Abstract
The recent emergence of highly virulent human adenoviruses (HAdVs) with new tissue tropisms underscores the need to determine their ontogeny. Here we report complete high quality genome sequences and analyses for all the previously unsequenced HAdV serotypes (n = 20) within HAdV species D. Analysis of nucleotide sequence variability for these in conjunction with another 40 HAdV prototypes, comprising all seven HAdV species, confirmed the uniquely hypervariable regions within species. The mutation rate among HAdV-Ds was low when compared to other HAdV species. Homologous recombination was identified in at least two of five examined hypervariable regions for every virus, suggesting the evolution of HAdV-Ds has been highly dependent on homologous recombination. Patterns of alternating GC and AT rich motifs correlated well with hypervariable region recombination sites across the HAdV-D genomes, suggesting foci of DNA instability lead to formulaic patterns of homologous recombination and confer agility to adenovirus evolution.
The evolution of any infectious organism represents a complex and dynamic transaction between pathogen and host. Evolution of viral pathogens may lead to altered virulence, enhanced transmission, altered tissue tropisms, and striking new disease manifestations. As a result, understanding viral evolution is vital to predict and prevent future disease outbreaks. Adenoviruses, due to their broad tropism and tractability, offer a useful model for studying the molecular evolution of DNA viruses. Adenoviruses have played an invaluable role in the study of human biology; current paradigms for RNA splicing and viral oncogenesis are two such examples1,2,3. Human adenoviruses (HAdVs) are also significant agents of disease, ranging in severity from mild, self-limited infections of mucosal surfaces, to severe, life threatening dissemination, particularly involving the respiratory tract4,5,6. Recently, outbreaks from evolving, novel HAdV types have been associated with fatal infections4,5.
There are currently over 60 HAdV types in seven species (human adenovirus A–G), with HAdV-D containing the most members, including a substantial number identified during the first two decades of the AIDS epidemic7. HAdVs have a linear, double stranded DNA genome that is 34–36 kb in size. Among HAdV-Ds, homologous recombination appears to play a major role in generating genome diversity4,5,8,9,10,11. In the past, a comprehensive investigation of their evolution was limited by a lack of cohort genome sequence data. To address this gap, we sequenced the complete genomes for all 20 previously unsequenced serotypes within HAdV-D, for which only limited nucleotide sequence data was previously available (Fig. 1A, and Supplemental Table 1), and herein present the first comprehensive analysis of the complete set of HAdV-D whole genomes.
Figure 1.
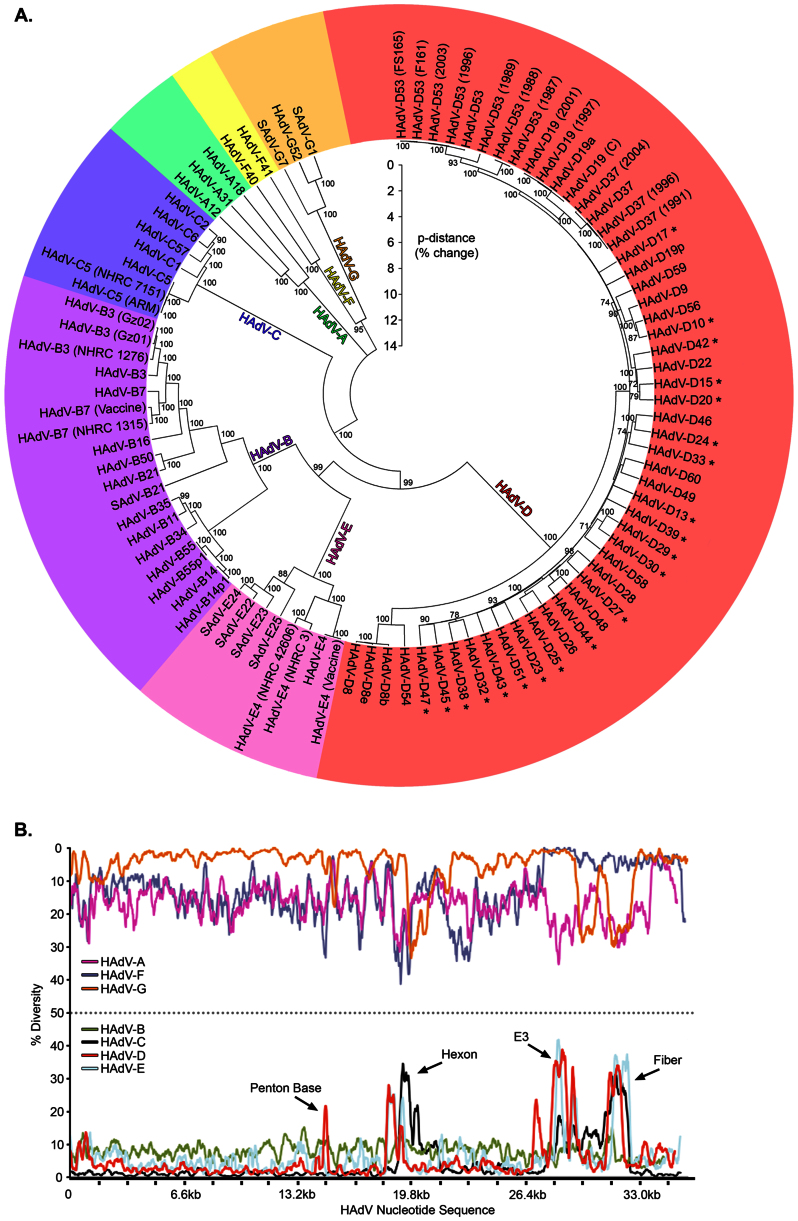
Human adenovirus diversity (A) Genome phylogenetic analysis of human adenoviruses is presented as a bootstrap-confirmed (500 replicates) neighbor-joining tree constructed with whole genome sequence from all known HAdV types.The evolutionary differences were computed using the p-distance method and represent the proportion of nucleotide differences. Genomes that are newly reported in this study are designated by a *. (B) Nucleotide diversity plots constructed using DnaSP v5 represent the average number of nucleotide differences per site between each type in every HAdV species. The % diversity is calculated on the y-axis and the x-axis illustrates the nucleotide position on the genome.
Results
HAdV-D genomes have focused genetic diversity located in hypervariable regions
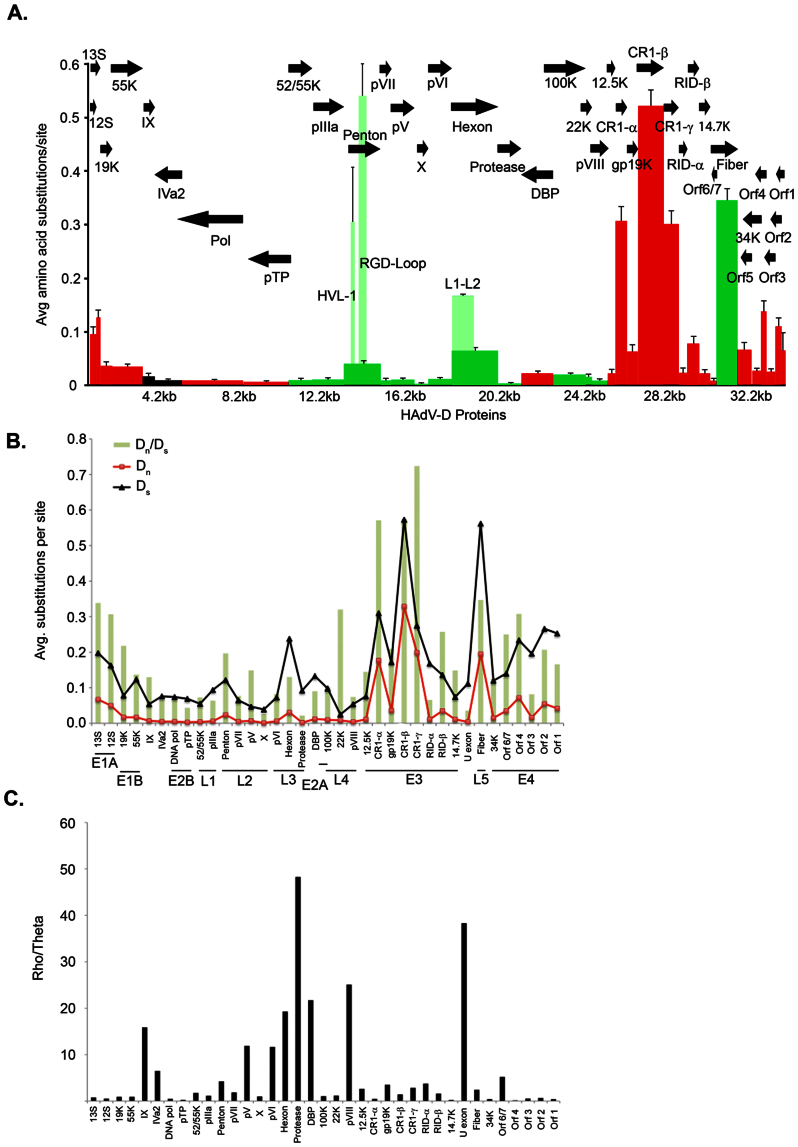
To place HAdV-Ds in context, genetic variation was compared across all HAdV species by constructing nucleotide diversity plots. While diversity varied between different HAdV species, HAdV-Ds were distinguished by a remarkable dichotomy between high nucleotide sequence conservation and stereotypical focal variation in regions including the hexon, fiber, and penton base genes, which encode for the three major structural proteins of the viral capsid, and the E3 transcription unit (Fig. 1B). HAdV-Cs showed a similar pattern but lacked variation in the penton base gene. As confirmation of the diversity plots, we inferred phylogenetic distances based on the average amino acid substitutions for each HAdV-D protein, validating higher average substitution rates in the hypervariable loops of the penton base and hexon proteins, the entire fiber protein, and the CR1-α, -β, and -γ genes within the E3 region. Amino acid substitutions were uncommon elsewhere, including for example, the highly conserved DNA polymerase (Fig. 2A). The ratios of nonsynonymous to synonymous nucleotide substitutions were also greatest for the variable regions (Fig. 2B). Therefore, each of these highly variable regions of HAdV-D genomes is likely under significant host immune pressure.
Figure 2. HAdV-D evolution.
(A) Amino acid diversity calculated in MEGA 4.02, measuring the average amino acid substitution for each HAdV-D protein. Each bar in the graph corresponds to a protein as represented by arrows. Red = early. Light green represents the hypervariable region of the hexon and penton base. Dark green = late genes. Black = intermediate genes. (B) Analysis of synonomous and non-synonomous mutations across the HAdV-D genome calculated using MEGA software. Synonomous (Ds) and non-synonomous (Dn) changes are represented in black and red lines, respectively. Green bars represent the ratio (Dn/Ds) for each gene. (C) Analysis of the rho (recombination) and theta (mutation) ratio as determined by DnaSP for each gene in the HAdV-D genome.
HAdV-D genomes may recombine more frequently than their species counterparts
Previously we have identified recombination in prototype and novel HAdV-D and HAdV-B types4,5,8,9,10. To identify recombination rates across HAdV species, genomes were parsed for recombination (rho) and mutation (theta) events12, for all HAdV types within each species. Across all HAdV species, rho/theta ratios ranged from 0.002 to 0.119 (Supplemental Table 2). Since the values for HAdV-D were lower than expected, we next examined each HAdV-D gene individually. We identified rho/theta ratios of >1 for many individual HAdV-D genes (Figs. 2C, S1A, and S1B), reflecting evolution through recombination. In contrast, HAdV-B, the second largest species after HAdV-D, showed greater amino acid variability across the genome (Figs. S2A and S2B), and rho/theta was ≤ 0.4 for every gene (Fig. S2C). Taken together this data suggests, in contrast to HAdV-B, the genomes of HAdV-D have evolved via recombination rather than by base substitution.
Proteotyping identifies recombination in every HAdV-D type
To examine potential recombination pairs within HAdV-D types, we applied a previously developed computational approach, known as proteotyping13. For the 38 HAdV-Ds analyzed, 28 unique hexon proteotypes were identified (Fig. 3A and Supplemental Table 3), of which 20 were unshared between viruses, and eight were shared. Among the shared hexon proteotypes, two were common to three virus members each, and six proteotypes to two virus members each. Thus, at least 10 out of the 38 HAdV-D types likely represent recombinants resulting from exchanges in the hexon coding region. Analysis of the fiber gene suggested 22 unique proteotypes among 38 viruses (Fig. 3B and Supplemental Table 3), of which 10 were shared between viruses. At least 16 fiber genes appear to have evolved as a result of homologous recombination.
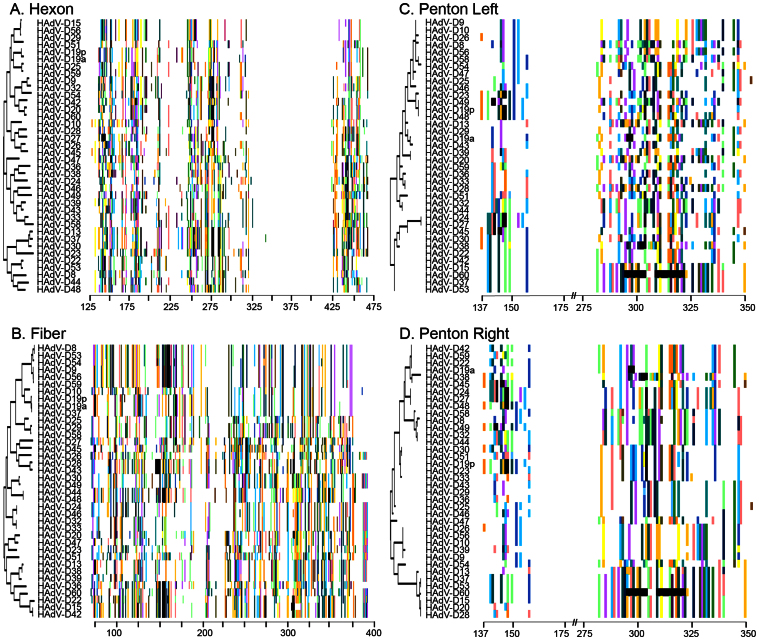
Figure 3. Proteotyping assignments for hypervariable HAdV-D proteins.
Neighbor-joining phylogenetic trees are shown on the left for each protein. The amino acid signatures are shown to the right. Each amino acid that was variable from consensus sequence was assigned a color. White regions represent sequence that are conserved and match consensus. (A) Hexon, (B) Fiber (C) Penton base, organized according to the HVL-1 based tree, (D) Penton base, organized according to the RGD loop (HVL-2) based tree.
In the penton base protein, there are two distinct hypervariable loops (HVLs) – separated by 123 amino acids – which may undergo recombination as separate segments9,14. The HVL-specific recombination of penton base proteotypes was examined further by generating one neighbor-joining tree based on HVL1, and one based on the arginine-glycine-aspartic acid (RGD) loop, also known as HVL2. As can be seen in Figs. 3C and 3D, the phylogenetic tree differs when sorting by HVL1 or HVL2, as does the overall proteotype pattern. Despite appearances, the genomic region encompassing both penton base HVLs statistically was more likely to recombine as a single unit than as two separate entities (p < .0001, Fisher's exact test, H0: independence). In contrast, two more distal regions of the genome – the hexon and fiber genes– were more likely to recombine independently of one another (p = .392), suggesting that proximity on the linear genome dictates the likelihood that two different hypervariable regions will undergo homologous recombination together. In summary, at least 24 of the penton base HVL1 and 28 of the RGD (HVL2) loops were the product or source of recombination with other HAdV-Ds (Supplemental Table 3). The E3 coding region in HAdV-Ds includes eight potential open reading frames15. Confirmed E3 gene products facilitate immune evasion by the virus16,17, but are not required for viral replication in vitro18. Our analysis of the putative gene product of E3 CR1-β19,20, chosen because of its remarkably high amino acid variability (Fig. 2A), showed 14 unique proteotypes (Figure S3A and Supplemental Table 3), with 10 shared and four unshared among viruses. At least 24 of 38 HAdV-Ds appeared to be recombinant for E3 CR1-β. In summary, proteotype analysis identified homologous recombination in at least two of the five examined hypervariable regions for every virus. As controls, the highly conserved DNA polymerase and DNA binding proteins were also analyzed, yielding only one proteotype for each gene (Figs. S3B and S3C).
HAdV-D recombination is found within GC/AT transition zones
GC-rich motifs have been associated with increased genomic stability and relative resistance to homologous recombination21. HAdV-D genomes are highly conserved, and also possess among the highest GC content among all HAdV species. Gene-by-gene analysis for GC content revealed regions of the HAdV-D genome most likely to undergo homologous recombination also demonstrate abrupt reductions in GC content (Fig. 4A). To analyze these areas we developed software (http://binf.gmu.edu/sequence_range) to identify potential regions responsible for homologous recombination in HAdV-Ds based on GC and AT content. As a control, SV40 (GenBank acc. no. NC_001526.2), known to readily undergo homologous recombination22, and HPV (GenBank acc. no. NC_001669.1), which does not regularly recombine23, were also subjected to similar analysis. Working empirically, we identified stereotypical GC/AT transition zones, 30–45 nucleotides in length, within conserved regions24 of HAdV-D DNA adjacent to all the observed hypervariable regions within the penton base, hexon, E3 CR1-β, and fiber genes (Fig. 4B and Supplemental Table 4). These represent potential sites for the initiation of homologous recombination of adjacent hypervariable regions. Analogous GC/AT transition zones were observed in SV40 but not in HPV (Lee and Chodosh, unpublished data).
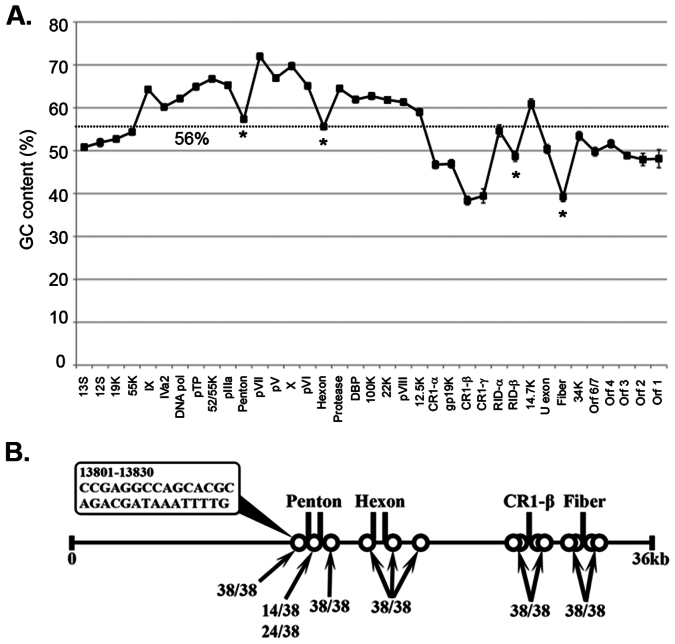
Figure 4. HAdV-D recombination analysis.
(A) Average % GC content per gene across the HAdV-D genome is presented. Error bars represent standard deviation. Dotted line represents the average % GC content across the whole genome. The penton base, hexon, and fiber genes all showed a significant decline in GC content compared to their nearest neighbor genes (*p < .0014). (B) Recombination hot spot analysis. A 15 bp sliding window was used to analyze GC to AT transition zones (10% threshold over mean % GC content). The HAdV-D genome is represented by a horizontal solid line. Vertical solid lines and circles indicate homologous regions and potential recombination hot spots. A penton base GC to AT transition zone example is presented in the bubble.
Discussion
This work presents whole genomic sequences for every previously unsequenced HAdV type, and publication of these sequences allows high-resolution analyses of HAdV evolution. To compare and contrast genomes among different HAdV species, we first set out to examine diversity within each species using nucleotide diversity plots. HAdV-D regions coding for the hexon, fiber, and penton base, as well as the E3 transcription unit were distinctly hypervariable. Each of the three major adenovirus capsid genes encodes at least one hypervariable component present on the exterior surface of the viral capsid25,26,27,28. For fiber and penton base proteins, amino acids within these hypervariable regions are responsible for host cell binding and internalization, respectively29. Confirmed E3 proteins facilitate immune evasion by the virus16,17. Looking broadly across the genome of HAdV-Ds, the degree of overall conservation punctuated by stereotypical regions of diversity appears in stark contrast to most other HAdV species. While the major capsid genes for all HAdV species demonstrate diversity, nucleotide sequence outside the capsid and E3 coding regions is more conserved in HAdV-Ds than any other species. This combination of high sequence conservation interspersed with formulaic areas of deviation is particularly characteristic of HAdV-Ds.
Homologous recombination is a critical mechanism for the maintenance of genome fitness and diversity30,31,32, and for HAdV-Ds may allow exchange of those regions of the genome most susceptible to immune pressures from the host. We examined the recombination potential among HAdV-D types. Comparison of recombination and mutation rates suggested mutation rather than recombination (rho/theta < 1) was the predominant evolutionary process driving genetic variation across HAdV-D genomes as a whole. Whole genome analysis of rho/theta for HAdV-Ds was low (0.05), but an averaging effect was suspected due to the high degree of sequence conservation across the majority of the genome. To test this, we performed the same analysis gene by gene, and identified many with rho/theta >1, consistent with evolution of those genes by recombination. To compare these results to another HAdV species, we also examined recombination and mutation rates in HAdV-B genomes, chosen because HAdV-B has the second largest number of unique types after HAdV-D. Unlike HAdV-Ds, recombination/mutation rate ratios across all genes of HAdV-Bs was < 1, suggesting that mutation rather than recombination plays a more important role in generating diversity within this species. Therefore, in comparison to HAdV-Bs, viruses within HAdV-D are more likely to recombine. It should be noted that these results could be biased by the comparatively fewer typed HAdV-Bs available for analysis (n = 10). However, this conclusion is consistent with the relatively greater degree of sequence homology – and therefore greater potential for homologous recombination – in HAdV-D genomes.
To identify the extent of HAdV-D recombination within the species, we applied a proteotyping method previously used to study the evolution of avian influenza virus13. Genome proteotyping examines molecular evolution by elucidating differences in predicted amino acid sequence that phylogenetic trees may fail to distinguish, and can differentiate shared or unshared “protein types” among a population of proteins coded for by the same gene. The results of this analysis suggest that recombination has occurred for at least two hypervariable regions of every known HAdV-D genome. Furthermore, by proteotyping, it was predicted that serum neutralization, which relies on the host's humoral immune response against hypervariable loops 1 and 2 on the hexon protein (ε determinant), can unequivocally identify only 53% (20/38) of the now fully characterized HAdV-D genomes.
HAdV-D genomes have one of the highest GC contents among HAdV species. Because GC content is associated with genome stability and resistance to recombination21, we examined the GC content of each gene across the genome. In areas prone to recombination, GC content was abruptly reduced. Notably, among HAdV-D genes judged most likely by proteotyping to have recombined, E3 CR1-β shows the lowest GC content of any open reading frame across the entire genome. A possible explanation for homologous recombination in dsDNA viruses involves the formation of hairpin loops in dissociated ssDNA during DNA replication33, mediated by a nucleotide region enriched with GC followed by one of equal length enriched for AT34,35. Further analysis identified stereotypical transition zones, GC-rich to AT-rich, 30–45 nucleotides in length, immediately preceding and following hypervariable nucleotide regions shown to be targets of homologous recombination. For example, highly similar GC/AT transition sequence was found in the conserved region between hypervariable loops 1 and 2 of the HAdV-D penton base gene, a site of certain homologous recombination9. Similar GC/AT transition zones were also identified in a SV40 genome but not in a HPV genome – only the former is known to recombine. These data suggest that abrupt transition in nucleotide sequence from GC-rich to AT-rich may be critical for recombination among HAdV-Ds (and also for other dsDNA viruses). GC/AT transition zones in HAdV-Ds permit rapid evolution by the virus in response to environmental bottlenecks and immune pressures. Further studies are in progress to directly test the role of GC/AT transition zones in homologous recombination among HAdV-Ds.
Our work completes the public collection of high quality reference sequences for all previously unsequenced HAdV serotypes, and provides a detailed foundation for the analysis of emerging viruses. In specific, HAdV-Ds appear to evolve through homologous sharing of specific genomic parts, containing hypervariable coding regions for surface epitopes or immune modulatory proteins. Therefore, existing HAdV-Ds represent an evolutionary sampling of potential diversity, and proteotype analysis defines the available recombination palette of hypervariable regions for the evolution of new HAdV-Ds. It is also important to note that the evolution of new HAdVs by homologous recombination requires co-infection in the same host cell of at least two unique genotypes from the same viral species36,37,38,39,40,41,42. The emergence of new HAdV-Ds during the AIDS epidemic suggests a role for persistence of multiple viruses under reduced immune surveillance43,44,45. Future experiments detailing the mechanism for molecular evolution of HAdVs will be vital as new viruses, some with altered tissue tropisms and increased virulence, emerge through homologous recombination.
Methods
Cells and virus
All viruses sequenced in this study were obtained from the American Type Culture Collection (ATCC, Manassas, VA) except for HAdV-D10, which was a kind gift from Dr. David Schnurr at the California Department of Public Health. Each viral stock was grown in A549 cells (CCL-185, ATCC) and purified by CsCl gradient. DNA was extracted using Roche MagnaPure (Branford, CT) and quantitated using a Nanodrop 8000 (Thermo Scientific, Wilmington, DE).
Genome sequencing and annotation
Purified DNA was sequenced on a Roche 454 DNA sequencer by Operon (Eurofins MWG Operon; Huntsville, Alabama), to at least 17-fold depth (Next Gen), with an accuracy of greater than 99% (Q20 or better) (Supplemental Table 1). The sequencing reads were assembled using CLC Genomics Workbench (http://www.clcbio.com/index.php?id=1240), with an N50 average of 5,260. HAdV-D13, D32, and D39 along with viral inverted terminal repeat regions were sequenced on an ABI 3730 XL (Applied Biosystems, Carlsbad, CA) to an 8-fold coverage. Annotation was performed using a custom annotation engine (Dyer and coworkers, unpublished) and the Genome Annotation Transfer Utility46, with confirmation from NCBI's open reading frame (ORF) finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). Artemis (http://www.sanger.ac.uk/resources/software/artemis/) was used to evaluate the data47,48. Open reading frames were BLAST-analyzed against GenBank sequences for confirmation and protein similarity. Splice sites were predicted using the GenScan web server at MIT (http://genes.mit.edu/GENSCAN.html). Quality control included sequence annotation and comparison with HAdV genome landmarks.
Sequence analysis
Sequences were aligned using the ClustalW49 option within the software Molecular Evolutionary Genetics Analysis (MEGA) 4.0.2 (http://www.megasoftware.net/)50. DnaSP v5.10.01 (http://www.ub.edu/dnasp/)51 was used to calculate nucleotide diversity across the whole genome. Whole genome alignments of each HAdV species were used to calculate the average number of nucleotide differences per site between the sequences. A window of 200 bps sliding window (20 bps) was used to create each plot. Recombination analysis was carried out using RDP3 program suite (http://darwin.uvigo.es/rdp/rdp.html)52. Phylogenetic analysis was preformed using bootstrap-confirmed neighbor-joining trees (500 replicates) also designed with MEGA 4.0.2 using the p-distance model. The average number of amino acid substitutions per site (amino acid diversity) between sequences was calculated using MEGA 4.0.2 using the Poisson correction model. MEGA was also used to estimate the number of synonymous substitutions per synonymous site (Ds) and the number of non-synonymous substitutions per non-synonymous site (Dn).
Proteotyping
Amino acid alignments were performed using ClustalW option and a maximum likelihood (ML) tree was created using MEGA software. From the amino acid alignment, a clade-guided consensus sequence was determined and each amino acid was assigned a unique, arbitrary color. Residues that matched the consensus were colored white and gaps in the alignment were colored black. Unique amino acids along with a 10% sequence divergence threshold were used to identify unique proteotypes.
Recombination hot spot analysis
Using C++, we developed a software program (http://binf.gmu.edu/sequence_range; username: GCATuser; password: GCATtransition) to identify possible homologous recombination hotspots over all 38 adenovirus genomes. The program uses a sliding and variable sized window to analyze GC and AT content by percentage. A 10% threshold was selected (above average GC content) to identify GC to AT transition sites. Among all “hits”, combinations showing GC-rich/GC-AT-moderate/AT-rich regions or GC-rich/AT-rich regions were selected as candidate regions for homologous recombination. Attention was directed to identification of such GC/AT transition zones adjacent to five hypervariable regions including both penton base hypervariable loops, hexon, fiber and E3 CR1-β genes.
Statistical methods
In proteotyping the hypervariable genes, Fisher's exact test was used to test the null hypothesis that recombination between any two genes occurred independently. Significance was predetermined as α = .05. To analyze whether Dn/Ds ratios differed across the genome, two approaches were taken. The first approach assumed normality of the logarithm of the ratios, and used the simple bootstrap to estimate their variances. This approach controlled the null hypothesis weakly, in that it is valid under the complete null hypothesis that all ratios are equal. While some of the unadjusted p-values were small, none met the threshold for significance at an overall 0.05 level once multiple comparison adjustments were applied. In a second approach, the equivalence of each log Dn/Ds ratio to the average of all remaining log ratios was tested. This vastly reduced the number of comparisons, but also the power. None of these p-values, while small without adjustment, met any threshold for significance at the α = .05 level.
GC nucleotide content differences were examined by assuming independence between adjacent genes, and for each triple, testing whether the difference between the first two of the three was negative, and the difference between the second two was positive. A single p value was calculated as the union of the two events that could make the outcome more extreme than what was observed, under the assumption that the two differences are independent; this is an upper bound when the magnitudes of the differences are positively associated. Correction was made for the 35 tests performed (p value threshold of 0.05/35 = 0.0014), such that p < .0014 was considered statistically significant.
Author Contributions
J.C., M.S.J., D.S., D.W.D., C.M.R., J.Y.L. and G.S. conceived and designed experiments. C.M.R., G.S., J.Y.L., S.D., J.R., E.L., M.A.Y. and R.A.B. performed the experiments and analyzed the data: C.M.R., J.C., M.S.J., D.S., D.W.D. and R.A.B. wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Data
Acknowledgments
Supported by NIH grants EY013124, EY021558, and EY014104, a Research to Prevent Blindness Senior Scientific Investigator Award (JC), the Massachusetts Lions Eye Research Fund, and with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, grant UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers).
References
- Chow L. T., Gelinas R. E., Broker T. R. & Roberts R. J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 (1977). [DOI] [PubMed] [Google Scholar]
- Berget S. M., Moore C. & Sharp P. A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A 74, 3171–5 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P. et al. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334, 124–9 (1988). [DOI] [PubMed] [Google Scholar]
- Robinson C. M. et al. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409, 141–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. P. et al. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J Clin Microbiol 48, 991–3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer J. C. Adenoviruses in the immunocompromised host. Clin Microbiol Rev 5, 262–74 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong J. C. et al. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J Clin Microbiol 37, 3940–5 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. P. et al. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4, e5635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C. M. et al. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J Virol 83, 8980–5 (2009).19553309 [Google Scholar]
- Singh G. et al. Over-reliance on the hexon gene leading to misclassification of human adenoviruses. J Virol 86, 4693–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H. et al. Recombination analysis of intermediate human adenovirus type 53 in Japan by complete genome sequence. J Gen Virol 92, 1251–9 (2011). [DOI] [PubMed] [Google Scholar]
- Morrell P. L., Toleno D. M., Lundy K. E. & Clegg M. T. Estimating the contribution of mutation, recombination and gene conversion in the generation of haplotypic diversity. Genetics 173, 1705–23 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer J. C. et al. Large-scale sequence analysis of avian influenza isolates. Science 311, 1576–80 (2006). [DOI] [PubMed] [Google Scholar]
- Al Qurashi Y. M., Alkhalaf M. A., Lim L., Guiver M. & Cooper R. J. Sequencing and phylogenetic analysis of the hexon, fiber, and penton regions of adenoviruses isolated from AIDS patients. J Med Virol 84, 1157–65 (2012). [DOI] [PubMed] [Google Scholar]
- Robinson C. M., Seto D., Jones M. S., Dyer D. W. & Chodosh J. Molecular evolution of human species D adenoviruses. Infect Genet Evol 11, 1208–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windheim M., Hilgendorf A. & Burgert H. G. Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr Top Microbiol Immunol 273, 29–85 (2004). [DOI] [PubMed] [Google Scholar]
- Mahr J. A. & Gooding L. R. Immune evasion by adenoviruses. Immunol Rev 168, 121–30 (1999). [DOI] [PubMed] [Google Scholar]
- Berkner K. L. & Sharp P. A. Generation of adenovirus by transfection of plasmids. Nucleic Acids Res 11, 6003–20 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blusch J. H. et al. The novel early region 3 protein E3/49K is specifically expressed by adenoviruses of subgenus D: implications for epidemic keratoconjunctivitis and adenovirus evolution. Virology 296, 94–106 (2002). [DOI] [PubMed] [Google Scholar]
- Windheim M. & Burgert H. G. Characterization of E3/49K, a novel, highly glycosylated E3 protein of the epidemic keratoconjunctivitis-causing adenovirus type 19a. J Virol 76, 755–66 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Moretto V., Ehrlich S. D., Duwat P. & Dabert P. GC-rich DNA sequences block homologous recombination in vitro. J Biol Chem 266, 6667–9 (1991). [PubMed] [Google Scholar]
- Jasin M., de Villiers J., Weber F. & Schaffner W. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell 43, 695–703 (1985). [DOI] [PubMed] [Google Scholar]
- Ostrow R. S., Zachow K. R. & Faras A. J. Molecular cloning and nucleotide sequence analysis of several naturally occurring HPV-5 deletion mutant genomes. Virology 158, 235–8 (1987). [DOI] [PubMed] [Google Scholar]
- Boursnell M. E. & Mautner V. Recombination in adenovirus: crossover sites in intertypic recombinants are located in regions of homology. Virology 112, 198–209 (1981). [DOI] [PubMed] [Google Scholar]
- Fuschiotti P. et al. Structure of the dodecahedral penton particle from human adenovirus type 3. J Mol Biol 356, 510–20 (2006). [DOI] [PubMed] [Google Scholar]
- Zubieta C., Schoehn G., Chroboczek J. & Cusack S. The structure of the human adenovirus 2 penton. Mol Cell 17, 121–35 (2005). [DOI] [PubMed] [Google Scholar]
- Crawford-Miksza L. & Schnurr D. P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol 70, 1836–44 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisch I. et al. Phylogenetic analysis and structural predictions of human adenovirus penton proteins as a basis for tissue-specific adenovirus vector design. J Virol 81, 8270–81 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E. & Nemerow G. R. Virus yoga: the role of flexibility in virus host cell recognition. Trends Microbiol 12, 162–9 (2004). [DOI] [PubMed] [Google Scholar]
- Szekvolgyi L. & Nicolas A. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J 277, 571–89 (2010). [DOI] [PubMed] [Google Scholar]
- White M. F. Homologous recombination in the archaea: the means justify the ends. Biochem Soc Trans 39, 15–9 (2011). [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T. & Eichler E. E. The evolution of human segmental duplications and the core duplicon hypothesis. Cold Spring Harb Symp Quant Biol 74, 355–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. D. & Bujarski J. J. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J Virol 69, 131–40 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. D. & Bujarski J. J. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J Virol 71, 3799–810 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K. et al. Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. J Gen Virol 88, 298–315 (2007). [DOI] [PubMed] [Google Scholar]
- McCarthy T., Lebeck M. G., Capuano A. W., Schnurr D. P. & Gray G. C. Molecular typing of clinical adenovirus specimens by an algorithm which permits detection of adenovirus coinfections and intermediate adenovirus strains. J Clin Virol 46, 80–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. et al. Resequencing microarray for detection of human adenoviruses in patients with community-acquired gastroenteritis: a proof-of-concept study. J Med Microbiol 59, 1387–90 (2010). [DOI] [PubMed] [Google Scholar]
- Cao Y. et al. Genotyping of human adenoviruses using a PCR-based reverse line blot hybridisation assay. Pathology 43, 488–94 (2011). [DOI] [PubMed] [Google Scholar]
- Al Qurashi Y. M., Guiver M. & Cooper R. J. Sequence typing of adenovirus from samples from hematological stem cell transplant recipients. J Med Virol 83, 1951–8 (2011). [DOI] [PubMed] [Google Scholar]
- Barrero P. R., Valinotto L. E., Tittarelli E. & Mistchenko A. S. Molecular typing of adenoviruses in pediatric respiratory infections in Buenos Aires, Argentina (1999–2010). J Clin Virol 53, 145–50 (2012). [DOI] [PubMed] [Google Scholar]
- Vora G. J. et al. Co-infections of adenovirus species in previously vaccinated patients. Emerg Infect Dis 12, 921–30 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzgar D. et al. PCR analysis of Egyptian respiratory adenovirus isolates, including identification of species, serotypes, and coinfections. J Clin Microbiol 43, 5743–52 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein J. M. & Dieterich D. T. The histopathology of 103 consecutive colonoscopy biopsies from 82 symptomatic patients with acquired immunodeficiency syndrome: original and look-back diagnoses. Arch Pathol Lab Med 125, 1042–6 (2001). [DOI] [PubMed] [Google Scholar]
- Yan Z., Nguyen S., Poles M., Melamed J. & Scholes J. V. Adenovirus colitis in human immunodeficiency virus infection: an underdiagnosed entity. Am J Surg Pathol 22, 1101–6 (1998). [DOI] [PubMed] [Google Scholar]
- Curlin M. E. et al. Frequent detection of human adenovirus from the lower gastrointestinal tract in men who have sex with men. PLoS ONE 5, e11321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherepanov V., Ehlers A. & Upton C. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7, 150 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K. et al. Artemis: sequence visualization and annotation. Bioinformatics 16, 944–5 (2000). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–8 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M. & Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24, 1596–9 (2007). [DOI] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–2 (2009). [DOI] [PubMed] [Google Scholar]
- Martin D. P. et al. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26, 2462–3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data