Abstract
Protamines are small, highly positively charged peptides used to package DNA to very high densities in sperm nuclei. Tight DNA packing is considered essential to minimize DNA damage by mutagens and reactive oxidizing species. A striking and general feature of protamines is the almost exclusive use of arginine over lysine for the positive charge to neutralize DNA. We have investigated whether this preference for arginine might arise from a difference in DNA condensation by arginine and lysine peptides. The forces underlying DNA compaction by arginine, lysine, and ornithine peptides are measured using the osmotic stress technique coupled with x-ray scattering. The equilibrium spacings between DNA helices condensed by lysine and ornithine peptides are significantly larger than the interhelical distances with comparable arginine peptides. The DNA surface-to-surface separation, for example, is some 50% larger with poly-lysine compared to poly-arginine. DNA packing by lysine rich peptides in sperm nuclei would allow much greater accessibility to small molecules that could damage DNA. The larger spacing with lysine peptides is due to both a weaker attraction and a stronger short ranged repulsion relative to the arginine peptides. A previously proposed model for poly-arginine and protamine binding to DNA provides a convenient framework for understanding the differences between the ability of lysine and arginine peptides to assemble DNA.
The compaction of DNA is a common theme in cell biology. The most tightly packed DNA is found in bacteriophages (1, 2) and vertebrate sperm nuclei (3, 4). In both cases, highly negatively charged DNA is found to be hexagonally organized with interhelical spacings of only 27–30 Å (or surface-to-surface separations of ~ 7–10 Å), corresponding to packing densities of 400 – 500 mg/ml. In these two cases, packing is accomplished quite differently. The DNA is actively transported into the bacteriophage capsid head using very strong molecular motors. Once in the head, the DNA is under tremendous pressure that is used to help eject the DNA into bacterial cells. In vertebrate sperm nuclei, DNA is spontaneously compacted, typically to 28–30 Å spacings by small, highly positively charged, arginine-rich peptides called protamines (5–8). The general consensus is that tight packing of DNA in sperm nuclei is necessary to limit DNA damage due to reactive oxidative species since DNA repair is absent (9–15).
The DNA condensation by cations has been studied for some time now (7, 16–24). The most extensively studied cations, Co(NH3)63+, Mn2+, and the biogenic amines spermidine3+ and spermine4+, condense DNA to 27–30 Å, spacings comparable to protamines (7, 21–24). Only if condensing ions bound to DNA are diluted with ions of lower charge or if excess counterion binding ‘overcharges’ DNA does the spacing between helices increase much beyond 30 Å (21, 25, 26). Only a narrow range of condensed DNA spacings have been observed. The almost exclusive use of arginine by protamines in preference to lysine is then somewhat puzzling. Of the 32 amino acids of salmon protamine, 21 are arginines with no lysines (27). Bovine protamine has 26 arginines out of 50 total amino acids again with no lysines (28). Humans have two protamines; P1 that has 50 amino acids with 24 arginines and no lysines and P2 that has 57 amino acids with 26 arginines and only 2 lysines (27). The N-terminal tails of histones are not as highly charged as protamines, but use both lysines and arginines to compact chromatin. We have previously characterized the packing of DNA by arginine peptides (18) and found that packing by salmon protamine can be quantitatively understood from analysis of the peptides (6). DNA is compacted by +3 and higher charged arginine peptides to the same 27 – 30 Å interhelical spacing range as for the several other DNA condensing ions. We now examine lysine peptides. Lysines are significantly worse than arginines for tightly compacting DNA. The DNA packing density decreases by at least ~ 25 % in comparing arginine and lysine peptides that have the same charge. Equivalently, the surface-to-surface separation between helices increases by some 50%. Ornithine that only differs from lysine by having one less methylene group in the side chain is only slightly better than lysine for compacting DNA, but still much worse than arginine.
We have been characterizing intermolecular forces for many biological macromolecules, including DNA, for many years now using osmotic stress coupled with x-ray scattering (29–31). As noted above, compaction of DNA by multi-charged cations does not result in anhydrous contact of helices, but is rather characterized by significant water left between helices, typically some 7–10 Å. This indicates that cation mediated attraction is balanced by a shorter ranged repulsive force. Our experiments have allowed us to separate the forces into their attractive and repulsive components. The attractive forces necessary for condensation arise from a correlation of positive charges of bound cations bound on one helix with the negative phosphate charges on another, (see, e.g., Kornyshev et al (32)). The spacing of positive and negative charges on apposing helices and helical fluctuations will modulate the attraction amplitude. Both attractive and repulsive forces can be described by exponential functions with ~2.5 and ~5 Å decay lengths for DNA condensed by a wide variety of cations: salmon protamine, Co(NH3)63+, Mn2+, spermidine3+, spermine4+, and several arginine peptides ranging in charge from +2 to +6 (7, 18, 22, 24, 31). Within this same framework, we find that the DNA interhelical forces mediated by lysine peptides have both a weaker attraction and a stronger short ranged repulsion than for the arginine peptides. Significantly, the particular double exponential form that describes the force curves for other condensing ions does fit the force data for lys+ and di-lys2+ and the ornithine peptides, but not for the tri-, tetra-, or hexa-lysine peptides. This can be explained if the mode of binding of lysine peptides to DNA changes as helices move closer. The model for poly-arginine or protamine binding to DNA from Hud et al (33) offers a possible way to account for the differences we observe between the arginine and lysine peptides.
Experimental Procedures
Materials
L-Arginine hydrochloride, l-lysine hydrochloride, l-ornithine hydrochloride, ammonium chloride, Putrescine dihydrochloride, dilysine dihydrochloride (di-lys2+), trilysine (tri-lys3+), tetralysine (tetra-lys4+), pentalysine (penta-lys5+), poly-l-arginine hydrochloride (P7662 15–70kDa, weight average Mw ~ 35,500), poly-l-lysine hydrochloride (P2658 15–30kDa) and poly-l-histidine hydrochloride (P9386 5–25kDa) were obtained from Sigma-Aldrich (St. Louis, MO). All Sigma-Aldrich chemicals were used without further purification. Di-arginine, di-ornithine, tri-ornithine, tetra-ornithine, hexa-lysine, hexa-arginine, and all mixed lys-arg peptides were custom synthesized and purified (>98%) by GenScript Corporation (Piscataway, NJ). The peptides were neutralized with Tris base and used without further purification. Bioultra grade polyethylene glycol (PEG), average molecular weight (MW) 8,000, was purchased from Fluka Chemical Company. High molecular weight DNA (MW > 5×106) was prepared and purified from adult chicken whole blood as described previously (7, 34) and dialyzed against 10 mM TrisCl (pH 7.5), 1mM EDTA.
Osmotic Stress
The method for direct force measurements by osmotic stress has been previously described (30). In brief, condensed macromolecular arrays, such as DNA, are equilibrated against a bathing polymer solution of known osmotic pressure. The polymer, typically polyethylene glycol (PEG), is too large to enter the condensed DNA phase and applies a direct osmotic pressure, Π, on the condensate. Water, salt, and other small solutes are free to exchange between the PEG and condensed DNA phases. After equilibration, the osmotic pressures in both phases are the same. Osmotic pressures of the PEG-salt solutions were measured directly using a Vapro Vapor Pressure Osmometer (model 5520, Wescor, Logan, UT). The interhelical spacing, Dint, can be determined from Bragg scattering of x-rays as a function of the applied PEG osmotic pressure.
For mono and divalent salts and peptides, i.e., non-condensing species, DNA pellets (~250 μg DNA) are prepared by precipitation with either ethanol or 5% PEG in appropriate salt solutions. Precipitates are then equilibrated against PEG-salt bath solutions in vast excess at a known PEG osmotic pressure. Pellets are typically equilibrated for 2 weeks with several changes of the bathing PEG-salt solution.
Since DNA spontaneously precipitates with +3 and higher charged cations, samples for x-ray scattering were prepared in one of two ways. Concentrated polycation solutions were added to 1 mg/mL chicken erythrocyte DNA (250 μg DNA) in 10 mM TrisCl (pH 7.5) in steps of ~0.2 mM. Each addition was thoroughly mixed before adding more condensing ions and continued until no DNA was in solution after centrifugation at ~ 10Kg for 10min. Typically, the lysine or arginine to DNA phosphate ratio was a 1 – 1.5 at the end point. Alternatively, condensing ions were added to DNA in a single aliquot to an equivalent final concentration. The resulting fibrous samples were centrifuged and the DNA pellets transferred to corresponding PEG-salt solutions containing condensing ion and 10 mM Tris (pH 7.5) and allowed to equilibrate for approximately 2 weeks. X-ray scattering profiles did not depend on the method used to prepare the DNA precipitate and did not change after 6 months.
Critical Concentrations
The critical concentration of each condensing cation (e.g. tri-lysine or tri-ornithine) for the precipitation of DNA from dilute solution was determined as described in Pelta et al., (19). A series of DNA samples were prepared with varied cation concentration in 10 mM Tris buffer. DNA concentration was ~15 μM base pairs in 1 mL total volume. After incubation at room temperature for ~ 1 hour, the solution was centrifuged at 16000 × g for 10 min and the DNA absorbance at 260 nm of the supernatant was measured. Critical concentrations were observed to decrease approximately by an order of magnitude for each additional charge as seen by others (35, 36). The cation concentration used in the bathing PEG-salt solution for the osmotic stress force measurements was 2 to 6 fold higher than the critical concentration. Over this range, the observed spacing between helices does not depend on cation concentration. The force curves for divalent ions (putrescine, di-lysine, di-ornithine and di-arginine) are insensitive to the ion concentration between 5 and 20 mM (data not shown).
X-ray Scattering
Ni-filtered Cu-Kα radiation from an UltraBright microfocus x-ray source from Oxford Instruments equipped with polycapillary focusing x-ray optics was used for the small angle x-ray scattering (SAXS) experiments. The primary beam was also collimated by a set of slits. Samples were sealed with a bath of equilibrating solution in the sample cell, and then mounted into a temperature-controlled holder at 20°C. The flight path between the sample and detector was helium filled. Typical exposure times were ~ 30 min. Diffraction patterns were recorded by direct exposure of Fujifilm BAS image plates and digitized with a Fujifilm FLA 3000 scanner. The images were analyzed using FIT2D (AP Hammersley, ESRF) and SigmaPlot 10.01 (SPSS) software programs. The sample to image plate distance was calibrated using silver behenate and found to be ~16.7 cm. Mean pixel intensities between scattering radii r − 0.05 mm and r + 0.05 mm averaged over all angles of the powder pattern diffraction, <I(r)>, were used to calculate integrated radial intensity profiles, 2πr<I(r)>. The strong scattering peaks correspond to interaxial Bragg diffraction from DNA helices packed in a hexagonal array. The Bragg spacing, DBr, and the actual distance between helices, Dint, are related by . For different samples equilibrated at the same PEG concentration, interaxial spacings are reproducible to within ~0.1 Å.
Overview of force analysis
Extensive osmotic pressure measurements on a wide variety of systems both charged and uncharged have been reported using the osmotic stress technique and are reviewed in Stanley and Rau (31) (and references therein). The common features observed allow a simple framework for fitting force curves. For univalent cation ion concentrations > 1 M and for higher valence cations, DNA osmotic pressures, Π, for interaxial spacings, Dint, out to ~ 40 Å can be described by two exponential forces with decay lengths that differ by a factor of two,
| (1) |
or
| (2) |
where R and A are pre-exponential osmotic pressure amplitudes. The longest decay length, λh, is ~ 5 Å. Since these forces are observed for interactions between both charged and uncharged molecules, we have concluded that they are due to changes in water structuring energies as surfaces approach, but an electrostatic explanation for the DNA forces is still possible. Both hydration forces and electrostatic theories of interacting helices predict a ratio of 2 between the decay length for attractive and repulsive interactions. The ~5 Å decay length force is the direct interaction either of charges on one helix with the charges on another in the electrostatic model and of hydration structures on adjacent helices for hydration forces. This force can be either repulsive or attractive depending on the positions of charges or hydration patches on adjacent helices. The 2.5 Å decay length force is always repulsive and is an image charge interaction for electrostatics, the interaction of charges on one helix with the low dielectric core of another. In the hydration model, the 2.5 Å decay length arises from the disruption of water structuring radiating from one helix by the presence of another. For DNA, at low univalent salt concentrations (<1 M) and spacings larger than ~ 30–35 Å, exponential electrostatic forces are observed with Debye-Hückel decay lengths, λD. At closer distances, the 2.5 Å decay length exponential force dominates or,
| (3) |
Many polyvalent counterions, including all the +3 and higher charged peptides examined here, cause DNA to spontaneously condense resulting in a finite equilibrium separation between the hexagonally packed DNA helices, Deq. For this case, A = −Re(−Deq/λh) and the osmotic stress force curves can be fit to,
| (4) |
with λh fixed at 5 Å.
The repulsive and attractive free energy contributions from the two individual forces per DNA base pair at a particular spacing D can be calculated by integrating osmotic pressure – volume work, Π dV, for each exponential from 8 to D assuming hexagonal packing.
| (5) |
and
| (6) |
where λh is 5 Å and b is the linear spacing between DNA base pairs, ~3.4 Å. At the equilibrium spacing, Deq, the depth of the free energy minimum is given,
| (7) |
Results
DNA packing and forces with arginine and lysine
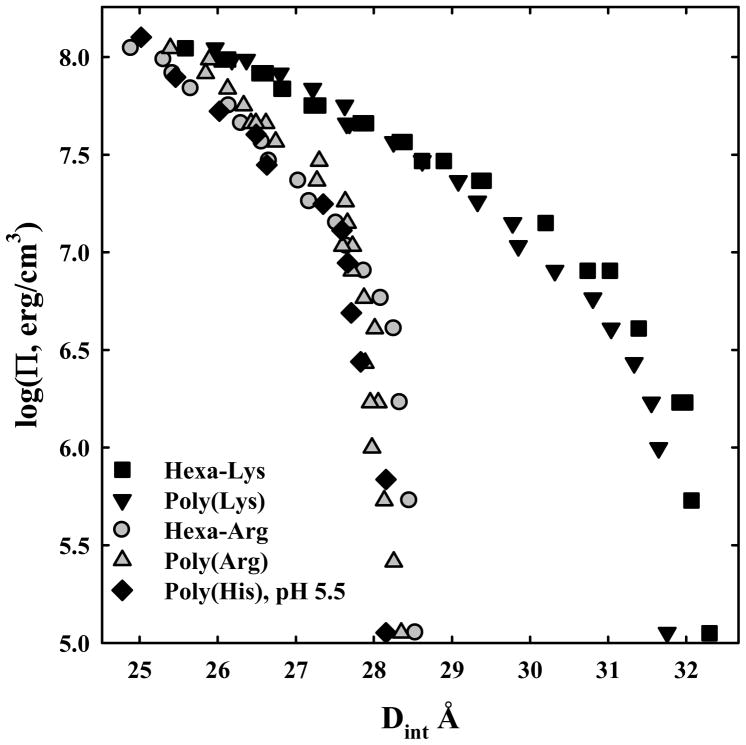
The almost exclusive use of arginine over lysine in sperm protamines might indicate a substantial difference in DNA packing densities with the amino acids. Figure 1 shows log osmotic pressure, Π, vs. DNA interaxial spacing, Dint, curves for hexa-arginine, poly-arginine, hexa-lysine, poly-lysine, and poly-histidine. The pH is 7.5 for the lysine and arginine peptides, but is pH 5.5 for poly-histidine in order to fully charge that peptide. The pKa of histidine is low enough that poly-histidine is not sufficiently charged at pH 7.5 to condense DNA. The peptide concentrations in the PEG stressing solutions are all high enough such that the forces are insensitive to peptide concentration over at least two-fold range. There is little difference between poly-histidine, hexa-arginine and poly-arginine and between hexa-lysine and poly-lysine. The arginine and histidine peptides condense DNA to equilibrium spacings that are within the 27 – 30 Å range that is characteristic of the several other DNA condensing ions that have been well characterized such as Co(NH3)63+, Mn2+, spermidine3+, and spermine4+. The lysine peptides stand out for their inability to densely pack DNA compared to arginine and charged histidine. The equilibrium spacing in the absence of osmotic pressure is 28.3 Å for hexa-arginine and 32.2 Å for hexa-lysine. Since the DNA diameter is ~ 20 Å, this difference corresponds to a 50% greater surface-to-surface separation for hexa-lysine - DNA compared to hexa-arginine - DNA arrays in the absence of osmotic pressure. Suwalsky and Traub (37) have previously reported 29.2 and 31.6 Å spacings for DNA condensed with poly-arginine and poly-lysine, respectively, at 100 % relative humidity. We also note that the apparent limiting slope at high osmotic pressures is observably larger for the lysine peptides than for the arginine peptides.
Figure 1.
Osmotic stress force curves for DNA with poly-arginine, hexa-arg6+, poly-lysine, hexa-lys6+, and poly-histidine. DNA pellets were made as described in Methods and Materials. The samples were then equilibrated against polyethylene glycol (PEG) solutions containing 10 mM Tris (pH 7.5) and 100 μM in monomer of poly-arginine
 , hexa-arg6+
, hexa-arg6+
 , poly-lysine ▼, and hexa-lys6+ ■ and containing 10 mM Tris (pH 5.5) and 100 μM in monomer of poly-histidine ◆. PEG is excluded from the DNA phase and applies an osmotic pressure Π on it. The spacing between helices, Dint, was measured by x-ray scattering. The points at log(Π) ~ 5 indicate the equilibrium distance between helices in the absence of PEG. Poly-arginine, hexa-arg6+, and poly-histidine all show similar DNA-DNA force curves. Poly-lysine and hexa-lys6+ are also similar but have equilibrium spacings much larger than the others.
, poly-lysine ▼, and hexa-lys6+ ■ and containing 10 mM Tris (pH 5.5) and 100 μM in monomer of poly-histidine ◆. PEG is excluded from the DNA phase and applies an osmotic pressure Π on it. The spacing between helices, Dint, was measured by x-ray scattering. The points at log(Π) ~ 5 indicate the equilibrium distance between helices in the absence of PEG. Poly-arginine, hexa-arg6+, and poly-histidine all show similar DNA-DNA force curves. Poly-lysine and hexa-lys6+ are also similar but have equilibrium spacings much larger than the others.
Lys+ and Orn+
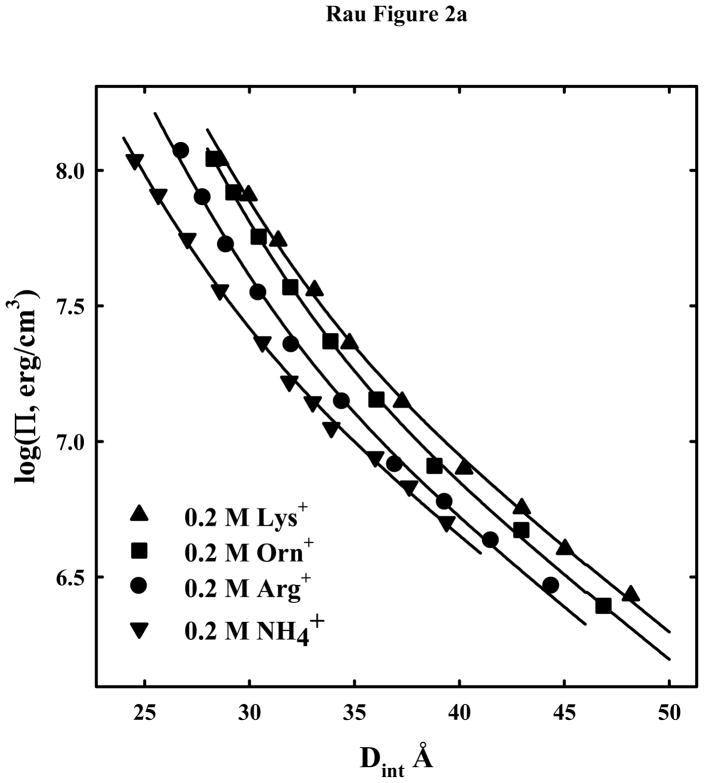
In order to further investigate the differences in DNA-DNA forces between lysine and arginine, we have measured force curves for a series of lysine and ornithine oligo-peptides to compare with arginine. Ornithine differs from lysine only by having one fewer methylene group in the side chain. Both arginine and ornithine have three methylene groups in the side chain before the end charged groups, guanidinium and amine, respectively. Figure 2a shows osmotic stress force curves for 0.2 M arg+, lys+, orn+, and a small univalent cation, NH4+, for comparison. At this salt concentration, the electrostatic repulsion between helices dominates at low pressures and larger spacings. At high pressures, the short decay length repulsive force dominates. The solid lines show fits of the data to equation (3) with λh = 5 Å and λD = 7 Å, the Debye-Hückel shielding length at 0.2 M univalent salt. The data can be fit quite well using only the force amplitudes as variables.
Figure 2.
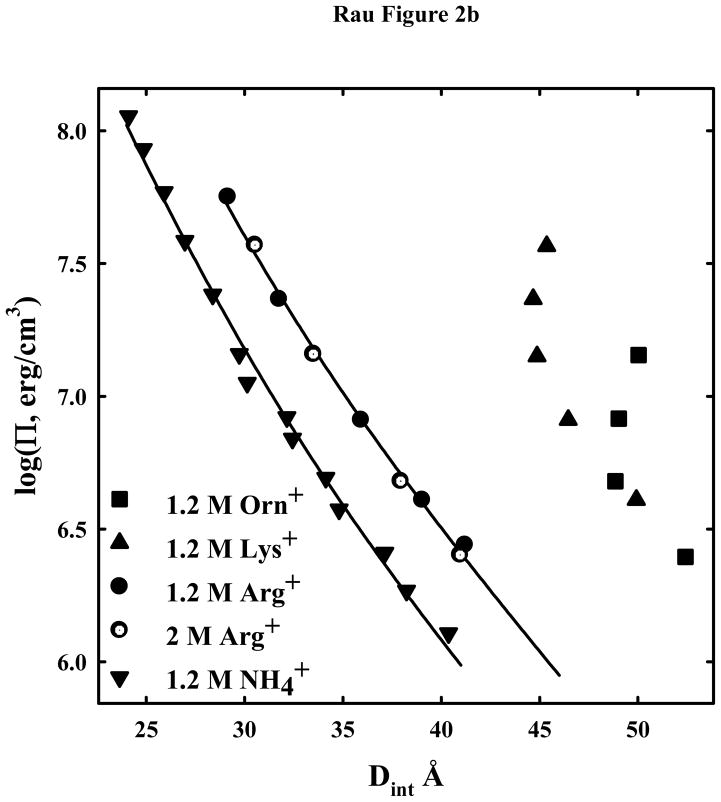
Osmotic stress force curves for DNA in ArgCl, LysCl, OrnCl, and NH4Cl. DNA condensates prepared by ethanol precipitation and equilibrated against PEG solutions containing 10 mM Tris (pH 7.5) and two concentrations of ArgCl ●, LysCl ▲, OrnCl ■, or NH4Cl ▼. (A): The salt concentration is 0.2M. The solid lines are fits of the data to equation (3) with λh = 5 Å and the Debye-Huckel shielding length at 0.2 M salt concentration λD = 7 Å. Electrostatics dominates at low pressures not hydration forces. (B) - The salt concentration is 1.2 M for arg+, lys+, orn+, and NH4+. Force measurements for 2 M arg+ (
 ) are also shown. The solid lines are fits of the arginine and ammonium data to a hydration force, equation (2) with λh = 5 Å. The overlap of the 1.2 and 2 M arg+ data indicates that the interactions are dominated by hydration forces at high salt concentrations, not the electrostatic repulsion seen at lower salt concentrations as in (A). The ornithine and lysine force curves show very different behavior from other univalent ions that have been examined.
) are also shown. The solid lines are fits of the arginine and ammonium data to a hydration force, equation (2) with λh = 5 Å. The overlap of the 1.2 and 2 M arg+ data indicates that the interactions are dominated by hydration forces at high salt concentrations, not the electrostatic repulsion seen at lower salt concentrations as in (A). The ornithine and lysine force curves show very different behavior from other univalent ions that have been examined.
Figure 2b shows osmotic stress force curves for 1.2 and 2 M arg+, 1.2 M lys+, 1.2 M orn+, and 1.2 M NH4+. At these high salt concentrations the direct hydration force with a decay length of ~ 5 Å dominates over electrostatics at the low osmotic pressures, hence the insensitivity to monovalent ion concentration for arg+. The fit to equation (3) with λh = 5 Å is quite good for both NH4+ and arg+. The amplitudes of the shorter ranged force are the same within experimental error for 0.2, 1.2 and 2 M arg+ and for 0.2 and 1.2 M NH4+. Both lys+ and orn+, however, show a very different and anomalous behavior at the higher concentration. At low pressures, a quite large limiting spacing, 45–50 Å, is reached. Weaker reflections are still observed in the 8–20 Å region that are typical of B-form DNA. The observed interaxial reflections are broad indicative of an array of spacings either due to fluctuations or to poor packing. At lower pressures than shown the DNA pellet dissolves. The insensitivity of the spacing to osmotic pressure indicates that the spacing is dominated by the packing of lysine and ornithine between helices, not water.
Di-arg2+, di-orn2+, di-lys2+
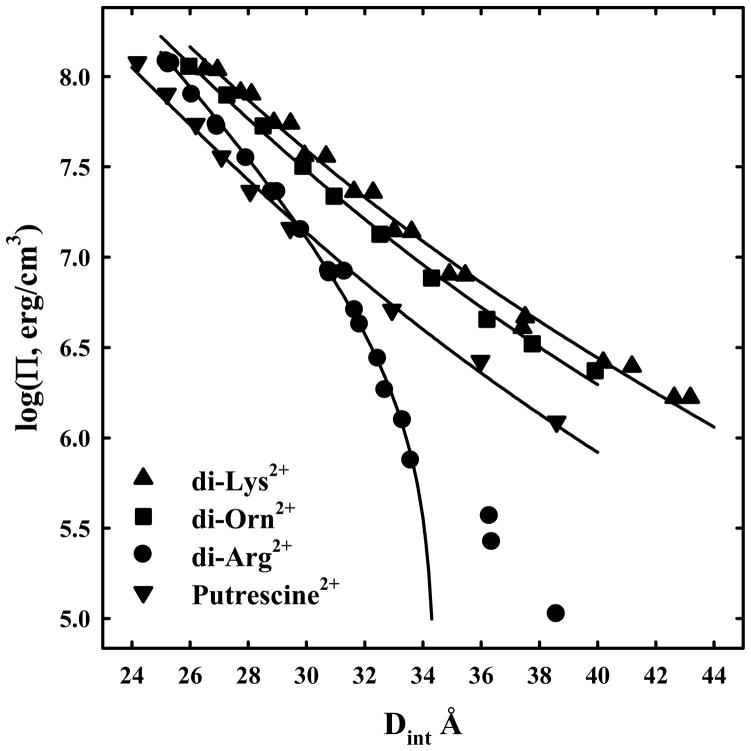
The remarkable ability of arginine to package DNA compared to lysine is clearly seen with the peptide dimers. Figure 3 shows DNA force curves in 10 mM di-arg2+, di-lys2+, di-orn2+, and, for comparison, the di-amine putrescine2+. At 10 mM concentration, force curves are insensitive to the divalent ion concentration over at least a change of a factor of two, indicating that the DNA is neutralized almost entirely by the divalent peptides. Di-arg2+ - DNA assemblies undergo a transition to net attraction at low pressures. We have only seen such a transition for divalent cations with Mn2+ and Cd2+. Other divalent cations, such as Mg2+, Ca2+, Zn2+, and putrescine2+, show strictly repulsive forces over the entire pressure range. The forces with di-orn2+ and di-lys2+ closely resemble other divalent cations as putrescine2+. Typically only the 2.5 and 5 Å decay length forces are seen for DNA with divalent cations. The solid lines are fits of the data for di-lys2+, di-orn2+, and putrescine to equation (2) with λh = 5 Å. Only the data after the low pressure transition for di-arg2+ is used for fitting to equation (4). The 5 Å decay length force is attractive for di-arg2+, but repulsive for di-lys2+ and di-orn2+. As with the 0.2 M univalent ions, the ornithine dimer is slightly less repulsive than the lysine dimer.
Figure 3.
Osmotic stress force curves for DNA in divalent di-arginine, di-lysine, di-ornithine, and putrescine, a divalent diamine. DNA condensates were prepared by ethanol precipitation and equilibrated against PEG solutions containing 10 mM Tris (pH 7.5) and 10 mM di-arg2+ ●, di-lys2+ ▲, di-orn2+ ■, or putrescine2+ ▼. The solid lines for di-lys, di-orn, and putrescine are fits to equation (2) with the decay length λh fixed at 5 Å. Di-arginine shows a transition at log(Π) ~ 5.7 from a repulsive to an attractive interaction. The solid line for di-arg is the best fit to equation (4) with the decay length λh fixed at 5 Å and using only the data after the transition.
Higher valence ornithine and lysine peptides
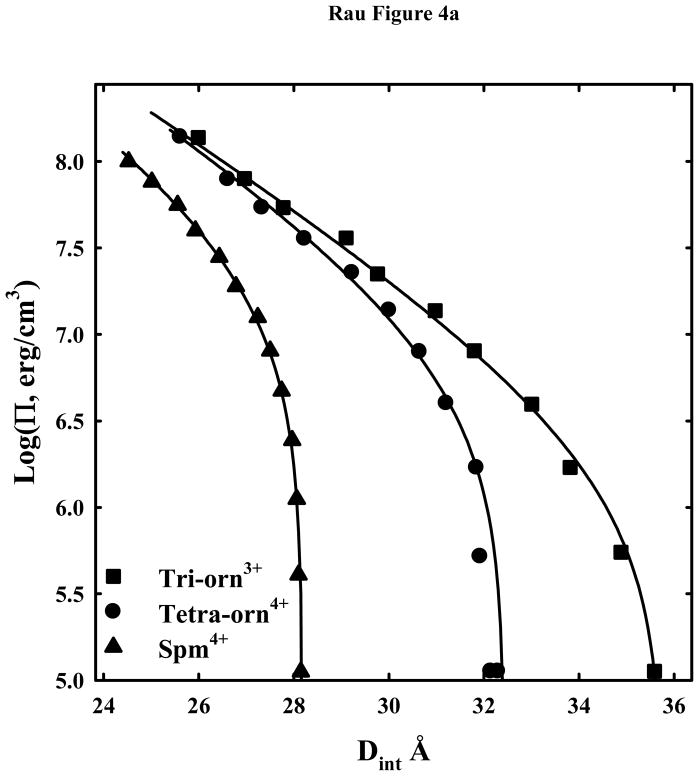
Figure 4a shows DNA osmotic stress force curves with tri-orn3+, tetra-orn4+, and, for comparison, the tetra-amine spermine4+. Both the ornithine peptides spontaneously assemble DNA in the absence of PEG. The solid lines are the fits to equation (4) with λh fixed at 5 Å. As we observed previously with the arginine peptides, Co(NH3)63+, spermidine3+, and spermine4+ this functional form gives a good description of the forces. The equilibrium spacings with tri-orn3+ and tetra-orn4+ with no applied PEG osmotic pressure are still significantly larger than for tri-arg3+, 30.1 Å, and tetra-arg4+, 29.6 Å (18).
Figure 4.
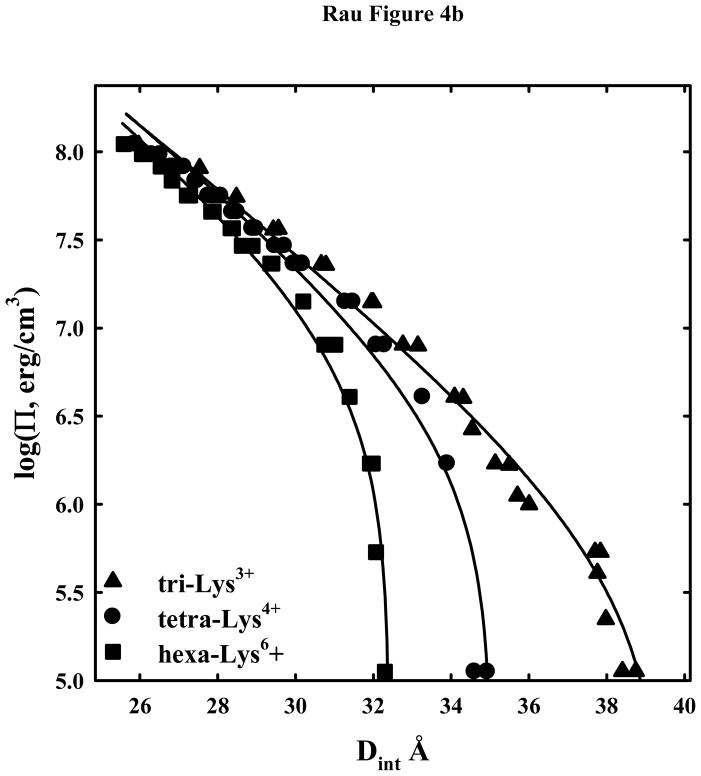
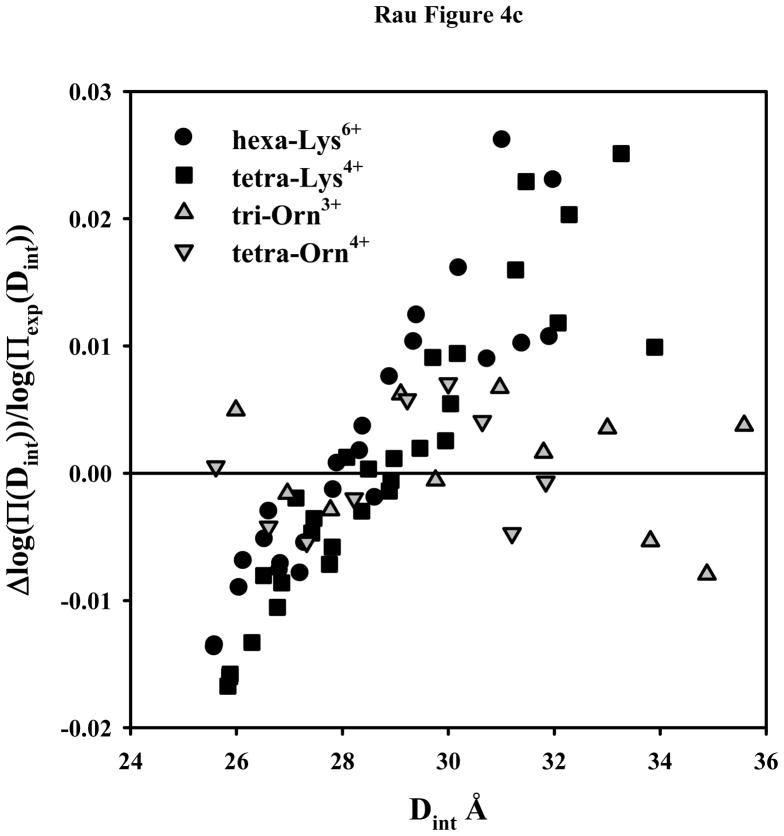
DNA-DNA interactions with +3 and higher charged lysine and ornithine peptides. (A) - Osmotic stress force curves for DNA in tri-ornithine3+, tetra-ornithine4+, and the tetra-amine spermine4+. DNA was precipitated by adding tri-orn3+, tetra-orn4+, or spermine4+ to a concentrated DNA solution in 10 mM Tris (pH 7.5) as described in Methods and Materials. The DNA pellets were then equilibrated against PEG solutions containing 10 mM Tris (pH 7.5) and 2 mM tri-orn ■, 0.5 mM tetra-orn ●, or 0.5 mM spermine ▲. The points at log(Π) ~ 5 are the equilibrium spacings, Deq, in the absence of PEG. The solid lines are fits of the data to equation (4) with the decay length λh fixed at 5 Å. Note the convergence of the ornithine force curves at high pressures. (B) - Osmotic stress force curves for DNA in tri-lysine3+, tetra-lysine4+, and hexa-lysine6+. DNA was precipitated by adding tri-lys3+, tetra-lys4+, or hexa-lys6+ to a concentrated DNA solution in 10 mM Tris (pH 7.5) as described in Methods and Materials. The DNA pellets were then equilibrated against PEG solutions containing 10 mM Tris (pH 7.5) and 2 mM tri-lys3+ ▲, 0.5 mM tetra-lys4+ ●, or 0.1 mM hexa-lys6+ ■. The points at log(Π) ~ 5 are the equilibrium spacings, Deq, in the absence of PEG. The solid lines are fits of the data to equation (4) with the decay length λh fixed at 5 Å. (C) – Normalized residuals for the osmotic stress force data shown in figures 4A and B, tetra-lys4+ ■, hexa-lys6+ ●, tri-orn3+
 , and tetra-orn4+
, and tetra-orn4+
 . Δlog(Π(Dint)) = log(Πexp(Dint)) − log(Πcalc(Dint)), where log(Πexp(Dint)) is the experimental log(Π) at Dint and log(Πcalc(Dint)) is the log(Π) at Dint calculated from the double exponential fit to the data. Note the systematic deviation of the lysine peptide data from the fit.
. Δlog(Π(Dint)) = log(Πexp(Dint)) − log(Πcalc(Dint)), where log(Πexp(Dint)) is the experimental log(Π) at Dint and log(Πcalc(Dint)) is the log(Π) at Dint calculated from the double exponential fit to the data. Note the systematic deviation of the lysine peptide data from the fit.
Figure 4b shows DNA osmotic stress force curves for tri-lys3+, tetra-lys4+, and hexa-lys6+. The equilibrium spacings in the absence of PEG osmotic pressure are more than 2 Å larger than for the corresponding ornithine peptide. Once again the solid lines are fits to equation (4) with λh fixed at 5 Å. In this case, however, the fits do not well describe the data.
Figure 4c show the normalized fitting residuals for peptides of figures 4a and 4b. Unlike the ornithine fits or the residuals for the set of arginine peptides reported previously, the lysine data shows large and systematic deviation from the double exponential form of equation (1). Either the decay lengths for the interactions mediated by the lysine peptides are different from the others or lysine peptides bound to DNA rearrange with increasing osmotic pressure to decrease the force significantly. If a factor of two is maintained between the short range repulsive force and the longer range attraction, then the best fitting decay lengths for the hexa-lys6+ force curve in figure 4b would be 3.5 and 7 Å.
Mixed lys/arg peptides
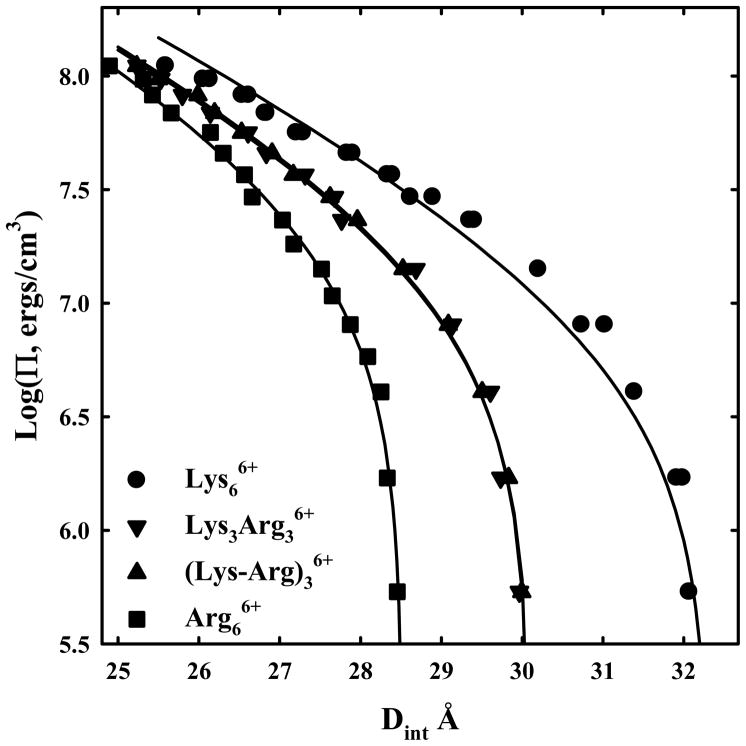
Given the large difference between lysine and arginine in packaging DNA, it is of interest to investigate mixed arg/lys peptides. Figure 5 shows force DNA curves for lys/arg hexamers, hexa-arg6+, hexa-lys6+, arg3lys36+ and arg-lys-arg-lys-arg-lys6+. The peptides with three arginines and three lysines are not distinguishable; the placement of the charged groups does not affect forces. The equilibrium spacing is almost at the midpoint between the hexa-arg6+ and hexa-lys6+ data. The solid lines are fits to equation (4) with λh fixed at 5 Å. The systematic deviation of the hexa-lys6+ data can be more clearly seen than figure 4b. The fit for hexa-arg6+ is very good and reasonable for the 3 arg/3 lys peptides.
Figure 5.
DNA-DNA forces with mixed arginine-lysine peptides. Osmotic stress force curves for DNA in hexa-arg6+, hexa-lys6+, lys3-arg36+, and (lys-arg)36+. DNA was precipitated by adding the +6 charged peptides to a concentrated DNA solution in 10 mM Tris (pH 7.5) as described in Methods and Materials. The DNA pellets were then equilibrated against PEG solutions containing 10 mM Tris (pH 7.5) and 100 μM hexa-lys6+ ●, hexa-arg6+ ■, lys3-arg36+ ▼, or (lys-arg)36+ ▲. The points at log(Π) ~ 5 are the equilibrium spacings, Deq, in the absence of PEG. The solid lines are fits of the data to equation (4) with the decay length λh fixed at 5 Å.
Discussion
The dense packing of DNA by protamines in sperm nuclei is considered necessary to prevent DNA damage due to mutagens and small reactive oxidative species (9–14). Osmotic stress force measurements indicate that salmon protamines compact DNA using the same forces as for much smaller cations with many fewer charges, as Co(NH3)63+, Mn2+, spermidine3+, spermine4+, and +2, +3, +4, and +6 arginine peptides (6, 7, 18, 22, 24). The same double exponential functional form of equation (1) with λh ~ 5 Å and variable amplitudes fits the osmotic stress force data well for all these different ions. The amplitudes of the attractive and short ranged repulsive forces of salmon protamine DNA condensates can be well understood from the amino acid composition (6). A striking feature of protamines is the almost exclusive use of arginine over lysine for the cationic charge (27, 38). Lewis et al (39) have shown that arginine rich protamines likely evolved from lysine rich histone H1, suggesting a functional preference for arginine. The results presented here demonstrate that arginine is much more effective in densely packing DNA than lysine. The equilibrium surface-to-surface distance between DNA helices increases by almost 50% in changing from poly-arginine or hexa-arginine to poly-lysine or hexa-lysine. Arginine rich peptides such as protamines would be highly favored if dense packaging of DNA in sperm nuclei is biologically important. DNA condensed with the lysine peptides have significantly larger equilibrium spacings than compacted not only with arginine peptides, but also with other condensing ions such as Co(NH3)63+, spermidine3+, spermine4+, or poly-histidine.
A striking feature of the osmotic stress force curves of DNA condensed with the lysine peptides that is not observed for the arginine and ornithine peptides is the inability of the double exponential form of equation (1) with λh ~ 5 Å decay lengths to fit the tri-lys3+, tetra-lys4+, and hexa-lys6+ data. The observations that a λh/2 ~ 2.5 Å exponential is able to fit the high pressure 0.2 M lys+ data and that the double exponential fit with λh ~ 5 Å is able to well describe the force curve with di-lys2+ and with the closely related ornithine peptides suggests that the underlying forces are the same for lysine as for arginine. The data can be rationalized if there are at least two distinct binding modes of the longer lysine peptides to DNA, one that optimizes binding energy to an isolated helix and another that optimizes the interaction between helices. As helices approach, lysine peptide binding would gradually shift from the first mode to the second. This will have the effect of decreasing the apparent limiting high pressure force from that expected if binding was unperturbed.
The double exponential curve fits allow us to separate the interhelical forces into their attractive and repulsive components. Rather than report force amplitudes, we calculate attractive or repulsive free energies (in units of kT per base pair) that are more meaningful. ΔG(Deq)/kT is the net free energy at the equilibrium spacing, i.e., at the free energy minimum, and is given by equation (7). ΔGA (25 Å)/kT and ΔGR (25 Å)/kT are the interaction energies per base pair calculated by integrating Π dV from ∞ to 25 Å for the λh and λh/2 decay length forces, respectively, and are given by equations (5) and (6).
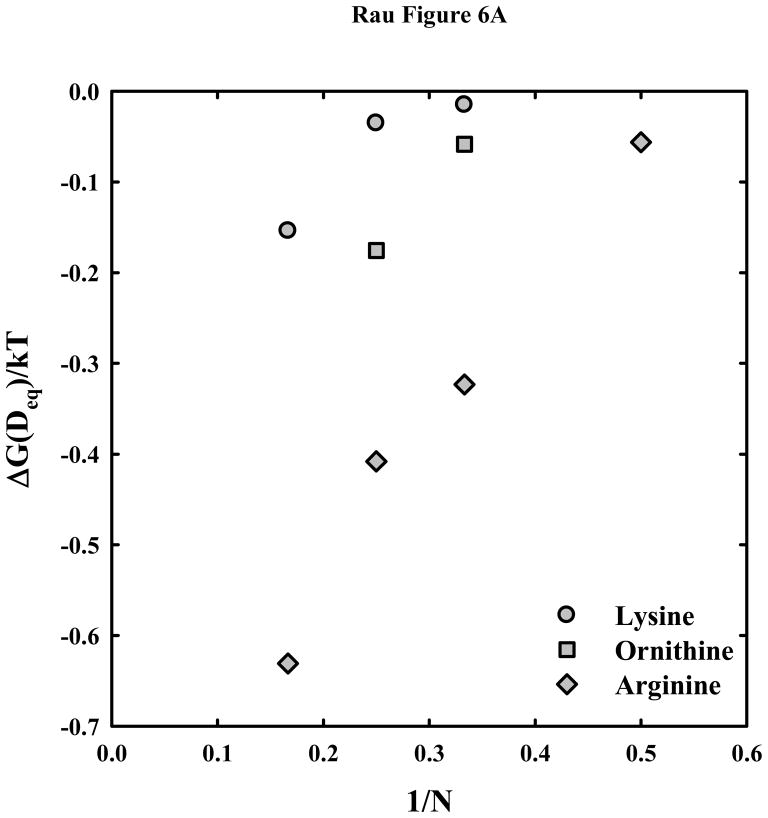
Figure 6a shows the free energy per base pair at the equilibrium spacing, ΔG(Deq)/kT, as dependent on the inverse of peptide length, 1/N, for arginine, ornithine, and lysine. The arginine data was taken from (18). We have previously seen that free energies vary approximately linearly with 1/N for the arginine peptides. The much weaker attraction with lysine and ornithine relative to arginine is readily apparent. The estimated free energy gained from attractive interhelical interactions with hexa-arg6+ is more than 5 times that with hexa-lys6+. Although, the forced fit of the lysine peptide to the λh ~ 5 Å double exponential function likely underestimates ΔG(Deq), the real value will still be less than for the ornithine peptides.
Figure 6.
Dependence of the interaction free energies on peptide length N. (A) – The free energy of interaction (in units of kT/bp) at the equilibrium spacing, Deq, is calculated from the double exponential fits to the osmotic stress data and equation (7) for the arginine
 , ornithine
, ornithine
 , and lysine
, and lysine
 peptides. The much weaker attraction between DNA helices with lysine and ornithine peptides compared with arginine is apparent. (B) – The attractive and repulsive free energy components at 25 Å (in units of kT/bp) of the interaction are calculated from the double exponential fits to the osmotic stress force curves and equations (5) and (6). The repulsive, ΔGR(25 Å)/kT, and attractive, ΔGA(25 Å)/kT, free energies, respectively, are shown for the arginine (
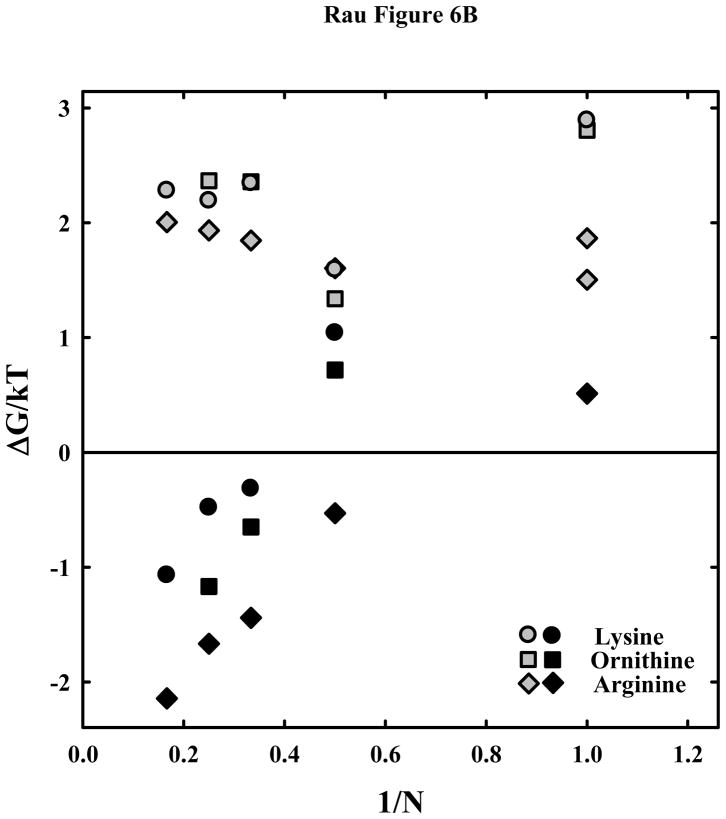
peptides. The much weaker attraction between DNA helices with lysine and ornithine peptides compared with arginine is apparent. (B) – The attractive and repulsive free energy components at 25 Å (in units of kT/bp) of the interaction are calculated from the double exponential fits to the osmotic stress force curves and equations (5) and (6). The repulsive, ΔGR(25 Å)/kT, and attractive, ΔGA(25 Å)/kT, free energies, respectively, are shown for the arginine (
 , ◆), ornithine (
, ◆), ornithine (
 , ■) and lysine (
, ■) and lysine (
 , ●) peptides.
, ●) peptides.
Figure 6b shows the attractive and repulsive force components for the arginine, lysine, and ornithine peptides as a function of the inverse length or charge of the peptide, N. Again the arginine data is taken from (18). The gray symbols are for the shorter ranged force that is always repulsive; the black symbols are for the λh decay length forces that can be either attractive or repulsive depending on the peptide length. Since we are forcing a fit of the tri-lys3+, tetra-lys4+, and hexa-lys6+ data to double exponential function with λh = 5 Å, the real value of ΔGR(25 Å)/kT is likely more repulsive and more attractive for ΔGA(25 Å)/kT due to the apparent shift in lysine peptide binding mode as helices approach. ΔGR(25 Å)/kT values for lys+ and orn+ are significantly larger than for arg+ or for almost any other univalent ion we have studied. This may be related to the quite unusual osmotic stress force curves observed with 1.2 M lys+ and orn+ (figure 2b). With such large repulsive energies, alternate structures and ligand binding modes that would ordinarily be energetically prohibited might now be accessible as helices are pushed close together. ΔGR(25 Å)/kT values for the peptides with charges ≥ +2 are not as different for the lysine, ornithine, and arginine peptides as for the +1 amino acids. The repulsive free energies of the lysine or ornithine peptides are, however, consistently larger than for arginine for charges +3 to +6.
The attractive free energies, ΔGA(25 Å)/kT, are in the order arginine > ornithine > lysine. Our results demonstrate that the observed large difference between equilibrium spacings for DNA compacted with lysine and arginine peptides is primarily due to a decreased attractive free energy, although the short ranged repulsion also contributes some to the much weaker net attraction between helices.
DNA assembly has also been characterized by determining the critical concentration of the multivalent ion necessary for precipitation. Both binding energies of these ions to isolated helices and the resultant net attractive energy between helices will contribute to the critical concentration. The binding data of Mascotti and Lohman (40, 41) indicate there is little difference in binding of a +4 lysine peptide and a +4 arginine peptide to isolated double stranded DNA at low salt concentrations (~ 100 mM). Leng and Felsenfeld (42) reported that poly-arginine precipitates DNA more readily than poly-lysine. Ando and coworkers (43, 44) have found that arginine peptides are much more efficient in precipitating DNA than the analogous ornithine peptides. At the low salt concentrations used here, 10 mM Tris, pH 7.5, the binding of most tetra- and higher valent ions is close to stoichiometric (35). None-the-less, Thomas and coworkers (36, 45) at similar low salt conditions have found that considerably more tetra-lys4+ was necessary for DNA condensation than spermine4+ and that significantly more penta-lys5+ was necessary than several +5 analogs of spermine. Brewer et al (46) reported that hexa-arg6+ is much more effective than hexa-lys6+ at condensing DNA. Combined, all these observations would suggest that the difference in critical concentration for precipitation is likely due to a difference in attractive interaction energies between helices as seen here rather than in binding energies to isolated helices.
The underlying reason for the weaker attraction with lysine peptides than with arginine peptides is unclear. It is likely not due to an intrinsic difference between charged amine and guanidinium groups for interhelical attraction. Biogenic amines such as spermidine3+ and spermine4+ are very efficient in condensing DNA resulting in equilibrium spacings of 29.7 and 28.2 Å, respectively, compared with 30.1 and 29.6 Å for tri-arg3+ and tetra-arg4+. The theoretical work of Kornyshev and coworkers (32, 47–49) seems to us best able to describe DNA-DNA interactions. Amplitudes of the ~5 Å decay length force will depend on the number, position, and relative correlation of charges on apposite helices. Attraction will result from a favorable juxtaposition of positive charges on one helix with negative charges on another. An increased distance separation of positive and negative charges on one helix will naturally increase the attraction of correlated charged on apposing helices. Thermal fluctuations of helices will decrease attraction. Cation charge is likely important for DNA compaction for, at least, two reasons. First, the net charge of DNA decreases as the charge of the cation increases by Manning condensation; the entropy loss from binding one +N ion is less than from binding N univalent ions. Second, the entropy loss from correlating one +N ion on a helix with phosphates on an adjacent helix is less than from correlating N univalent ions. The difference in the strength of attraction between the lysine peptides and the other condensing ions of the same charge is likely due to a difference in the specific binding interactions of lysine peptides with DNA compared to the others, particularly the relative positions of the cationic positive charges and DNA phosphates.
One proposed arrangement of DNA and cation charges that can result in favorable correlations is that cations bind in DNA grooves. Adjacent helices can position themselves for a favorable interaction of the positively charged groove of one helix with the negatively charged backbone of another (49). Since the major groove of B-form DNA is significantly wider than the minor groove, the positive-negative charge separation and, hence, the attractive force could be greater for cations that bind in the major groove compared to the minor groove. The difference between the DNA–DNA forces with lysine and arginine peptides most likely is the result of a difference in details in the binding mode. The model of Hud et al (33) for poly-arginine binding provides another possible framework for considering these correlations. In this model, the neutral peptide backbone binds in the major groove of DNA, allowing the side chain guanidinium groups to reach the sugar-phosphate backbone and to hydrogen bond to neighboring phosphates along one strand. The result is an alternating, equally spaced arrangement of positive and negative charges along the backbone. Adjacent helices can arrange for a favorable interaction of these separated charges along the backbone. The Hud et al model may result in weaker attraction compared with major groove placement of the cation charge since the ~7 Å separation between phosphates along the backbone is significantly less than the ~ 20 Å across the major groove. On the other hand, the tight binding of an arginine to two phosphates along the backbone may suppress helical fluctuations and increase attraction.
If we assume a similar binding mode for the lysine peptide with the backbone in the major groove, the side chain amine of lysine is not large enough to span the distance between two phosphates and will likely only interact closely with one, unlike arginine. The ability of arginine to hydrogen bond to two phosphates along the backbone and of lysine to only one is consistent with the observations of Mascotti and Lohman (41) who observed that the binding of oligo-arginines to double stranded DNA is enthalpically favored compared to oligo-lysine binding. If the amine of lysine only interacts with a single DNA phosphate, then the alternating sequence of charges along the backbone will not be equally spaced, helical fluctuations will not be suppressed by cross-linking adjacent phosphates, and the attraction will be weaker. The osmotic pressure dependent repositioning of lysines may be a movement of a lysine amine that is closely associated with one phosphate to a position intermediate between phosphates along the backbone or to the major groove. Lys+ and di-lys2+ may not be long enough to stabilize the specific groove binding over other binding locations. The histidine side chain is certainly too short to reach the phosphate backbone and the histidine charge would remain primarily in the major groove and would be expected to mediate a larger DNA-DNA attractive force than lysine as seen in figure 1. Ornithine with one less methylene in the side chain than lysine might not be long enough to reach and hydrogen bond to the phosphate backbone. As a result, the ornithine peptides could still be well fit by the double exponential form with λh ~ 5 Å.
Conclusion
The difference in the ability of arginine and lysine peptides to compact DNA is quite substantial. In general, the lysine peptides show both weaker attraction force and stronger short-ranged repulsion which lead to significantly larger spacing between DNA helices for lysine peptide condensation compared to arginine peptides. If tight packing of DNA in sperm nuclei is critical for protection from damage, there would be a strong bias for replacing lysine with arginine as protamines evolved. The large difference in forces between structurally similar arginine and lysine peptides emphasizes the importance of the conformation of the bound cation in creating interhelical correlations. The Hud et al (33) model for arginine binding to DNA extended to lysine is consistent with our observations.
Acknowledgments
Funding: This work was supported by the Intramural Research Program of the National Institutes of Health, NICHD (Eunice Kennedy Shriver National Institute of Child Health and Human Development).
Abbreviations
- PEG
polyethylene glycol
- arg
arginine
- lys
lysine
- orn
ornithine
- Π
PEG osmotic pressure
Contributor Information
Jason DeRouchey, Email: derouchey@uky.edu, Department of Chemistry, University of Kentucky, Lexington, KY 40506; Phone: (859)323-2827; Fax (859)323-1069.
Brandon Hoover, Email: brandon.hoover@nih.gov, The Program in Physical Biology, The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892; Phone: 301-402-4698.
Donald C Rau, Email: raud@mail.nih.gov, The Program in Physical Biology, The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892; Phone: 301-402-4698.
References
- 1.Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Current opinion in structural biology. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Nurmemmedov E, Castelnovo M, Catalano CE, Evilevitch A. Biophysics of viral infectivity: matching genome length with capsid size. Quarterly reviews of biophysics. 2007;40:327–356. doi: 10.1017/S0033583508004666. [DOI] [PubMed] [Google Scholar]
- 3.Blanc NS, Senn A, Leforestier A, Livolant F, Dubochet J. DNA in human and stallion spermatozoa forms local hexagonal packing with twist and many defects. Journal of Structural Biology. 2001;134:76–81. doi: 10.1006/jsbi.2001.4365. [DOI] [PubMed] [Google Scholar]
- 4.Hud NV, Allen MJ, Downing KH, Lee J, Balhorn R. Identification of the elemental packing unit of DNA in mammalian sperm cells by atomic force microscopy. Biochemical and biophysical research communications. 1993;193:1347–1354. doi: 10.1006/bbrc.1993.1773. [DOI] [PubMed] [Google Scholar]
- 5.Brewer L. Deciphering the structure of DNA toroids. Integr Biol. 2011;3:540–547. doi: 10.1039/c0ib00128g. [DOI] [PubMed] [Google Scholar]
- 6.DeRouchey JE, Rau DC. Role of amino acid insertions on intermolecular forces between arginine peptide condensed DNA helices: implications for protamine-DNA packaging in sperm. The Journal of biological chemistry. 2011;286:41985–41992. doi: 10.1074/jbc.M111.295808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rau DC, Parsegian VA. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices - Evidence for long-range attractive hydration forces. Biophys J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toma AC, de Frutos M, Livolant F, Raspaud E. DNA Condensed by Protamine: A “Short” or “Long” Polycation Behavior. Biomacromolecules. 2009;10:2129–2134. doi: 10.1021/bm900275s. [DOI] [PubMed] [Google Scholar]
- 9.Braun RE. Packaging paternal chromosomes with protamine. Nature Genetics. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- 10.Carrell DT, Emery BR, Hammoud S. The aetiology of sperm protamine abnormalities and their potential impact on the sperm epigenome. International journal of andrology. 2008;31:537–545. doi: 10.1111/j.1365-2605.2008.00872.x. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Marin C, Gosalvez J, Roy R. Types, causes, detection and repair of DNA fragmentation in animal and human sperm cells. International journal of molecular sciences. 2012;13:14026–14052. doi: 10.3390/ijms131114026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muratori M, Marchiani S, Maggi M, Forti G, Baldi E. Origin and biological significance of DNA fragmentation in human spermatozoa. Frontiers in bioscience: a journal and virtual library. 2006;11:1491–1499. doi: 10.2741/1898. [DOI] [PubMed] [Google Scholar]
- 13.Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- 14.Oliva R. Protamines and male infertility. Human Reproduction Update. 2006;12:417–435. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- 15.Aitken RJ, De Iuliis GN. On the possible origins of DNA damage in human spermatozoa. Molecular human reproduction. 2010;16:3–13. doi: 10.1093/molehr/gap059. [DOI] [PubMed] [Google Scholar]
- 16.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 17.DeRouchey J, Netz RR, Radler JO. Structural investigations of DNA-polycation complexes. European Physical Journal E. 2005;16:17–28. doi: 10.1140/epje/e2005-00003-4. [DOI] [PubMed] [Google Scholar]
- 18.DeRouchey J, Parsegian VA, Rau DC. Cation charge dependence of the forces driving DNA assembly. Biophysical Journal. 2010;99:2608–2615. doi: 10.1016/j.bpj.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelta J, Livolant F, Sikorav JL. DNA aggregation induced by polyamines and cobalthexammine. J Biol Chem. 1996;271:5656–5662. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 20.Raspaud E, de la Cruz MO, Sikorav JL, Livolant F. Precipitation of DNA by polyamines: A polyelectrolyte behavior. Biophysical Journal. 1998;74:381–393. doi: 10.1016/S0006-3495(98)77795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raspaud E, Durand D, Livolant F. Interhelical spacing in liquid crystalline spermine and spermidine-DNA precipitates. Biophysical Journal. 2005;88:392–403. doi: 10.1529/biophysj.104.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rau DC, Parsegian VA. Direct measurement of temperature-dependent solvation forces between DNA double helices. Biophys J. 1992;61:260–271. doi: 10.1016/S0006-3495(92)81832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schellman JA, Parthasarathy N. X-ray diffraction studies in cation-collapsed DNA. J Mol Biol. 1984;175:313–329. doi: 10.1016/0022-2836(84)90351-6. [DOI] [PubMed] [Google Scholar]
- 24.Todd BA, Parsegian VA, Shirahata A, Thomas TJ, Rau DC. Attractive forces between cation condensed DNA double helices. Biophys J. 2008;94:4775–4782. doi: 10.1529/biophysj.107.127332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeRouchey JE, Rau DC. Salt effects on condensed protamine-DNA assemblies: anion binding and weakening of attraction. The journal of physical chemistry B. 2011;115:11888–11894. doi: 10.1021/jp203834z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Rau DC. Incomplete ion dissociation underlies the weakened attraction between DNA helices at high spermidine concentrations. Biophys J. 2005;89:1932–1940. doi: 10.1529/biophysj.105.065060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis JD, Song Y, de Jong ME, Bagha SM, Ausio J. A walk though vertebrate and invertebrate protamines. Chromosoma. 2003;111:473–482. doi: 10.1007/s00412-002-0226-0. [DOI] [PubMed] [Google Scholar]
- 28.Queralt R, Adroer R, Oliva R, Winkfein RJ, Retief JD, Dixon GH. Evolution of protamine P1 genes in mammals. Journal of molecular evolution. 1995;40:601–607. doi: 10.1007/BF00160507. [DOI] [PubMed] [Google Scholar]
- 29.Leikin S, Parsegian VA, Rau DC, Rand RP. Hydration Forces. Annual Review of Physical Chemistry. 1993;44:369–395. doi: 10.1146/annurev.pc.44.100193.002101. [DOI] [PubMed] [Google Scholar]
- 30.Parsegian VA, Rand RP, Fuller NL, Rau DC. Osmotic-Stress For The Direct Measurement of Intermolecular Forces. Methods in Enzymology. 1986;127:400–416. doi: 10.1016/0076-6879(86)27032-9. [DOI] [PubMed] [Google Scholar]
- 31.Stanley C, Rau DC. Evidence for water structuring forces between surfaces. Curr Opin Colloids Interface Sci. 2011;16:551–556. doi: 10.1016/j.cocis.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornyshev AA, Lee DJ, Leikin S, Wynveen A. Structure and interactions of biological helices. Reviews of Modern Physics. 2007;79:943–996. [Google Scholar]
- 33.Hud NV, Milanovich FP, Balhorn R. Evidence of novel secondary structure in DNA-bound protamine is revealed by raman-spectroscopy. Biochem. 1994;33:7528–7535. doi: 10.1021/bi00190a005. [DOI] [PubMed] [Google Scholar]
- 34.McGhee JD, Wood WI, Dolan M, Engel JD, Felsenfeld G. A 200 base pair region at the 5′ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981;27:45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- 35.Korolev N, Berezhnoy NV, Eom KD, Tam JP, Nordenskiold L. A universal description for the experimental behavior of salt-(in)dependent oligocation-induced DNA condensation. Nucleic Acids Research. 2009;37:7137–7150. doi: 10.1093/nar/gkp683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nayvelt I, Thomas T, Thomas TJ. Mechanistic differences in DNA nanoparticle formation in the presence of oligolysines and poly-L-lysine. Biomacromolecules. 2007;8:477–484. doi: 10.1021/bm0605863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwalsky M, Traub W. A comparative X-ray study of nucleoprotamine and DNA complexes with polylysine and polyarginine. Biopolymers. 1972;11:2223–2231. doi: 10.1002/bip.1972.360111103. [DOI] [PubMed] [Google Scholar]
- 38.Balhorn R. The protamine family of sperm nuclear proteins. Genome Biology. 2007;8:227.1–227.8. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis JD, Saperas N, Song Y, Zamora MJ, Chiva M, Ausio J. Histone H1 and the origin of protamines. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4148–4152. doi: 10.1073/pnas.0308721101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mascotti DP, Lohman TM. Thermodynamics of single-stranded RNA and DNA interactions with oligolysines containing tryptophan. Effects of base composition, Biochemistry. 1993;32:10568–10579. doi: 10.1021/bi00091a006. [DOI] [PubMed] [Google Scholar]
- 41.Mascotti DP, Lohman TM. Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry. 1997;36:7272–7279. doi: 10.1021/bi970272n. [DOI] [PubMed] [Google Scholar]
- 42.Leng M, Felsenfeld G. The preferential interactions of polylysine and polyarginine with specific base sequences in DNA. Proceedings of the National Academy of Sciences of the United States of America. 1966;56:1325–1332. doi: 10.1073/pnas.56.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawashima S, Ando T. Interaction of basic oligo-L-amino acids with deoxyribonucleic acids. Oligo-L-arginines of various chain lengths and herring sperm DNA. Journal of biochemistry. 1978;84:343–350. doi: 10.1093/oxfordjournals.jbchem.a132134. [DOI] [PubMed] [Google Scholar]
- 44.Kawashima S, Inoue S, Ando T. Interaction of basic oligo-L-amino acids with deoxyribonucleic acid. Oligo-L-ornithines of various chain lengths and herring sperm DNA. Biochimica et biophysica acta. 1969;186:145–157. doi: 10.1016/0005-2787(69)90497-3. [DOI] [PubMed] [Google Scholar]
- 45.Vijayanathan V, Lyall J, Thomas T, Shirahata A, Thomas TJ. Ionic, structural, and temperature effects on DNA nanoparticles formed by natural and synthetic polyamines. Biomacromolecules. 2005;6:1097–1103. doi: 10.1021/bm0493234. [DOI] [PubMed] [Google Scholar]
- 46.Brewer L, Corzett M, Lau EY, Balhorn R. Dynamics of protamine 1 binding to single DNA molecules. The Journal of biological chemistry. 2003;278:42403–42408. doi: 10.1074/jbc.M303610200. [DOI] [PubMed] [Google Scholar]
- 47.Cherstvy AG, Kornyshev AA, Leikin S. Torsional deformation of double helix in interaction and aggregation of DNA. The journal of physical chemistry B. 2004;108:6508–6518. doi: 10.1021/jp0380475. [DOI] [PubMed] [Google Scholar]
- 48.Kornyshev AA, Leikin S. Electrostatic interaction between helical macromolecules in dense aggregates: An impetus for DNA poly- and mesomorphism. Proc Natl Acad Sci USA. 1998;95:13579–13584. doi: 10.1073/pnas.95.23.13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kornyshev AA, Leikin S. Electrostatic zipper motif for DNA aggregation. Physical Review Letters. 1999;82:4138–4141. [Google Scholar]