruber et al., 2012). Mutations were also observed in the ATRX and DAXX genes, encoding proteins responsible for loading of the H3.3 variant into the telomere region (Schwartzentruber et al., 2012). Pancreatic neuroendocrine tumors (PanNETs) with ATRX and DAXX mutations also exhibit ALT (Heaphy et al., 2011). Since this phenotypic effect is associated with a H3.3 loading defect, the G34 mutations may also interfere with H3.3 loading. In contrast, tumors with K27M mutations did not display ALT, and these mutations may instead mimic dimethylated lysine 27, a repressive Polycomb mark, given that methionine is a natural mimic of this epigenetic mark (Hyland et al., 2011). H3.3 G34R mutations have also been reported in primitive neuroectodermal tumors of the CNS (Gessi et al., 2013), mirroring the defect in the ATRX-DAXX-H3.3 axis in other brain tumors and PanNETs.
Mutations in HIST1H3B and HIST1H1C have been found in diffuse large B-cell lymphoma (DLBCL), although the mutations do not occur in clusters (Lohr et al., 2012; Morin et al., 2011). These might be functionally different from the hotspot mutations seen in brain tumors. Focal deletion of a histone gene cluster at 6p22 is seen in near-haploid cases of acute lymphoblastic leukemia (Holmfeldt et al., 2013).
Histone Methylation Writers
The MLL gene, encoding one of the H3K4 methyltransferases has over 50 translocation fusion partners in different lineages of leukemia. These rearrangements account for 80% of the cases of infant leukemia and 5–10% of adult leukemia cases, and are generally associated with poor prognosis (Tan et al., 2011a). The primary mechanism has been attributed to the recruitment of inappropriate epigenetic factors to MLL targets, by fusions between recruitment proteins and the DNA-binding N-terminus of MLL. Target genes for these recruited complexes include the HOX genes, particularly HOXA9, whose upregulation is a key feature of MLL leukemia. MLL regulates the expression of HOX genes in normal pluripotent cells, but the oncogenic fusion proteins keep them from being turned off during differentiation and therefore impart stem-cell like properties. Targeted therapeutic strategies are emerging for AML with MLL fusions, including inhibition of menin (encoded by MEN1), DOT1L, PRMT1, the histone acetylation reader BRD4, and LSD1 (Zeisig et al., 2012). In addition to the translocations, loss-of-function mutations of MLL-MLL3 have been reported in many different types of cancer, including AML - possibly another way of disturbing the temporal control at promoters associated with pluripotency. MLL2 is mutated at very high frequency in B-cell follicular lymphoma and diffuse large B-cell lymphoma, consistent with the gain-of-function mutations of EZH2 in the same tumor types.
While menin is critical to the oncogenic effects of MLL fusion proteins in AML (Yokoyama and Cleary, 2008), loss of function mutations have been found in PanNETs (Jiao et al., 2011), consistent with a tumor-suppressor role, suggesting that cellular context is important.
The recurrent t(5;11)(q35;p15.5) translocation in AML results in the fusion of the H3K36 methyltransferase NSD1 to nucleoporin-98 (NUP98), with elevated levels of H3K36me3 levels at HOXA genes and accompanying transcriptional activation. Translocations involving another dedicated H3K36 methyltransferase WHSC1/MMSET/NSD2 are seen in 20% of multiple myelomas. Another H3K36 methyltransferase, SETD2 is recurrently mutated in clear cell renal cell carcinomas (ccRCC) (Dalgliesh et al., 2010).
A recent study reconstructed the phylogenetic structure of molecular events in ccRCC with multiple spatially separated samples from the same tumors (Gerlinger et al., 2012). In both of the two patients studied, distinct SETD2 inactivating mutations were found in different parts of the same tumor. Immunohistochemistry staining confirmed H3K36me3 loss in all the mutant tumors. This convergent somatic evolution indicates that failure to establish H3K36 methylation marks provides a strong selective advantage relatively late in ccRCC progression. A similar molecular convergence was found for KDM5C, an H3K4 demethylase, in one of the two patients. This, together with recurrent mutations in other epigenetic regulators, shows that epigenetic dysregulation, often mediated by genetic events, is important in advanced ccRCCs.
EZH2, the writer for the H3K27 methylation mark associated with Polycomb repression, has long been viewed as an oncogene in cancer. Indeed, gain-of-function mutations are seen in lymphomas. However, loss-of-function mutations in this gene have recently been described in other cancers. We discuss these divergent effects of EZH2 mutations and other alterations to this pathway in more detail later (Figure 4).
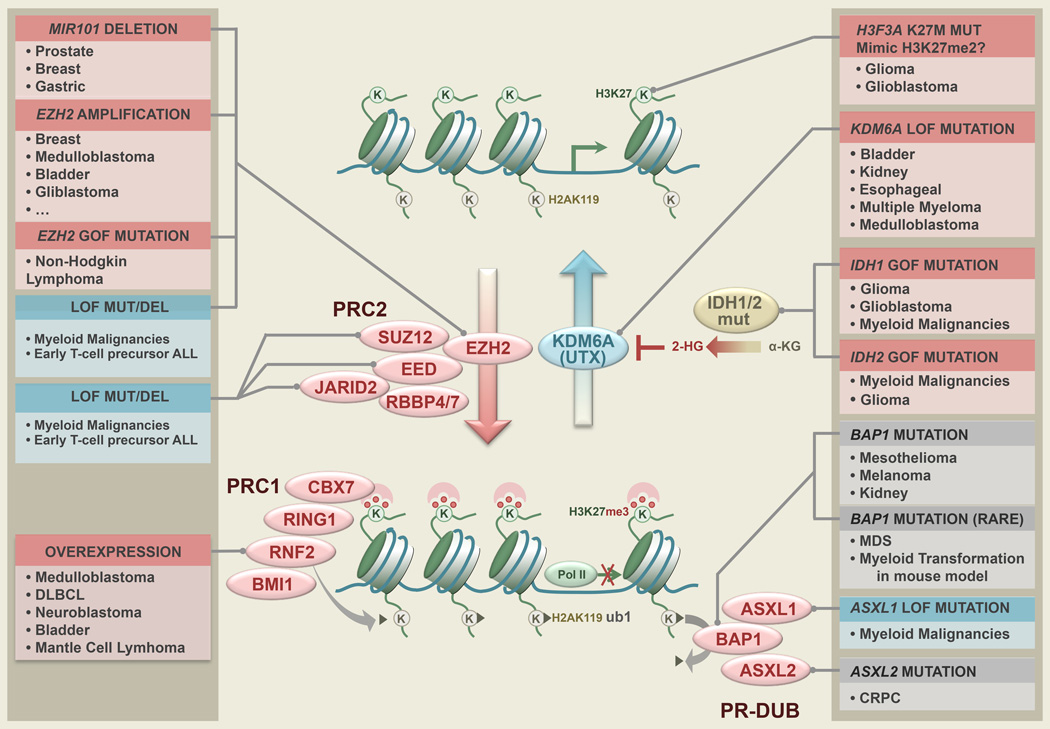
Figure 4. Genetic Disruption of Epigenetic Control at H3K27 in Cancer.
The counteracting writer EZH2 and eraser KDM6A/UTX form a pair in regulating an important epigenetic mark, methylation at H3 lysine 27. EZH2 catalyzes the methylation process with help from other components in the Polycomb Repressive Complex 2 (PRC2), while KDM6A, part of the Trithorax complex, removes this repressive mark. The K27me3 mark attracts another Polycomb complex, PRC1, which ubiquitinates H2AK119, and thereby blocks PolII elongation. Another Polycomb complex, PR-DUB is also critical to the maintainance of the repression at a subset of the Polycomb genes, although it removes the H2AK119ub mark and thus counteracts PRC1 in that regard. Mutations and genetic alterations spanning a wide spectrum of human cancers hit this epigenetic pathway. Solid tumors show possibly neomorphic histone K27 mutations (mimicking H3K27me2), UTX mutation, EZH2 amplification and/or overexpression due to genomic loss of the repressive microRNA miR101, as well as amplification/overexpression of the PRC1 member BMI1, and lymphoma exhibits gain-of-function mutations of EZH2, consistent with a gain of Polycomb repression (red boxes) in the affected malignancies. In contrast, myeloid malignancies and ALL, particularly early T-cell precursor ALL show mutations that could sabotage Polycomb repression (blue boxes). Mouse models show that loss of BAP1, the enzymatic unit of PR-DUB, leads to myeloid transformation, although BAP1 mutation in MDS is rare. Gray boxes indicate that the effect on H3K27me3 is not clear.
Histone Methylation Erasers
Consistent with EZH2 overexpression in various solid tumors, the corresponding eraser KDM6A/UTX is mutated in more than a dozen tumor types, with the highest frequency in bladder (Gui et al., 2011; van Haaften et al., 2009). The H3K9 demethylase KDM4C/GASC1 is amplified in breast cancer and has been shown to drive transformation (Liu et al., 2009; Rui et al., 2010). Ectopic expression of this putative oncogene in vitro causes an efficient decrease of H3K9me3 (Cloos et al., 2006). Its co-amplification with JAK2 (Rui et al., 2010) -which phosphorylates H3Y41 and prevents binding of H3K9 methylation reader HP1 to the H3K9 methylation mark - in lymphoma makes for an interesting example of a single genetic event hitting two possible epigenetic regulators. Inhibiting the two co-amplified and cooperating gene products is efficient at killing these lymphoma cells.
Histone Acetylation Writers and Erasers
The counteracting histone acetyltransferases (HATs) and deacetylases (HDACs) are considered to be promiscuous, and often have important non-histone substrates such as TP53. There are three major families of HATs, namely the CBP/P300, GNAT, and the MYST families. CREBBP is mutated at high frequency in follicular lymphoma and diffuse large-cell B-cell lymphoma (DLBCL) (Morin et al., 2011; Pasqualucci et al., 2011) and in ALL (Mullighan et al., 2011), particularly relapsed hyperdiploid ALL (Inthal et al., 2012). Its paralogue EP300 also undergoes frequent mutation (Gayther et al., 2000; Gui et al., 2011; Pasqualucci et al., 2011; Zhang et al., 2012) and loss of heterozygosity (LOH) in many different epithelial cancers.
HATs have also been implicated in gene fusions. The t(8;16)(p11;p13) translocation in AML fuses the N-terminal part of MOZ, the founding member of the MYST HAT family, to the major part of the CBP gene containing the acetylase domain. MOZ has also been found to be involved in fusions with EP300 (Yang, 2004). These translocations generating chimeric oncoproteins with the DNA-binding domain of MOZ and the transcription-activating domain of another coactivator are associated with AML M5/M4. These results suggest that both disruption and redirection of HAT could contribute to cancer.
Reports of mutations in HDACs are rare. Rather, HDACs are often co-opted by other genetic alterations. A prime example is the PML-RARα translocation, responsible for 95% of the AML FAB-M3 (APL, Acute promyelocytic leukemia) cases. The leukemogenetic effect of this translocation is primarily mediated through aberrant recruitment of N-CoR/HDAC repressor complexes (Minucci and Pelicci, 2006). The retinoic acid receptor-alpha (RARα) part binds to retinoic acid-responsive elements (RAREs), while the PML moiety recruits the HDAC-containing repressive complex. All-trans retinoic acid (ATRA) targets RARα, dissociates these repressor complexes and effectively induces differentiation of the leukemic promyelocytes. Combination therapy of ATRA and arsenic trioxide shows excellent clinical response, and turned APL into a highly curable disease (Wang and Chen, 2008). Another fusion protein PLZF-RARα blocks differentiation by a similar mechanism. However, APL with this translocation is ATRA-resistant due to the higher affinity of the PLZF moiety to the N-CoR complex, but a combination of ATRA with HDAC inhibitors can fully reverse the transcriptional repression and induce terminal differentiation for this type of AML (Wang and Chen, 2008). Similarly, the RUNX1-ETO fusion, the AML1-ETO fusion, and the CBF-MYH11 protein from inv(16), all recruit HDACs, and efficacy of HDAC inhibitors has been demonstrated for all three (Zeisig et al., 2012).
Epigenetic Readers
The epigenetic readers add another layer of control to the epigenetic state, by serving as interpreters of the epigenetic state and relaying epigenetic signals. Many of the epigenetic writer/eraser/remodelers have intrinsic reader domains, or interact with dedicated readers to sense the presence or absence of particular epigenetic marks. Translocations joining BRD4 or occasionally BRD3, both readers of the BET Bromodomain-containing family, to almost the entire length of the NUT gene define a lethal, poorly differentiated pediatric tumor, NUT midline carcinoma (NMC) (French et al., 2008). The BET family members targetable by BET inhibitors (Filippakopoulos et al., 2010), are lysine acetylation readers that bind transcriptionally active chromatin as acetylated lysine readers, and are targetable by BET inhibitors (Filippakopoulos et al., 2010). Functional studies show that the BRD-NUT fusion oncoprotein binds avidly to acetylated histones, resulting in a differentiation block, potentially by interfering with transcriptional programs driving differentiation (French et al., 2008). BRD3 is significantly mutated in lung adenocarcinomas (Imielinski et al., 2012), and BRD8 in liver cancer (Fujimoto et al., 2012). In addition, the plant homeodomain (PHD)-domain containing gene PHF6 is recurrently mutated in AML (Van Vlierberghe et al., 2011) and overall loss of this gene (mutation and/or deletion) is observed in TALL (Van Vlierberghe et al., 2010).
Chromatin Remodelers
A large number of SWI/SNF complexes exist in mammals and contribute to lineage- and tissue-specific gene expression (Wilson and Roberts, 2011). The two major types of SWI/SNF complexes are BAF (BRM-containing, or SWI/SNF-A) and PBAF (BRG1 containing, or SWI/SNF-B), defined by a core enzymatic unit being either SMARCA2 (BRM) or SMARCA4 (BRG1). Those complexes also contain other core units, such as SMARCB1/SNF5 and ARID1A/B, unique to BAF, and PBRM1 and BRD7, unique to PBAF. Truncating mutations in the SMARCB1 gene are very common in malignant rhabdoid tumours (RTs) (Versteege et al., 1998), a rare yet lethal tumor diagnosed in children. The biallelic nature of the inactivation fits with a tumor suppressor role for SMARCB1. Familial cases of RTs are associated with inheritance of one defective SMARCB1 allele. SMARCB1 is also mutated in a few other cancers (Figure 1). SMARCA4 mutation is also seen in familial cases of RT (Schneppenheim et al., 2010), indicating that it is indeed SWI/SNF dysfunction that is responsible for RT development. SMARCA4 is also mutated Burkitt’s Lymphoma, in a mutually exclusive manner with ARID1A mutations (Love et al., 2012), again suggesting a driver role for SWI/SNF mutations. Recurrent SMARCA4 mutation is also seen in lung cancer (Imielinski et al., 2012) and medulloblastoma (especially the WNT subtype) (Jones et al., 2012; Parsons et al., 2011; Pugh et al., 2012; Robinson et al., 2012).
ARID1A mutations have been found in more than ten different tumor types, with the highest rate in the clear cell subtype of ovarian cancer, where it is mutated in more than half of the tumors (Jones et al., 2010; Wiegand et al., 2010). ARID1A is also mutated in the endometrioid subtypes of ovarian (Wiegand et al., 2010) and endometrial cancers (Guan et al., 2011). ARID1B and ARID2 mutations are also seen in various cancer types, including liver cancer (Fujimoto et al., 2012) and melanoma (Hodis et al., 2012), among others. In addition, the polybromo-containing PBRM1 in the PBAF complex was recently found to be the second most mutated gene in clear cell renal cell carcinomas (Varela et al., 2011). Another SWI/SNF gene, SMARCE1, is highly recurrently mutated in clear cell meningiomas (Smith et al., 2013). The exceptionally high mutation rate of SWI/SNF member in clear cell tumors from different tissues (ovary, kidney, and meninges) highlights an interesting possible link between clear cell tumors and SWI/SNF dysfunction.
ATRX, responsible for H3.3 incorporation at telomeres and pericentric heterochromatin is often mutated in pancreatic neuroendocrine tumours (PanNETs), the second most common malignancy of the pancreas (Jiao et al., 2011). Interestingly, there are also recurrent mutations in the associated chaperone DAXX in the same cancer type, and the two mutations are mutually exclusive. Mutation of these two genes are also found in GBM where they are mutually exclusive with the H3F3A mutations described earlier, with any of these three genes mutated in almost half of the tumors studied (Schwartzentruber et al., 2012). These mutations all lead to alternative lengthening of telomeres associated with increased genomic instability (Heaphy et al., 2011; Schwartzentruber et al., 2012). With the possible exception of ATRX and DAXX, most of the mutations in the SWI/SNF family members are not associated with genomic instability (Wilson and Roberts, 2011). Rather, perturbed differentiation may be the major mechanism, as cells from different lineages co-exist within an individual rhabdoid tumor.
The CHD family chromatin remodelers can be divided into three classes. Class I (CHD1/2), Class II (CHD3/4, in the NuRD/Mi-2/CHD complex), and Class III (CHD5–9). The NuRD/Mi-2/CHD complexes are unique in that they have core enzymatic subunits with at least two distinct functions: ATP-dependent remodeling (CHD3 and CHD4), as well as histone deacetylase (HDAC1 and HDAC2) functions (Lai and Wade, 2011), and therefore couple two epigenetic processes in one complex for transcriptional repression. Their MBD and MTA subunits target the complex to different parts of the genome by binding methylated DNA (MBD) or other transcription factors (MTA), in physiological and pathological conditions. For example, the MTA-2 containing NuRD complex associates with TWIST in breast cancer, represses genes such as E-cadherin (CDH1), and contributes to EMT. CHD4 is mutated in 17% of serous endometrial cancer (Le Gallo et al., 2012). Of the other CHDs, CHD1 is the second most-frequently deleted gene in prostate cancer, defining an ETS-negative subtype (Grasso et al., 2012), with mutations reported as well (Berger et al., 2011).
Epigenetic Insulators
CTCF is located in 16q22.1 with LOH in breast and prostate cancers (Filippova et al., 1998) and Wilms’ tumors (Mummert et al., 2005). CTCF mutation has also been reported in breast cancer (Filippova et al., 2002), and this mutation is found to be significant in a cohort of 510 tumors (TCGA, 2012c). Rare mutations in this gene have also been reported for prostate cancer (Filippova et al., 2002), Wilms’ tumor (Filippova et al., 2002), AML (Dolnik et al., 2012), ALL (Mullighan et al., 2011; Zhang et al., 2012)), and endometrial cancers (Le Gallo et al., 2012). The functional implications of CTCF deletion/mutation, especially given the low frequency, have not been fully delineated yet, but abrogation of proper insulation might be one mechanism.
Genetic Disruption of a Central Epigenetic Control Circuit
Figure 4 illustrates the diverse ways in which a central epigenetic control circuit can be impacted in cancer. EZH2 catalyzes methylation at H3K27, as part of Polycomb Repressive Complex 2 (PRC2). Two other core subunits of PRC2 are EED and SUZ12, and other components such as JARID2 can be part of a PRC2 complex too. EZH2 has long been thought to be oncogenic since it is overexpressed as a result of amplification of EZH2 in breast, bladder and other cancers (Bracken et al., 2003), as well as genetic loss of miR101, which represses EZH2 in prostate cancer (Varambally et al., 2008). In line with this view, gain-of-function hotspot mutations (Y641 and A677,) in the SET-domain of EZH2 have been found in a significant portion of lymphomas (Lohr et al., 2012; Morin et al., 2010; Morin et al., 2011; Pasqualucci et al., 2011). More convincingly, EZH2 amplification and overexpression in two of the largest subgroups of medulloblastoma is mutually exclusive with mutation of the H3K27 demethylase KDM6A, suggesting that accumulation of H3K27me3 is a key step in these tumors (Robinson et al., 2012).
On the other hand, loss of function mutations of EZH2 have also been found in a series of myeloid malignancies, including MDS, Multiple Myeloma, MPN and MDS/MPN, as well as in head-and-neck squamous cell carcinomas (HNSCCs) (Ernst et al., 2010; Nikoloski et al., 2010; Stransky et al., 2011), suggesting that PRC2 can also act as tumor suppressors. Aside from the myeloid malignancies, mutually exclusive recurrent deletion and loss of function mutations of EZH2 and SUZ12 have also been found in T-lineage acute lymphoblastic leukemia (T-ALL) (Ntziachristos et al., 2012) or of all three PRC2 subunits in early-T-cell-precursor ALL (Zhang et al., 2012) further substantiate a tumor suppressor role. Indeed, disruption of EZH2 is sufficient to induce T-ALL in mice (Simon et al., 2012). PRC2 component mutations are much more common in early T-cell precursor ALL, a lymphoblastic leukemia with myeloid features. The Polycomb repressive deubiquitinase (PR-DUB) component ASXL1 is also mutated in myeloid malignancies, and ASXL1 mutation mediates myeloid transformation through loss of PRC2 repression (Abdel-Wahab et al., 2012), in line with the observed loss of function of PRC2 members in myeloid disorders. Mouse models also show that loss of BAP1, the enzymatic unit of PR-DUB, lead to myeloid transformation (Dey et al., 2012). This, together with a BAP1 catalytic mutation found in a MDS patient lacking other MDS mutations (Dey et al., 2012) further lends credibility to the idea that loss of Polycomb repression drive myeloid disorders, while in B-cell lymphoma and solid tumors gain of Polycomb repression seem important.
EZH2 also exhibits PRC2-independent oncogenic activities. For example, in castration-resistant prostate cancer Akt-mediated phosphorylation of EZH2 at S21 can shift EZH2 from PRC2-dependent promoters to EZH2 ‘solo’ promoters. This EZH2 activity is often associated with androgen receptor (AR), and activates gene expression at these loci (Xu et al., 2012).
These complex ways in which the H3K27me3 axis is disrupted in cancer suggest differential therapeutic approaches should be developed for 1) myeloid malignancies and early T-cell ALL, 2) lymphomas and some solid tumors, and possibly 3) hormone-associated cancers. In particular, caution should be used when considering EZH2 inhibitors for the first group of tumors, which feature genetic lesions leading to loss of PRC2 repression.
Conclusions
It is clear that the cancer genome and epigenome influence each other in a multitude of ways (Figure 5). They offer complementary mechanisms to achieve similar results, such as the inactivation of tumor-suppressor genes such as BRCA1, CDKN2A, VHL, and RB1 by either deletion or epigenetic silencing, and they can work cooperatively, as in the case of CIMP and BRAF mutation in colorectal cancer, where CIMP appears to create a permissive context for BRAF mutation as early as in the precursor lesion (Hinoue et al., 2009; Yamamoto et al., 2012).
Figure 5. Interplay Between the Cancer Genome and Epigenome.
The genome and epigenome influence each other, as the genome provides the primary sequence information and encodes regulators of epigenetic states, while the epigenome controls the accessibility and interpretation of the genome. Changes in one can influence the other, forming a partnership in producing genetically or epigenetically encoded phenotypic variation subject to Darwinian selection for growth advantage, and thus eventually achieving the hallmarks of cancer (Hanahan and Weinberg, 2011). Genetic instability and mutation, and epigenomic disruption can be considered enabling characteristics of cancer cells.
Many questions and challenges remain. For example, the explosion in the number of epigenetic regulator mutations identified in human cancer has underscored the importance of epigenetic control in tumor suppression, but the phenotypic consequences of these mutations remain largely uncharacterized. However, this plethora of newly identified mutations in epigenetic regulators opens entirely new avenues of therapeutic attack (Dawson and Kouzarides, 2012).
Another major question remaining in the field is the mechanistic basis for some well-recognized examples of disruption of epigenetic control, such as CIMP in colorectal cancer. Even in the case of G-CIMP in gliomas and its very tight association with IDH1 mutation, it is not clear what confers the gene specificity of the hypermethylation events. Correlated events such as CIMP create other challenges as well. In contrast to most mutations, CIMP-associated DNA methylation events are highly correlated, with a large number of recurrent alterations that appear to be passenger events without functional contribution to the cancer process. This high degree of correlation precludes the straightforward use of recurrence frequency among different tumors as a main filter criterion in the identification of functionally relevant epigenetic driver events. Therefore, the identification of epigenetic drivers must rely more on the analysis of transcriptional consequences, mutual exclusivity with other events in the same pathway within a tumor, and complementary mechanisms of inactivation of the same gene in other tumors, and most importantly, functional experimental validation of an impact of the epigenetic gene inactivation on cellular proliferation, immortality, angiogenesis, cell death, invasion or metastasis.
Cancer epigenetics and genetics may in orm each other. enetics can shed light on the identity of epigenetic drivers by revealing mutual exclusivity with genetic aberrations in the same gene or pathway. Epigenetics may also provide insight into genetic drivers in a similar fashion. In addition, the understanding of epigenetic networks provides a framework to interpret the functional significance of lower-frequency drivers in the same pathway. The high frequency of epigenetic regulator mutations seen in various cancers, the hotspot nature of some mutations found, mutual exclusivity between different mechanisms affecting the same genes/pathways, clonal analysis highlighting convergent evolution, and validations in experimental systems all attest to the importance of mutations in epigenetic regulators in cancer, and strengthen the concept that disruption of epigenetic control is a common enabling characteristic of cancer cells.
Acknowledgements
This work was supported by NIH grants R01-170550, R01-CA157918, R01-DA030325, and U24 CA143882 (PWL). We would like to thank Peter Jones, Benjamin Berman and Timothy Triche Jr. for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, Caricasole A, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- Baer C, Claus R, Plass C. Genome-Wide Epigenetic Regulation of miRNAs in Cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Hoppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986;46:2917–2922. [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature Genetics. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Boyarchuk E, Montes de Oca R, Almouzni G. Cell cycle dynamics of histone variants at the centromere, a model for chromosomal landmarks. Curr Opin Cell Biol. 2011;23:266–276. doi: 10.1016/j.ceb.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis- erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16:R88–R95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Cohen NM, Kenigsberg E, Tanay A. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell. 2011;145:773–786. doi: 10.1016/j.cell.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Coolen MW, Stirzaker C, Song JZ, Statham AL, Kassir Z, Moreno CS, Young AN, Varma V, Speed TP, Cowley M, et al. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat Cell Biol. 2010;12:235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- Craene BD, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2012;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diala ES, Hoffman RM. Hypomethylation of HeLa cell DNA and the absence of 5-methylcytosine in SV40 and adenovirus (type 2) DNA: analysis by HPLC. Biochem Biophys Res Commun. 1982;107:19–26. doi: 10.1016/0006-291x(82)91663-1. [DOI] [PubMed] [Google Scholar]
- Dion V, Lin Y, Hubert L, Jr., Waterland RA, Wilson JH. Dnmt1 Deficiency Promotes CAG Repeat Expansion in the Mouse Germline. Hum Mol Genet. 2008;5:5. doi: 10.1093/hmg/ddn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, Mauch J, Kelkenberg-Schade S, Haldemann B, Fries T, Kronke J, Kuhn MW, Paschka P, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood. 2012;120:e83–e92. doi: 10.1182/blood-2011-12-401471. [DOI] [PubMed] [Google Scholar]
- Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Encode-Project-Consortium. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Estecio MR, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, Oki Y, Kondo Y, Jelinek J, Shen L, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010;20:1369–1382. doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000a;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, Baylin SB, Herman JG. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000b;60:2368–2371. [PubMed] [Google Scholar]
- Fabbri M, Calin GA. Epigenetics and miRNAs in human cancer. Adv Genet. 2010;70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Fenouil R, Cauchy P, Koch F, Descostes N, Cabeza JZ, Innocenti C, Ferrier P, Spicuglia S, Gut M, Gut I, et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, Doggett NA, Lobanenkov VV. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- Filippova GN, Qi CF, Ulmer JE, Moore JM, Ward MD, Hu YJ, Loukinov DI, Pugacheva EM, Klenova EM, Grundy PE, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard JK. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, Kucera KS, Willard HF, Myers RM. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS genetics. 2011;7:e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessi M, Gielen GH, Hammes J, Dorner E, Muhlen AZ, Waha A, Pietsch T. H3.3 G34R mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: possible diagnostic and therapeutic implications? J Neurooncol. 2013 doi: 10.1007/s11060-012-1040-z. [DOI] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin 5' CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Mao TL, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL, Shih Ie M. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. Increased methylation variation in epigenetic domains across cancer types. Nature genetics. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome research. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Rapkins RW, Kwok CT, Srivastava S, Wong JJ, Khachigian LM, Polly P, Goldblatt J, Ward RL. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5'UTR. Cancer Cell. 2011;20:200–213. doi: 10.1016/j.ccr.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 2011;12:756–766. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013 doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussin J, Sinnett D, Casals F, Idaghdour Y, Bruat V, Saillour V, Healy J, Grenier JC, de Malliard T, Busche S, et al. Rare allelic forms of PRDM9 associated with childhood leukemogenesis. Genome Res. 2013 doi: 10.1101/gr.144188.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland EM, Molina H, Poorey K, Jie C, Xie Z, Dai J, Qian J, Bekiranov S, Auble DT, Pandey A, et al. An evolutionarily 'young' lysine residue in histone H3 attenuates transcriptional output in Saccharomyces cerevisiae. Genes & development. 2011;25:1306–1319. doi: 10.1101/gad.2050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inthal A, Zeitlhofer P, Zeginigg M, Morak M, Grausenburger R, Fronkova E, Fahrner B, Mann G, Haas OA, Panzer-Grumayer R. CREBBP HAT domain mutations prevail in relapse cases of high hyperdiploid childhood acute lymphoblastic leukemia. Leukemia. 2012;26:1797–1803. doi: 10.1038/leu.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Dent SYR. Chromatin: Receiver and Quarterback for Cellular Signals. Cell. 2013;152 doi: 10.1016/j.cell.2013.01.017. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nature Genetics. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr., Vogelstein B, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence- dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Kim M, Trinh BN, Long TI, Oghamian S, Laird PW. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic acids research. 2004;32:5742–5749. doi: 10.1093/nar/gkh912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:E1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nature reviews Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nature reviews Genetics. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, et al. Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet. 2012;8:e1002692. doi: 10.1371/journal.pgen.1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nature Genetics. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, Yang ZQ. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene. 2009;28:4491–4500. doi: 10.1038/onc.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughery JE, Dunne PD, O'Neill KM, Meehan RR, McDaid JR, Walsh CP. DNMT1 deficiency triggers mismatch repair defects in human cells through depletion of repair protein levels in a process involving the DNA damage response. Human molecular genetics. 2011;20:3241–3255. doi: 10.1093/hmg/ddr236. [DOI] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2012 doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci U S A. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummert SK, Lobanenkov VA, Feinberg AP. Association of chromosome arm 16q loss with loss of imprinting of insulin-like growth factor-II in Wilms tumor. Genes Chromosomes Cancer. 2005;43:155–161. doi: 10.1002/gcc.20176. [DOI] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat Rev Genet. 2012;14:62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk A, Sansom O, Selfridge J, Caballero IM, Salozhin S, Aithozhina D, Cerchietti L, Meng FG, Augenlicht LH, Mariadason JM, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208. doi: 10.1128/MCB.26.1.199-208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Raddatz G, Gao Q, Bender S, Jaenisch R, Lyko F. Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation. PLoS Genet. 2012;8:e1003146. doi: 10.1371/journal.pgen.1003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, Wright GW, Lenz G, Ngo VN, Shaffer AL, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Berger J, Bishop SM, Hendrich B, Bird A, Clarke AR. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat Genet. 2003;34:145–147. doi: 10.1038/ng1155. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Schneppenheim R, Fruhwald MC, Gesk S, Hasselblatt M, Jeibmann A, Kordes U, Kreuz M, Leuschner I, Martin Subero JI, Obser T, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS genetics. 2011;7:e1001286. doi: 10.1371/journal.pgen.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkins SB, Bocker T, Swisher EM, Mutch DG, Gersell DJ, Kovatich AJ, Palazzo JP, Fishel R, Goodfellow PJ. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet. 1999;8:661–666. doi: 10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- Smith MJ, O'Sullivan J, Bhaskar SS, Hadfield KD, Poke G, Caird J, Sharif S, Eccles D, Fitzpatrick D, Rawluk D, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013 doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
- Sproul D, Kitchen RR, Nestor CE, Dixon JM, Sims AH, Harrison DJ, Ramsahoye BH, Meehan RR. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol. 2012;13:R84. doi: 10.1186/gb-2012-13-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011a;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011b;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, O'Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA, C.G.A.R.N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA, C.G.A.R.N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012a;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA, C.G.A.R.N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012b;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA, C.G.A.R.N. Comprehensive molecular portraits of human breast tumours. Nature. 2012c;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome research. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, Cox NJ, Dogan AU, Pass HI, Trusa S, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H. Epigenetic drivers of genetic alterations. Adv Genet. 2010;70:309–323. doi: 10.1016/B978-0-12-380866-0.60011-3. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B, Philippe J, Gonzalez-Garcia S, Toribio ML, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, Nicolas C, Payer AR, Fernandez HF, Tallman MS, et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25:130–134. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Wang RY, Kuo KC, Gehrke CW, Huang LH, Ehrlich M. Heat-and alkali-induced deamination of 5-methylcytosine and cytosine residues in DNA. Biochim Biophys Acta. 1982;697:371–377. doi: 10.1016/0167-4781(82)90101-4. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature Genetics. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, Eder C, Dicker F, Fasan A, Haferlach C, Haferlach T, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26:934–942. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nature Genetics. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, Windpassinger C, Wackernagel W, Loy S, Wolf I, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Suzuki H, Yamano HO, Maruyama R, Nojima M, Kamimae S, Sawada T, Ashida M, Yoshikawa K, Kimura T, et al. Molecular dissection of premalignant colorectal lesions reveals early onset of the CpG island methylator phenotype. Am J Pathol. 2012;181:1847–1861. doi: 10.1016/j.ajpath.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell. 2003;4:121–131. doi: 10.1016/s1535-6108(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Yamazaki J, Estecio MR, Lu Y, Long H, Malouf GG, Graber D, Huo Y, Ramagli L, Liang S, Kornblau SM, et al. The epigenome of AML stem and progenitor cells. Epigenetics. 2013;8:92–104. doi: 10.4161/epi.23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature genetics. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig BB, Kulasekararaj AG, Mufti GJ, So CW. SnapShot: Acute myeloid leukemia. Cancer Cell. 2012;22:698–698. e691. doi: 10.1016/j.ccr.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]