Abstract
The emergence of multi- and extensively drug-resistant tuberculosis (MDR-TB and XDR-TB, respectively) has intensified the critical public health implications of this global disease. The fitness of Mycobacterium tuberculosis (M.tb.) strains exhibiting MDR and XDR phenotypes is of fundamental importance in predicting whether the MDR-/XDR-TB epidemic will be sustained across the human population. Here we describe a potential mechanism of M.tb. resistance to the TB drug isoniazid conferred by loss of a sigma factor, SigI. We demonstrate that the gain of isoniazid resistance in the M.tb. ΔsigI mutant might not diminish the organism’s fitness for causing disease. These findings have significant implications when considering the ability of drug resistant M.tb. strains to initiate untreatable TB epidemics, as it is possible that loss or alteration of SigI function could play a role in the generation of MDR and XDR M.tb. strains of suitable fitness to spread in a community setting.
Mycobacterium tuberculosis (M.tb.) is an ancient pathogen that has coevolved with humans for tens of thousands of years1, and the discovery of antibiotics effective against this organism was a monumental breakthrough in the treatment of tuberculosis (TB) patients. However, as has been the case for many other pathogenic microorganisms, drug-resistant strains of M.tb. have arisen through natural selection within humans receiving antimicrobials. While drug resistance-conferring mutations provide the microbe with a selective advantage in the presence of the drug, these genetic alterations may be associated with a loss of fitness in the drug-free environment, either through less efficient binding of a natural substrate, wasted metabolic energy producing resistance proteins, or the sacrifice of a pathway involved in the drug’s action2. Despite this, multi- and extensively drug-resistant (MDR and XDR, respectively) M.tb. strains have spread beyond populations in which they were selected by inappropriate antimicrobial therapy and into treatment-naive patients, where the strains have shown considerable pathogenicity even among non-immunocompromised hosts, indicating that many drug-resistant M.tb. strains are of sufficient fitness to spread person-to-person3-5.
Isoniazid (INH) is a bactericidal first-line TB drug with an extremely high potency for members of the M. tuberculosis complex, but not other non-tuberculous mycobacteria6. Upon entering tubercle bacilli, INH is activated to form an isonicotinyl free radical by the bacterium’s own peroxidase/catalase enzyme encoded by katG7. The INH free radical reacts with NAD to form an INH-NAD adduct that is a highly potent inhibitor of the essential enzyme InhA, an enoyl ACP-reductase required for mycolic acid biosynthesis8-10. Surveys of clinical isolates that demonstrate INH-resistance reveal that approximately 50% have alterations in katG (including complete deletion of the gene), and an additional 25% contain compensatory promoter mutations that upregulate the expression of the inhA gene11. Mutations in a small number of other genes encoding activators or regulators of NADH, such as mshA or ndh, have also been described and may account for some clinically-observed INH resistance12. Thus, about 25% of INH-resistant M.tb. isolates are resistant through undetermined mechanisms.
In this study we describe a novel mechanism of INH resistance in M.tb. We identify a regulatory gene, sigI, that when deleted confers INH-resistance by reducing the transcription levels of katG. We further show that M.tb. lacking SigI is not attenuated, but rather hypervirulent, compared to wild-type M.tb. in a mouse model of TB, demonstrating that this SigI-related mechanism of INH resistance is not associated with a loss of fitness in M.tb.
Results
Identification of the SigI regulon
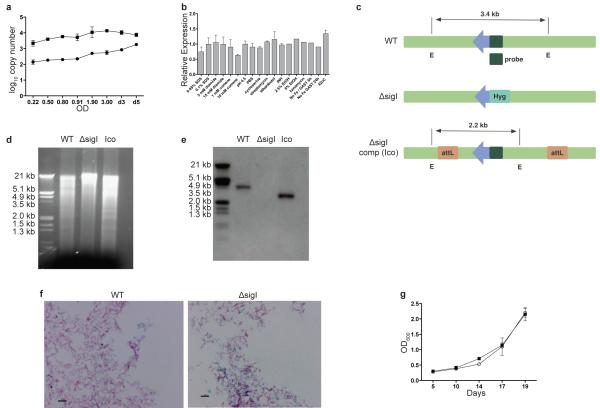
The M.tb. genome encodes 13 sigma factors, including one principal sigma factor (sigA), two principal-like sigma factors (sigB, sigF), and 10 extracytoplasmic function sigma factors (sigC-sigM)13. Previous reports on 12 of these sigma factors (sigA-sigH, sigJ-sigM) have revealed the basic features of their regulons and how they are utilized by M.tb. to adjust to various environmental conditions, including macrophage and mouse infection models, with most sigma factors involved in the regulation of virulence programs13. Therefore, we investigated the role of the relatively uncharacterized sigma factor SigI in the regulation of M.tb. gene expression and virulence phenotypes. Using quantitative, real-time reverse transcription polymerase chain reaction (qRT-PCR), sigI gene expression was examined under various growth phases and stress conditions. sigI transcript levels were increased during stationary phase (Fig. 1a) as well as during heat shock (Fig. 1b). However, little variation in sigI expression was observed in response to other stress conditions.
Figure 1. sigI expression and validation of M.tb. ΔsigI.
(a) Transcript copy numbers of sigI (closed circle) and sigA (closed square) during M.tb. growth in culture. (b) Using copy number, sigI transcription was analyzed under stress conditions (relative to no stress conditions). The following stress conditions were analyzed: detergent (SDS), disulfide (diamide), oxidative (cumene), acid (pH 4.5), antibiotic (cycloserine, streptomycin, ethambutol, INH, kanamycin), ethanol (EtOH), low iron (No Fe GAST) and heat shock (42°C). . (c) Diagram of the restriction map for Southern blotting of wild-type, ΔsigI and the sigI-complemented (Ico) M.tb. strains. Arrow indicates sigI gene. E: EcoRI restriction site. Hyg: hygromycin resistance gene. (d) Agarose gel of EcoRI-digested genomic DNA for Southern blotting. (e) Southern blot with probe (primers described in Supplementary Table S2). (f) Acid-fast staining of wild-type and ΔsigI mutant M.tb. strains. Scale bar: 5 μm. (g) Growth curves for wild-type (open circle) and ΔsigI (closed square) M.tb. strains. Three biological replicates of all experiments were performed, and error bars represent standard deviation.
To examine the role of sigI more closely, we constructed a sigI deletion mutant and the corresponding complemented strain (Fig. 1c), both of which were confirmed by Southern blot (Fig. 1d, e). The sigI mutant displayed normal cellular morphology as observed by acid-fast staining (Fig. 1f) and growth rate in liquid culture (Fig. 1g). To identify the SigI regulatory network, we compared global gene expression patterns between the ΔsigI mutant and the parent wild-type strain using microarray (Supplementary Table S1). As the microarray analyses served as a screening tool for the identification of SigI-regulated genes, the complemented strain was not included in the microarray studies. The SigI regulon identified included an ATP synthase (Rv1304), heat shock proteins (Rv0440, Rv0350, and Rv3417), and katG. These data supported the role for SigI during heat shock (Fig. 2a). In addition to its ability to activate INH, the KatG catalase-peroxidase has been reported to play a detoxification role during oxidative stress, and has been found to be essential for full virulence in macrophage and mouse infection models14-17. This manuscript describes the follow-up validation and further characterization of the relationship between sigI and katG as suggested in our microarray screen.
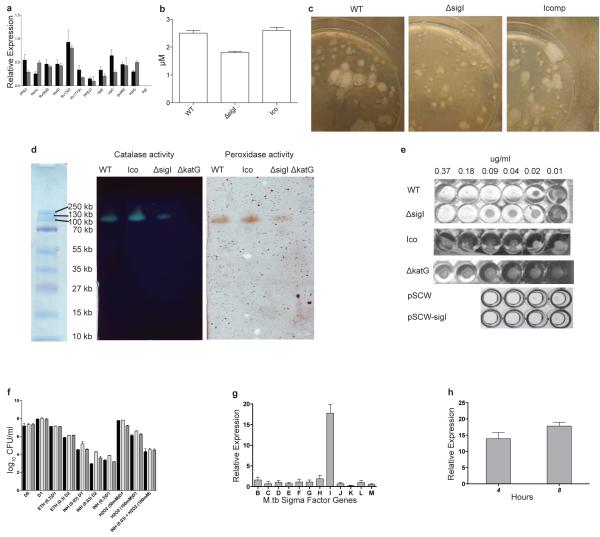
Figure 2. in vitro characterization of M.tb. ΔsigI.
(a) The down-regulation of genes, including katG, which were identified by microarray in the ΔsigI mutant, was confirmed by qRT-PCR. Data represent gene expression (based on copy number) of the mutant compared to wild-type. (b) Catalase activity within total cell lysates from wild-type M.tb., the ΔsigI mutant, and the complemented strain (Ico). (c) Catalase activity from bacteria growing on a 7H10 agar plate was detected with 10% H2O2. (d) Bacterial lysates were resolved by native PAGE. Catalase activity was visualized using 1% ferric chloride and 1% potassium ferricyanide. Peroxidase activity was visualized using a solution of 0.5 mg/ml p-diaminobenzidine, 3mM H2O2. (e) INH MIC assay results for wild-type and ΔsigI M.tb. strains, as well as the sigI overexpressing strain (pSCW-sigI) and its empty vector control strain (pSCW). (f) Wild-type (black bars), ΔsigI mutant (light gray bars), and complement (dark gray bars) strains were exposed to INH, ETH, H2O2 and INH + H2O2 for 1 or 2 days and then plated on 7H10 agar for CFU enumeration. The combined INH and H2O2 exposure was for 1 day. The ETH and INH concentrations are in ug/mL, and the H2O2 concentration is in mM. (g) Expression levels of M.tb. sigB-sigM following 4-hours of acetamide-induced overexpression of sigI (relative to expression prior to acetamide exposure). (h) M.tb. katG expression after 4 and 8 hours of acetamide-induced expression of sigI. Three biological replicates of all experiments were performed, and error bars represent standard deviation.
Catalase production is decreased in the M.tb. ΔsigI mutant
To determine whether the association between sigI deficiency and decreased katG expression was functionally significant, we examined the catalase-peroxidase activity of the ΔsigI mutant upon exposure to hydrogen peroxide. As shown in Fig. 2b, c, this activity was decreased in the mutant compared to wild-type bacteria, as well as in the bacterial lysates (Fig. 2d). The ΔkatG mutant was devoid of detectable catalase activity (Fig. 2d), as has been well-documented17. These data indicated that SigI plays a role in the regulation of katG expression, but that other transcriptional mechanisms maintain a baseline level of katG expression.
The M.tb. ΔsigI mutant exhibits resistance to INH
In view of the fact that KatG is necessary for the activation of INH12, we investigated whether SigI-mediated expression of katG affected the organism’s resistance to this antibiotic. We first determined the minimal inhibitory concentration-90% (MIC90) of INH for the ΔsigI mutant, wild-type and complemented strains, and the well-characterized INH resistant mutant ΔkatG17. The ΔkatG mutant lacks catalase-peroxidase activity altogether and is highly resistant to INH with an MIC90 of 10 μg/ml12 (Fig. 2e). As shown in Fig. 2e, the MIC90 of the ΔsigI mutant was 0.18 μg/ml, four times higher than the wild-type strain (MIC90 = 0.04 μg/ml), while the complement strain exhibited similar INH susceptibility as wild-type M.tb.
Since ethionamide (ETH) is an anti-TB drug which inhibits the product of inhA without a need for KatG-dependent activation8, we also tested the susceptibility of the ΔsigI mutant and related strains to ETH in order to evaluate if the ΔsigI mutant’s INH resistance was mediated by katG- or inhA-dependent pathways. Bacteria were treated with INH, ETH, and H2O2 at various concentrations, and bacterial survival was determined after 24 and 48 hours. We observed no difference in survival of ETH-treated bacteria compared to wild-type, whereas the ΔsigI mutant exhibited higher survival when exposed to INH (Fig. 2f). These data indicate that the M.tb. ΔsigI mutant’s resistance to INH is due to reduced SigI-dependent katG expression, rather than from inhA-related gene regulation. An interesting additional finding was the observation that the ΔsigI mutant expressed sufficient catalase/peroxidase activity to protect from H2O2 stress to near wild-type levels (Fig. 2d, f). The wild-type and ΔsigI bacteria had similar survival when exposed to H2O2 and INH simultaneously, again indicating equivalent responses to H2O2. This may be due to a mechanism where the ΔsigI mutant expresses sufficient catalase/peroxidase activity to protect from H2O2 oxidative damage despite compromised ability to activate INH.
Overexpression of sigI results in increased susceptibility to INH
To differentiate whether SigI directly regulates katG transcription or if katG expression is regulated by other SigI-dependent co-factors, we constructed a conditionally overexpressing sigI merodiploid strain in which the second copy of sigI was expressed from the acetamide-inducible promoter Pace (Fig. 2g). Chemically-induced overexpression of sigI led to enhancement of katG expression, suggesting a direct transcriptional effect (Fig. 2h). We also measured the MIC to INH for the sigI over-expression strain, and this strain exhibited a lower MIC (0.02 μg/ml) than either the wild-type or control strain carrying the empty vector (Fig. 2e). These data further indicate a specific role for SigI in the bacterial response to INH.
SigI directs transcription of katG
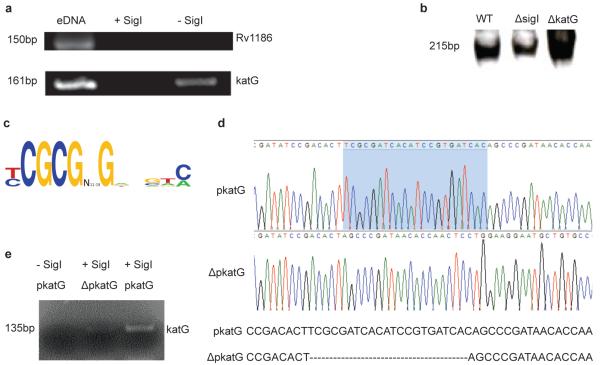
Transcription of M.tb. katG has been shown to occur from two different promoters: one upstream of the furA gene, which itself is upstream of katG, and also one directly upstream of katG, between furA and katG18. We utilized an in vitro transcription (IVT) assay to test directly whether SigI could direct transcription of katG from either of these promoter sequences. We expressed and purified recombinant SigI protein to >98% purity for use in the assembly of an RNA polymerase holoenzyme; purified recombinant SigI was combined with the E. coli RNA core polymerase (i.e., RNA polymerase lacking a sigma factor component). This SigI-containing enzyme complex was then incubated in a reaction buffer containing a DNA template comprising the katG gene, including the promoter sequence 379 bp up-stream. Any transcripts generated by the IVT reaction were then amplified and evaluated by RT-PCR. Although the SigI-containing RNA polymerase holoenzyme was unable to direct transcription from the non-SigI-regulated gene Rv1186, SigI-directed transcription was detected from the katG promoter, providing direct evidence that SigI can direct transcription from the katG promoter located 379 bp upstream of the gene (Fig. 3a).
Figure 3. SigI transcribes the katG gene.
(a) SigI-containing RNA polymerase holoenzyme was able to direct transcription from the promoter region 379 bp upstream of the katG gene. After transcription by SigI, the gene product was amplified by RT-PCR. The ORF region Rv1186 was used as a negative control for IVT, as SigI does not direct transcription of this gene. (b) Total bacterial lysates from wild-type and ΔsigI strains were utilized as RNA polymerase component in an IVT assay in which the promoter region 33bp upstream of katG was used as the DNA template. (c) Putative SigI binding motif as identified from upstream sequences of the genes identified by microarray to be a part of the SigI regulon. (d) Nucleotide sequence of the katG promoter; the shaded region indicates the deletion. (e) SigI cannot direct transcription from an IVT template in which the putative SigI binding motif has been deleted (from promoter region 33bp upstream of katG, ΔpkatG). Three biological replicates of all experiments were performed.
Next, we evaluated the transcriptional activities present in the cytoplasm of the ΔsigI mutant compared to wild-type. Bacterial lysates prepared from wild-type M.tb., the ΔsigI mutant, as well as the ΔkatG mutant, were used for IVT experiments using a katG template with the promoter located 33 bp upstream of the gene. As presented in Fig. 3b, lysates from the ΔsigI mutant were associated with significantly decreased katG transcription, further demonstrating a direct SigI transcriptional effect in expression of katG. These transcription-based assays thus indicate that SigI transcribes katG from both of its known promoters18. Based on this information and the upstream sequences of the genes comprising the putative SigI regulon (Fig. 2a), we identified a possible SigI binding motif (Fig. 3c). To validate this putative SigI binding motif, we next performed an IVT assay using a katG template with this motif deleted from the promoter sequence immediately upstream of the katG gene (Fig. 3d). Removal of this short sequence nearly abolished the ability of SigI to direct transcription of katG (Fig. 3e), thus confirming SigI-mediated transcription of katG. Taken together with our results, these findings indicate a possible role for SigI-mediated katG expression in the context of INH treatment.
M.tb. ΔsigI is not attenuated in cell culture infections
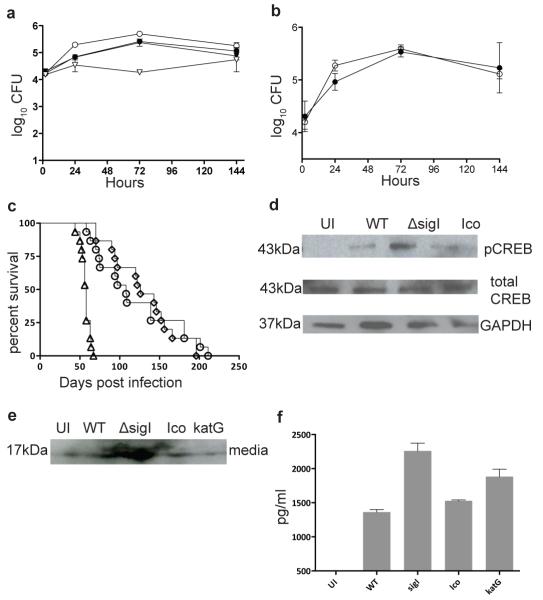
M.tb. strains lacking a functional KatG enzyme are severely attenuated in virulence in both macrophage and mouse infection models15, 17, suggesting that a decrease in the SigI-dependent expression of katG could be associated with defective growth and virulence in these model systems. Therefore, we analyzed the ability of the ΔsigI mutant to survive within J774A.1 murine macrophages. Activated cells were infected with wild-type M.tb. or the ΔsigI or ΔkatG mutants, and intracellular bacterial loads were determined after 24 hours, 3 days and 6 days post-infection. As shown in Fig. 4a, survival of the ΔsigI strain was similar to wild-type, while the ΔkatG mutant was attenuated in growth, indicating that the ΔsigI mutant was not associated with a decrease in virulence. Furthermore, when J774A.1 cells were co-infected with both wild-type and ΔsigI M.tb., the strains grew equally well and with the same growth kinetics as in singly infected cells, again indicating that the ΔsigI strain was not attenuated for growth in murine macrophage-like cells (Fig.4b).
Figure 4. M.tb. ΔsigI mutant strain is not attenuated in cells or mice.
(a) J774A.1 murine macrophages were infected with wild-type M.tb. (closed circle), the ΔsigI mutant (open circle), the complemented strain (closed triangle), and the ΔkatG mutant (open triangle). (b) J774A.1 cells were co-infected with wild-type M.tb. (closed circle) and the ΔsigI mutant (open circle). (c) Time-do-death for three groups each of 20 Balb/c mice that were infected with an inoculum that implanted ~ 3.7 log10 bacteria per lung by aerosol; wild type (open circle), sigI (open triangle), and complement strain (open diamond). (d) CREB phosphorylation was measured by Western blot in M.tb.-infected J774A.1 cells at 12h. (e) TNF-α production was measured in infected J774A.1 by Western blot. (e) TNF-α production was measured in infected J774A.1 by ELISA. The time-to-death experiment was performed once. Three biological replicates of all pther experiments were performed, and error bars represent standard deviation.
M.tb. ΔsigI is hypervirulent in BALB/c mice
We next evaluated whether this trend would extend to in vivo infections. Groups of 20 female BALB/c mice were infected by the aerosol route with wild type M.tb., the ΔsigI mutant, or the complemented ΔsigI strain and monitored for time to-death; day 1 CFU counts for each strain were 3.75, 3.86 and 3.75 log10CFU, respectively. Interestingly, compared to the wild-type and complemented strains, infection with the ΔsigI mutant was associated with more rapid death of the mice, with half of the ΔsigI-infected mice dying by day 58 post infection, while the corresponding value for wild-type and complement straininfected mice was day 139 and day 146, respectively (Fig. 4c). Thus, the M.tb. ΔsigI mutant strain displayed a hypervirulent phenotype in this time-to-death mouse infection model.
Because the in vivo data were contrary to our hypothesis that the ΔsigI mutant would exhibit attenuated growth and virulence in our model systems, we examined an additional parameter associated with M.tb. virulence, namely CREB phosphorylation and TNF-α production. Recent reports from our laboratory and others have indicated that upon infection, an initial increase in TNF-α levels benefit these organisms, and that virulent mycobacteria actively modulate host signaling to increase the expression of this pro-inflammatory cytokine19-21. The ΔsigI mutant, in contrast to the wild-type or complemented strains, induced increased CREB phosphorylation within infected J774A.1 cells (Fig. 4d), as well as increased TNF-α secretion (Fig. 4e, f). These data are in agreement with the in vivo results, indicating that the ΔsigI mutant is not attenuated for growth or virulence within these model systems, despite exhibiting KatG-associated INH resistance. The infection with the ΔkatG mutant may have also induced increased TNF-α secretion from these cells, as we observed an increase in TNF-α in the culture supernatant as measured by ELISA (Fig. 4f) that was not observed in Western blot analysis (Fig. 4e). Thus, from our data, the role of TNF-α is unclear in its relationship to the attenuated ΔkatG mutant.
Lack of sigI confers low-level resistance to INH in vivo
To address the relevance of sigI deletion in M.tb. resistance to INH, we infected female BALB/c mice with wild-type or ΔsigI M.tb. (with day 1 log10CFU counts of 3.44 and 3.71, respectively), and treatment with INH was initiated at 14 days post infection. For each strain, INH was administered by oral gavage at 0.5, 1.5 and 6.0 mg/kg/day, and lung CFU counts were obtained at 28 and 56 days post infection (i.e., after 14 and 42 days, respectively, of INH treatment). All ΔsigI-infected mice that did not receive INH or were treated at the lowest dose (0.5 mg/kg/day) died before the end of the experiment (Fig. 5a), again indicating increased virulence of this strain compared to wild-type, even in the presence of low-level INH. In addition, ΔsigI-infected mice receiving either 1.5 or 6.0 mg/kg/day of INH had significantly higher lung CFU counts at both day 28 and day 56 post infection compared to mice infected with wild-type M.tb. (Fig. 5b). We hypothesize that the increased survival of the ΔsigI strain is related to the low-level INH resistance displayed by this strain in vitro; however, it is possible that this observation is due to the increased virulence of the strain. Taken together, these data confirm the virulent phenotype of the ΔsigI mutant strain and demonstrate that lack of SigI function is associated with increased survival in the presence of INH.
Figure 5. The ΔsigI mutant strain is resistant to INH. in vivo.
Two groups of 50 BALB/c mice each were infected with wild-type and ΔsigI M.tb. with an inoculum that implanted ~ 3.5 log10 bacteria per lung by aerosol. Fourteen days post-infection, INH was administered to the mice daily (5 days/week) by oral gavage at doses of 0.5, 1.5 and 6.0 mg/kg/day. (a) Time-do-death analysis of infected mice treated with 0.5 mg/kg/day INH (blue line) or not treated (red line). Solid lines: wild-type. Dashed lines: ΔsigI. Triangle: sham treatment (no INH). Square: 1.5 mg/kg/day INH. Circle: 6.0 mg/kg/day INH. (b) CFU counts in mouse lungs for mice treated untreated or with INH at 1.5 and 6.0 mg/kg/day. All mice infected with the ΔsigI strain died before the day 56 time point. This experiment was performed once, with multiple mice per group. Error bars represent standard deviation.
Discussion
Mathematical models have indicated that the relative fitness of drug resistant M.tb. strains is the most important determinant when predicting the spread and epidemic potential of MDR- and XDR-TB2, 22. Here, we have presented a possible mechanism of INH resistance, i.e., loss of sigI and the associated SigI-dependent katG expression. The ΔsigI mutant did not exhibit diminished growth or virulence within mouse infection models, and in fact our data demonstrate that this mutant strain was more virulent than wild-type M.tb. under the conditions tested. Furthermore, we have shown that the loss of sigI results in increased bacterial survival in the presence of INH, indicating an in vivo role for SigI in INH resistance. These findings could have significant implications for the spread of drug resistant M.tb. strains.
Recently, Ando and colleagues have characterized INH-resistant human clinical isolates of M.tb. in Japan23. In their analysis of 108 INH-resistant isolates, they identified three unique mutations in the intergenic region upstream of katG in 4% of the isolates. Three of these isolates did not contain additional mutations in the examined sequences (katG, inhA promoter, aphC, ndh or kasA-kasB), while one isolate also contained a mutation in the inhA promoter region. These recent findings indicate that changes in the katG promoter sequence can affect resistance to INH, and that this mode of INH resistance is relevant in human TB. While the putative SigI binding site that we identified was not mutated in these INH-resistant strains, this work by Ando et al. indicates that alterations in the katG promoter region influence susceptibility to INH. Thus, these findings support our hypothesis that SigI-mediated transcription of katG may be is associated with M.tb. susceptibility to INH.
Although M.tb. lacking KatG function exhibits attenuated fitness for growth compared to wild-type M.tb., a second compensatory mutation has been identified which results in a regain of fitness, namely mutations in the ahp gene, which encodes a peroxidase12. Therefore, the generation of INH-resistance through this mechanism can be thought of as a two-step process whereby the initial katG mutation is associated with a fitness loss, followed by a secondary, compensatory mutation in ahp, which may increase the ability of the drug-resistant strain to survive in the community. In this communication, we have described an alternative pathway towards INH resistance in which a step towards drug (or multidrug) resistance, i.e., mutation of sigI, is not associated with a fitness cost.
Methods
Construction of mutant strains
The ΔsigI mutant was created using a suicide vector approach using the plasmid pCK068624. The upstream flanking region of the sigI gene was amplified by PCR using primers sigIP1 and sigIP2 (Supplementary Table S2) and directionally cloned into pCK0686 using NdeI and SpeI restriction sites. Similarly, the downstream flanking region of the sigI gene was amplified by PCR using primers sigIP3 and sigIP4 and then directionally cloned into pCK0686 using PacI and XhoI restriction sites to create pKOsigI. The suicide vector pKOsigI was then transformed into electrocompetent M.tb. using a Bio-Rad E. coli Pulser, and transformants were selected using 7H10 plates containing Hygromycin B (50 μg/mL). Double crossover events leading to a proper gene disruption were isolated by selection on plates containing Hygromycin B (50 μg/mL) and 5% sucrose. A complemented strain was constructed using the integrating plasmid pMH94. A 4.1 kb region containing the sigI gene was amplified by PCR using primers psigIcomp5 and psigIcomp6 (Supplementary Table S2), and the resulting amplicon was cloned into pMH94 using the XbaI restriction site. The resulting complementation plasmid was transformed into electrocompetent ΔsigI cells, and transformants were selected using both Kanamycin (10 μg/mL) and Hygromycin B (50 μg/mL).
Purification of recombinant SigI
Recombinant SigI was purified as C-terminal His-tagged recombinant protein. The entire ORF of the M.tb. sigI gene was PCR amplified using primers pETsigI1 and pETsigI2 (Supplementary Table S2). The amplicon was digested with NdeI and HindIII, and ligated to NdeI and HindIII digested pET22b (+) (Novagen). The resulting vector, pET-sigI, was then transformed into E. coli BL21(DE3) strain. SigI overexpression was induced using 1mM IPTG and after 3 hours of induction, the cells were harvested and disrupted by sonication in binding buffer (50mM Tris-Cl, 0.5M NaCl, 10mM Imidazol, 8M urea). The lysate was applied to a Histrap FF crude column (GE Healthcare) and purified using an FPLC system (Amersham). The purity of the protein was visualized using SDS-PAGE, and protein concentration was determined using the Bradford method.
Conditionally overexpressing sigI
Previously, we constructed sigF and sigB overexpression vectors pSCW35 and pSCW40 containing an acetamide promoter-sigF or sigB fusion gene respectively, as well as a control vector pSCW38, containing only the acetamide promoter25. To overexpress sigI in M. tuberculosis, the entire sigI gene was amplified by PCR using primers pACEI1 and pACEI2 and directionally cloned into pSCW35 using NdeI and PacI restriction sites. The resulting vector, pSCW-sigI, was transformed into electrocompetent M. tuberculosis and transformants were isolated on 7H10 plates containing kanamycin (10μg/ml).
Microarray
Total RNA was extracted from wild-type M.tb. and ΔsigI mutant at both OD600=1 and OD600=2. One microgram of RNA was reverse-transcribed with random hexamers (Invitrogen), and the resulting cDNA was labeled with Cy3-dCTP or Cy5-dCTP and competitively hybridized to whole genome arrays. Arrays were scanned using an Axon 4000B scanner and images were analyzed using GenePix pro 4.0 software. The resulting data were normalized to the total integrated intensity across both channels, and the ratio of Cy5 and Cy3 was compared and calculated. Q value was obtained using SAM (Significance Analysis of Microarrays). The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus26 and are accessible through GEO series accession number GSE35231.
Real-time RT-PCR
The ΔsigI mutant was confirmed by real-time RT-PCR from RNA isolated from M. tuberculosis CDC1551. Bacterial cells were centrifuged, washed with PBS and resuspended in Trizol reagent (Invitrogen) followed by bead beating with 0.1mm diameter silica beads at 5000 rpm. After centrifugation at 10,000 × g for 5 min at 4°C, the supernatants were collected, treated with chloroform and centrifuged again to collect the aqueous phase. Finally, RNA was precipitated with 1 volume of isopropyl alcohol, washed with 75% ethanol and dried. About 1μg of DNase I-treated RNA was transcribed with Superscript II Reverse Transcriptase (Invitrogen) using random primers (Invitrogen). Real-time PCR was carried out using IQ SYBR Green I PCR kit (Bio-Rad). The cDNA and RT primer set (Supplementary Table S2) were added to each reaction and specific amplification was detected at the extension cycle. The threshold cycle (CT) was converted to copy numbers using a DNA standard curve.
in vitro Transcription assay
The DNA templates including the putative katG promoter regions were prepared by PCR using primers katG1 and katG2 (Supplementary Table S2). Two picomoles of E. coli RNA core polymerase (Epicentre) were incubated with 20 pmol of purified sigma factor protein for 30 min in transcription assay buffer (10mM Tris, 50mM KCl, 10mM MgCl2, 0.1mM EDTA, 0.25μg/ul BSA), then 0.09 μg of template DNA was added and incubated with 0.35mM 11-biotin UTP, 1mM ATP, 1mM CTP, 1mM GTP and 0.65mM UTP mixture 25. The template with the putative SigI-binding motif deleted was generated using the mega-primer method27.
Survival of sigI mutant with several antibiotics and determine the MIC
Survival of ΔsigI mutant with INH, ETH, and H2O2 was analyzed by the addition of INH (0.03 μg/ml and 0.3 μg/ml), ETH (0.3 μg/ml) and H2O2 (50mM and 100mM) to cultures for 24h and 48h; aliquots were plated onto 7H10 agar. The MIC of INH for sigI mutant was also checked using 24-0.01μg/ml INH. INH was added to the media and the growth of wild-type, ΔsigI mutant, the sigI complemented strain, and the katG deletion mutant strain were checked after 7 and 14 days.
Catalase activity assay
Whole cell lysates from wild-type and the ΔsigI deletion mutant were prepared using a bead beater method. Ten micrograms of total protein was used to measure the catalase activity using a catalase assay kit (Calbiochem). Catalase and peroxidase activity were also measured using a gel staining method where the cell lysate was separated using a 10% non-denaturing PAGE gel. After electrophoresis, the gel was incubated in 0.03% H2O2 for 5 min and washed. Catalase activity was visualized by soaking the gel in a solution containing 1% (w/v) ferric chloride and 1% (w/v) potassium ferricyanide. Peroxidase activity was detected by incubating in detection solution (0.5 mg/ml p-diaminobenzidine, 3mM H2O2).
Macrophage infection
J774A.1 cells were cultivated in cRPMI (2mM glutamine, 10% FBS) and activated by incubating with interferon-γ (500U/ml) for 12hrs, followed by incubation with lipopolysaccharide (200ng/ml) for 3h before infection. After washing the macrophages 3 times with cRPMI, 105 bacteria were used to infect macrophage culture at an MOI of 1:1. After 2hrs of infection, macrophages were washed 5 times with RPMI media, and cultivated with cRPMI with 5% CO2 at 37°C. Macrophages were harvested, washed 3 times with RPMI and then lysed using 0.1% triton X-100 in PBS at each time point (2 hrs, 3 days, and 6 days after infection). Macrophage lysate was plated on 7H10 plates for enumeration of bacterial CFU.
TNF-α and CREB measurement
TNF-α production was measured using the DuoSet mouse TNF-α ELISA kit (R&D Biosystems) and by western blot using anti-mouse TNF-α antibody with biotin conjugate(Invitrogen), diluted 1:1000. CREB phosphorylation was measured by western blot using phospho-CREB antibody at 1:2000, total CREB antibody at 1:3000 (both from Cell Signaling), and GAPDH antibody at1:35000 (Sigma).
Virulence assessment of ΔsigI mutant
Three groups of 20 randomized six-week old female BALB/c mice (Charles River) were infected with either wild-type, ΔsigI, or the complemented strain using a Middlebrook inhalation exposure system (Glas-Col) with 10 mL of a diluted mid-log phase M.tb. culture intended to implant approximately 1000 bacilli in the lungs. One day post-infection, 3 mice from each group were sacrificed in order to determine the implanted dose. Mice were observed until death, with the time of each death recorded. Mice were maintained in isolator cages, with no more than 5 mice per cage, in the Johns Hopkins Center for Tuberculosis Research Animal Biosafety Level 3 facility. All animal experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and all procedures were approved by the Johns Hopkins University Animal Care and Use Committee.
Bacterial survival in vivo during INH treatment
Two groups (50 mice per group) of randomized 6-week old female BALB/c mice (Charles River) were infected by aerosol as described above with either wild-type or ΔsigI M.tb. with implants of approximately 3.5 log10 bacilli into the lung. Five mice from each group were sacrificed on day 1 to determine implant dose, and an additional 5 mice from each group were sacrificed on day 14 to determine lung bacterial burden prior to initiating treatment. Starting on day 14 post-infection, the remaining mice were given INH five days per week via oral gavage (200 μL total volume) at either 0, 0.5, 1.5 or 6.0 mg/kg/day (INH was dissolved in water). Four mice from each group were sacrificed at day 28, and all remaining mice were sacrificed at day 56 post infection. Lung homogenates were plated on selective 7H11 agar plates for CFU determination, and on days 28 and 56, lung homogenates were also plated on selective 7H11 agar plates containing 0.1 μg/mL.
Supplementary Material
Acknowledgements
We thank André Kübler and Mariama Maiga for their assistance in processing experimental samples. We also thank Peter McCaffrey for his assistance in assembling the figures. Support from the Howard Hughes Medical Institute and the National Institutes of Health (awards AI036973, AI037856 and AI079590) is gratefully acknowledged.
Footnotes
Author contributions. J-H.L. designed the study, performed most of the experiments and co-wrote the manuscript. N.C.A. contributed to study design, performance of experiments and wrote the manuscript. S.N. and D.E.G. assisted with performance of experiments. S.L. and H.G. contributed to study design, performance of experiments and data analysis. W.R.B. designed experiments, supervised the study and co-wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
Accession codes: The microarray data have been deposited in the Gene Expression Omnibus under accession number GSE35231.
REFERENCES
- 1.Wirth T, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 2009;13:1456–1466. [PubMed] [Google Scholar]
- 3.Blower SM, Chou T. Modeling the emergence of the ‘hot zones’: tuberculosis and the amplification dynamics of drug resistance. Nat. Med. 2004;10:1111–1116. doi: 10.1038/nm1102. [DOI] [PubMed] [Google Scholar]
- 4.Gandhi NR, et al. Extensively drug-resistant tuberculosis as a cause of death in patients coinfected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 5.Palmero D, et al. Infectiousness and virulence of multidrug-resistant and drug susceptible tuberculosis in adult contacts. Medicina (B. Aires) 2002;62:221–225. [PubMed] [Google Scholar]
- 6.Kourbeti IS, Maslow MJ. Nontuberculous mycobacterial infections of the lung. Curr. Infect. Dis. Rep. 2000;2:193–200. doi: 10.1007/s11908-000-0035-7. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee A, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 9.Dessen A, Quemard A, Blanchard JS, Jacobs WR, Jr, Sacchettini JC. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science. 1995;267:1638–1641. doi: 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- 10.Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Jr, Sacchettini JC. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 2009;13:1320–1330. [PubMed] [Google Scholar]
- 12.Vilcheze C, Jacobs WR., Jr. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu. Rev. Microbiol. 2007;61:35–50. doi: 10.1146/annurev.micro.61.111606.122346. [DOI] [PubMed] [Google Scholar]
- 13.Sachdeva P, Misra R, Tyagi AK, Singh Y. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J. 2010;277:605–626. doi: 10.1111/j.1742-4658.2009.07479.x. [DOI] [PubMed] [Google Scholar]
- 14.Collins DM. In search of tuberculosis virulence genes. Trends Microbiol. 1996;4:426–430. doi: 10.1016/0966-842x(96)10066-4. [DOI] [PubMed] [Google Scholar]
- 15.Manca C, Paul S, Barry CE, 3rd, Freedman VH, Kaplan G. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 1999;67:74–79. doi: 10.1128/iai.67.1.74-79.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol. Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 17.Pym AS, Saint-Joanis B, Cole ST. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 2002;70:4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Master S, Zahrt TC, Song J, Deretic V. Mapping of Mycobacterium tuberculosis katG promoters and their differential expression in infected macrophages. J. Bacteriol. 2001;183:4033–4039. doi: 10.1128/JB.183.13.4033-4039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal N, Lamichhane G, Gupta R, Nolan S, Bishai WR. Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature. 2009;460:98–102. doi: 10.1038/nature08123. [DOI] [PubMed] [Google Scholar]
- 20.Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volkman HE, et al. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen T, Murray M. Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nat. Med. 2004;10:1117–1121. doi: 10.1038/nm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ando H, et al. Downregulation of katG expression is associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 2011;79:1615–1628. doi: 10.1111/j.1365-2958.2011.07547.x. [DOI] [PubMed] [Google Scholar]
- 24.Kaushal D, et al. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8330–8335. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, Karakousis PC, Bishai WR. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J. Bacteriol. 2008;190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyagi R, Lai R, Duggleby RG. A new approach to ‘megaprimer’ polymerase chain reaction mutagenesis without an intermediate gel purification step. BMC Biotechnol. 2004;4:2. doi: 10.1186/1472-6750-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.