Abstract
The rearrangement of T and B lymphocyte antigen receptor loci occurs within a highly complex chromosomal environment and is orchestrated through complex mechanisms. Over the past decade, a large body of literature has highlighted the significance of chromatin architecture at antigen receptor loci in supporting the genomic assembly process: in preparation for recombination, these loci tend to contract and form multiple loops that shorten the distances between gene segments and facilitate recombination events. A CCCTC binding factor, CTCF, has received much attention in this regard since it has emerged as an important regulator of chromatin organization and transcription. In this review, we will summarize recent work outlining conformational dynamics at antigen receptor loci during lymphocyte development and we will discuss the role of CTCF in antigen receptor locus conformation and repertoire development.
Introduction
Adaptive immunity in jawed vertebrates is mediated by T and B lymphocytes that express highly diverse and clonally distributed antigen receptors. The diversity of lymphocyte antigen receptors is generated primarily by the assembly of variable (V), diversity (D) and joining (J) gene segments at T cell receptor (TCR) and immunoglobulin (Ig) loci (1). This process, known as V(D)J recombination, is catalyzed by the recombination-activating gene-1 and -2 proteins (RAG-1 and RAG-2, hereafter referred to as RAG). The RAG proteins recognize recombination signal sequences (RSSs)3 that flank all V, D and J gene segments, and with two RSSs held in a synaptic complex, create double-strand breaks that can be rejoined to assemble V, D and J gene segments with tremendous combinatorial diversity. Antigen receptor loci undergo recombination in a manner that is regulated according to cell lineage and developmental stage in T and B lymphocyte precursors (2, 3). During T lymphocyte development in the thymus, Tcrb, Tcrd and Tcrg genes recombine at the CD4−CD8− double negative (DN) stage, whereas Tcra genes recombine at the CD4+CD8+ double positive (DP) stage. B lymphocyte development in the bone marrow is similarly characterized by developmentally-staged recombination of Ig heavy (Igh) and light (Igk and Igl) chain genes, with Igh recombination in pro-B cells and Igk and Igl recombination in pre-B cells. Recombination events are also regulated within individual loci; for example, D-to-J recombination precedes V-to-DJ recombination at both Igh and Tcrb, and proximal V-to-J recombination precedes distal V-to-J recombination at Tcra (2, 3).
The V(D)J recombination programs at TCR and Ig loci are controlled at multiple levels (3–5). Most fundamental is the restriction imposed by RAG protein expression, which, except in special circumstances (6), is limited to developing pro- and pre-B cells and DN and DP thymocytes (7). Beyond that, the individual antigen receptor loci carry complex arrays of cis-regulatory elements, including enhancers and promoters, that dictate regional changes to transcription and chromatin structure that, in turn, allow RAG proteins to access particular RSSs at the appropriate developmental stage (3–5). In many instances, functional communication between enhancers and promoters must occur over long distances across a landscape that includes many such elements, raising questions as to how these functional interactions are targeted and regulated (8). Most recently, the spatial dynamics of antigen receptor loci within immature lymphocyte nuclei has also emerged as a critical aspect of their regulation (4, 9, 10). The seminal observation relied on three dimensional fluorescence in situ hybridization (3D-FISH) to visualize immunoglobulin loci in cell nuclei: Igh and Igk moved away from the nuclear periphery and the two ends of the Igh locus were less separated in nuclei of pro-B cells as compared to other cells (11). Subsequent studies revealed “contraction” to be a general property of antigen receptor loci that occurs during the developmental stage in which V segments undergo recombination, with “decontraction” occurring subsequently (12–16). Because contraction and decontraction can be detected even in recombinase-deficient nuclei, they are thought to set the stage for and to terminate long-distance recombination events, respectively, during lymphocyte development. Since RAG proteins bind preferentially to J or D-J clusters to form “recombination centers”, contraction and decontraction would move distant V gene segments into or out of these recombination centers to allow regulated assembly of a V gene repertoire (4).
Chromatin architecture, CTCF and cohesin
Eukaryotic genomes are packaged at multiple levels to solve a critical “space” problem in the nucleus while simultaneously facilitating transcription and replication of the DNA template (17). Recent technological advances in the analysis of genomic spatial relationships have allowed insights into the principles of higher-order chromatin organization (18). These technologies are all derivative of the chromosome conformation capture technique (3C), in which interacting DNA fragments are initially trapped by chemical crosslinking; following a restriction enzyme digest and addition of DNA ligase under conditions favoring intramolecular ligation, any two interacting DNA fragments can then be identified and quantified by PCR using oligonucleotide primers specific for the two fragments (19). In a version of this technology known as 4C, circular DNA resulting from ligation of both ends of two interacting fragments is amplified by inverse PCR using two primers in a “bait” or “viewpoint” sequence, and the entire universe of sequences interacting with the bait is determined by microarray or deep sequencing (20, 21). In a further adaptation of this technology known as Hi-C, there is no defined “bait”; rather, deep sequencing is used to identify the entire genomic universe of interacting sequences (22). Over the last several years, 4C and Hi-C analysis has revealed segregation of the genome into discrete spatial compartments of up to a few megabases in length that correspond to domains of active or inactive chromatin. Long-distance interactions are more frequent within a domain but also occur between domains, with active chromatin domains interacting more frequently with other active domains, and inactive chromatin domains interacting more frequently with other inactive domains (20, 22–24). The data support a “fractal globule” model of the genome in which locally looped and packaged globular units are then further assembled to package the entire genome.
CTCF (CCCTC-binding factor) is a highly conserved, ubiquitously expressed transcription factor that binds a GC-rich consensus sequence (25). CTCF has been attributed multiple functions, including transcriptional activation, transcriptional repression and transcriptional insulation (the ability to block an enhancer from activating a promoter when CTCF is situated between the two elements). In addition, CTCF has been shown to function as a chromatin organizer that mediates long-distance looping interactions in the genome (25). The ability of CTCF to mediate such loops may underlie some or all of the above activities; CTCF-mediated loops could activate transcription by bringing together enhancer and promoter elements that are associated with nearby CTCF sites, or could insulate or suppress transcription by segregating enhancers and promoters that have intervening CTCF sites. Indeed, there are tens of thousands of CTCF binding sites distributed in a range of contexts across mammalian genomes (26–30). As might be expected for a role in insulation, the boundaries between topologically and functionally defined chromatin domains are highly enriched for CTCF sites. However, only 15% of all CTCF sites are located at these boundaries (24); other CTCF sites are distributed at enhancers, promoters, and other genic and intergenic sites and mediate interactions between these sites (24, 30–32). Recent analysis of the pro-B cell genome indicated that long-distance interactions within chromatin spatial compartments were associated with CTCF binding, whereas those between spatial compartments were associated with the binding of lineage-specific transcription factors (23).
Cohesin is a multi-subunit protein complex that is well-known for its role in mediating sister-chromatid cohesion during cell division; it is thought to function by forming a closed ring around the two newly replicated DNA double strands (33). Cohesin is now also appreciated to play important roles in gene expression and chromatin organization in interphase cells: notably, it binds in a CTCF-dependent fashion to about 70% of CTCF binding sites genome-wide (34–36) and is necessary for CTCF-dependent insulation and looping (37–39). Although in this manner CTCF and cohesin function together at many sites in the genome, they can also interact with other transcription factors and can function independent of each other (40, 41).
ChIP-chip and ChIP-Seq analyses have revealed antigen receptor loci to be particularly enriched in binding sites for CTCF and cohesin (26, 42–47). As a result, both proteins have drawn attention as potential regulators of locus conformation, transcriptional activity and V(D)J recombination. We focus below on recent studies of the Igh, Igk and Tcra/Tcrd loci. Much less is known about Tcrb locus architecture and there have been no studies of the Tcrg and Igl loci.
The studies of locus architecture and transcription discussed below have generally been conducted using immature cell populations isolated from recombinase-deficient mice, because this eliminates the confounding effects of changes in spatial relationships that are a consequence of V(D)J recombination. That said, it is an assumption that the RAG proteins themselves would not substantially impact the parameters being measured; this could be addressed by analyzing cells isolated from mice expressing a catalytically inactive RAG protein complex (48).
Igh locus
The murine Igh locus on chromosome 12 contains nearly 200 VH gene segments spanning 2.7 Mb of DNA, followed by 10–13 DH, four JH, and eight CH gene segments (2) (Fig. 1A). Igh locus V(D)J recombination occurs in a strictly ordered fashion, with DH-to-JH recombination preceding VH-to-DHJH recombination in pro-B cells. The VH-to-DHJH step is further regulated to support allelic exclusion, as it is terminated as a consequence of feedback inhibition in pre-B cells. Notably, each Igh allele generally undergoes VH-to-DHJH recombination only once, since this rearrangement eliminates all unrearranged DH gene segments. Therefore, although several DH-proximal VH gene segments do rearrange at elevated frequencies, DH-distal VH gene segments must be able to compete effectively with these proximal VH gene segments to allow the assembly of a diverse Igh repertoire.
Figure 1.
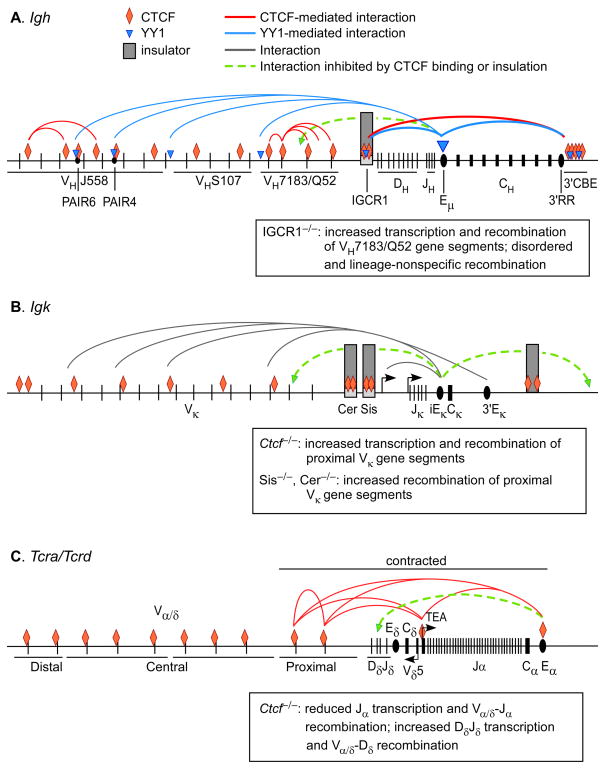
Long-distance interactions at antigen receptor loci. Long-distance DNA contacts are depicted for the contracted conformation of the Igh locus in pro-B cells (A), the contracted conformation of the Igk locus in pre-B cells (B) and the 3′ contracted but 5′ extended conformation of the Tcra/Tcrd locus in DP thymocytes (C). Interactions known or presumed to be mediated by CTCF or YY1 are indicated. Transcription and recombination phenotypes of various genetic models are summarized (boxes). Igk interactions involving Sis (45) are not depicted.
3D-FISH data have shown that the Igh locus contracts specifically in pro-B cells (11, 12, 14, 49) and that this conformation brings all VH gene segments into proximity of the DHJH cluster (13). Spatial-distance measurements and computer modeling suggested that in its extended conformation in pre-pro B cells, the Igh locus is organized into at least three rosette-like compartments, each composed of multiple DNA loops, and that these compartments merge in pro-B cells (13). Unrearranged Igh loci then appear to decontract in pre-B cells, correlating with the suppression of VH-to-DH recombination associated with allelic exclusion (14). Molecular mechanisms of Igh contraction and decontraction are only partly understood. Transcription factors Pax5, Yin-Yang 1 (YY1) and Ikaros have been shown to be essential for complete Igh locus contraction as well as normal frequencies of distal VH rearrangement (12, 50, 51). Since distal VH segments appear to be “accessible” in Pax5- or YY1-deficient pro-B cells based on their germline transcription and histone modifications, it has been inferred that distal VH-to-DHJH rearrangement relies on Igh locus contraction (12, 50). Elimination of the Igh intronic enhancer (Eμ), located between JH and Cμ segments, also causes a loss of Igh contraction (52). YY1 may directly regulate Igh contraction by binding to Eμ (50, 52) and to other sites in the VH array (53). However, it may also influence Igh contraction indirectly by regulating the expression of Pax5 (50, 53). Igh locus contraction was also reduced by CTCF knockdown, but the effect was rather modest (54). The data suggest that multiple factors cooperate to shape the contracted configuration of the Igh locus in pro-B cells.
The Igh locus contains 85 or more CTCF sites, the majority of which are distributed across the VH region (43, 54) (Fig. 1A). CTCF sites in the proximal portion of the VH array are located immediately downstream of VH RSSs, whereas those in the distal portion are intergenic (42, 44). Included among the latter are CTCF sites at a series of 14 homologous Pax5-activated intergenic repeat (PAIR) elements, some of which express Pax5-dependent and pro-B cell-specific antisense transcripts (43, 53, 54). Only a few CTCF binding sites are found outside of the VH region. Two are located ~3.2 and 5.6 kb upstream of DFL16.1, the most 5′ DH gene segment, in a region called 5′DFL16 or intergenic control region 1 (IGCR1); nine others are densely arrayed in an area called the 3′ CTCF-binding element (3′CBE) that flanks the 3′ regulatory region (3′ RR) downstream of the CH gene segments (42, 54–56). The majority of Igh locus CTCF sites also bind cohesin (54). Notably, neither CTCF nor cohesin bind to Eμ (52, 54).
Long-distance DNA interactions across the Igh locus have recently been probed at high resolution by 3C and 4C (52–54, 56) (Fig. 1A). These studies have described a 300 kb, multiple-loop structural domain at the 3′ end of the Igh locus, defined by interactions among IGCR1, Eμ, 3′RR, and 3′CBE, that are present in both pre-pro-B cells and pro-B cells. The interaction between IGCR1 and 3′CBE is CTCF- and cohesin-dependent (54), whereas Eμ interactions with IGCR1 and 3′CBE are CTCF- and cohesin-independent (52, 54). Eμ interactions with these sites may be mediated by YY1, since this factor binds not only to Eμ, but in one study was found to bind to IGCR1 and 3′CBE as well (52). Further 4C analyses conducted using either a proximal VH site or a distal VH site as bait described two additional multiple-loop regions, each spanning several hundred kilobases. Most, but not all, interacting sequences bound CTCF; however, the CTCF-dependency of these interactions requires further study (52). Because the two multiple-loop regions do not interact with each other, they were hypothesized to reside in distinct structural domains (52). Nevertheless, more comprehensive studies of long-distance interactions will be needed for a better picture of Igh domain structure in pre-pro- and pro-B cells.
In addition to the relatively local interactions described above, Eμ was shown to interact with two distant sites, one in the proximal portion of the VH array (near the 5′ end of the VH7183 gene segments) and one in the distal portion of the VH array (near the 3′ end of the VHJ558 gene segments) (Fig. 1A). These long-distance interactions are Eμ-dependent and may also involve YY1, since YY1 binding was detected at both sites (52). It was also recently shown that Eμ interacts across 1.8–2.0 Mb with two PAIR elements (PAIR4 and PAIR6) that are major sites of intergenic antisense transcription in the distal portion of the VH array (53). These long-distance interactions are pro-B cell-specific and YY1-dependent; hence, they correlate with, and may reflect, Igh locus contraction. Notably, long-distance Eμ-PAIR interactions do not reflect a functional role for Eμ in PAIR activation, since PAIR antisense transcription is Eμ-independent. Nevertheless, PAIR transcription, like Igh contraction, is YY1-dependent (53). Therefore, it was suggested that PAIR and Eμ elements might interact because they are independently recruited into the same transcription factory and that this co-recruitment may be the basis for Igh locus contraction (44, 53, 57). Consistent with this, knockdown of CTCF in pro-B cells caused increased PAIR transcription and increased PAIR-Eμ interactions (53, 54). However, this observation is inconsistent with the notion that CTCF promotes Igh locus contraction; the modest Igh conformational change detected by 3D-FISH in CTCF knockdown pro-B cells (54) might reflect more local changes in loop organization within Igh domains, rather than decontraction per se.
How do changes in CTCF-mediated loop structures impact Igh recombination? Deletion of the 4kb IGCR1 region or mutation of its two CTCF binding sites led to dramatic dysregulation of the VH repertoire, with substantially elevated transcription and recombination of the most DH-proximal VH gene segments (56) (Fig. 1A). Remarkably, proximal VH gene segments were also found to rearrange to DH gene segments prior to DH-JH joining and to DH or DHJH segments even in thymocytes, and they were not subject to feedback inhibition from productively assembled Igh alleles (56). Deletion of the IGCR1 region also disrupted the IGCR1-3′CBE loop, as well as Eμ interactions with both elements. However, since the functional dysregulation requires only mutation of the IGCR1 CTCF binding sites, it may specifically reflect the loss of the CTCF-dependent IGCR1-3′CBE loop (54, 56). The data suggest that CTCF plays a critical role in insulating the proximal VH gene segments from the influence of Eμ, perhaps a direct result of CTCF-mediated looping between the IGCR1 and 3′CBE regions. Consistent with this interpretation, deletion of hs5-7, containing 7 of 9 CTCF sites in the 3′CBE region, partially reduced the IGCR1-3′RR/3′CBE interaction and caused a mild increase in proximal VH usage (58). Moreover, a dysregulation similar to that documented for IGCR1 deletion was observed for a distal VH gene segment that was repositioned between IGCR1 and the DH cluster on an otherwise wild-type Igh allele (59).
If IGCR1 functions as an insulator, it might be expected to suppress physical interactions between Eμ and upstream sites in the Igh locus. However, it is apparent that Eμ can interact with numerous distant sites in the locus in pro-B cells, even in the presence of IGCR1 and an IGCR1-3′CBE loop (52, 53). Therefore, the dynamics of IGCR1-mediated looping and the basis for its impacts on long-distance DNA contacts, transcriptional activation and V(D)J recombination will be important areas for future study.
A note of caution that applies to the interpretation of all studies involving knockout or knockdown of pleiotropic transcription or architectural proteins is that it is difficult to know whether observed effects are direct (as is often assumed) or indirect. Studies involving binding site mutations, as in the example of IGCR1, can be particularly revealing in this regard.
Igk locus
Studies of the murine Igk locus are yielding a picture rather similar to that of the Igh locus. The murine Igk locus on chromosome 6 contains 96 functional Vκ gene segments that are distributed across 3.2 Mb and are situated upstream of four Jκ gene segments and Cκ (Fig. 1B). Although Igk alleles can undergo secondary Vκ-to-Jκ recombination to replace an initial VκJκ rearrangement that is out-of-frame or that provokes autoreactivity, opportunities for secondary rearrangements are limited by the small number of Jκ segments. Therefore, Igk conformational features should be important to foster the development of a broad Vκ repertoire by providing Jκ-proximal and -distal Vκ gene segments similar opportunities for rearrangement. Based on 3D-FISH, the Igk locus was reported to undergo contraction in pre-B cells to facilitate Vκ-to-Jκ recombination at this stage (14). However, a recent Hi-C study documented an extensive, locus-wide network of physical interactions involving Vκ segments and the intronic κ enhancer (iEκ, situated in the Jκ-Cκ intron) that, although absent in pre-pro-B cells, is apparent in pro-B cells well before the initiation of Igk recombination (23). The presence of this interaction network in pro-B cells suggests that Igk contraction may occur earlier during B cell development than previously thought.
The Igk locus contains approximately 60 sites to which CTCF binds in pre-B cells; binding was found to be lower in pro-B cells (42, 45). Strong CTCF binding was detected at the silencer in the intervening sequence (Sis), a recombination silencer in the intergenic region between Vκ and Jκ segments (60, 61), at the newly described contracting element for recombination (Cer) immediately upstream of Sis (62), and at the 5′ and 3′ boundaries of the locus (42, 45) (Fig. 1B). Other CTCF sites are distributed at intergenic locations across the 3.2 Mb Vκ array. A functional role for CTCF in Igk locus recombination was evaluated in mb1-cre Ctcff/f mice supplied with a pre-rearranged Igμ transgene (45). The pre-B cell Vκ repertoire in these mice was strongly biased towards usage of proximal Vκ gene segments. Consistent with this, germline transcription of proximal Vκ gene segments was substantially increased, as were interactions of iEκ and the 3′ κ enhancer (3′Eκ) with sites distributed across the proximal 1 Mb of the Vκ array (45). The enhancers also displayed increased interactions with sites outside of the Igk locus. The data suggest that CTCF plays a role in insulation, and that dysregulation of the Vκ repertoire is a consequence of hyper-activation of proximal Vκ segments by the Igk enhancers in the absence of CTCF.
Sis-deficient mice also displayed a proximally biased Vκ repertoire, indicating that the Sis CTCF site may contribute to insulation (61) (Fig. 1B). Cer deletion imparted an even stronger proximal bias to Vκ rearrangement and also allowed Vκ-Jκ rearrangement in thymocytes (62). Thus, Cer may contribute to insulation as well. However, Cer (unlike Sis) deletion modestly reduced Igk locus contraction, suggesting locus conformation as a potential explanation for the proximal Vκ bias (62). Additional experiments will be required to determine whether the conformational change and proximal Vκ bias reflect lost binding of CTCF or of another factor to Cer, and analysis on a recombinase-deficient background may be needed to fully evaluate effects of Sis and Cer on transcription and long-distance Eκ interactions. Although Sis and Cer have both been attributed other functions (60–62), their potential roles as insulators separating Vκ segments from the Eκ-regulated Jκ domain appear similar to that of IGCR1 in the Igh locus.
CTCF likely contributes to the network of long-distance interactions at the Igk locus detected by Hi-C in pro-B cells (23). However, many of the described iEκ interactions mapped to sites of E2A occupancy. This suggests the possibility that E2A may drive the clustering of widely distributed Vκ gene segments with iEκ and may in this way facilitate contraction and Vκ-to-Jκ rearrangement (23). This possibility warrants further study.
Tcra/Tcrd locus
As compared to either Igh or Igk, the Tcra/Tcrd locus on murine chromosome 14 (Fig. 1C) boasts a more nuanced program of conformational states and distinct regulatory functions for CTCF. These features are thought to facilitate a complex developmental program that transitions from Tcrd gene assembly in DN thymocytes to Tcra gene assembly in DP thymocytes (3). The locus contains approximately one hundred V genesegments that are distributed across 1.5Mb. A small subset of these V segments rearrange to two Dδ and Jδ gene segments to assemble a Tcrd repertoire. In contrast, most V segments can rearrange to a large array of 61 Jα gene segments to assemble a Tcra repertoire.
Notably, similar to Igh recombination, complete V-to-Dδ-to-Jδ recombination can only occur once per allele due to elimination of Dδ gene segments. In contrast, the large arrays of Vα and Jα gene segments can support multiple rounds of Vα-to-Jα recombination on each allele, allowing thymocytes multiple chances to assemble a Tcra gene that can promote positive selection (3). It is well established that initial (primary) rearrangements in early DP thymocytes are targeted to Jα segments at the extreme 5′ end of the Jα array by the activity of the T early α (TEA) and Jα49 promoters, and that as a function of DP thymocyte lifespan, subsequent (secondary) rearrangements are targeted to progressively more 3′ Jα segments by the activities of the introduced Vα gene segment promoters (63–66). This 5′-to-3′ Jα progression is coupled with an inevitable, reciprocal 3′-to-5′ progression V gene segment utilization, but there is controversy over the extent to which this progression is regulated and coordinated with the Jα progression. PCR analysis of thymic Vα-to-Jα rearrangements has shown that the most Jα-proximal Vα gene segments are biased to rearrange to the most 5′ Jα gene segments, that the most Jα-distal Vα gene segments are biased to rearrange to substantially more 3′ Jα gene segments, and that broadly distributed multi-member Vα families tend to rearrange to the entire set of Jα gene segments (67–69). This has been interpreted as a regulated and coordinated progression of Vα gene segment availability. Recent deep-sequencing analysis of Tcra transcripts in peripheral CD8+ T cells painted a picture that was generally consistent with the observations outlined above (70). However, features of the observed combinatorial diversity conflicted with the notion of coordinated Vα and Jα progressions, leading the authors to propose that following primary rearrangements between 3′ Vα and 5′ Jα gene segments, the remaining Vα segments simultaneously become available for recombination to Jα gene segments (70). Further analysis may be required to conclusively resolve this issue. For example, the study discussed above analyzed a peripheral CD8 T cell repertoire that had been shaped by thymic selection (70), rather than the complete spectrum of recombination events occurring in DP thymocytes. Moreover, additional factors, including duplicated or triplicated portions of the V array featuring nearly identical V gene segments, make analysis of the Tcra/Tcrd locus repertoire particularly challenging.
Analysis of Tcra/Tcrd locus conformation by 3D-FISH revealed that, as compared to control B cells, the locus is fully contracted in DN thymocytes but undergoes 5′ end extension to adopt a unique 3′ contracted and 5′ decontracted configuration in DP thymocytes (16) (Fig. 1C) (although an earlier study had reached different conclusions for unknown reasons (15)). What would be the rationale for this behavior? In DN thymocytes the locus is limited to a single round of Tcrd gene recombination per allele and both proximal and distal Vδ gene segments serve as recombination substrates (reviewed in (71)); this Igh-like behavior is best facilitated by a fully contracted configuration of the locus. In contrast, in DP thymocytes the locus undergoes multiple rounds of recombination with primary recombination biased towards use of 3′ Vα and 5′ Jα gene segments. A 3′ contracted and 5′ decontracted configuration would favor the initial usage of 3′ Vα gene segments and thereby save more 5′ Vα gene segments for subsequent rounds of recombination (16). It should be noted that the conformations described above are adopted by the unrearranged Tcra/Tcrd locus in recombinase-deficient thymocytes. Whether and how locus conformation in DP thymocytes adjusts to prior Tcrd gene or primary Tcra gene recombination is not known.
Recent 3C studies have documented a network of interactions within the 3′-contracted domain of the Tcra/Tcrd locus that forms in DP thymocytes (Fig. 1C). The Tcra enhancer (Eα) becomes active in DP thymocytes and is known to activate promoters distributed across 500 kb (71), a region that may correspond to the contracted 3′ domain. Included among these promoters are the TEA promoter associated with 5′ Jα gene segments and the promoters of proximal V gene segments. Eα was shown to contact individual V and J promoters and to bring these promoters in contact with each other to form a “chromatin hub” (47). Because RAG proteins preferentially bind to 5′ Jα gene segments to form a recombination center in DP thymocytes (48), hub formation would bring proximal V gene segments into this recombination center and would facilitate the synapsis of V and J RSSs to support primary Tcra recombination. Notably, although Eα is required to establish this network of DNA contacts within the contracted portion of the Tcra/Tcrd locus, it is not required for 3′ end contraction per se (16). This differs from the reported role for Eμ in Igh locus contraction (52). CTCF also plays no role in 3′ end contraction in DP thymocytes (47). Hence, the molecular basis for Tcra/Tcrd locus contraction is unknown.
As is the case for Igh and Igk, there are many binding sites for CTCF and cohesin in the Tcra/Tcrd locus (46, 47) (Fig. 1C). However the distribution of binding sites is strikingly different than for Igh and Igk, since in the Tcra/Tcrd locus, CTCF sites generally mark cis-regulatory elements, including most V gene segment promoters, the TEA promoter and Eα (47). Conditional knockout of Ctcf or Rad21 (which encodes a cohesin subunit) caused reduced interactions between Eα and the TEA promoter, reduced TEA transcription, and reduced 5′ Jα accessiblity (46, 47). Loss of CTCF also partially disrupted interactions between Eα and proximal Vα gene segments and between Vα and Jα gene segments (47). As a result, CTCF- and cohesin-deficiency were associated with reduced Vα-to-Jα rearrangement (46, 47). Surprisingly, CTCF-deficiency, or deletion of the TEA promoter and its associated CTCF site, caused increased interaction between Eα and the DδJδCδ cluster and increased transcription and recombination of Dδ and Jδ gene segments. Because the DδJδCδ cluster lacks CTCF sites, it was suggested that CTCF normally synergizes with Eα-bound factors to specify CTCF-marked promoters (eg., TEA) as Eα targets, and that this suppresses Eα interactions with suboptimal, CTCF-free targets (eg. Dδ and Jδ promoters). With CTCF eliminated, this specificity is lost, leading to reduced Tcra gene rearrangement and increased Tcrd gene rearrangement in DP thymocytes (47). CTCF-dependent long-distance interactions are thought to serve primarily a targeting function at the Tcra/Tcrd locus, rather than an insulating function as at Igh and Igk, because Tcra/Tcrd locus CTCF sites are positioned at, rather than between, critical cis-regulatory elements.
Conclusions
Antigen receptor locus conformation is manipulated at multiple levels to support gene assembly by V(D)J recombination. The evidence suggests at least two layers of organization: multiple-loop rosette-like structures span hundreds of kilobases and interactions between such structures mediate global spatial relationships. Long-distance interaction networks appear to be orchestrated by ubiquitous and lineage-specific transcription factors (YY1, Pax5 and E2A) as well as chromatin architectural proteins (CTCF and cohesin). However, there appears not to be a single paradigm that fits all antigen receptor loci. CTCF-mediated looping can suppress or stimulate V(D)J recombination through effects on RSS accessibility, depending on whether those loops insulate enhancers from promoters (eg. Igh and Igk) or target enhancers to promoters (eg. Tcra). CTCF-mediated looping may also regulate V(D)J recombination by influencing RSS synapsis. Synapsis may be facilitated when RSSs are positioned near cis-regulatory elements brought into contact by CTCF (eg. Tcra). Synapsis was also shown to be inhibited when accessible RSSs are segregated into different chromatin loops by CTCF (72), although this result is difficult to reconcile with the obvious need for RSS synapsis between loops at antigen receptor loci. It seems clear that the development of antigen receptor repertoires must occur as an exceedingly complex function of conformational states and spatial relationships dictated by the distribution of binding sites for CTCF and other factors. It will take additional time and effort to make sense of it all.
Footnotes
This work was supported by National Institutes of Health Grant R37 GM41052 (to M.S.K.).
Abbreviations used in this paper: 3C, chromosome conformation capture; 3′CBE, 3′ CTCF binding element; 3D-FISH, three-dimensional fluorescence in situ hybridization; 3′RR, 3′ regulatory region; Cer, contracting element for recombination; CTCF, CCCTC binding factor; DN, double negative; DP, double positive; Eα, Tcra enhancer; Eμ, intronic Igh enhancer; iEκ, intronic Igk enhancer; IGCR1, intergenic control region 1; PAIR, Pax5 activated intergenic repeat; RSS, recombination signal sequence; Sis, silencer in the intervening sequence; TEA, T early α; YY1, Yin-Yang 1
References
- 1.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 2.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 3.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 5.Osipovich O, Oltz EM. Regulation of antigen receptor gene assembly by genetic-epigenetic crosstalk. Sem Immunol. 2010;22:313–322. doi: 10.1016/j.smim.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale JS, Ames KT, Boursalian TE, Fink PJ. Cutting Edge: Rag deletion in peripheral T cells blocks TCR revision. J Immunol. 2010;184:5964–5968. doi: 10.4049/jimmunol.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo TC, Schlissel MS. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr Opin Immunol. 2009;21:173–178. doi: 10.1016/j.coi.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev. 2012;22:79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossen C, Mansson R, Murre C. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol. 2012;30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 11.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 12.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 16.Shih HY, Krangel MS. Distinct contracted conformations of the Tcra/Tcrd locus during Tcra and Tcrd recombination. J Exp Med. 2010;207:1835–1841. doi: 10.1084/jem.20100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubner MR, Eckersley-Maslin MA, Spector DL. Chromatin organization and transcriptional regulation. Curr Opin Genet Dev. 2012 doi: 10.1016/j.gde.2012.11.006. (in press) http://dx.doi.org/10.1016/j.gde.2012.11.00618. [DOI] [PMC free article] [PubMed]
- 18.de Wit E, de Laat W. A decade of 3C technologiesinsights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 20.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 22.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips JE, V, Corces G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic acids research. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 36.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk DA, Kong X, Ball AR, Jr, Calof AL, Lander AD, Groudine MT, Yokomori K. Cohesin mediates chromatin interactions that regulate mammalian beta-globin expression. J Biol Chem. 2011;286:17870–17878. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Lucas JS, Bossen C, Murre C. Transcription and recombination factories: common features? Curr Opin Cell Biol. 2011;23:318–324. doi: 10.1016/j.ceb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ribeiro de Almeida C, Stadhouders R, de Bruijn MJ, Bergen IM, Thongjuea S, Lenhard B, van Ijcken W, Grosveld F, Galjart N, Soler E, Hendriks RW. The DNA-binding protein CTCF limits proximal Vκ recombination and restricts κ enhancer interactions to the immunoglobulin κ light chain locus. Immunity. 2011;35:501–513. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih HY, Verma-Gaur J, Torkamani A, Feeney AJ, Galjart N, Krangel MS. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci U S A. 2012;109:E3493–3502. doi: 10.1073/pnas.1214131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayegh CE, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynaud D, I, Demarco A, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A. 2012;109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3′ end of the Igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol Cell Biol. 2005;25:1511–1525. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Volpi SA, Verma-Gaur J, Hassan R, Ju Z, Roa S, Chatterjee S, Werling U, Hou H, Jr, Will B, Steidl U, Scharff M, Edelman W, Feeney AJ, Birshtein BK. Germline deletion of Igh 3′ regulatory region elements hs 5, 6, 7 (hs5-7) affects B cell-specific regulation, rearrangement, and insulation of the Igh locus. J Immunol. 2012;188:2556–2566. doi: 10.4049/jimmunol.1102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bates JG, Cado D, Nolla H, Schlissel MS. Chromosomal position of a VH gene segment determines its activation and inactivation as a substrate for V(D)J recombination. J Exp Med. 2007;204:3247–3256. doi: 10.1084/jem.20071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Z, Widlak P, Zou Y, Xiao F, Oh M, Li S, Chang MY, Shay JW, Garrard WT. A recombination silencer that specifies heterochromatin positioning and Ikaros association in the immunoglobulin κ locus. Immunity. 2006;24:405–415. doi: 10.1016/j.immuni.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 61.Xiang Y, Zhou X, Hewitt SL, Skok JA, Garrard WT. A multifunctional element in the mouse Igk locus that specifies repertoire and Ig loci subnuclear location. J Immunol. 2011;186:5356–5366. doi: 10.4049/jimmunol.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang Y, Park SK, Garrard WT. Vκ gene repertoire and locus contraction are specified by critical DNase I hypersensitive sites within the Vκ-Jκ intervening region. J Immunol. 2013;190:1819–1826. doi: 10.4049/jimmunol.1203127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 64.Hawwari A, Krangel MS. Role for rearranged variable gene segments in directing T cell receptor α recombination. Proc Natl Acad Sci U S A. 2007;104:903–907. doi: 10.1073/pnas.0608248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mauvieux L, Villey I, de Villartay JP. T early alpha (TEA) regulates initial TCRVAJA rearrangements and leads to TCRJA coincidence. Eur J Immunol. 2001;31:2080–2086. doi: 10.1002/1521-4141(200107)31:7<2080::aid-immu2080>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 66.Villey I, Caillol D, Selz F, Ferrier P, de Villartay JP. Defect in rearrangement of the most 5′ TCR-Jα following targeted deletion of T early α (TEA): implications for TCR α locus accessibility. Immunity. 1996;5:331–342. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- 67.Pasqual N, Gallagher M, Aude-Garcia C, Loiodice M, Thuderoz F, Demongeot J, Ceredig R, Marche PN, Jouvin-Marche E. Quantitative and qualitative changes in V-J alpha rearrangements during mouse thymocytes differentiation: implication for a limited T cell receptor alpha chain repertoire. J Exp Med. 2002;196:1163–1173. doi: 10.1084/jem.20021074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thuderoz F, Simonet MA, Hansen O, Pasqual N, Dariz A, Baum TP, Hierle V, Demongeot J, Marche PN, Jouvin-Marche E. Numerical modelling of the V-J combinations of the T cell receptor TRA/TRD locus. PLoS Comput Biol. 2010;6:e1000682. doi: 10.1371/journal.pcbi.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jouvin-Marche E, Fuschiotti P, Marche PN. Dynamic aspects of TCRα gene recombination: qualitative and quantitative assessments of the TCRα chain repertoire in man and mouse. Adv Exp Med Biol. 2009;650:82–92. doi: 10.1007/978-1-4419-0296-2_7. [DOI] [PubMed] [Google Scholar]
- 70.Genolet R, Stevenson BJ, Farinelli L, Osteras M, Luescher IF. Highly diverse TCRα chain repertoire of pre-immune CD8+ T cells reveals new insights in gene recombination. EMBO J. 2012;31:1666–1678. doi: 10.1038/emboj.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hawwari A, Krangel MS. Regulation of TCR δ and α repertoires by local and long-distance control of variable gene segment chromatin structure. J Exp Med. 2005;202:467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shrimali S, Srivastava S, Varma G, Grinberg A, Pfeifer K, Srivastava M. An ectopic CTCF-dependent transcriptional insulator influences the choice of Vβ gene segments for VDJ recombination at TCRβ locus. Nucleic Acids Res. 2012;40:7753–7765. doi: 10.1093/nar/gks556. [DOI] [PMC free article] [PubMed] [Google Scholar]