ABSTRACT
Taphrina deformans is a fungus responsible for peach leaf curl, an important plant disease. It is phylogenetically assigned to the Taphrinomycotina subphylum, which includes the fission yeast and the mammalian pathogens of the genus Pneumocystis. We describe here the genome of T. deformans in the light of its dual plant-saprophytic/plant-parasitic lifestyle. The 13.3-Mb genome contains few identifiable repeated elements (ca. 1.5%) and a relatively high GC content (49.5%). A total of 5,735 protein-coding genes were identified, among which 83% share similarities with other fungi. Adaptation to the plant host seems reflected in the genome, since the genome carries genes involved in plant cell wall degradation (e.g., cellulases and cutinases), secondary metabolism, the hallmark glyoxylate cycle, detoxification, and sterol biosynthesis, as well as genes involved in the biosynthesis of plant hormones. Genes involved in lipid metabolism may play a role in its virulence. Several locus candidates for putative MAT cassettes and sex-related genes akin to those of Schizosaccharomyces pombe were identified. A mating-type-switching mechanism similar to that found in ascomycetous yeasts could be in effect. Taken together, the findings are consistent with the alternate saprophytic and parasitic-pathogenic lifestyles of T. deformans.
IMPORTANCE
Peach leaf curl is an important plant disease which causes significant losses of fruit production. We report here the genome sequence of the causative agent of the disease, the fungus Taphrina deformans. The genome carries characteristic genes that are important for the plant infection process. These include (i) proteases that allow degradation of the plant tissues; (ii) secondary metabolites which are products favoring interaction of the fungus with the environment, including the host; (iii) hormones that are responsible for the symptom of severely distorted leaves on the host; and (iv) drug detoxification enzymes that confer resistance to fungicides. The availability of the genome allows the design of new drug targets as well as the elaboration of specific management strategies to fight the disease.
Introduction
Taphrina deformans is the agent of peach leaf curl, a disease that affects orchards throughout the temperate regions of the world. Hosts include peach (Prunus persica) and, to a lesser extent, almond trees (Prunus dulcis). T. deformans is the most intensively studied species of the genus due to its importance as a plant pathogen worldwide, all known peach and nectarine cultivars being susceptible to infection (1). The economic burden of peach leaf curl is variable, depending on the areas of production. The disease causes $2.5 to $3 million in losses in the United States annually (2). In northern Italy, it represents an important threat to the tree and can affect 60 to 90% of shoots (3). One of the most significant symptomatic effects occurs on leaves that become severely distorted and reddish (4). These symptoms are usually attributed to the production of indole-3-acetic acid (IAA) by T. deformans (5).
T. deformans is a fungus belonging to the subphylum Taphrinomycotina, a basal lineage of the phylum Ascomycota. The genus Taphrina belongs to the order Taphrinales in the class Taphrinomycetes. The Taphrinomycotina subphylum also contains the fission yeasts of the genus Schizosaccharomyces and the animal parasites of the genus Pneumocystis (6). T. deformans is dimorphic, i.e., it has a biotrophic hyphal state which is parasitic-pathogenic on plant tissues and a saprophytic yeast state that may overwinter on plant surfaces. Only the yeast state can be grown on conventional culture media in vitro (7).
We report here the sequence of the genome of T. deformans strain PYCC 5710, as well as comparative genomic analysis of its gene repertoire. The genome carries key genes for plant invasion and colonization.
RESULTS
Nuclear and mitochondrial genomes.
The T. deformans strain PYCC 5710 genomic DNA sequencing yielded a 13.3-Mb genome assembly with a 29.3-fold coverage (Table 1). This corresponds to 394 scaffolds, 50% of which are longer than 72 kb. The completeness of the genome was estimated to be 98.7%, and 99% of the 3,923 publicly available expressed sequence tags (ESTs) could be mapped on the assembly. A single copy of the nuclear rRNA operon is present in a single scaffold (scaffold 361). The 18S rRNA gene displays 100% identity with the previously reported allele of this gene (8), confirming the identity of the strain sequenced here. Telomeric sequences presumably corresponding to the reported eight subtelomeres are located in 14 scaffolds (Table 1; see Text S1 in the supplemental material). No obvious toxin biosynthesis or surface antigen gene clusters are present in the putative subtelomeric regions, as has been observed in other pathogens (9, 10). Antigen gene clusters are also not present in other parts of the genome. However, as is discussed below, secondary metabolite gene clusters, which may be involved in toxin production, were found elsewhere in this genome.
TABLE 1 .
T. deformans nuclear genome statistics and comparison to those of other members of the Taphrinomycotina subphylum
| Characteristic | T. deformans | P. jiroveciia | S. pombeb |
|---|---|---|---|
| Assembly size (Mb) | 13.3 | 8.1 | 12.5 |
| Average GC (%) | 49.5 | 29.1 | 36.0 |
| No. of scaffolds | 394 | 356c | 3d |
| Scaffold N50 (kb) | 71.9 | 41.6c | NAe |
| Telomere repeat motif | TTAGGG | TTAGGG | TTAC(A)(C)G(1–8) |
| Repeat (%) | 1.5f | 9.8 | 9.4 |
| No. of protein-coding genes | 5,735 | 3,898 | 5,124 |
| Gene density (genes per Mb) | 431 | 481 | 554 |
| Average gene length (ntg) | 1,492 | 1,472 | 1,446 |
| Mean exon per gene | 2.1 | 3.7 | 2.0 |
| Mean intron length (nt) | 78 | 61 | 81 |
| No. of tRNAs | 169 | 71 | 171 |
These data are from reference 18. P. jirovecii infects specifically humans.
These statistics are from http://www.pombase.org (73).
Contigs.
Chromosomes.
NA, not applicable or not available from the website
Detailed description of T. deformans repeated elements is shown in Table S2 in the supplemental material.
nt, nucleotide.
The T. deformans genome includes a low number of repeated transposable elements (TEs) (see Table S1 in the supplemental material) and a relatively high GC content compared to those of other Taphrinomycotina members (Table 1). We therefore investigated the presence of genome defense mechanisms that might be responsible for the underrepresentation of TEs. These include the repeat induced point mutation (RIP; a mechanism that limits the spreading of TEs in fungi [11, 12]), RNA interference (RNAi) (13), and the meiotic silencing by unpaired DNA (MSUD) (14). We screened the whole genome for the RIP footprints and found no evidence of regions with typical RIP bias (see Fig. S1). This is consistent with the fact that the RIP seems restricted to a few fungal species, including the model fungus Neurospora crassa (12). On the other hand, most of the genes involved in RNAi and MSUD are present in the genome (see Table S2), suggesting that the spreading and silencing of TEs might be mediated by these mechanisms.
The mitochondrial genome is 30 kb in size, which makes it larger than the 20-kb mitochondrial genome of Schizosaccharomyces pombe and the 23-kb mitochondrial genome of Pneumocystis carinii, the species infecting specifically humans (see Table S3 in the supplemental material). Analysis revealed a 624-bp group I intron inside the cytochrome c oxidase subunit gene 1 (cox1) gene, an 813-bp group I intron inside the cytochrome b (cob) gene, and an 840-bp group II intron inside the cox2 gene. In S. pombe, the cox1 gene contains two group I introns (15), the cob gene contains one group II intron (16), and the cox2 gene contains a group II intron (17). The T. deformans mitochondrial genome carries an additional truncated nonfunctional cox1 gene. The same feature has been reported in Ustilago maydis (J. C. Kennell and C. Boehmer, unpublished data), but the biological implication of this is not known.
Gene content.
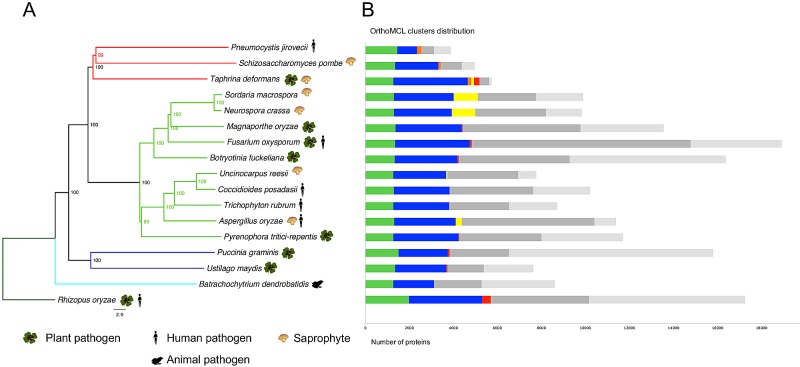
We identified 5,735 protein-coding genes, 44% of which are supported by ESTs and 83% by similarity to proteins found in other fungi (see Table S4 in the supplemental material). About 30% of the total genes were manually curated, including all those which are discussed in detail in the text. To better understand the gene content of T. deformans, we compared its proteome to those of 16 relevant fungi, including two plant-nonpathogenic Taphrinomycotina members, seven plant pathogens, four plant saprophytes, two human pathogens, and one animal pathogen. These fungi were selected to reflect different host infection mechanisms. The maximum likelihood phylogeny derived from alignment of 227 single-copy orthologs is consistent with previous phylogenies (6, 18, 19), placing T. deformans in the basal Taphrinomycotina subphylum (Fig. 1A).
FIG 1 .
Phylogeny and ortholog distribution in T. deformans and other fungi. (A) The phylogeny was inferred by the maximum likelihood method using RAxML (72) from concatenated alignment of 227 single-copy orthologs conserved across species. The scale bar represents 2 amino acid changes per site, and the bootstrap values are indicated on nodes. Different subphylum branches are highlighted as follows: Taphrinomycotina, red; Pezizomycotina, light green; Basidiomycota, dark blue; Chytridiomycota, light blue; Mucormycotina, dark green. (B) The colored bars represent orthologous proteins identified using OrthoMCL (69) in the fungi shown in panel A. The bars are divided into seven categories: present in all species (green), present in T. deformans and at least one other species (blue), Taphrinomycotina specific (orange), present in T. deformans and plant saprophytes (yellow), present in T. deformans and plant fungal pathogens (red), other proteins (dark gray), and species specific (light gray).
Ortholog clustering identified 13,566 protein groups containing at least one protein from T. deformans (Fig. 1B). Out of the 5,735 T. deformans proteins, 22.3% are included in clusters present in all species (shown as green strips in Fig. 1B); 58.4% are shared, i.e., present in T. deformans and at least one other species (shown as blue strips); and 4.4% are present only in Taphrinomycotina members (shown as orange strips).
To identify genes that might be responsible for T. deformans pathogenicity, we classified organisms according to their lifestyle. Fungi can be free-living as saprophytes recycling nutrients and/or can establish interactions with a host (20). These interactions are diverse and can vary from biotrophy (i.e., the fungus is able to live within its host and cannot be cultured in vitro in the case of obligate biotrophy) to necrotrophy (i.e., the fungus kills its host to access nutrients). Here, we deliberately simplified the problem by considering an organism to be a pathogen if it can cause plant disease and to be a saprophyte if it can live on dead plant material (Fig. 1). We then conducted a search for T. deformans genes that are shared only with saprophytes and genes that are shared only with pathogens. We excluded genes that show similarity in S. pombe and Pneumocystis jirovecii proteomes, which are not plant pathogens. These genes were excluded because, despite its saprophytic lifestyle, S. pombe is not able to infect plants or grow on dead plant material. A small proportion of T. deformans proteins (2.2%, n = 126) are shared exclusively with saprophytes (shown as yellow strips in Fig. 1B). Only 52% of this subset of genes had a functional annotation, and this portion is enriched (P < 0.05) in functional categories such as “nucleic acid binding,” “carbohydrate metabolic process,” and “calcium ion transport” (see Table S5 in the supplemental material). Another 4.5% of T. deformans proteins (n = 259) are shared exclusively with plant pathogens (shown as red strips in Fig. 1B), and 76% of them had a functional annotation. These proteins are enriched (P < 0.05) in the functional categories “lipid metabolic process,” “cutinase activity,” and “response to oxidative stress” (see Table S6). Among the proteins shared with plant pathogens are (i) cutinases and lipases, which have a suspected role in fungal pathogenesis as they are important for penetration into both host cell and intercellular space (21, 22), and (ii) 24 putative secreted effector proteins, which are known to be crucial for plant-fungus interaction (23). We also found genes that might be involved in the transport and metabolism of specific compounds such as toxins, drugs, or hormones. These include an auxin efflux carrier and a particular drug resistance protein. The latter is a transporter that was shown to confer resistance to benomyl and methotrexate in Candida albicans (24). Taken together, these results are consistent with the fact that T. deformans has both saprophytic and parasitic-pathogenic lifestyles.
Host-pathogen interaction.
During its life cycle, T. deformans enters and damages plant tissues. The genome encodes a full repertoire of carbohydrate-active enzymes, including those required for degrading the plant cell wall (see Table S7 in the supplemental material). Additionally, the genome encodes 70 proteases, among which 21 appear to be secreted (see Table S8 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm). Although T. deformans possesses cellulases and xylanases that allow the degradation of cellulose and hemicellulose, it lacks lignin-degrading enzymes necessary for the degradation of lignocellulose and lignin components (FOLymes). These enzymes are also absent in other fungi (i.e., U. maydis, Cryptococcus neoformans, N. crassa, and Saccharomyces cerevisiae) (25). Other plant fungal parasites, such as the white rot fungus Schizophyllum commune, possess a full repertoire of FOLymes (25). After entering into its host organism, T. deformans is able to cause stable infection using different mechanisms and effectors. Among them, the most important are the following.
(i) Plant hormones and carotenoids.
The production of the plant auxin hormone indole-3-acetic acid (IAA) by T. deformans is generally considered to be responsible for the formation of characteristic leaf deformation symptoms (5, 26). In U. maydis, the tryptophan aminotransferases (encoded by Tam genes) and indole-3-acetaldehyde dehydrogenases (encoded by Iad genes) are required for production of IAA (27). As in U. maydis, two orthologs of each Tam gene (locus tags TAPDE_001394 and TAPDE_004347) and Iad gene (locus tags TAPDE_005337 and TAPDE_000671) are present in T. deformans, suggesting a similar route of IAA biosynthesis.
The carotenoid pigments are responsible for the characteristic pink coloration of T. deformans yeast colonies (7). The T. deformans strain PYCC 5710 sequenced here is among the highest carotenoid accumulators among Taphrinomycotina members (28). Accordingly, all genes necessary for carotenoid biosynthesis are present in the assembly, including the geranylgeranylpyrophosphate synthase (locus tag TAPDE_002204), the lycopene cyclase/phytoene synthase (locus tag TAPDE_005571), and the phytoene dehydrogenase (locus tag TAPDE_002896). Carotenoids are important protective compounds in fungi, safeguarding against UV light and other oxidative stresses (29). This role may be of particular importance in T. deformans, which lives exposed in the high-light environment at or near the plant surface. In U. maydis, carotenoids can also function as precursors for the production of retinal, a molecule which acts as a chromophore in the photoactive protein opsin (29). Similarly, the genome of T. deformans carries three opsin genes (locus tags TAPDE_003195, TAPDE_001673, and TAPDE_005609), as well as a single copy of the carotenoid cleavage oxygenase (locus tag TAPDE_002691), an enzyme which cleaves β-carotene into two molecules of opsin. The role of carotenoids and their derivatives in stress biology and photobiology is not characterized in T. deformans. In plants, carotenoids are important as precursors of the plant stress hormone abscisic acid (ABA). In T. deformans, we identified genes orthologous to those involved in ABA biosynthesis of the plant parasite fungus Botrytis cinerea, which are distinct from those of plants (30). The ABA biosynthesis cluster includes a cytochrome P450 monooxygenase similar to B. cinerea ABA1 and ABA2 (locus tag TAPDE_000660), an ortholog of ABA3 (locus tag TAPDE_004842), and several short-chain dehydrogenases/reductases similar to ABA4 (locus tags TAPDE_000889, TAPDE_001904, TAPDE_002246, TAPDE_002541, and TAPDE_000083). Additionally, the T. deformans genome harbors putative orthologs of the plant ABA biosynthesis genes, i.e., zeaxanthin epoxidase (locus tag TAPDE_003610), xanthoxin dehydrogenase (locus tag TAPDE_000917), molybdenum cofactor sulfurase (locus tag TAPDE_004842), 9-cis-epoxycarotenoid dioxygenase (locus tag TAPDE_002691), and abscisic-aldehyde oxidase (locus tag TAPDE_004279). These findings suggest the possibility of ABA production, which has not been previously reported or tested for in Taphrina, although observed in the ascomycetous plant pathogen B. cinerea (30). However, expression studies would be required to determine if these genes are activated upon stress or under other conditions. Our phylogenetic analyses did not provide conclusive evidence of the transfer of the plant-like ABA biosynthesis gene cluster from plant to T. deformans (data not shown).
(ii) Mating genes.
In the T. deformans genome, several locus candidates for putative MAT cassettes akin to those of S. pombe were identified, namely, potential orthologs of matmc and matpi genes, as well as of genes involved in mating type switching (e.g., swi1, swi5, swi6, and swi10). Many other genes described as sex related in S. pombe seem to be also present (31), e.g., those encoding the components of the pheromone-activated mitogen-activated protein kinase (MAPK) signaling cascade (see Table S9 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm). Some of the genes involved in mating-time switching (sap1, swi2, and msh3), meiosis (e.g., mei2), or silencing (clr1) and one of the pheromone receptor genes (mam2) are either absent or appear to be truncated. Others, like a putative dcr1 homolog, lack relevant domains, in this case the helicase at the N terminus, but conserve all the others.
(iii) Secondary metabolism.
Secondary metabolites are small active molecules that are produced by many microorganisms, including bacteria, fungi, and plants. They are usually not essential but can confer advantages when microbes are interacting with the environment (32). Four genes that define putative secondary metabolite biosynthesis gene clusters have been identified in the T. deformans genome. Three of them bear a similarity to nonribosomal peptide synthases (NPRS), and one appears to be a hybrid NPRS/polyketide synthase (PKS) (see Table S10 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm). Since these biosynthetic gene clusters are not present in other Taphrinomycotina members (i.e., Schizosaccharomyces spp. and Pneumocystis spp.), T. deformans appears to have a unique capacity for the production of secondary metabolites.
Drug targets.
The genome of T. deformans encodes targets for known antifungal drugs. Despite its apparent resistance to azole treatment (33), we identified the gene encoding the lanosterol 14-alpha-demethylase which is targeted by the phytosanitary antifungal molecules difenoconazole, tebuconazole, and myclobutanil (34, 35). Moreover, most of the genes involved in the biosynthesis of different sterols (brassicasterol, ergosterol, and cholesterol) (see Table S11 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm) are conserved. The apparent resistance to azoles might be explained by the fact that T. deformans uses brassicasterol as a major sterol in its membranes, instead of ergosterol (36). Genes encoding β-1,3 glucan synthase, which is targeted by the antifungal echinocandins, and cytochrome bc1, which is targeted by trifloxystrobin, were also identified. Additionally, T. deformans possesses a homolog of the Bcmfs1 gene, which was shown to be crucial for the resistance of Botryotinia fuckeliana to the fungicides fludioxonil, camptothecin, and cercosporin (37).
Metabolic capabilities.
The T. deformans yeast state is able to grow on conventional fungal or yeast media (7). Consistently, its genome encodes most of the respiratory and biosynthetic pathways (e.g., citric acid cycle, glycolysis, and amino acid synthesis). Nevertheless, several genes involved in the metabolism of galactose and other sugars are lacking (see Table S12 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm). This is consistent with the fact that T. deformans has a relatively narrow spectrum of assimilation of carbon and nitrogen sources. Indeed, sugars utilized include cellobiose, d-xylose, and d-glucose but not galactose, sucrose, maltose, sorbose, or trehalose (7). Additionally, T. deformans has been reported to be auxotrophic for thiamine and biotin (38), and consistently, genes specific to the biosynthesis of these vitamins are missing in the genome (see Table S12 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm). T. deformans may possess a functional glyoxylate cycle since the hallmark genes, i.e., isocitrate lyase and malate synthase, are present in its genome (see Table S12 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm), and the activity of the latter gene is supported by EST data (NCBI EST locus tag TDE00002395). This cycle allows the synthesis of glucose from lipids in the absence of a carbohydrate source (39).
DISCUSSION
The T. deformans nuclear genome is larger and harbors a larger number of genes than those of P. jirovecii and S. pombe. Its mitochondrial genome is similar in size to that of P. jirovecii but larger than that of S. pombe. The nuclear genome presents a low content of repeated elements and the same telomere repeat as that in plants, mammals, and filamentous fungi (18, 40). The apparently low content of transposable elements seems not to be correlated with RIP activity, which is also absent in other Taphrinomycotina members. A single copy of the ribosomal DNA unit has been identified in the genome. To date, this characteristic was observed only in Pneumocystis species (41, 42); it is generally believed to be associated with a low replication rate and is compatible with the notably slow growth of the yeast state of T. deformans in culture (7). The genome also carries genes for important mechanisms such as respiratory and biosynthetic pathways, plant cell and tissue degradation, sterol biosynthesis, and mating genes. This gene content allows this organism to be both a plant pathogen and saprophyte.
Despite its relatively reduced genome compared to those of other fungal plant pathogens (e.g., 13 versus 59.9 Mb for Fusarium oxysporum), about 5% of T. deformans genes are conserved only in fungal plant pathogens. These genes are likely to constitute the genetic basis of its virulence. Enrichment analysis indicated that many of these genes are involved in cutinase activity, lipid metabolism, and response to oxidative stress activity, which are all key factors for successful invasion of plant cells (21, 22). The cutinases hydrolyze the cutin, which protects the surface of the plant leaves. Consequently, cutinases are predicted to be involved in the early phase of the infection. The lipid metabolism is crucial for the maintenance of cell wall integrity and the signaling network and thus might be important for virulence. The genes involved in the response to oxidative stress might also provide protection against stress resulting from exposure to high-intensity sunlight and/or plant defense mechanisms.
Two genomic features make T. deformans unique among Taphrinomycotina members. First, it harbors four putative secondary metabolite gene clusters, although such clusters were lost during the evolution of other ascomycetes such as S. pombe (43), S. cerevisiae (43), and P. jirovecii (18). However, their functionality and final products in T. deformans remain to be determined. Second, the genome carries genes putatively involved in the biosynthesis of plant hormones (IAA) that may be responsible for the formation of characteristic leaf deformation symptoms. It has been suggested that hormone production capabilities as well as secondary metabolite gene clusters in other fungi could have been acquired via horizontal gene transfer (44, 45). However, the hormone biosynthetic pathways differed between fungi and plants in all cases examined so far (30, 46). This is consistent with the view that plant hormone biosynthesis results rather from a convergent evolution involving different metabolic pathways or enzymes, which produce similar compounds in plants and fungi (47).
This fungus is homothallic since single haploid cells give rise to dikaryotic hyphae and asci on plant tissue (48). In contrast to U. maydis, conjugation of cells heterozygous at the mating type (MAT) locus does not occur. However, since heterozygosity of paired nuclei in the dikaryotic hyphae is often required for pathogenic development, a mating-type-switching mechanism similar to that found in ascomycetous yeasts could be in effect. Many locus candidates for the expected sex-related genes and MAT cassettes were identified in the T. deformans genome. The MAT cassettes are notoriously difficult to characterize due to the short lengths of their coding sequences (CDSs) and their low degree of sequence conservation. Two of the three putative cassettes seem incomplete, lacking one of the expected genes, and some genes involved in mating type switching are either absent or truncated. The missing cassette and genes may have gone undetected or are present in gaps of the present genome sequence.
MATERIALS AND METHODS
Strain and culture conditions.
The T. deformans strain PYCC 5710 (=CBS 356.35) cells were grown in liquid YEPD medium (1% [wt/vol] Difco yeast extract, 2% Difco peptone, 2% glucose) at 25°C with rotary shaking (150 × g). Cells were harvested by centrifugation at 6,000 rpm for 1 min, washed twice with 30 ml of EDTA (20 mM, pH 5.8), aliquoted (5 g per aliquot), and stored at −20°C until use.
Genomic DNA isolation, sequencing, and assembly.
Genomic DNA from a single aliquot was isolated using the protocol of Lachance (49) and used for whole-shotgun sequencing. Data were generated by a combination of both mate-paired and single-end Roche 454 GS FLX sequencing with Titanium chemistry. Raw data statistics can be found in Table S13 at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm. Vectors and other contaminants (7.3% of total reads) were removed using Phred and Cross match processing tools (50, 51) with the UniVec database (available at ftp://ftp.ncbi.nih.gov/pub/UniVec/) and a custom collection of bacterial genomes. The mitochondrial genome reads were isolated (ca. 4.3% of total reads), and assembled separately (see Text S2 in the supplemental material). The nuclear genome was assembled using the remaining filtered sequences (88.4%) with Newbler (version 2.6). The genome completeness was estimated using the CEGMA pipeline (52) and alignment of 3,923 ESTs (BLASTn cutoff, ≤10−5).
Gene prediction and functional annotations.
The ab initio gene finder GeneMark-ES (53) was self-trained on the genome sequence assembly. Augustus (54) and SNAP (55) gene finders were trained on 3,923 expressed sequence tags (ESTs) available at http://www.ncbi.nlm.nih.gov/nucest/?term=Taphrina%20deformans). Final gene models were produced using Maker pipeline (v 2.10) (56) based on trained predictors, 3,923 ESTs, and a set of 36,252 reference proteins corresponding to the proteomes of S. pombe, S. cerevisiae, N. crassa, and Neosartorya fischeri and five T. deformans proteins (available from http://www.uniprot.org/; release 2012_07) (57). Gene structures were visualized and manually corrected if necessary using Apollo (v.1.11.7) (58). tRNAs and other noncoding RNAs were predicted using Infernal (v.1.0.2) (59), tRNA-scan-SE (v.1.21) (60), and Rfam (v.10.1) (61). Mapping of coding sequences (CDSs) to KEGG biochemical pathways was performed using Priam (62). Carbohydrate active enzymes were further documented according to the CAZy database (available at http://www.cazy.org/) (62, 63). Functional enrichment tests were performed using BLAST2GO (64). Proteases and secreted proteins were annotated using InterPro v.40, SIMAP, and SignalP 4.0 (65–67). Secreted proteins were identified using InterPro and correspond to proteins bearing a characteristic signal peptide and having no transmembrane domain. The telomere regions were annotated using in-house methods (see Text S1 in the supplemental material). Repeats were identified using TransposonPSI (available at http://transposonpsi.sourceforge.net/) and RepeatMasker (v.3.2.8) (available at http://www.repeatmasker.org) with RepBase (v.16.02) (68). The repeat point mutation (RIP) indexes were calculated according to the method of Lewis et al. (12), using Perl scripts available at https://github.com/hyphaltip/fungaltools.
Gene family analysis and phylogeny.
Gene families for T. deformans and 15 other fungi were built using OrthoMCL with a Markov inflation of 1.5 and a maximum E value of 10−5 (69). The fungal proteomes used were those of S. pombe, Magnaporthe oryzae, Fusarium oxysporum, Botryotinia fuckeliana, Puccinia graminis, Ustilago maydis, Rhizopus oryzae, Neurospora crassa, Aspergillus oryzae, Sordaria macrospora, Uncinocarpus reesii, Pyrenophora tritici-repentis, Trichophyton rubrum, Coccidioides posadasii, Batrachochytrium dendrobatidis (available at http://www.uniprot.org/; release 2012_09), P. jirovecii (18), and T. deformans (this study). Single-copy orthologs were identified using OMA (standalone version 0.99q) (70), concatenated, and aligned using MAFFT with the L-INS-i method (71). Phylogeny was inferred using RAxML (v.7.2.8) (72) with 100 bootstrap replicates and BLOSUM62 as model.
Additional supplemental material.
Additional supplemental material can be found at http://www.chuv.ch/imul/imu_home/imu_recherche/imu_recherche_hauser/imu-phauser-suppldata.htm.
Nucleotide sequence accession numbers.
The whole-genome sequence has been deposited at ENA-EMBL under accession no. CAHR00000000. Raw reads have been deposited at ENA SRA under accession no. ERP001279.
SUPPLEMENTAL MATERIAL
Telomeres. Download
Mitochondrial genome assembly and annotation. Download
Composite RIP indexes (CRI) in Taphrina deformans and Neurospora crassa genomes. (A) No scaffold in the T. deformans exhibits a large AT-rich region with typical RIP bias (CRI index value, >0). The example of scaffold 5 is shown. All 394 scaffolds of the genome assembly were subjected to the same analysis with similar results. (B) The linkage group I sequence from N. crassa which has been reported to be subjected to the RIP (45) was used as a control. Download
Repeated elements in T. deformans genome.
T. deformans genes involved in RNA-mediated silencing.
T. deformans mitochondrial genome statistics and comparison to those of other Taphrinomycotina members.
Gene model support.
Overrepresented Gene Ontology (GO) categories of genes present in T. deformans and plant fungal saprophytes.
Overrepresented Gene Ontology (GO) categories of genes present in T. deformans and plant fungal pathogens.
Carbohydrate-active enzyme genes in T. deformans and selected fungi.
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation grant 310030-124998 to P.M.H. and M.P. and by an Academy of Finland Fellowship to K.O. (decision no. 251397 and 256073). The computations were performed at the Vital-IT Center for high-performance computing (http://www.vital-it.ch) of the SIB (Swiss Institute of Bioinformatics). SIB receives financial supports from the Swiss Federal Government through the State Secretariat for Education and Research (SER).
We thank Thomas Bernard for help with enzyme annotation and helpful discussions. We thank Cláudia Carvalho (PYCC, Portugal) for preparing genomic DNA.
Footnotes
Citation Cissé OH, Almeida JMGCF, Fonseca Á, Kumar AA, Salojärvi J, Overmyer K, Hauser PM, Pagni M. 2013. Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. mBio 4(3):e00055-13. doi:10.1128/mBio.00055-13.
REFERENCES
- 1. Ogawa JM, Zehr EI, Bird GW, Ritchie K, Uriu K, Uyemoto JK. 1995. Compendium of stone fruit diseases. APS Press, St. Paul, MN. [Google Scholar]
- 2. Chester KS. 1947. Nature and prevention of plant diseases, 2nd ed. The Blakiston Company, Philadelphia, PA. [Google Scholar]
- 3. Rossi V, Bolognesi M, Giosuè S, Mazzini F, Ponti I, Spada G. 2005. Biologia ed epidemiologia dell’agente della bolla del pesco. Inf. Agrario 10:50–67 [Google Scholar]
- 4. Kern H, Naef Roth S. 1975. Zur Bilding von Auxinen und Cytokininen durch Taphrina-Arten. Phytopathology 83:193–222 [Google Scholar]
- 5. Yamada T, Tsukamoto H, Shiraishi T, Nomura T, Oku H. 1990. Detection of indoleacetic acid biosynthesis in some species of Taphrina causing hyperplastic diseases in plants. Ann. Phytopathol. Soc. Jpn. 56:532–540 [Google Scholar]
- 6. Sugiyama J, Hosaka K, Suh SO. 2006. Early diverging Ascomycota: phylogenetic divergence and related evolutionary enigmas. Mycologia 98:996–1005 [DOI] [PubMed] [Google Scholar]
- 7. Fonseca Á, Rodrigues MG. 2011. Taphrina fries, p 823–858 In Kurtzman CP, Fell JW, Boekhout T, The yeasts, a taxonomic study, 5th ed, vol 2 Elsevier, Amsterdam, The Netherlands: . http://dx.doi.org/10.1016/B978-0-444-52149-1.00073-2 [Google Scholar]
- 8. Bacigalova K, Lopandic K, Rodrigues M, Fonseca A, Herzberg M, Pinsker W, Prillinger H. 2002. Phenotypic and genotypic identification and phylogenetic characterisation of Taphrina fungi on alder. Mycol. Prog. 2:179–196 [Google Scholar]
- 9. Keely SP, Renauld H, Wakefield AE, Cushion MT, Smulian AG, Fosker N, Fraser A, Harris D, Murphy L, Price C, Quail MA, Seeger K, Sharp S, Tindal CJ, Warren T, Zuiderwijk E, Barrell BG, Stringer JR, Hall N. 2005. Gene arrays at Pneumocystis carinii telomeres. Genetics 170:1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Lafton A, Latgé JP, Li W, Lord A, Lu C, Majoros WH, May GS, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 11. Selker EU. 2002. Repeat-induced gene silencing in fungi. Adv. Genet. 46:439–450 [DOI] [PubMed] [Google Scholar]
- 12. Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schübeler D, Selker EU. 2009. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 19:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fulci V, Macino G. 2007. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr. Opin. Microbiol. 10:199–203 [DOI] [PubMed] [Google Scholar]
- 14. Shiu PK, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–916 [DOI] [PubMed] [Google Scholar]
- 15. Lang BF. 1984. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe: highly homologous introns are inserted at the same position of the otherwise less conserved cox1 genes in Schizosaccharomyces pombe and Aspergillus nidulans. EMBO J. 3:2129–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lang BF, Ahne F, Bonen L. 1985. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe. The cytochrome b gene has an intron closely related to the first two introns in the Saccharomyces cerevisiae cox1 gene. J. Mol. Biol. 184:353–366 [DOI] [PubMed] [Google Scholar]
- 17. Schäfer B, Kaulich K, Wolf K. 1998. Mosaic structure of the cox2 gene in the petite negative yeast Schizosaccharomyces pombe: a group II intron is inserted at the same location as the otherwise unrelated group II introns in the mitochondria of higher plants. Gene 214:101–112 [DOI] [PubMed] [Google Scholar]
- 18. Cissé OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4(1):e00428-00412 http://dx.doi.org/10.1128/mBio.00428-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y, Leigh JW, Brinkmann H, Cushion MT, Rodriguez-Ezpeleta N, Philippe H, Lang BF. 2009. Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Mol. Biol. Evol. 26:27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lowe RG, Howlett BJ. 2012. Indifferent, affectionate, or deceitful: lifestyles and secretomes of fungi. PLoS Pathog. 8:e1002515 http://dx.doi.org/10.1371/journal.ppat.1002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolattukudy P. 1985. Enzymatic penetration of the plant cuticle by fungal pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:4080–40877753774 [Google Scholar]
- 22. Voigt CA, Schäfer W, Salomon S. 2005. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 42:364–375 [DOI] [PubMed] [Google Scholar]
- 23. Stergiopoulos I, de Wit PJ. 2009. Fungal effector proteins. Annu. Rev. Phytopathol. 47:233–263 [DOI] [PubMed] [Google Scholar]
- 24. Fling ME, Kopf J, Tamarkin A, Gorman JA, Smith HA, Koltin Y. 1991. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol. Gen. Genet. 227:318–329 [DOI] [PubMed] [Google Scholar]
- 25. Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kües U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FW, vanKuyk PA, Horton JS, Grigoriev IV, Wösten HA. 2010. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 28:957–963 [DOI] [PubMed] [Google Scholar]
- 26. Perley JE, Stowe BB. 1966. On the ability of Taphrina deformans to produce indoleacetic acid from tryptophan by way of tryptamine. Plant Physiol. 41:234–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reineke G, Heinze B, Schirawski J, Buettner H, Kahmann R, Basse CW. 2008. Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol. Plant Pathol. 9:339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Eijk GW, Roeymans HJ. 1982. Distribution of carotenoids and sterols in relation to the taxonomy of Taphrina and Protomyces. Antonie Van Leeuwenhoek 48:257–264 [DOI] [PubMed] [Google Scholar]
- 29. Estrada AF, Brefort T, Mengel C, Díaz-Sánchez V, Alder A, Al-Babili S, Avalos J. 2009. Ustilago maydis accumulates beta-carotene at levels determined by a retinal-forming carotenoid oxygenase. Fungal Genet. Biol. 46:803–813 [DOI] [PubMed] [Google Scholar]
- 30. Siewers V, Kokkelink L, Smedsgaard J, Tudzynski P. 2006. Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl. Environ. Microbiol. 72:4619–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Müller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C. 2011. Comparative functional genomics of the fission yeasts. Science 332:930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fraenkel GS. 1959. The râisons d’être of secondary plant substances; these odd chemicals arose as a means of protecting plants from insects and now guide insects to food. Science 129:1466–1470 [DOI] [PubMed] [Google Scholar]
- 33. Sancholle M, Weete JD, Montant C. 1984. Effects of triazoles on fungi: I. Growth and cellular permeability. Pestic. Biochem. Physiol. 21:31–44 [Google Scholar]
- 34. Follas G, Welsh RD. 1993. Control of leaf curl in stone fruit with difenoconazole. Proc. NZ Plant Protect. Conf. 46:18-20 [Google Scholar]
- 35. Rekanovic E, Mihajlovic M, Potocnik I. 2010. In vitro sensitivity of Fusarium graminearum (Schwabe) to difenoconazole, prothioconazole and thiophanate-methyl. Pestic. Fitomed. 25:325–333 [Google Scholar]
- 36. Weete JD, Montant C. 1983. Effects of triazoles on fungi. II. Lipid composition of Taphrina deformans Biochim. Biophys. Acta 752:19–29 [Google Scholar]
- 37. Hayashi K, Schoonbeek HJ, De Waard MA. 2002. Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol. 68:4996–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laaser G. 1989. Vergleichende systematische Studien an Basidiomycetenhefen unter besonderer Berücksichtigung der Hefestadien. Bibl. Mycol. 130:1–325 [Google Scholar]
- 39. Kornberg HL, Beevers H. 1957. The glyoxylate cycle as a stage in the conversion of fat to carbohydrate in castor beans. Biochim. Biophys. Acta 26:531–537 [DOI] [PubMed] [Google Scholar]
- 40. Underwood AP, Louis EJ, Borts RH, Stringer JR, Wakefield AE. 1996. Pneumocystis carinii telomere repeats are composed of TTAGGG and the subtelomeric sequence contains a gene encoding the major surface glycoprotein. Mol. Microbiol. 19:273–281 [DOI] [PubMed] [Google Scholar]
- 41. Nahimana A, Francioli P, Blanc DS, Bille J, Wakefield AE, Hauser PM. 2000. Determination of the copy number of the nuclear rDNA and beta-tubulin genes of Pneumocystis carinii f. sp. hominis using PCR multicompetitors. J. Eukaryot. Microbiol. 47:368–372 DOI: 10.1111/j.1550-7408.2000.tb00062.x. PubMed [DOI] [PubMed] [Google Scholar]
- 42. Tang X, Bartlett MS, Smith JW, Lu JJ, Lee CH. 1998. Determination of copy number of rRNA genes in Pneumocystis carinii f. sp. hominis. J. Clin. Microbiol. 36:2491–2494 PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. U. S. A. 100:15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chapman DJ, Regan MA. 1980. Evolution of a biochemical pathway: evidence from comparative biochemistry. Annu. Rev. Plant Physiol. 31:639–645 [Google Scholar]
- 45. Nowrousian M, Stajich JE, Chu M, Engh I, Espagne E, Halliday K, Kamerewerd J, Kempken F, Knab B, Kuo HC, Osiewacz HD, Pöggeler S, Read ND, Seiler S, Smith KM, Zickler D, Kück U, Freitag M. 2010. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 6:e1000891 http://dx.doi.org/10.1371/journal.pgen.1000891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher-plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35:155–189 [Google Scholar]
- 47. Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B. 2001. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J. Plant Growth Regul. 20:319–331 [DOI] [PubMed] [Google Scholar]
- 48. Kramer CL. 1960. Morphological development and nuclear behavior in the genus Taphrina. Mycologia 52:295–320 [Google Scholar]
- 49. Lachance MA. 1985. Current views on the yeast species. Microbiol. Sci. 2:122–126 [PubMed] [Google Scholar]
- 50. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 51. Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 52. Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067 [DOI] [PubMed] [Google Scholar]
- 53. Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18:1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stanke M, Schöffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62 http://dx.doi.org/10.1186/1471-2105-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59 http://dx.doi.org/10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18:188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. UniProt Consortium 2012. Reorganizing the protein space at the universal protein resource (UniProt). Nucleic Acids Res. 40:D71–D75 http://dx.doi.org/10.1093/nar/gks060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lewis SE, Searle SM, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglir L, Birney E, Crosby MA, Kaminker JS, Matthews BB, Prochnik SE, Smithy CD, Tupy JL, Rubin GM, Misra S, Mungall CJ, Clamp ME. 2002. Apollo: a sequence annotation editor. Genome Biol. 3:RESEARCH0082 http://dx.doi.org/10.1186/gb-2002-3-12-research0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res. 37:D136–D140 http://dx.doi.org/10.1093/nar/gkp725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Claudel-Renard C, Chevalet C, Faraut T, Kahn D. 2003. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31:6633–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 http://dx.doi.org/10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36:3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJ, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. 2012. Interpro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40:D306–D312 http://dx.doi.org/10.1093/nar/gkr948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 67. Rattei T, Tischler P, Götz S, Jehl MA, Hoser J, Arnold R, Conesa A, Mewes HW. 2010. SIMAP—a comprehensive database of pre-calculated protein sequence similarities, domains, annotations and clusters. Nucleic Acids Res. 38:D223–D226 http://dx.doi.org/10.1093/nar/gkp949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110:462–467 [DOI] [PubMed] [Google Scholar]
- 69. Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Roth AC, Gonnet GH, Dessimoz C. 2008. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics 9:518 http://dx.doi.org/10.1186/1471-2105-9-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39–64 [DOI] [PubMed] [Google Scholar]
- 72. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 73. Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bähler J, Kersey PJ, Oliver SG. 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40:D695–D699 http://dx.doi.org/10.1093/nar/gkr853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Telomeres. Download
Mitochondrial genome assembly and annotation. Download
Composite RIP indexes (CRI) in Taphrina deformans and Neurospora crassa genomes. (A) No scaffold in the T. deformans exhibits a large AT-rich region with typical RIP bias (CRI index value, >0). The example of scaffold 5 is shown. All 394 scaffolds of the genome assembly were subjected to the same analysis with similar results. (B) The linkage group I sequence from N. crassa which has been reported to be subjected to the RIP (45) was used as a control. Download
Repeated elements in T. deformans genome.
T. deformans genes involved in RNA-mediated silencing.
T. deformans mitochondrial genome statistics and comparison to those of other Taphrinomycotina members.
Gene model support.
Overrepresented Gene Ontology (GO) categories of genes present in T. deformans and plant fungal saprophytes.
Overrepresented Gene Ontology (GO) categories of genes present in T. deformans and plant fungal pathogens.
Carbohydrate-active enzyme genes in T. deformans and selected fungi.