SUMMARY
The specification of neuronal subtypes in the cerebral cortex proceeds in a temporal manner; however, the regulation of the transitions between the sequentially generated subtypes is poorly understood. Here, we report that the forkhead-box transcription factor Foxg1 coordinates the production of neocortical projection neurons through the global repression of a default gene program. The delayed activation of Foxg1 was necessary and sufficient to induce deep-layer neurogenesis, followed by a sequential wave of upper-layer neurogenesis. A genome-wide analysis revealed that Foxg1 binds to mammalian-specific non-coding sequences to repress over 12 transcription factors expressed in early progenitors, including Ebf2/3, Dmrt3, Dmrta1 and Eya2. These findings reveal an unexpected prolonged competence of progenitors to initiate corticogenesis at a progressed stage during development and identify Foxg1 as a critical initiator of neocorticogenesis through spatiotemporal repression, a system that balances the production of non-radially and radially migrating glutamatergic subtypes during mammalian cortical expansion.
INTRODUCTION
The functional integrity of mammalian brain systems depends on the precisely coordinated production of diverse neuron populations during development. Specifically, in the cerebral cortex, distinct neuronal subtypes are produced in a stereotypical temporal order (Angevine and Sidman, 1961). In recent years, considerable progress has been made in the identification of genes that control the differentiation of each neuronal type in the neocortex (Fame et al., 2011; Leone et al., 2008). In contrast, little is known about the regulation of the transitions between the sequentially generated subtypes.
Interestingly, although most cortical glutamatergic neurons arise from local progenitors that migrate radially and differentiate into projection neurons, some exceptions exist, in which early-born neurons originate within the surrounding pallial progenitors and invade the neocortex through a distinct migration mode. These cells have both mitogenic and patterning effects on late-born projection neurons and are unique to mammalian vertebrates (Borello and Pierani, 2010; Puelles, 2011). By far the most characterized neurons, Cajal-Retzius (CR) cells, which express the glycoprotein Reelin (Reln), have emerged rapidly both in number and molecular diversity over the course of mammalian evolution (Meyer, 2010; Pollard et al., 2006). Functionally, this is not surprising, given the specialized roles of these cells in regulating both the radial migration and areal expansion of later-born projection neurons, which are unique to the laminated neocortex system. Mechanistically, the regulation of the switch from the production of early signaling cells to radially migrating projection neurons requires a developmental process in the broader context of cortical evolution, which ultimately balances the numbers of these two functionally distinct subtypes. Hence, such mechanisms must utilize a system adaptable to changes in cortical size during mammalian evolution.
Both mouse and human cortical progenitors faithfully recapitulate in vitro the sequential generation of principal glutamatergic subtypes in vivo: preplate (ppl), deep-layer (DL), and upper-layer (UL) neurons (Eiraku et al., 2008; Gaspard et al., 2008; Shi et al., 2012). These studies imply that a common intrinsic program regulating progenitor cell competence might regulate transitions between non-radially and radially migrating mammalian cortical subtypes. Indeed, CR cells, which represent the earliest glutamatergic cell lineage in the developing neocortex (Hevner et al., 2003b), differentiate prior to all projection neuron subtypes in vitro (Eiraku et al., 2008; Gaspard et al., 2008). The unique differentiation capacity of CR cells raises the intriguing hypothesis that these progenitors represent a default progenitor state prior to commitment to a radially migrating neuron production program.
Foxg1, a member of the forkhead-box family of transcription factors (TFs), is one of the earliest TFs expressed in the anterior neural plate. Extensive studies in Xenopus, zebrafish, chick, and mice have shown that Foxg1 plays evolutionarily conserved roles in telencephalic growth (Ahlgren et al., 2003; Hanashima et al., 2002; Regad et al., 2007), cell migration (Tian et al., 2012; Miyoshi and Fishell, 2012) and patterning (Manuel et al., 2010; Roth et al., 2010), in part by antagonizing TGFβ/Smad pathways (Seoane et al., 2004) and repressing p27Kip1 (Hardcastle and Papalopulu, 2000) and Wnt8b (Danesin et al., 2009) expression. Recently, however, mutations in human FOXG1 have been associated with a congenital form of neurodegenerative diseases, namely Rett syndrome and West syndrome (Naidu and Johnston, 2011; Striano et al., 2011). Furthermore, the inactivation of Foxg1 during the early production period of mouse neocortical projection neurons alters the neurogenesis process to the earliest CR cells without altering the BMP/Wnt signaling pathway (Hanashima et al., 2007). Collectively, these observations raise an intriguing hypothesis that Foxg1 might have functions beyond its evolutionarily conserved roles to mediate the transition from non-radially to radially migrating neurogenesis.
To test this hypothesis, we used an in vivo reversible gene expression system to synchronously manipulate Foxg1 expression in cortical progenitor cells. By activating Foxg1 expression after its prolonged inactivation, we demonstrate that expression of Foxg1 is necessary and sufficient to switch from the production of earliest CR to DL projection neurons. We further show that Foxg1 binds to mammalian-specific non-coding sequences to repress the expression of multiple TFs. These observations define Foxg1 as a key coordinator of the early transcriptional network, identifying a novel regulatory system to balance the number of functionally unique glutamatergic subtypes during the course of mammalian cortical development.
RESULTS
Cortical Progenitors Exhibit Restricted Spatiotemporal Competence for CR Cell Production upon Foxg1 Inactivation
To determine the role of Foxg1 in regulating the early competence of cortical progenitors, we first assessed the temporal and spatial capacity for neurogenesis upon Foxg1 inactivation. Within the cortex, Reln-expressing CR cells are the earliest differentiating neurons and migrate tangentially to form a preplate at embryonic day (E) 11.5 (Figure 1A). At E13.5, DL (layers V/VI) projection neurons, as indicated through Ctip2 expression (Arlotta et al., 2005), migrate radially into the cortical plate (CP; Figure 1B). At E18.5, Brn2-expressing neurons in layers II/III (McEvilly et al., 2002) migrate and differentiate in the upper CP (Figure 1D). In Foxg1−/− mice, neither Ctip2+ nor Brn2+ cells were detected in the cortex, whereas the number of Reln+ CR cells was increased at respective stages (Figures 1E–H). To assess the migration patterns of these neurons, we introduced pCAGGS-GFP constructs into E14.5 Foxg1+/− and Foxg1−/− cortices using electroporation and examined the neuronal distribution and morphology at E18.5 (Figures 1I and 1J). In contrast to control GFP+ neurons, showing characteristic leading processes oriented towards the pia (Figures 1I’), these processes were randomly oriented showing no coordinated migration in Foxg1−/− neurons (Figures 1J’). These data show that CR cells are generated at the expense of the subsequent generation of radially migrating projection neurons in the absence of Foxg1 (Figure 1K).
Figure 1. Temporal Competence of Cortical Progenitor Cells Upon Foxg1 Inactivation.
(A–H) Coronal sections of E11.5 to E18.5 Foxg1+/− and Foxg1−/− cortices indicate expression of Reln (green), Ctip2 (red), and Brn2 (blue). vz, ventricular zone; ppl, preplate; cp, cortical plate; mz, marginal zone; iz, intermediate zone.
(I and J) Migration of E14.5 pCAGGS-GFP-electroporated neurons in E18.5 Foxg1+/− and Foxg1−/−. (I′,J′) Enlarged views of the boxed regions in (I,J).
(K) Schematic model of neurogenesis. The large circles indicate progenitors, and the small circles indicate post-mitotic neurons. The arrows indicate a transition in cell competence or neuronal differentiation. CR, Cajal-Retzius progenitors; DL, deep-layer progenitors; UL, upper-layer progenitors.
(L and M) Foxg1 (red) and DAPI staining (blue) of E13.5 (L) and E15.5 (M) wild-type cortices.
(N) Schematic diagram of the Foxg1tetOFoxg1 line. Foxg1 transgene expression is repressed in the presence of doxycycline (Dox).
(O–V) Foxg1 and Reln expression in Foxg1tetOFoxg1 mice or Foxg1tTA/+ control littermates. Dox was administered at E13 or E15, and the embryos were harvested at E16.5 and E18.5, respectively.
(W and X) Schematic diagram of temporal competence of cortical progenitors upon Foxg1 inactivation. UL progenitors do not acquire a CR cell fate upon Foxg1 inactivation (X). Whether these progenitors retain the UL cell identity is under investigation. KO, knockout.
Scale bar: 100 µm.
See also Figure S1.
We next assessed the spatial competence of CR cell production in the Foxg1−/− cortex. CR cells represent a heterogeneous population derived from spatially discrete sources: cortical hem, pallial-subpallial boundary (PSB), septum, choroid plexus, and thalamic eminences. The former three major subtypes can be further identified through the combinatorial expression of common and specific markers: p73 in septal- and cortical hem–derived CR cells, calretinin in early septal- and PSB-derived CR cells, and ER81 in septal-derived CR cells (Griveau et al., 2010; Yoshida et al., 2006; Zimmer et al., 2010). We observed increased numbers of both p73+/calretinin−/Reln+ (cortical hem identity; Figure S1B′) and p73−/calretinin+/Reln+ (PSB identity; Figure S1C′) CR cells in the E12.5 Foxg1−/− cortex. In contrast, ER81+ CR cells were not detected in the Foxg1−/− cortex across rostrocaudal positions at E11.5 or E12.5 (Figures S1A′–C′ and data not shown). These data demonstrate that the loss of Foxg1 results in the overproduction of most CR cell subtypes, except those of rostral identity.
We next assessed the temporal competence window for CR cell production within neocortical progenitor cells. Our previous experiments demonstrated that the conditional removal of Foxg1 expression during the DL production period results in the reversion of DL progenitors to CR cells (Hanashima et al., 2004). We observed that Foxg1 is expressed in both DL (Figure 1L) and UL progenitors (E15.5; Figure 1M). Therefore, we used Foxg1tetOFoxg1 conditional knockout mice (Hanashima et al., 2007) to assess whether UL progenitors retain the competence to differentiate into CR cells upon inactivation of Foxg1 at E15 (Figure 1N). In contrast to DL progenitors, from which ectopic CR cells were induced (Figure 1T), UL progenitors did not adopt the CR cell identity when Foxg1 was inactivated at E15 (Figure 1V). These results show that cortical progenitors undergo a progressive competence restriction during the DL-to-UL transition and that UL progenitors no longer require Foxg1 to repress the earliest CR cell identity (Figures 1W and 1X).
Foxg1 Induction Initiates DL Projection Neuron Production
The progressive restriction of CR cell generation further suggested that cortical progenitors utilize an intrinsic program to regulate transitions between cortical subtype identities. Thus, we assessed whether the manipulation of Foxg1 expression could shift the timing of cortical neuron production through the regulation of the temporal competence of cortical progenitors. For this task, we designed a reversible Foxg1 expression experiment in which Foxg1 was inactivated during DL neuron production and subsequently re-expressed during UL neuron production (Figure 2A). Based on previous results, we predicted two opposing scenarios in which the progression for competence of cortical progenitor cells proceeds in the absence of Foxg1: (a) a progressive intrinsic clock, in which UL neurons are produced according to their normal birthdate following CR cell production, or (b) a resetting of temporal competence, in which DL neurons are generated after a prolonged period of CR cell genesis. To examine these possibilities, we took advantage of a tTA-mediated gene expression system to reversibly express Foxg1 in vivo after its initial repression (Figure 1N). To circumvent early developmental defects resulting from the loss of Foxg1, doxycycline (Dox) was administered starting at E9.5 (Figure 2A), when the size differences between the Foxg1 heterozygote and homozygote cortex are minimal (Xuan et al., 1995).
Figure 2. DL Neurons are Produced After Prolonged CR Cell Production Upon Foxg1 Induction.
(A) Models for the progression of temporal competence in the absence of Foxg1. (a) In this progressive intrinsic clock model, the repression of Foxg1 during the period of DL production does not affect the timing of UL neuron production. (b) According to this model, Foxg1 re-expression after prolonged inactivation initiates DL neurogenesis at E14.5. Each circle represents progenitor state.
(B–Q) Foxg1 and Reln (B–M) and Ctip2 and DAPI (N and O) immunohistochemistry and Fezf2 in situ hybridization (P and Q) of coronal sections from Foxg1tTA/+ controls and Foxg1tetOFoxg1 [E9.5–14.5off] mice. (B–G) Dox was administered from E9.5 to E14.5, at which point the cortices were analyzed; (H–Q) Dox was administered from E9.5 to E14.5 and replaced with H2O from E14.5 to E18.5, at which point the cortices were analyzed. Scale bars: 100 µm (B-M); 50 µm (all others).
(R) Experimental design and summary.
(S–Y) In utero electroporation of pCAGGS-GFP (S–S''', U–U''',X) or pCAGGS-GFP and pCAGGS-Foxg1 (T–T''',V–V''',Y) into E14.5 Foxg1−/− cortex and analyzed at E18.5. (S'–V''') indicate enlarged views of the boxed regions shown in (S–V).
See also Figures S2 and S3.
We first repressed Foxg1 expression from E9.5 through E14.5 with Dox administration (referred to as Foxg1tetOFoxg1 [E9.5–14.5off] mice) and examined neurogenesis at E14.5 (Figures 2B–G). As predicted, we observed excessive numbers of Reln+ neurons in the CP of Foxg1tetOFoxg1 mice (Figure 2G), consistent with the requirement for Foxg1 in suppressing CR cell identity in DL progenitors (Figure 1W). To further validate whether Foxg1-lineage progenitor cells retained the capacity to differentiate into CR neurons at this stage, we isolated cortical progenitors utilizing the LacZ reporter introduced into the Foxg1 locus (Figure 1N) and CD133 expression (which marks cortical progenitor cells). Dissociated cells from E14.5 Foxg1tetOFoxg1 [E9.5–14.5off] cortices were labeled with fluorescein di-β-galactopyranoside and CD133-APC, and FACS-sorted progenitors were differentiated for 24 hr in vitro (Figures S2A and S2B). Nearly all FACS-purified cells (98.1%) were positive for LacZ (Figure S2C). In addition, Reln+ and Tuj1+ post-mitotic neurons were selectively eliminated, confirming progenitor purification (Figure S2C). To assess CR cell differentiation, we utilized previously reported CR neuron markers (Yamazaki et al., 2004). These results revealed a greater than 2-fold up-regulation of 22 CR cell marker genes (Figure S2D), whereas cortical progenitor marker expression was decreased (Figure S2E). Thus, the increase in CR cell marker gene expression reflected LacZ+ progenitor differentiation rather than the expansion of LacZ− CR precursors after sorting. Together, these results demonstrate that Foxg1-lineage progenitors retain the capacity to differentiate into CR cells after Foxg1 inactivation from E9.5 to E14.5 in vivo.
Next, we removed Dox treatment at E14.5, the transition stage from DL to UL neuron production during normal development (Hevner et al., 2003a). Low levels of Foxg1 protein were readily detected within the SOX2+ progenitors in E15.5 Foxg1tetOFoxg1 [E9.5–14.5off] cortices (Figure S3A), and further examination at E18.5 revealed numerous post-mitotic cells expressing Foxg1 (Figure 2K), which were located under the supernumerary CR cell layer (Figure 2M). Notably, we observed that many of these neurons expressed Ctip2. (Figure 2O). To further validate that up-regulated Ctip2 expression represented a transition in cell identity, we assessed the expression of Fezf2, a gene that is both required and sufficient for the specification of DL subcortical projection neurons (Molyneaux et al., 2005). Indeed, these neurons expressed Fezf2 mRNA (Figure 2Q).
As Foxg1 plays prominent roles in the cell cycle regulation of cortical progenitors (Hanashima et al., 2002), we assessed whether the activation of the cell cycle upon Foxg1 induction is responsible for the CR to DL transition (Figure S4). We observed no reduction of the cell cycle length in E15.5 Foxg1tetOFoxg1 [E9.5–14.5off] progenitors (Figures S4T and S4V) compared with E12.5 Foxg1tetOFoxg1 [E9.5–12.5off] progenitors (Figures S4K and S4U); rather, the cell cycle length was increased (Figure S4W).
To further confirm that Foxg1 directs temporal identity transition through alterations in cell competence, we manipulated Foxg1 expression in a restricted number of cortical progenitors (Figure 2W). The co-electroporation of pCAGGS-Foxg1 and pCAGGS-GFP constructs into E14.5 Foxg1−/− constitutive knockout cortices was sufficient to induce Ctip2+ neurons within, but not outside, the GFP+ cells (Figures 2T’–T”’ and 2V’–V”’) and Fezf2 expression (Figures 2Y). Together, these data demonstrate that cortical progenitor cells switch their intrinsic competence to adopt a DL neuron fate upon Foxg1 re-expression even after a prolonged period of CR cell production in vivo.
The Onset of Foxg1 Expression Triggers Sequential Neurogenesis in the Neocortex
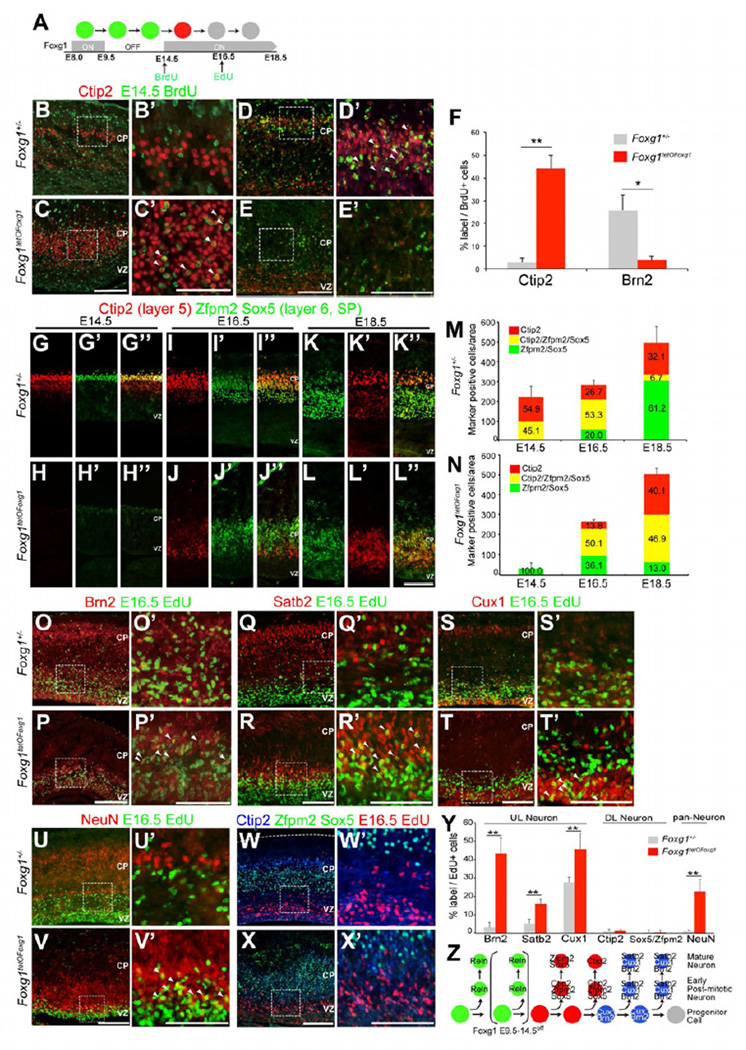
The induction of DL neurons did not distinguish whether (a) Foxg1 expression is required solely for the switch from CR cells to DL neuron production or (b) Foxg1 induction is sufficient to trigger the production of the full complement of the radially migrating projection neuron program. To address this issue, we administered pulses of bromodeoxyuridine (BrdU) and ethynyluridine (EdU) at E14.5 and E16.5, respectively, to label temporal cohorts of cortical neurons born after Foxg1 re-expression (Figure 3A). We first examined the fate of E14.5 BrdU-labeled cells (Figures 3B–F). Consistent with previous reports (Arlotta et al., 2005), we observed virtually no Ctip2-labeled BrdU+ cells in the control cortices at E18.5 (3.0 ± 8%; Figures 3B′ and 3F), indicating that the majority of Ctip2+ cells were generated prior to E14.5. In contrast, many BrdU+ neurons were co-labeled with Ctip2 in the Foxg1tetOFoxg1 [E9.5–14.5off] cortex (44.1 ± 6.0%; Figures 3C′ and 3F). Labeling using Brn2 in the controls, showed that 25.8 ± 68% of BrdU+ cells expressed Brn2 (Figures 3D’ and 3F), indicating a shift from DL to UL neurogenesis during normal development. Interestingly, in the Foxg1 mutants, we detected a low number of Brn2+ neurons located near the ventricular zone (VZ) (Figure 3E); however, the majority of these cells lacked BrdU label (Figure 3F). This observation suggests that UL neurons may be generated later than E14.5 in Foxg1 mutants.
Figure 3. The Onset of Foxg1 Triggers Sequential DL and UL Neurogenesis.
(A) Schematic diagram of the birthdating studies.
(B–E) BrdU and Ctip2 (B and C) or Brn2 (D and E) immunohistochemistry in coronal sections of E18.5 Foxg1tetOFoxg1 [E9.5–14.5off] mice and Foxg1tTA/+ littermates. (B′–E′) Enlarged views of the boxed regions shown in (B–E). The arrowheads indicate cells that are double-labeled with Ctip2 and BrdU, or Brn2 and BrdU. Scale bars: 100 µm (B–E), 50 µm (B'–E').
(F) Quantitative analysis of the percentage of BrdU+ cells that expresses Ctip2 and Brn2. *P < 0.05 and **P < 0.01.
(G–L) Ctip2 (red) and Zfpm2/Sox5 (merged in green) immunohistochemistry in E14.5, E16.5 and E18.5 Foxg1tetOFoxg1 [E9.5–14.5off] and Foxg1tTA/+ cortices. Scale bars: 50 µm (G–L'').
(M,N) Quantitative analysis of DL cells expressing Ctip2, Zfpm2 and Sox5. The y-axes indicate the total number of neurons per unit area that expressed any of the three markers. Colored bars represent the relative proportion of Ctip2+ only (red), Zfpm2/Sox5-positive only (green) and Ctip2+ cells that also expressed Zpfm2/Sox5 (yellow) of total DL cells.
(O–X) Double-detection of EdU and respective markers: UL neurons; Brn2, Satb2, Cux1 (O-T), mature neurons; NeuN (U,V), DL neurons; Ctip2, Zpfm2/Sox5 (W,X) in E18.5 Foxg1tetOFoxg1 [E9.5–14.5off] mice and Foxg1tTA/+ littermates. (O′–X′) Enlarged views of the boxed regions shown in (O–X). The arrowheads indicate cells that were double-labeled with the indicated markers and EdU. Scale bars: 100 µm (O–X), 50 µm (O'-X').
(Y) Quantitative analysis of the percentage of EdU+ cells that were co-labeled with the respective markers. **P < 0.01.
(Z) Schematic diagram of neurogenesis upon Foxg1 expression. Cux1 and Brn2 are expressed in both progenitors and neurons, whereas the others are expressed in post-mitotic neurons. Zpfm2/Sox5 and Ctip2 are co-expressed in early post-mitotic neurons but are later differentially expressed in layers V and VI/SP neurons. The grey circle indicates potential glial progenitors.
See also Figure S4.
We next assessed the generation of DL subtypes using combinatorial markers. During early corticogenesis (E14.5), the majority of DL neurons in the CP have been shown to co-express Ctip2 and Sox5/Zfpm2 (Kwan et al., 2008) (Figure 3G’’). However, by E18.5, the expression of Ctip2 is downregulated in layer VI and SP neurons, and is maintained at high levels only in layer V neurons (Figures 3B and 3K’’) (Kwan et al., 2008). In Foxg1tetOFoxg1 [E9.5–14.5off] mutants, the number of Ctip2 single-positive cells increased from E16.5 to 18.5; however, even at E18.5, 46.9% of the total number of DL neurons continued to expressed both Ctip2 and Zfpm2/Sox5, indicating a delayed DL subtype segregation consistent with late DL neuron production onset. The total number of DL neurons in the E18.5 Foxg1 mutants was comparable to that of the controls (509.3 ± 81.8 and 495.7 ± 180 cells/unit area, respectively). Collectively, these data indicate that not only the production but also the segregation timing between DL subtype markers (Ctip2 and Sox5/Zfpm2) was shifted concomitantly with the extended window of CR cell production.
We further assessed the fate of neurons born at E16.5 by examining EdU-labeled cells. Consistent with previous studies, only a fraction of Brn2+ cells were generated at this late period during normal development (3.0 ± 2.8% Brn2+/EdU+cells; Figures 3O′ and 3Y) (Hevner et al., 2003a). Other UL neuron markers, including Satb2 (5.2 ± 2.4%), Cux1 (27.8 ± 2.7%) and the mature neuron marker NeuN (0.8 ± 0.6%; Figures 3Q’, 3S’, 3U’ and 3Y), were also detected at low abundance, implying a transition from neurogenesis to gliogenesis (Seuntjens et al., 2009). In contrast, a significantly higher proportion of EdU+ cells in the Foxg1 mutants expressed Brn2 (43.2 ± 8.4%), Satb2 (15.8 ± 2.6%), Cux1 (45.6 ± 8.8%) and NeuN (22.6 ± 6.9%) (Figures 3P’, 3R’, 3T’, 3V’ and 3Y) but not DL neuron markers Ctip2 (1.0 ± 0.8%) or Sox5/Zfpm2 (0.1 ± 1.4%), implying that E16.5 progenitors primarily contribute to UL neurons in Foxg1 mutants. Together, these data suggest that corticogenesis in the Foxg1tetOFoxg1 [E9.5–14.5off] mutants proceed normally after a prolonged period of CR cell production, albeit with a temporal shift.
Temporal Transcriptome Analysis Reveals a Switch in Early Transcriptional Network upon Foxg1 Induction
The reversible Foxg1 expression system enabled the in vivo synchronization of the corticogenesis program, which provided a unique opportunity to explore the molecular logic underlying the temporal competence shift from non-radially to radially migrating glutamatergic subtypes. Importantly, the level of Foxg1 expression in the absence of Dox in the Foxg1tetOFoxg1 cortex was between the levels observed in the heterozygote and wild type (Figure S3), implying that the phenotype achieved through Foxg1 induction reflects the progression of the endogenous gene program within its lineage, rather than overexpression. We proposed that this reversible expression system would allow us to identify physiologically relevant targets of Foxg1 required for this early identity transition.
Therefore, we used FACS to isolate cortical progenitors from E14.5, E15.0, E15.5, and E16.5 Foxg1tetOFoxg1 [E9.5–14.5off] cortices (Figure 4A). E15.5 Foxg1tetOFoxg1 [E9.5–15.5off] cortices (i.e., Dox administered from E9.5 to E15.5) were used as Foxg1 non-induced controls. Quantitative RT-PCR (qPCR) indicated that Foxg1 mRNA levels were increased at E15.0 and restored at E16.5 (Figure S3D). Both immunoblotting (Figure S3E) and immunohistochemistry (Figure S3F) results indicated that Foxg1 expression was detected at E15.5 and increased at E16.5. These data suggest that the earliest downstream genes might respond to Foxg1 at approximately 24 hr after Dox removal. Total RNA prepared from FACS-sorted progenitors was reverse-transcribed, labeled, and hybridized to Affymetrix Genechip microarrays. To identify genes regulated in a Foxg1-dependent manner, we applied stringent filtering steps to detect the significant differential expression of genes without potential biases (Table S1). First, microarray datasets from the five experimental conditions were subjected to an analysis of variance (ANOVA), and the significant differential expression of transcripts was clustered into 30 groups (3408/45038 transcripts; Figure 4B). Among these, Wnt8b, a previously identified Foxg1-repressed target (Danesin et al., 2009), was clustered in the early down-regulated gene group (group II), validating the microarray analysis. Notably, multiple CR-specific genes (Ebf2/3, Lhx9 and Zic3) (Inoue et al., 2008; Yamazaki et al., 2004) were also among the down-regulated gene cluster (group III). These results imply that Foxg1 might switch early cell identity through the repression of multiple CR-specific genes.
Figure 4. Temporal Transcriptome Analysis of Foxg1-induced Cortical Progenitors In Vivo.
(A) Experimental design.
(B) Heatmap representing ANOVA cluster analysis. A total of six groups are indicated (groups I–VI). Datasets were obtained from two independent analyses of each experimental condition. E15.5 (Dox+) represents non-induced negative controls. Representative genes within each cluster are depicted on the right; blue indicates a previously reported Foxg1 target gene (Wnt8b), and red indicates reported CR cell markers.
(C) Analysis of transcript response to Foxg1 induction. The response time (x axis) was calculated as the time required to reach a half-maximum response at E16.5 (inset). Response magnitude (y axis) is represented by fold change (log2). Red indicates transcription factors; green indicates Wnt8b.
See also Tables S1 and S2.
Although ANOVA enables the statistical assessment of differentially expressed transcripts across multiple experimental samples, estimating the precise time and magnitude of the responses to predict the genes and events downstream of Foxg1 required further time-dependent criteria. Therefore, we subjected the six clusters (constitutively up- or down-regulated groups: 682/3408 transcripts) to a fold-change analysis, and genes that were up- or down-regulated by more than 2-fold from E14.5 to E16.5 were depicted (206/682 transcripts). Next, we independently measured the response of each transcript to Foxg1 induction by calculating the time-response as indicated by the half-maximal response time relative to 48 hr (x) and the response magnitude as indicated by the fold change in the transcript expression level at 48 hr (y; Figure 4C, Table S2). This analysis also revealed Wnt8b among the earliest genes to respond to Foxg1 (response time: 20.2 hr; Figure 4C). Furthermore, we identified multiple probe sets obtained from the same genes (three probes each for Ebf2, Ebf3, and Nr4a2) that responded similarly (Figure 4C), further validating the microarray study.
Two noticeable patterns stood out from these findings. First, most down-regulated genes responded between 20 and 35 hr, whereas the up-regulated genes had a broader range of response times, from 10 to 45 hr (Figure 4C). Second, more TFs were present among down-regulated genes than among up-regulated genes (19/59 [32.2%] down-regulated transcripts, 14/147 [9.5%] up-regulated transcripts). These results imply that up-regulated genes that respond with a longer delay (≥35 hr) might include genes that are indirect targets of Foxg1.
To identify the gene network downstream of Foxg1, we further analyzed the TFs because they can directly regulate the gene expression responsible for the early CR-to-DL transition (13 down-regulated TFs [19 transcripts], 12 up-regulated TFs [14 transcripts]). Candidate TF expression was assessed using qPCR for progenitors isolated from E14.5, E15.5, and E16.5 Foxg1tetOFoxg1 [E9.5–14.5off] cortices. The expression trend detected in the microarray studies was validated, except for Mapk8 (Figure S5A). Next, we induced Foxg1 expression for 24 hr in Foxg1tetOFoxg1 mice treated with Dox from E9.5 to 13.5 (E14.5 Foxg1tetOFoxg1 [E9.5–13.5off]). We expected bona fide Foxg1 targets to respond in a manner similar to that of E15.5 Foxg1tetOFoxg1 [E9.5–14.5off] in these one-day-early Foxg1-induced progenitors, whereas developmental stage–dependent genes would respond differently. According to these criteria, most candidate down-regulated TFs (11/12) responded identically to E13.5 Foxg1 induction (Figure S5B). However, 3/3 early-up-regulated TFs (Sdpr, Cebpd, Sox8; genes that responded within 24 hr in Figure S5A) were induced in the presence and absence of Dox (Figure S5B). These TFs were eliminated from further analysis.
Foxg1 Binds to Highly Mammalian Conserved Sequences to Regulate Global Gene Expression In Vivo
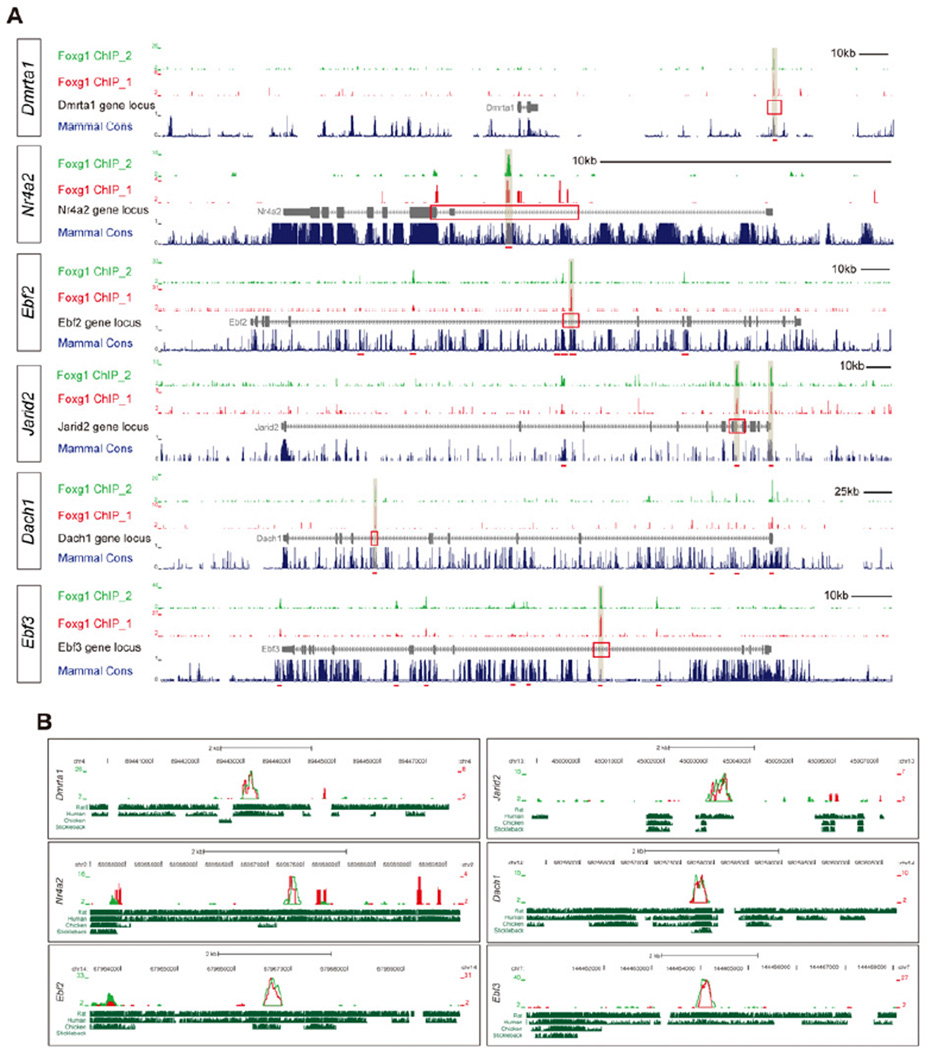
The transcriptome datasets obtained from complementary microarray and qPCR analyses identified highly Foxg1-responsive TFs within cortical progenitors. To verify whether any of these candidates are potential direct targets of Foxg1, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq). A comparison of Foxg1 and control datasets with the Model-based Analysis of the ChIP-Seq (MACS) peak-calling algorithm (Zhang et al., 2008) identified 5817 overlapping peaks between the two Foxg1 ChIP-Seq replicates (see Experimental Procedures). An examination of the binding site distribution in the gene loci revealed the preferential recruitment of Foxg1 to intronic sequences within the down-regulated TFs (41.7%, 5/12 genes) but not up-regulated TFs (0%, 0/7 genes) (Figure 5A and Table S3). To assess whether the Foxg1-bound sequences outside the gene body were potential regulatory elements, we cloned these Foxg1 binding sites upstream of the luciferase reporter gene and assessed transcription. The forced expression of Foxg1 in P19 cells significantly repressed the activity of these binding sites (4/5 genes; p<0.05)(Figure S6). Notably, these Foxg1-bound non-coding sequences were highly conserved in mammals but were underrepresented in chicks and teleosts (Figure 5B).
Figure 5. ChIP-seq Analysis of Foxg1-repressed Gene Loci.
(A) Views of entire gene loci for the indicated genes. The data are presented from two independent ChIP-seq analyses (Foxg1 ChIP_1 and Foxg1 ChIP_2). The genes are listed in order of response to Foxg1. The red underline indicates MACS peaks and the light-brown bars mark MACS peaks with highest fold enrichment within the indicated region.
(B) Enlarged views of the red-boxed regions in (A) and conservation between mouse and rat, human, chicken, and stickleback.
Finally, all 19 TFs were analyzed through in situ hybridization (ISH) to determine their spatiotemporal expression patterns in E11.5 wild type, E14.5 control, and E14.5 and E16.5 Foxg1tetOFoxg1 [E9.5–14.5off] cortices (Figure 6 and data not shown). The results revealed three noticeable trends. (1) At E11.5, down-regulated TFs exhibited bimodal expression patterns, either caudal-to-rostral gradient expression within the neuroepithelium (Dmrt3, Eya2, Nr4a2) or restricted expression in early preplate resembling migrating CR cells (Rgmb, Ebf2/3, Lhx9). (2) Most of these genes were down-regulated or remained weakly expressed in presumptive CR cells in E14.5 control cortices. (3) In Foxg1tetOFoxg1 [E9.5–14.5off] cortices, all genes were up-regulated in either the VZ or CP at E14.5, whereas expression was significantly reduced in both cortical VZ and CP at E16.5. Thus, the ISH expression pattern recapitulated the temporally shifted repression according to the Foxg1 induction time scale. Collectively, these data define Foxg1 as a key regulator of the early transcriptional network in switching cell identity in the developing cerebral cortex.
Figure 6. Q-PCR and ISH Analysis of Foxg1-repressed Transcription Factors.
(Left column) qPCR data of E14.5, E15.5, and E16.5 Foxg1tetOFoxg1 [E9.5–14.5off] cortical progenitors. The values are relative to GAPDH expression. (Right columns) The representative ISH images from E11.5 wild type (sagittal), E14.5 control, E14.5 and E16.5 Foxg1tetOFoxg1 [E9.5–14.5off] (coronal) cortices. The figures are shown in order of response to Foxg1 (early to late), with the exception of Wnt8b, which is a positive control. Scale bars: 100 µm (E14.5, E16.5); 200 µm (E11.5).
See also Figures S5 and Table S4.
DISCUSSION
Foxg1 Regulates Early Cortical Gene Networks
Here, we demonstrate that cortical progenitors retain an unexpected prolonged competence to initiate corticogenesis at a progressed stage during development upon Foxg1 induction. Although specific TFs induce the production of neuronal subtypes beyond their normal birthdates (Molyneaux et al., 2005), the genes that directly shift temporal competence in the mammalian central nervous system have not been reported. Our reversible Foxg1 expression system enabled the in vivo synchronization of corticogenesis and provided a unique opportunity to explore the molecular logic underlying the shift in temporal competence of cortical progenitors. Although such strategies have successfully identified gene regulatory networks in embryonic stem cell differentiation, whether they can be applied to an advanced stage of cell lineage (e.g., cortical progenitor cells) had not been previously assessed in vivo. By extending clustering-based methods, our time-response detection algorithm enabled the investigation of gene expression dynamics in a time-sensitive manner. The data revealed specific patterns in response timing among the genes regulated through a single TF. Consistent with previous reports that Foxg1 can act as a transcriptional repressor (Yao et al., 2001), we observed an increased representation of TFs among the down-regulated genes (Figure 4) and showed that Foxg1 binds to these down-regulated genes in vivo (Figure 5). Interestingly, these TFs might not only be selectively expressed, but may also be required for CR cell development (Chiara et al., 2012; Inoue et al., 2008). These results indicate that the transition from early CR cell to the projection neuron production program involves the rapid repression of multiple TFs (≥20 hr), followed by delayed induction of up-regulated TFs (≥30 hr). These observations are consistent with previous reports that developmental cell fate decisions favor the utilization of repressor cascades, which are more robust to noise in protein-production rates than activator cascades (Jacob et al., 2008; Rappaport et al., 2005). Intriguingly, repressor networks are also the major regulatory cascades responsible for the segregation of subtype identities within neocortical DLs. Fezf2, Tbr1 and Satb2 are expressed in corticospinal, corticothalamic and corticocortical projection neurons, respectively, and the loss of any one of these genes results in a switch to alternative subtype identities (Alcamo et al., 2008; Chen et al., 2008; Han et al., 2011; McKenna et al., 2011). Notably, in these mutants, the timing and total number of DL cells generated appear grossly normal. Together with our current data, the specification of cortical projection neuron subtypes likely involves two critical steps: (1) the suppression of a default identity and commitment to projection neuron fate through Foxg1-mediated TF cascade and (2) the cross-regulatory determination within projection neurons through subtype-specific TFs. Whether similar repression cascades account for temporal identity transitions during later steps of cortical neurogenesis remains unknown.
Role of Foxg1 in Cortical Specification
Our study identified Foxg1 as a key coordinator that initiates cortical neurogenesis. Interestingly, it has been shown that neocortical specification requires the repression of cortical hem and PSB identity through the expression of another TF, Lhx2 (Chou et al., 2009; Mangale et al., 2008). Foxg1 and Lhx2 are expressed in the neuroepithelium as early as E8.0 and E8.5, respectively (Mangale et al., 2008; Tao and Lai, 1992). Both knockout and chimera studies have revealed that, within the cortex, Foxg1−/− cells express Lhx2 caudally, whereas Lhx2−/− progenitors retain Foxg1 expression laterally (Mangale et al., 2008; Muzio and Mallamaci, 2005). These studies imply that Foxg1 and Lhx2 might function cooperatively, but independently, to establish neocortical identity. Indeed, their temporal requirements for cortical specification are distinct. The removal of Foxg1 at E13 is sufficient to revert DL cells to CR cell identity, but the conditional removal of Lhx2 exhibits an early (~E11.5) competence window for neocortex-to-paleocortex transition (Chou et al., 2009). Additionally, because Foxg1 is essential for establishing ventral telencephalic identity, the absence of obvious PSB expansion in Foxg1−/− is likely secondary to the loss of ventral gene expression. These observations also imply that the primary targets of Foxg1 and Lhx2 might be largely non-redundant.
Interestingly, genetic fate-mapping studies have indicated that the caudal telencephalon, including the future archipallium, is derived from a lineage that is distinct from that of a more rostral telencephalic compartment. The compound loss of Emx2 and Pax6 results in the loss of archipallial territories without affecting anterior FGF8 or Foxg1 expression (Kimura et al., 2005). Within the caudal forebrain, Emx2 and Pax6 are expressed in dorsomedial and ventrolateral regions, respectively, where Foxg1 delineates boundaries at a cellular resolution (Figure S7 and data not shown). These areas consist of the cortical hem, PSB, choroid plexus and thalamic eminence, all of which are presumptive CR cell sources (Bielle et al., 2005; Imayoshi et al., 2008; Tissir et al., 2009; Yoshida et al., 2006) (Figure 7B). Our ISH studies revealed that the primary repressed Foxg1 target TFs also exhibit caudal-to-rostral gradient expression within the early telencephalon (Figure 6). Thus, the onset of Foxg1 expression in the anterior neural ridge induced through FGF8 might switch early cell competence in an opposed rostral-to-caudal gradient (Shimamura and Rubenstein, 1997) (Figure 7).
Figure 7. Proposed model for the switch in neurogenesis in the cerebral cortex.
(A) Foxg1 (red) is induced in the anterior neural ectoderm through rostral Fgf8 expression (yellow) and expands caudally in the neural plate.
(B) After neural tube closure, Foxg1 shifts the rostral limit of caudal telencephalic gene expression within the neuroepithelium (indicated in green) and initiates projection neuron production in the dorsal progenitors. Expression of these genes is only observed rostrally in migrating CR neurons. Note that the cortical hem corresponds only to the dorsal part of the CR cell competent region (green) in the sagittal section. Ventrally, the caudal limits of Foxg1 expression are the PSB and thalamic eminence (Pax6+ and Sfrp2+ region in Figure S7). PSB, pallium-subpallium boundary; CPe, choroid plexus; ThE, thalamic eminence.
See also Figure S7.
Consistent with specialized roles, it has been proposed that the wide distribution of CR cells in the marginal zone is limited to mammals and is further elaborated in both number and molecular diversity in humans (Medina and Abellan, 2009; Pollard et al., 2006). CR cells are under-represented in chicks, whereas zebrafish lack obvious CR cells and Reln and Foxg1 expression domains largely overlap (Costagli et al., 2002; Nomura et al., 2008). Our ChIP-seq data revealed that Foxg1-bound non-coding sequences within early-repressed TFs are highly conserved in mammals but are largely absent in chicks and teleosts (Figure 5B). The expansion of mammalian cortical size during evolution may have co-opted efficient default-mode compensatory mechanisms to generate sufficient numbers of early signaling cells prior to the onset of corticogenesis, which involves a novel projection neuron migration mode.
EXPERIMENTAL PROCEDURES
Mice
Foxg1lacZ/+ mice (Xuan et al., 1995) were maintained on a CD1 background and intercrossed to obtain Foxg1lacZ/lacZ null embryos. Foxg1 conditional mutants (Foxg1tetOFoxg1) were generated by crossing Foxg1tTA/+ mice with Foxg1lacZ/+;tetOFoxg1IRESlacZ double-heterozygous mice (Hanashima et al., 2007). Animals were housed in the Animal Housing Facility of the RIKEN CDB according to institute guidelines.
In Situ Hybridization and Immunohistochemistry
Embryos were dissected, and the brains were fixed in 4% paraformaldehyde (PFA) for 1 hr. For advanced-stage embryos, brains were perfused with PBS and 4% PFA prior to fixation. Following 30% sucrose replacement, fixed brains were embedded in OCT compound, and 12 µm slices were cut on a cryostat. In situ hybridization and immunohistochemistry was performed as previously described (Hanashima et al., 2007). For details, see the Extended Experimental Procedures.
In Utero Electroporation
Pregnant dams from Foxg1+/− intercrosses were anesthetized by intraperitoneal injection with Nembutal sodium solution (Lundbeck Inc.). The electroporation was performed on E14.5 embryonic brains using an electroporator (CUY21E; Nepa Gene). The procedural details are provided in Extended Experimental Procedures.
FACS
The cerebral cortices of E14.5 mouse embryos were dissociated using a Neural Tissue Dissociation Kit (Sumilon). For FDG labeling, pre-warmed 5 mM FDG (Sigma) was added dropwise to dissociated cells (3×106) and incubated at 37°C for 1 min. For APC-conjugat ed IgG or CD133 labeling, dissociated cells (3×106) were incubated with APC-conjugated IgG or CD133 on ice for 30 min. FACS was performed using FACSAria II and analyzed using FACSDiva 6.1 software (Becton Dickinson). The procedural details are provided in Extended Experimental Procedures.
GeneChip Analysis
Total RNA was prepared using an RNeasy Mini Kit (QIAGEN), and the quality was assessed with an Agilent 2100 Bioanalyzer (Agilent Technology). The cDNA synthesis and cRNA labeling reactions were performed using the 3’ IVT-Express Kit according to the manufacturer’s instructions (Affymetrix). High-density oligonucleotide arrays for Mus musculus (Mouse Genome 430 2.0), containing 45038 probes, were performed according to the Expression Analysis Technical Manual (Affymetrix).
Temporal transcriptome analysis
To identify differentially expressed genes, one-way ANOVAs with post hoc tests were performed. Multiple comparisons were corrected with false discovery rates (FDRs), and an FDR of less than 0.05 was chosen as significant. For probes with a ratio ≥2.0, the time required to reach 50% expression relative to E16.5 was designated as the response time (Figure 4C inset). The procedural details are described in Table S1 and Extended Experimental Procedures.
Quantitative RT-PCR
Quantitative RT-PCR was performed using SYBR green labeling (SYBR Premix Ex Taq II; Takara) and a 7900 Fast Real Time PCR System (Applied Biosystems). The quantitative analysis was performed using the delta-delta Ct method with GAPDH as an internal control. The primers used for qPCR are listed in Table S4.
ChIP-seq
Dissected cerebral cortices from E14.5 wild-type embryos were subjected to the ChIP assay using Foxg1 antibodies (Neuracell). The resulting ChIPed DNAs from two independent ChIP experiments and input DNAs were sequenced (pair-end, 50-mer) on an Illumina Hiseq2000 platform. For details, see the Extended Experimental Procedures.
Statistical Analysis
The quantitative data are presented as the mean ± standard error of mean (S.E.M) from representative experiments (n≥3). For the statistical analysis, the data were evaluated with a Student’s t-test. P-values < 0.05 or 0.1 were considered significant.
Supplementary Material
HIGHLIGHTS.
Foxg1 directs projection neuron production onset in the neocortex
Foxg1 acts cell autonomously to switch mammalian cortical subtypes
Progenitors retain a prolonged competence to initiate corticogenesis in vivo
Ebf2/3, Dmrta1, Eya2 are mammalian-specific repression targets of Foxg1
ACKNOWLEDGMENTS
We thank P. Arlotta for critical reading of the manuscript; S. Garel, T. Bullmann and Y. Gonda for valuable discussions; Y. Sasai for the Foxg1 antibody; S. Garel, J. Aruga, and T. Curran for plasmids; and Y. Wada, R. Oda, and C. Kumamoto for excellent technical assistance. This work was supported through a Grant-in-Aid for Scientific Research on Innovative Areas “Neural Diversity and Neocortical Organization” from the MEXT of Japan to C.H. and a grant from the NIMH (R01MH094589) to B.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahlgren S, Vogt P, Bronner-Fraser M. Excess FoxG1 causes overgrowth of the neural tube. J. Neurobiol. 2003;57:337–349. doi: 10.1002/neu.10287. [DOI] [PubMed] [Google Scholar]
- Alcamo E, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell S. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux B, Chen J, Inoue J, Kominami R, Macklis J. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Bielle F, Griveau A, Narboux-Neme N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat. Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- Borello U, Pierani A. Patterning the cerebral cortex: traveling with morphogens. Current Opin. Genet. Dev. 2010;20:408–415. doi: 10.1016/j.gde.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz L, McConnell S. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang S, Hattox A, Rayburn H, Nelson S, McConnell S. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara F, Badaloni A, Croci L, Yeh ML, Cariboni A, Hoerder-Suabedissen A, Consalez GG, Eickholt B, Shimogori T, Parnavelas JG, Rakic S. Early B-cell factors 2 and 3 (EBF2/3) regulate early migration of Cajal-Retzius cells from the cortical hem. Dev. Biol. 2012;365:277–289. doi: 10.1016/j.ydbio.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Perez-Garcia C, Kroll T, O'Leary D. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costagli A, Kapsimali M, Wilson SW, Mione M. Conserved and divergent patterns of Reelin expression in the zebrafish central nervous system. J. Comp. Neurol. 2002;450:73–93. doi: 10.1002/cne.10292. [DOI] [PubMed] [Google Scholar]
- Danesin C, Peres JN, Johansson M, Snowden V, Cording A, Papalopulu N, Houart C. Integration of telencephalic Wnt and hedgehog signaling center activities by Foxg1. Dev. Cell. 2009;16:576–587. doi: 10.1016/j.devcel.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Fame RM, MacDonald JL, Macklis JD. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- Griveau A, Borello U, Causeret F, Tissir F, Boggetto N, Karaz S, Pierani A. A novel role for Dbx1-derived Cajal-Retzius cells in early regionalization of the cerebral cortical neuroepithelium. PLoS Biol. 2010;8:e1000440. doi: 10.1371/journal.pbio.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan K, Shim S, Lam M, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Fernandes M, Hebert J, Fishell G. The role of Foxg1 and dorsal midline signaling in the generation of Cajal-Retzius subtypes. J. Neurosci. 2007;27:11103–11111. doi: 10.1523/JNEUROSCI.1066-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Li S, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hanashima C, Shen L, Li S, Lai E. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J. Neurosci. 2002;22:6526–6536. doi: 10.1523/JNEUROSCI.22-15-06526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle Z, Papalopulu N. Distinct effects of XBF-1 in regulating the cell cycle inhibitor p27(XIC1) and imparting a neural fate. Development. 2000;127:1303–1314. doi: 10.1242/dev.127.6.1303. [DOI] [PubMed] [Google Scholar]
- Hevner R, Daza R, Rubenstein J, Stunnenberg H, Olavarria J, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev. Neurosci. 2003a;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Hevner R, Neogi T, Englund C, Daza R, Fink A. Cajal-Retzius cells in the mouse: transcription factors, neurotransmitters, and birthdays suggest a pallial origin. Brain Res. Dev. Brain Res. 2003b;141:39–53. doi: 10.1016/s0165-3806(02)00641-7. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135:2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- Inoue T, Ogawa M, Mikoshiba K, Aruga J. Zic deficiency in the cortical marginal zone and meninges results in cortical lamination defects resembling those in type II lissencephaly. J. Neurosci. 2008;28:4712–4725. doi: 10.1523/JNEUROSCI.5735-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Maurange C, Gould A. Temporal control of neuronal diversity: common regulatory principles in insects and vertebrates? Development. 2008;135:3481–3489. doi: 10.1242/dev.016931. [DOI] [PubMed] [Google Scholar]
- Kimura J, Suda Y, Kurokawa D, Hossain Z, Nakamura M, Takahashi M, Hara A, Aizawa S. Emx2 and Pax6 function in cooperation with Otx2 and Otx1 to develop caudal forebrain primordium that includes future archipallium. J. Neurosci. 2005;25:5097–5108. doi: 10.1523/JNEUROSCI.0239-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K, Lam M, Krsnik Z, Kawasawa Y, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone D, Srinivasan K, Chen B, Alcamo E, McConnell S. The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale V, Hirokawa K, Satyaki P, Gokulchandran N, Chikbire S, Subramanian L, Shetty A, Martynoga B, Paul J, Mai M, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M, Martynoga B, Yu T, West J, Mason J, Price D. The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development. 2010;137:487–497. doi: 10.1242/dev.039800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvilly R, de Diaz M, Schonemann M, Hooshmand F, Rosenfeld M. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J. Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G. Building a human cortex: the evolutionary differentiation of Cajal-Retzius cells and the cortical hem. J Anat. 2010;217:334–343. doi: 10.1111/j.1469-7580.2010.01266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. Dynamic FoxG1 expression coordinates the integration of multipolar pyramidal neuron precursors into the cortical plate. Neuron. 2012;74:1045–1058. doi: 10.1016/j.neuron.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux B, Arlotta P, Hirata T, Hibi M, Macklis J. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Muzio L, Mallamaci A. Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J. Neurosci. 2005;25:4435–4441. doi: 10.1523/JNEUROSCI.4804-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu S, Johnston MV. Neurodevelopmental disorders: Clinical criteria for Rett syndrome. Nat. Rev. Neurol. 2011;7:312–314. doi: 10.1038/nrneurol.2011.64. [DOI] [PubMed] [Google Scholar]
- Nomura T, Takahashi M, Hara Y, Osumi N. Patterns of neurogenesis and amplitude of Reelin expression are essential for making a mammalian-type cortex. PloS one. 2008;3:e1454. doi: 10.1371/journal.pone.0001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K, Salama S, Lambert N, Lambot M, Coppens S, Pedersen J, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Puelles L. Pallio-pallial tangential migrations and growth signaling: new scenario for cortical evolution? Brain Behav. and Evol. 2011;78:108–127. doi: 10.1159/000327905. [DOI] [PubMed] [Google Scholar]
- Rappaport N, Winter S, Barkai N. The ups and downs of biological timers. Theor. Biol. Med. Model. 2005;2:22. doi: 10.1186/1742-4682-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T, Roth M, Bredenkamp N, Illing N, Papalopulu N. The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat. Cell Biol. 2007;9:531–540. doi: 10.1038/ncb1573. [DOI] [PubMed] [Google Scholar]
- Roth M, Bonev B, Lindsay J, Lea R, Panagiotaki N, Houart C, Papalopulu N. FoxG1 and TLE2 act cooperatively to regulate ventral telencephalon formation. Development. 2010;137:1553–1562. doi: 10.1242/dev.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le H, Shen L, Anderson S, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave K, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat. Neurosci. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 2012;15:477–486. S471. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura K, Rubenstein J. Inductive interactions direct early regionalization of the mouse forebrain. Development. 1997;124:2709–2718. doi: 10.1242/dev.124.14.2709. [DOI] [PubMed] [Google Scholar]
- Striano P, Paravidino R, Sicca F, Chiurazzi P, Gimelli S, Coppola A, Robbiano A, Traverso M, Pintaudi M, Giovannini S, et al. West syndrome associated with 14q12 duplications harboring FOXG1. Neurology. 2011;76:1600–1602. doi: 10.1212/WNL.0b013e3182194bbf. [DOI] [PubMed] [Google Scholar]
- Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- Tian C, Gong Y, Yang Y, Shen W, Wang K, Liu J, Xu B, Zhao J, Zhao C. Foxg1 has an essential role in postnatal development of the dentate gyrus. J. Neurosci. 2012;32:2931–2949. doi: 10.1523/JNEUROSCI.5240-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Ravni A, Achouri Y, Riethmacher D, Meyer G, Goffinet A. DeltaNp73 regulates neuronal survival in vivo. Proc Natl Acad Sci USA. 2009;106:16871–16876. doi: 10.1073/pnas.0903191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S, Baptista C, Balas G, Tao W, Soares V, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Sekiguchi M, Takamatsu M, Tanabe Y, Nakanishi S. Distinct ontogenic and regional expressions of newly identified Cajal-Retzius cell-specific genes during neocorticogenesis. Proc Natl Acad Sci USA. 2004;101:14509–14514. doi: 10.1073/pnas.0406295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Lai E, Stifani S. The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Mol Cell Biol. 2001;21:1962–1972. doi: 10.1128/MCB.21.6.1962-1972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Assimacopoulos S, Jones K, Grove E. Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development. 2006;133:537–545. doi: 10.1242/dev.02209. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Lee J, Griveau A, Arber S, Pierani A, Garel S, Guillemot F. Role of Fgf8 signalling in the specification of rostral Cajal-Retzius cells. Development. 2010;137:293–302. doi: 10.1242/dev.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.