ABSTRACT
Anthrax toxin proteins from Bacillus anthracis constitute a highly efficient system for delivering cytotoxic enzymes to the cytosol of tumor cells. However, exogenous proteins delivered to the cytosol of cells are subject to ubiquitination on lysines and proteasomal degradation, which limit their potency. We created fusion proteins containing modified ubiquitins with their C-terminal regions fused to the Pseudomonas exotoxin A catalytic domain (PEIII) in order to achieve delivery and release of PEIII to the cytosol. Fusion proteins in which all seven lysines of wild-type ubiquitin were retained while the site cleaved by cytosolic deubiquitinating enzymes (DUBs) was removed were nontoxic, apparently due to rapid ubiquitination and proteasomal degradation. Fusion proteins in which all lysines of wild-type ubiquitin were substituted by arginine had high potency, exceeding that of a simple fusion lacking ubiquitin. This variant was less toxic to nontumor tissues in mice than the fusion protein lacking ubiquitin and was very efficient for tumor treatment in mice. The potency of these proteins was highly dependent on the number of lysines retained in the ubiquitin domain and on retention of the C-terminal ubiquitin sequence cleaved by DUBs. It appears that rapid cytosolic release of a cytotoxic enzyme (e.g., PEIII) that is itself resistant to ubiquitination is an effective strategy for enhancing the potency of tumor-targeting toxins.
IMPORTANCE
Bacterial toxins typically have highly efficient mechanisms for cellular delivery of their enzymatic components. Cytosolic delivery of therapeutic enzymes and drugs is an important topic in molecular medicine. We describe anthrax toxin fusion proteins containing ubiquitin as a cytosolic cleavable linker that improves the delivery of an enzyme to mammalian cells. The ubiquitin linker allowed modulation of potency in cells and in mice. This effective strategy for enhancing the intracellular potency of an enzyme may be useful for the cytosolic delivery and release of internalized drugs.
Introduction
The therapeutic benefit of drugs depends on achieving high potency toward the target (an enzyme, cell, bacterium, parasite, virus, etc.) while avoiding damage to the host organism. One approach to killing tumor cells has been to use highly potent bacterial and plant toxins that act catalytically in the cytosol of the targeted cells. Most commonly, specificity has been sought by linking these toxins chemically or genetically to antibodies that bind to cell surface materials enriched on tumor cells. These proteins, initially termed “immunotoxins” and now more generally referred to as “targeted toxins” (TTs), have been under development for decades, but few have reached clinical use (1–3). This appears to be due to inadequate specificity for the tumor versus the host (i.e., a low therapeutic index), low efficiency of delivery to the cytosol, and other factors. Several approaches have been explored for improving the therapeutic indices of TTs (reviewed in reference 2) or to increase the uptake of TTs into the cytosol of tumor cells (4).
Our research group has used a different approach to achieve tumor cell specificity. This approach exploits the fact that anthrax toxin from Bacillus anthracis activity depends on proteolytic activation of the receptor-bound protective antigen (PA) protein by cell surface proteases (5–7). Replacing the site normally cleaved by furin and related proteases with sequences recognized by matrix metalloproteases or urokinase plasminogen activator has yielded potent agents having high specificity and efficacy in mouse tumor models. The protease-activated PA assembles into an oligomeric-protein-conducting channel that efficiently delivers the anthrax toxin catalytic effector proteins to endosomes and then translocates them to the cytosol. The native anthrax effector proteins lethal factor (LF) and edema factor can be replaced with a fusion protein containing the N-terminal 254 amino acids of anthrax toxin lethal factor (lethal factor N terminus [LFn]) and the Pseudomonas aeruginosa exotoxin A (PE) catalytic domain (PEIII). Once in the cytosol, PEIII will transfer ADP-ribose to eukaryotic elongation factor 2 (eEF2), resulting in protein synthesis inhibition and cell death. This system is highly effective in terms of cytosolic delivery and tumor-specific activation. It has been tested successfully on a number of tumor types (8) and is expected to be active on nearly all types of solid tumors.
One factor that affects the potency of all TTs but that has received limited attention is the issue of the stability of the effector proteins once they have reached the cytosol. It was noted in 1989 that many protein toxins have a strong bias against the presence of lysine residues in their catalytic domains (9). In retrospect, it is now evident that this feature limits the attachment of ubiquitin and the resulting proteasomal degradation of toxins (10). The cytosolic stability and resulting potencies of several toxins have been shown to depend on the N-end rule, which specifies that the N-terminal amino acid of a polypeptide determines the efficiency with which side chain lysine residues are ubiquitinated for proteasomal targeting (11). The N-end rule applies to LF and LFn-based fusion proteins (12, 13), indicating that it will impact the efficacy of anthrax toxin-based TTs.
Ubiquitin is a small eukaryotic protein that plays a major role in signal transduction and many other processes in addition to its role in protein degradation. Ubiquitin contains a diglycine motif at its C terminus that is conjugated to the epsilon amine of a lysine within the target protein. According to the N-end rule noted above, ubiquitination occurs on proteins with specific destabilizing N-terminal residues (11), and ubiquitination may occur on several sites within one protein. Ubiquitinated proteins are targeted for degradation by the 26S proteasome system (14). Ubiquitin itself may be ubiquitinated after its conjugation to a target protein, leading to creation of polyubiquitin chains. These chains may be built upon any of the seven lysine residues within ubiquitin by ubiquitin ligases, although Lys48 is most often used (15). Furthermore, polyubiquitination of Lys63 instead of Lys48 seems to result in less protein degradation (16). Ubiquitination is a balanced process with deubiquitinating enzymes (DUBs) counteracting ubiquitination. The DUBs recognize the C-terminal diglycine motif of ubiquitin and release ubiquitin from labeled proteins. This specific release of cargo from ubiquitin fusions is described by Varshavsky as the ubiquitin fusion technique (17), which can be used to conditionally stabilize or destabilize a protein.
In this study, we examined how the insertion of ubiquitin variants within the TT LFn-PEIII fusion protein altered enzymatic activity, cytotoxicity, and stability of the TTs. We used ubiquitin variants that allowed us to study the accessibility of the TTs to DUBs and polyubiquitination. The results obtained indicate that intracellular release of the catalytic PEIII domain is achievable and that ubiquitination of the TTs controls their persistence in the cytosol and thus determines their potency.
RESULTS
Design of TTs.
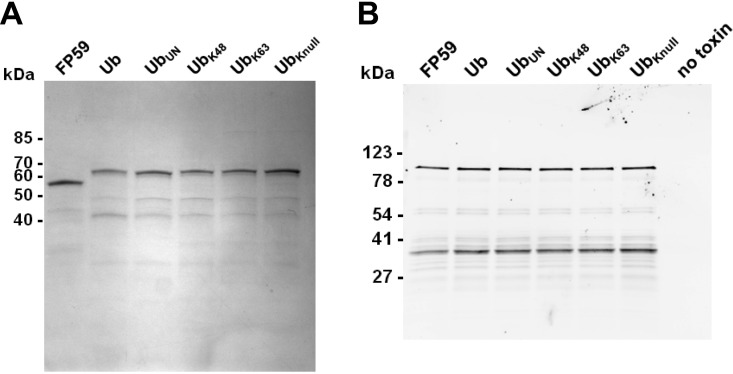
Six different TTs were constructed and analyzed in this study (Fig. 1A). The five ubiquitin-containing TTs are based on the TT anthrax fusion toxin FP59 that contains LFn at the N terminus and PEIII at its C terminus. An alignment of the amino acid sequences shows the differences in the ubiquitin fusion proteins (Fig. 1B). The TT designated “Ub” (Fig. 1A) contains the human wild-type ubiquitin with the C-terminal diglycine motif that is specifically recognized by DUBs. The TT UbUN has the same sequence but with three glycines (including the diglycine motif) replaced by a sequence of three alanines, which renders the sequence uncleavable by DUBs. The UbK48 and UbK63 mutants have all lysine residues of ubiquitin replaced by arginine residues except for one lysine residue, either Lys48 or Lys63, respectively. UbKnull contains ubiquitin with all seven lysines replaced by arginine. UbK48, UbK63, and UbKnull all retain the intact C-terminal diglycine motif. All proteins were successfully purified in yields of at least 1.5 mg per liter of culture (Fig. 2A). The molecular masses of all six proteins were confirmed by electrospray ionization mass spectrometry. Since the accessibility of PEIII within a larger polypeptide or toxin may limit its activity (18), we confirmed the catalytic activity of the five TTs containing ubiquitin or variants of ubiquitin in comparison to the fusion protein FP59 (Fig. 2B). All samples show a band for biotin-containing ADP-ribosylated eEF2 at a molecular mass of 100 kDa and an additional band of the same intensity at around 40 kDa. The latter is a degradation product of eEF2, an artifact of the purification of eEF2 from Saccharomyces cerevisiae yeast. This fragment contains the site of ADP-ribosylation of eEF2 and is thus likewise ADP-ribosylated and detected (19).
FIG 1 .
Domain structures and sequences of ubiquitin fusion proteins. Six different TTs were used for the study on the effects of ubiquitin. (A) LFn-PEIII (FP59) is the basic TT with the PA-binding LFn domain directly fused to the catalytic domain PEIII. All other TTs contain ubiquitin between these two domains, with either an uncleavable alanine sequence (UbUN) or with several or all of the lysines of ubiquitin mutated to arginines (UbK48, UbK63, and UbKnull). All the ubiquitin mutants have the cleavable diglycine motif. (B) The amino acid sequences of the variable portions of the proteins are shown. All PEIII domains end in the native PE sequence REDLK (not shown).
FIG 2 .
Analysis of purified TTs. (A) All TTs were analyzed on a Tris-glycine polyacrylamide gel by Coomassie blue staining. Each lane contained 1 µg of the indicated protein. (B) The enzymatic activity of PEIII within all TTs was studied by in vitro ADP-ribosylation. Each TT (20 ng) was incubated with eEF2, and biotin-labeled NAD+ and the ADP-ribosylated eEF2 were detected after Western blotting by streptavidin detection. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gels.
In vitro deubiquitination of TTs.
In order to analyze and compare the cleavability of the ubiquitin-containing TTs, the fusion proteins were incubated with HN6 cells (a human head and neck cancer cell line) lysates. The lysates contain active DUBs that are expected to cleave the C-terminal diglycine motif of the ubiquitin (20). All TTs containing ubiquitin with the natural C-terminal diglycine sequence were cleaved within the 2-h reaction time (Fig. 3A, B, and C). The cleavage product, LFn-ubiquitin, has an expected molecular mass of 41 kDa and is detected as a band migrating between the 50-kDa and 36-kDa marker bands. FP59, which lacks ubiquitin, showed no cleavage within 2 h (Fig. 3C). Furthermore, UBUN, containing three alanines instead of the diglycine motif, remained intact (Fig. 3C). All TTs were also exposed to lysates preincubated with the DUB inhibitor ubiquitin aldehyde. Pretreatment of the lysates with the inhibitor prevented cleavage of the fusion proteins, demonstrating the specificity of the cleavage. In order to see the effect of purified DUBs, the TTs were incubated with either Ubiquitin carboxyl-terminal hydrolase isozyme L3 (UCH-L3) or Otubain-1 (Otub1). UCH-L3 incubation resulted in low-efficiency cleavage, while Otub1 did not achieve any detectable cleavage (data not shown). These DUBs are apparently not primarily responsible for the intracellular cleavage of the TTs or require further components in the cytosol to achieve efficient cleavage.
FIG 3 .
Deubiquitinating enzyme (DUB) cleavage of TTs. (A to C) Each TT (150 ng) was incubated with HN6 cell lysate for 0, 10, and 120 min. Additional samples were incubated for 120 min in lysates that were preincubated with the DUB inhibitor ubiquitin aldehyde (ub ald). Samples were analyzed by simultaneous anti-LF and anti-PE immunodetection following Western blotting.
Deubiquitination of TTs in cells.
Studies on deubiquitination of TTs within cells were done using CHO TEM8 T4 cells, Chinese hamster ovary cells stably transfected with tumor endothelial marker 8 (TEM8) that overexpress the TEM8 anthrax toxin receptor and internalize more TT, thereby facilitating its detection in the cytosolic fractions. All full-length TTs were detected in the cytosol after 1-h toxin exposure and Western blotting with anti-LF and anti-PE combined (Fig. 4A to C, top black arrow). Mutant PAΔFF (PA with Phe314 and Phe315 deleted) binds and delivers LFn and TTs to endosomes but cannot support their translocation to the cytosol (21). The presence of small amounts of full-length TTs is probably an indication that the cytosolic extracts are contaminated with some endosomal contents. The bigger cleavage product, LFn-ubiquitin, with an apparent molecular mass of 41 kDa, was most likely detected as a very weak band in all fractions (white arrowhead), even in the PAΔFF samples and in the uncleavable UBUN fractions. However, PEIII, the released catalytic domain of the TTs, is clearly detected in the cytosol of cells incubated with UbK48, UbK63, and UbKnull (Fig. 4B and C, bottom black arrow with dashed line at an apparent molecular mass of 24 kDa). The corresponding band for the Ub TT is very weak in comparison, and the UbUN sample has no band for PEIII (Fig. 4A). The data demonstrate that DUB processing of the susceptible TT occurs in the cytosol to release the free PEIII domain. The ubiquitination inhibitor Pyrazone-41 (PYR-41) did not increase the amounts of any of the proteins detected, neither the uncleaved TTs nor the cleaved LFn-ubiquitin or PEIII. The FP59 lanes showed only the expected band for full-length FP59.
FIG 4 .
DUB cleavage of TTs in cells. (A to C) Each TT (0.25 µg/ml) was incubated in the presence of 1 µg/ml PA on CHO TEM8 T4 cells for 0 h and 1 h. Additional samples were preincubated for 1 h with 50 µM PYR-41 or incubated with PAΔFF (ΔFF) instead of PA. Thirty-five nanograms of Ub, UbK48, and UbKnull were loaded as controls. Cytosolic fractions were isolated and analyzed by simultaneous anti-LF and anti-PE immunodetection following Western blotting. The band marked by a black pound symbol indicates a cytosolic protein of CHO cells stained by the polyclonal LF antibody without any relevance for the detection of the fusion proteins.
Cytotoxicity of TTs.
Cytotoxicity analyses employed HN6 cells. This human cell line was chosen as a model cell line for proof of principle, since this cell line is a suitable model for human head and neck cancer when transplanted into nude mice. A 48-h toxin exposure resulted in dose-dependent cytotoxicity with cytotoxicity in the order UbKnull > UbK63 > FP59 > UbK48 >> Ub > UbUN (Fig. 5A). The 50% survival index (SI50) values determined for the TTs are shown in Table 1. The observed SI50 values range from 615 pM (UbUN) to 3.7 pM (UbKnull). These values are 0.02-fold and 3-fold changes of the SI50 of the ubiquitin-free FP59 (FP59 SI50, 11 pM), respectively. A shorter (2-h) period of toxin exposure reduced the observed cytotoxicities for all TTs (Fig. 5B). However, the reductions in potency were about the same for all TTs, since the relative SI50 values (compared to FP59 value) remained similar for all TTs (Table 1). Additionally, FP59-sensitive mouse RAW264.7 cells (murine leukemic monocytes/macrophages) were preincubated with the E1 ubiquitin-activating enzyme inhibitor PYR-41 (22) and subsequently with the different TTs for a further 18 h before cell survival was measured. RAW264.7 cells were used, since they die within 18 h of continuous toxin exposure, while other cell lines need up to 48 h and are thus affected by PYR-41 toxicity (data not shown). PYR-41 increased the cytotoxicities of UbK63, UbKnull, and FP59 only slightly (see Fig. S1D to F in the supplemental material), while increasing the toxicities of UbK48, and especially Ub and UBUN, to larger, if still modest extents (Fig. S1A to C), consistent with the expectation that the latter TTs are more susceptible to ubiquitination and inactivation.
FIG 5 .
Cytotoxicity of TTs on HN6 human head and neck cancer cells. HN6 cells (10,000 cells/well) were exposed to different concentrations of TTs for 48 h (A) or 2 h (B). All samples contained a fixed concentration of 250 ng/ml PA and various TT concentrations. Viable cells were quantitated in an assay employing 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). Relative survival was calculated as the percentage of living cells after treatment in relation to untreated cells. Error bars indicate standard errors of the means (SEM) of 6 (A) and 4 (B) independent experiments performed in triplicate.
TABLE 1.
Cytotoxicity of the targeted toxins on HN6 cellsa
| Targeted toxin | 48-h toxin exposure |
2-h toxin exposure |
||||
|---|---|---|---|---|---|---|
| SI50 (pM) | P valueb | Factor of enhancementc | SI50 (pM) | P value | Factor of enhancement | |
| UbUN | 615 | 0.052 | 0.02 | NDd | 0.066 | ND |
| Ub | 100 | 0.045 | 0.11 | 773.0 | 0.053 | 0.03 |
| UbK48 | 014 | 0.093 | 0.79 | 79 | 0.012 | 0.34 |
| FP59 | 011 | 027 | ||||
| UbK63 | 006.2 | 0.051 | 1.8 | 018 | 0.496 | 1.5 |
| UbKnull | 003.7 | 0.038 | 3 | 011 | 0.114 | 2.5 |
SI50 values, P values, and factors of enhancement for the cytotoxicity of the TTs on HN6 cells obtained from Fig. 5.
The P value was calculated for the FP59 value versus the values of all TTs. Bold P values are below 0.05, which is defined to be a significant difference.
The factor of enhancement was calculated by comparison of the SI50 value of FP59 to those of all other TTs.
ND, not determined.
Tumor treatment.
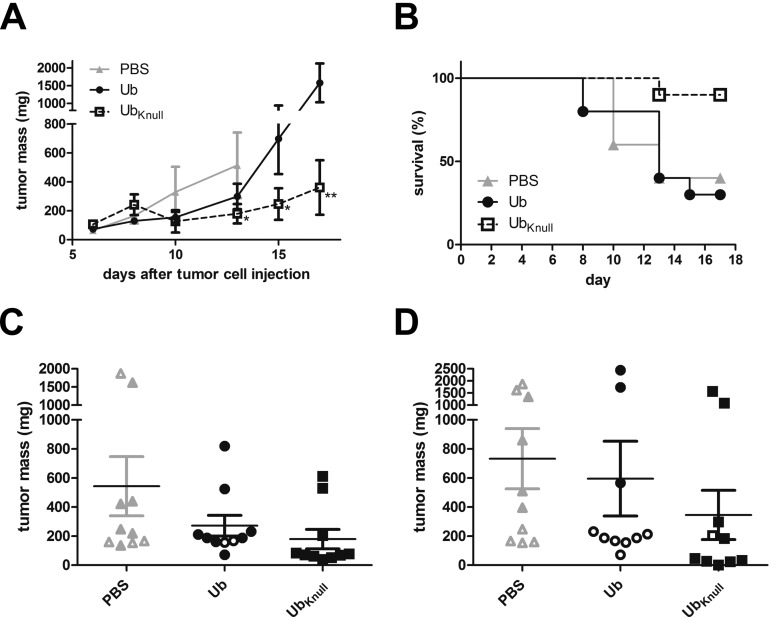
The Ub and UbKnull fusion proteins as well as the control FP59 protein were administered with the matrix metalloprotease-activated PA-L1 to mice with implanted LL3 mouse melanoma cell tumors. Treatment with PA-L1 plus Ub or UbKnull resulted in strong tumor growth inhibition (Fig. 6A). UbKnull significantly reduced tumor growth compared to the group treated with phosphate-buffered saline (PBS group) (the control group) on day 13 (P = 0.05). Due to rapid tumor growth and ulceration in the PBS group, several mice were euthanized before the end of the experiment. Therefore, only the Ub and UbKnull groups were compared at days 15 and 17; UbKnull treatment was significantly better than Ub treatment (P = 0.04 and P = 0.01, respectively). UbKnull treatment resulted in the development of inflammation and edema after the third injection, while Ub treatment did not result in any observable side effects. More aggressive tumor growth in the PBS and Ub groups resulted in a higher number of mice being euthanized before the end of the experiment (Fig. 6B). Individual tumor masses at day 13 and day 17 demonstrate a very good antitumor effect of the UbKnull treatment (Fig. 6C and D, respectively). Eight of 10 mice showed a strong response to the treatment, and one mouse was tumor-free. Treatment with PA-L1 plus FP59 resulted in severe inflammation and edema after the first injection, and no further injections were possible on day 8 or 10. One mouse was euthanized due to lethargy, most likely related to the FP59 treatment. Even at the end of the experiment, 7 of 10 mice showed symptoms like those following the first treatment. Due to the toxic effects of this treatment, the FP59 group was not further analyzed for tumor growth-inhibitory effects.
FIG 6 .
Tumor treatment by TTs in a mouse model. Ten C57BL/6 mice per group were intradermally injected with LL3 mouse melanoma tumor on day 0. Intratumoral injection of 50 µl TT was performed on days 6, 8, and 10 using either PBS (gray triangles), 5 µg PA-L1 plus 1.15 µg Ub (black circles), or 5 µg PA-L1 plus 1.15 µg UbKnull (squares). (A) Mean tumor masses until day 17. Tumor mass was calculated from measurements of the tumor width, depth, and height. PBS group values are displayed only until day 13 because of decreased group size due to tumor ulcerations. Significant decreases in tumor volume in the UbKnull group compared to the PBS group (day 13) and to the Ub group are indicated by asterisks. One asterisk indicates P smaller or equal to 0.05 and two asterisks indicate P smaller or equal to 0.01. Error bars indicate the SEM. (B) Mouse survival. Tumor ulcerations and excessive tumor sizes were premature endpoints and resulted in group size reductions before the end of the experiment on day 17. (C and D) Individual tumor masses as calculated for day 13 (C) and day 17 (D) data. Open symbols indicate tumor masses from animals euthanized before the analyzed time point. Horizontal lines depict the mean tumor masses for the different groups (including animals euthanized at earlier time points), and error bars indicate the SEM.
DISCUSSION
TTs containing bacterial toxin catalytic domains possess high potency, at least in theory, but this high potency requires that the TT be targeted with high specificity to avoid damage to nontarget cells. A high inherent potency allows lower doses to be used, which can help to limit the inevitable damage that comes from clearance of administered proteins to, and accumulation in, the liver and kidneys (23). However, for low protein doses to be effective, the protein must be efficiently bound and internalized and translocated to the cytosol, and once there, it must have sufficient stability to inactivate its target. The studies described here focus on the latter steps in TT action. The modified anthrax toxin used in our study utilizes a highly efficient delivery system which can deliver a number of different “payloads” in addition to the natural toxin lethal factor, as it was shown that the system is able to deliver PEIII (24), diphtheria toxin A chain, Shiga toxin catalytic domain (25), Bcl-XL protein (26), as well as the reporter beta-lactamase (27, 28).
A key feature of the TTs described here is the insertion of a ubiquitin domain between the targeting domain (LFn) and the catalytic payload (PEIII). A prerequisite for success of this design is that the ubiquitin domain not limit the efficiency of translocation of the enlarged, 3-domain polypeptides to the cytosol. Previous studies of the translocation process show that polypeptides must completely unfold to pass through the narrow lumen of the oligomeric PA channel (13, 29). Proteins that are tightly folded, either naturally or due to ligand binding, will not be translocated, although it is possible that chaperones may facilitate translocation of certain proteins (30). Ubiquitin is a tightly folded protein (31), and it was not certain that it would readily translocate. However, the fact that several of the proteins characterized here had potencies very similar to that of the FP59 protein indicate that ubiquitin unfolding occurred readily, at least in the context of these LFn fusion proteins and in the HN6 cells.
A second prerequisite for success of the strategy described here is that cytosolic DUBs be able to cleave the fusion proteins at an adequate rate. Here, the strategy is aided by the great diversity of DUBs present in cells (32). It was evident from the cleavages we observed (Fig. 3 and 4) and the pattern of toxicities (Fig. 5 and Table 1) that at least a few of the DUBs recognized the ubiquitin and the modified ubiquitins in the context of the 3-domain fusions. Experiments with purified DUBs showed that UCH-L3 could be one of the DUBs involved to some degree in cleavage of the TTs, while Otub1 does not recognize the ubiquitin-containing TTs as the substrates. Even replacement of all lysines with arginine (in the UbKnull protein) did not greatly diminish the ability of the DUBs to cleave the fusion proteins.
The release of a free catalytic domain (e.g., PEIII) in the cytosol has a number of potential and real advantages, including the opportunity to increase its stability and potency. Typically, it can be expected that any polypeptide used in the delivery of the catalytic domain will contain a number of lysines that provide targets for ubiquitination. This is clearly the case with LFn fusion proteins, since 32 of the 254 residues of LFn are lysine. Thus, an LFn-PEIII fusion protein (e.g., FP59) might be expected to be rapidly degraded. In fact, we and others have shown that LF and LFn are subject to the N-end rule (12, 13) and therefore that the side chain lysines are ubiquitinated, provided the N-terminal region has a destabilizing residue under the N-end rule. In retrospect, it is rather surprising that LF and LFn fusion proteins possess high potency. Thus, the introduction of a ubiquitin domain into a TT provides a way to cause release of the catalytic domain, freeing it from other domains (usually in the N-terminal region) that would promote its degradation. In the case of the PEIII fusions described here, the catalytic domain contains only two lysines, and therefore is inherently resistant to proteasomal degradation.
Another advantage of the strategy described here is the ability to release the TT catalytic domain having a specific, desirable N-terminal residue. This objective was one impetus to the original development of the ubiquitin fusion method (17). In the case of TTs, it is often desirable to generate a free catalytic domain having an N-terminal residue that is an N-end rule stabilizing residue.
Introducing ubiquitin as a linker between the two domains of FP59 to produce the TT designated “Ub” decreased its potency nearly 10-fold (Table 1). The introduction of ubiquitin targets the fusion protein to the proteasome after polyubiquitination (33) and prevented efficient accumulation of PEIII in the cytosol of cells (Fig. 4). Even though in vitro cleavage was as efficient as for the other cleavable TTs (Fig. 3), release of PEIII may be too slow to save most of the fusion protein from degradation. The uncleavable UbUN showed no cleavage and was apparently degraded too fast for the PEIII to efficiently inhibit protein synthesis (Fig. 4). Only the DUB-cleavable TTs having a reduced number of Lys residues demonstrated successful accumulation of PEIII in the cytosol and thus much higher cytotoxicities. The release of PEIII from the rest of the molecule helps PEIII to persist longer in the cytosol, which is a requisite for high cytotoxicity (34). UbKnull, which is resistant to ubiquitination of the inserted ubiquitin domain (35), is even more toxic than FP59 and presents the lowest SI50 of all TTs studied here (Table 1). Its high cytotoxicity is apparently due to the release of PEIII at rates well above those at which ubiquitination of the lysines on the LFn domain targets the intact fusion protein to the proteasome. Due to severe side effects of the FP59 treatment, UbKnull could not be compared directly to FP59 in the tumor model. However, lower side effects combined with a strong antitumor effect indicate the advantageous effect of the UbKnull insertion in FP59. The comparison to Ub in the tumor experiment clearly shows the importance of mutating the lysines of ubiquitin for increasing the protein’s stability (Fig. 6A). In cell culture experiments, the UbK63 mutant was slightly more cytotoxic than the UbK48 mutant (Table 1), consistent with prior evidence that ubiquitination of these two lysines leads to different outcomes. Thus, Jacobson et al. described differences for the proteasomal processing of polyubiquitin chains built on lysine 48 versus lysine 63, with faster deubiquitination of lysine 63-linked polyubiquitin chains and less proteasomal accessibility (16). This would explain the lower SI50 value for UbK63 compared to UbK48. UbK63 would persist longer in the cytosol before it is degraded and counteract the lower release of PEIII detected (Fig. 3C). Thus, the different ubiquitin variants allow the modulation of PEIII release and protein stability so as to achieve different levels of cytotoxicity.
The effect of the ubiquitination inhibitor PYR-41 was rather weak in the cytotoxicity studies (see Fig. S1 in the supplemental material). Higher concentrations are typically needed to obtain sufficient inhibition of ubiquitin activation (22). However, these concentrations were toxic for the cell lines used here. Furthermore, there was no increased stability of the TTs observed due to preincubation of PYR-41 (Fig. 4). As Fig. S1 shows, the effects of PYR-41 are rather weak, probably due to the instability of PYR-41.
Cleavable sequences have been introduced in other TTs for separation of the different moieties. Thus, Heisler et al. introduced furin- and cytosolic-protease-cleavable peptide sequences in a fusion of epidermal growth factor and the plant toxin saporin to release the toxin in the cytosol (36). Furin cleavage sites were also introduced into TTs by other groups to achieve increased cytotoxicity (37, 38). Fusions to ubiquitin have more commonly been used to decrease the stability of proteins, including green fluorescent protein (39) and beta-lactamase (33). In an approach to enhance TT potency more like that described here, Tcherniuk et al. fused ubiquitin to saporin, a plant protein toxin. In that study (40), the DUB cleavage sequence of ubiquitin was exchanged with a prostate-specific antigen cleavage site. Thus, the authors intended to separate ubiquitin and saporin only in the vicinity of tumor cells expressing prostate-specific antigen, following the same idea used for the activation of anthrax toxin PA by urokinase plasminogen activator or matrix metalloproteases (6). In contrast to PA, where the proteolytic cleavage results in activation of PA, prostate-specific antigen cleavage would separate ubiquitin and saporin with the goal of enrichment of ubiquitin-free saporin in the vicinity of tumor cells without including a strategy for achieving binding and delivery of the payload to the cytosol, and no gain in potency was achieved.
The data presented here demonstrate the value of introducing ubiquitin to the anthrax toxin delivery system (PA and LFn) in order to efficiently deliver enzymes to the cytosol. Variants with the lysines of ubiquitin replaced by arginines have improved stability and significant antitumor effects. Thus, the ubiquitin system for intracellular release provides an effective system for the fine-tuning of toxin uptake, half-life, nonspecific toxicity, and overall efficacy. Future studies on this system will hopefully provide further drugs capable of the safe and efficient elimination of tumor cells and, eventually, may warrant clinical trials in humans.
MATERIALS AND METHODS
Cloning of ubiquitin-containing targeted toxins.
All TTs used for this study are based on the anthrax fusion toxin FP59, consisting of LFn-PEIII (5) and were modified by inserting different ubiquitin variants between LFn and PEIII. LFn consists of the N-terminal 254 residues of anthrax toxin lethal factor (Fig. 1) and has the sequence AGG…QEINL (but in FP59 includes an additional N-terminal HM added due to cloning). PEIII consists of the C-terminal 216-residue catalytic domain of PE and has the sequence AEFL…REDLK. Details on cloning, expression, and purification of the ubiquitin-containing targeted toxins are described in the supplemental information (see Text S1 in the supplemental material). PA and the mutant PAΔFF (PA with Phe314-Phe315 deleted) were expressed as described earlier (21, 41).
Enzymatic activity of TTs.
The enzymatic activities of PEIII within all TTs were measured as described previously (42). Incubation of the TTs with purified eEF2 and biotinylated NAD+ allows the detection of ADP-ribosylated eEF2 by using streptavidin to detect biotin on eEF2 after SDS-PAGE and Western blotting.
Cell culture.
Cell culture experiments were performed on HN6 cells (a human head and neck cancer cell line) (43), CHO TEM8 T4 cells (Chinese hamster ovary cells stably transfected with anthrax receptor tumor endothelial marker 8) (41, 44), and RAW264.7 cells (murine leukemic monocytes/macrophages). See Text S1 in the supplemental material for details on cell culture and cytotoxicity assays.
Cleavage by deubiquitinating enzymes.
Lysates of HN6 cells were obtained after growing 6 × 106 HN6 cells overnight, washing them twice with phosphate-buffered saline (PBS) (150 mM NaCl, 8.3 mM Na2HPO4, 1.7 mM KH2PO4 [pH 7.4]), and incubating them with 300 µl of PBS supplemented with 1% Triton X-100. After 30 min at 4°C on a rotary shaker, cells were resuspended and centrifuged (30 min, 4°C, 16,000 × g). The supernatant (cell lysates) were used for in vitro cleavage of ubiquitin fusion toxins by incubation of 75 ng of the TTs for up to 120 min at 37°C with 8 µl of the cell lysate supplemented with 2 mM dithiothreitol (DTT) in a total volume of 9 µl. Additional experiments were performed with ubiquitin aldehyde (16) to inhibit deubiquitinating activity. Ubiquitin aldehyde (0.5 µl; final concentration, 2.5 µM) was incubated with the lysate 5 min at 37°C prior to TT addition.
Following incubation with the lysate, samples were separated by SDS-PAGE and Western blotted using the iBlot system (Invitrogen, Life Technologies, Grand Island, NY). TTs were detected by a polyclonal rabbit anti-LF serum and infrared dye-conjugated secondary antibodies on the Odyssey imager infrared detection system (LI-COR, Lincoln, NE).
Samples for detection of intracellular cleavage were separated by SDS-PAGE and Western blotted using the iBlot system and the Western blot signal enhancer kit (Thermo, Waltham, MA) for signal enhancement. TTs were detected by polyclonal rabbit anti-LF serum, polyclonal rabbit anti-PE (anti-Pseudomonas exotoxin A) serum (Sigma-Aldrich, St. Louis, MO), and infrared dye-conjugated secondary antibodies (Rockland Immunochemicals, Gilbertsville, PA) on the Odyssey imager infrared detection system (LI-COR, Lincoln, NE).
Detection of intracellular cleavage was performed on CHO TEM8 T4 cells (1 × 106 cells overnight in 12-well plates). The cells were incubated with 1 µg/ml PA or PAΔFF (PA with Phe314 and Phe315 deleted, a mutant that fails to deliver LF to the cytosol [21]), and 1 µg/ml of the TTs in 0.5 ml medium for 1 h or washed off immediately (0 h). Further samples were preincubated with 50 µM PYR-41 (22) for 1 h at 37°C before the addition of PA and the TTs at the indicated concentrations for 1 h. All cells were washed twice with PBS and incubated with trypsin-EDTA at 37°C until all cells could be transferred into new tubes for centrifugation (5 min, 4°C, 1,000 × g). For cytosol isolation, the cells were resuspended in 125 µg/ml saponin (Sigma-Aldrich, St. Louis, MO) in PBS, supplemented with protease inhibitor cocktail (Roche), and incubated for 10 min on ice (45). The complete supernatants after centrifugation (30 min, 4°C, 16,000 × g) were analyzed by SDS-PAGE and Western blotting.
Animal experiments.
All animal experiments were performed under protocols approved by the NIAID Animal Care and Use Committee. Female C57BL/6 mice (Jackson Labs, Bar Harbor, ME) were injected with 0.8 × 106 LL3 mouse melanoma cells intradermally in the neck on day 0. After 6 days (all tumors had a width of at least 4 mm), mice were randomly assigned to four groups of 10 mice each and injected with 50 µl sterile PBS with different drug combinations intratumorally every other day (day 6, day 8, and day 10). Mice were treated with PBS, 5 µg PA-L1 (PA with a mutated furin cleavage site to achieve tumor-selective cleavage and activation of PA by matrix metalloproteinase 2 [5]) plus 1 µg FP59 in PBS, 5 µg PA-L1 plus 1.15 µg LFnUbPEIII in PBS, or 5 µg PA-L1 plus 1.15 µg LFnUbKnullPEIII in PBS). Tumors were measured every other day with a caliper, and tumor mass was calculated [tumor mass = (width × depth × height)/2, with tumor mass measured in milligrams and width, depth, and height all measured in millimeters] with a final measurement on day 17. Mice with tumors with one diameter exceeding 20 mm or ulceration were euthanized.
SUPPLEMENTAL MATERIAL
Additional detailed Materials and Methods. Download
Cytotoxicity of TTs on RAW264.7 cells in the presence of the E1 ubiquitin-activating enzyme inhibitor PYR-41. RAW264.7 cells (15,000 cells/well) were preincubated with 21 µM PYR-41 or without PYR-41 for 1 h and further exposed to the TTs for 18 h. TTs were added to the cells in various concentrations with a fixed concentration of 250 ng/ml PA. Relative survival was calculated as described in the legend to Fig. 5. Error bars indicate SEM of two independent experiments performed in triplicate. Each panel presents the relative survival data for the indicated TT. Download
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD.
We thank J. Eric Anderson for mass spectrometric analysis of the proteins described here.
Footnotes
Citation Bachran C, Morley T, Abdelazim S, Fattah RJ, Liu S, Leppla SH. 2013. Anthrax toxin-mediated delivery of the Pseudomonas exotoxin A enzymatic domain to the cytosol of tumor cells via cleavable ubiquitin fusions. mBio 4(3):e00201-13. doi:10.1128/mBio.00201-13
REFERENCES
- 1. Alfano RW, Leppla SH, Liu S, Bugge TH, Duesbery NS, Frankel AE. 2008. Potent inhibition of tumor angiogenesis by the matrix metalloproteinase-activated anthrax lethal toxin: implications for broad anti-tumor efficacy. Cell Cycle 7:745–749 [DOI] [PubMed] [Google Scholar]
- 2. Hetzel C, Bachran C, Tur MK, Fuchs H, Stöcker M. 2009. Improved immunotoxins with novel functional elements. Curr. Pharm. Des. 15:2700–2711 [DOI] [PubMed] [Google Scholar]
- 3. Weldon JE, Pastan I. 2011. A guide to taming a toxin—recombinant immunotoxins constructed from Pseudomonas exotoxin A for the treatment of cancer. FEBS J. 278:4683–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachran C, Dürkop H, Sutherland M, Bachran D, Müller C, Weng A, Melzig MF, Fuchs H. 2009. Inhibition of tumor growth by targeted toxins in mice is dramatically improved by saponinum album in a synergistic way. J. Immunother. 32:713–725 [DOI] [PubMed] [Google Scholar]
- 5. Liu S, Netzel-Arnett S, Birkedal-Hansen H, Leppla SH. 2000. Tumor cell-selective cytotoxicity of matrix metalloproteinase-activated anthrax toxin. Cancer Res. 60:6061–6067 [PubMed] [Google Scholar]
- 6. Liu S, Redeye V, Kuremsky JG, Kuhnen M, Molinolo A, Bugge TH, Leppla SH. 2005. Intermolecular complementation achieves high-specificity tumor targeting by anthrax toxin. Nat. Biotechnol. 23:725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu S, Wang H, Currie BM, Molinolo A, Leung HJ, Moayeri M, Basile JR, Alfano RW, Gutkind JS, Frankel AE, Bugge TH, Leppla SH. 2008. Matrix metalloproteinase-activated anthrax lethal toxin demonstrates high potency in targeting tumor vasculature. J. Biol. Chem. 283:529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abi-Habib RJ, Singh R, Liu S, Bugge TH, Leppla SH, Frankel AE. 2006. A urokinase-activated recombinant anthrax toxin is selectively cytotoxic to many human tumor cell types. Mol. Cancer Ther. 5:2556–2562 [DOI] [PubMed] [Google Scholar]
- 9. London E, Luongo CL. 1989. Domain-specific bias in arginine/lysine usage by protein toxins. Biochem. Biophys. Res. Commun. 160:333–339 [DOI] [PubMed] [Google Scholar]
- 10. Falnes PO, Olsnes S. 1998. Modulation of the intracellular stability and toxicity of diphtheria toxin through degradation by the N-end rule pathway. EMBO J. 17:615–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varshavsky A. 2011. The N-end rule pathway and regulation by proteolysis. Protein Sci. 20:1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta PK, Moayeri M, Crown D, Fattah RJ, Leppla SH. 2008. Role of N-terminal amino acids in the potency of anthrax lethal factor. PLoS One 3:e3130 http://dx.doi.org/10.1371/journal.pone.0003130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wesche J, Elliott JL, Falnes PO, Olsnes S, Collier RJ. 1998. Characterization of membrane translocation by anthrax protective antigen. Biochemistry 37:15737–15746 [DOI] [PubMed] [Google Scholar]
- 14. Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. 2007. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282:17375–17386 [DOI] [PubMed] [Google Scholar]
- 16. Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW. 2009. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 S proteasome. J. Biol. Chem. 284:35485–35494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Varshavsky A. 2005. Ubiquitin fusion technique and related methods. Methods Enzymol. 399:777–799 [DOI] [PubMed] [Google Scholar]
- 18. Leppla SH, Martin OC, Muehl LA. 1978. The exotoxin of P. aeruginosa: a proenzyme having an unusual mode of activation. Biochem. Biophys. Res. Commun. 81:532–538 [DOI] [PubMed] [Google Scholar]
- 19. Bär C, Zabel R, Liu S, Stark MJ, Schaffrath R. 2008. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol. Microbiol. 69:1221–1233 [DOI] [PubMed] [Google Scholar]
- 20. Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. 2010. Potent delivery of functional proteins into mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem. Biol. 5:747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh Y, Klimpel KR, Arora N, Sharma M, Leppla SH. 1994. The chymotrypsin-sensitive site, FFD315, in anthrax toxin protective antigen is required for translocation of lethal factor. J. Biol. Chem. 269:29039–29046 [PubMed] [Google Scholar]
- 22. Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li CC, Kenten JH, Beutler JA, Vousden KH, Weissman AM. 2007. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 67:9472–9481 [DOI] [PubMed] [Google Scholar]
- 23. Soler-Rodriguez AM, Uhr JW, Richardson J, Vitetta ES. 1992. The toxicity of chemically deglycosylated ricin A-chain in mice. Int. J. Immunopharmacol. 14:281–291 [DOI] [PubMed] [Google Scholar]
- 24. Arora N, Leppla SH. 1993. Residues 1–254 of anthrax toxin lethal factor are sufficient to cause cellular uptake of fused polypeptides. J. Biol. Chem. 268:3334–3341 [PubMed] [Google Scholar]
- 25. Arora N, Leppla SH. 1994. Fusions of anthrax toxin lethal factor with Shiga toxin and diphtheria toxin enzymatic domains are toxic to mammalian cells. Infect. Immun. 62:4955–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu XH, Collier RJ, Youle RJ. 2001. Inhibition of axotomy-induced neuronal apoptosis by extracellular delivery of a Bcl-XL fusion protein. J. Biol. Chem. 276:46326–46332 [DOI] [PubMed] [Google Scholar]
- 27. Hobson JP, Liu S, Rønø B, Leppla SH, Bugge TH. 2006. Imaging specific cell-surface proteolytic activity in single living cells. Nat. Methods 3:259–261 [DOI] [PubMed] [Google Scholar]
- 28. Hu H, Leppla SH. 2009. Anthrax toxin uptake by primary immune cells as determined with a lethal factor-beta-lactamase fusion protein. PLoS One 4:e7946 http://dx.doi.org/10.1371/journal.pone.0007946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thoren KL, Krantz BA. 2011. The unfolding story of anthrax toxin translocation. Mol. Microbiol. 80:588–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamayo AG, Slater L, Taylor-Parker J, Bharti A, Harrison R, Hung DT, Murphy JR. 2011. GRP78(BiP) facilitates the cytosolic delivery of anthrax lethal factor (LF) in vivo and functions as an unfoldase in vitro. Mol. Microbiol. 81:1390–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ralat LA, Kalas V, Zheng Z, Goldman RD, Sosnick TR, Tang WJ. 2011. Ubiquitin is a novel substrate for human insulin-degrading enzyme. J. Mol. Biol. 406:454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reyes-Turcu FE, Ventii KH, Wilkinson KD. 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78:363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stack JH, Whitney M, Rodems SM, Pollok BA. 2000. A ubiquitin-based tagging system for controlled modulation of protein stability. Nat. Biotechnol. 18:1298–1302 [DOI] [PubMed] [Google Scholar]
- 34. Falnes PO, Ariansen S, Sandvig K, Olsnes S. 2000. Requirement for prolonged action in the cytosol for optimal protein synthesis inhibition by diphtheria toxin. J. Biol. Chem. 275:4363–4368 [DOI] [PubMed] [Google Scholar]
- 35. Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. 2005. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25:2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heisler I, Keller J, Tauber R, Sutherland M, Fuchs H. 2003. A cleavable adapter to reduce nonspecific cytotoxicity of recombinant immunotoxins. Int. J. Cancer 103:277–282 [DOI] [PubMed] [Google Scholar]
- 37. Goyal A, Batra JK. 2000. Inclusion of a furin-sensitive spacer enhances the cytotoxicity of ribotoxin restrictocin containing recombinant single-chain immunotoxins. Biochem. J. 345:247–254 [PMC free article] [PubMed] [Google Scholar]
- 38. Wang F, Ren J, Qiu XC, Wang LF, Zhu Q, Zhang YQ, Huan Y, Meng YL, Yao LB, Chen SY, Xu YM, Yang AG. 2010. Selective cytotoxicity to HER2-positive tumor cells by a recombinant e23sFv-TD-tBID protein containing a furin cleavage sequence. Clin. Cancer Res. 16:2284–2294 [DOI] [PubMed] [Google Scholar]
- 39. Deichsel H, Friedel S, Detterbeck A, Coyne C, Hamker U, MacWilliams HK. 1999. Green fluorescent proteins with short half-lives as reporters in Dictyostelium discoideum. Dev. Genes Evol. 209:63–68 [DOI] [PubMed] [Google Scholar]
- 40. Tcherniuk SO, Chroboczek J, Balakirev MY. 2005. Construction of tumor-specific toxins using ubiquitin fusion technique. Mol. Ther. 11:196–204 [DOI] [PubMed] [Google Scholar]
- 41. Liu S, Leung HJ, Leppla SH. 2007. Characterization of the interaction between anthrax toxin and its cellular receptors. Cell. Microbiol. 9:977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bachran C, Sutherland M, Bachran D, Fuchs H. 2007. Quantification of diphtheria toxin mediated ADP-ribosylation in a solid-phase assay. Clin. Chem. 53:1676–1683 [DOI] [PubMed] [Google Scholar]
- 43. Yeudall WA, Crawford RY, Ensley JF, Robbins KC. 1994. MTS1/CDK4I is altered in cell lines derived from primary and metastatic oral squamous cell carcinoma. Carcinogenesis 15:2683–2686 [DOI] [PubMed] [Google Scholar]
- 44. Liu S, Leppla SH. 2003. Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J. Biol. Chem. 278:5227–5234 [DOI] [PubMed] [Google Scholar]
- 45. Newman ZL, Leppla SH, Moayeri M. 2009. CA-074Me protection against anthrax lethal toxin. Infect. Immun. 77:4327–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional detailed Materials and Methods. Download
Cytotoxicity of TTs on RAW264.7 cells in the presence of the E1 ubiquitin-activating enzyme inhibitor PYR-41. RAW264.7 cells (15,000 cells/well) were preincubated with 21 µM PYR-41 or without PYR-41 for 1 h and further exposed to the TTs for 18 h. TTs were added to the cells in various concentrations with a fixed concentration of 250 ng/ml PA. Relative survival was calculated as described in the legend to Fig. 5. Error bars indicate SEM of two independent experiments performed in triplicate. Each panel presents the relative survival data for the indicated TT. Download