Abstract
Simian virus 40 (SV40) isolates differ in oncogenic potential in Syrian golden hamsters following intraperitoneal inoculation. Here we describe the effect of intravenous exposure on tumor induction by SV40. Strains SVCPC (simple regulatory region) and VA45-54(2E) (complex regulatory region) were highly oncogenic following intravenous inoculation, producing a spectrum of tumor types. Three lymphoma cell lines were established; all expressed SV40 T-antigen, were immortalized for growth in culture, and were tumorigenic following transplantation in vivo. New monoclonal antibodies directed against hamster lymphocyte surface antigens are described. The cell lines expressed MHC class II and macrophage markers and were highly phagocytic, indicating a histiocytic origin. Many hamsters that remained tumor-free developed SV40 T-antigen antibodies, suggesting that viral replication occurred. This study shows that route of exposure influences the pathogenesis of SV40-mediated carcinogenesis, that SV40 strain VA45-54(2E) is lymphomagenic in hamsters, that hamster lymphoid cells of histiocytic origin can be transformed in vivo and established in culture, and that reagents to hamster leukocyte differentiation molecules are now available.
Keywords: SV40, T-antigen, Polyomavirus, Hamsters, Lymphomas, Viral strains, Anti-hamster lymphocyte antibodies, Tumorigenicity, Phagocytosis
Introduction
Polyomavirus simian virus 40 (SV40) is an oncogenic DNA virus that was an inadvertent contaminant of early poliovirus vaccines that were prepared using cultures of primary monkey kidney cells (Butel and Lednicky, 1999; Stratton et al., 2003). It is estimated that millions of people in the US and worldwide received one or more doses of contaminated vaccines between 1955 and 1963. Exposures may have occurred beyond that time period, as the USSR oral poliovirus vaccine may have been contaminated as late as 1978 (Cutrone et al., 2005). SV40 has been found to be associated with selected human cancers at varying frequencies, including non-Hodgkin’s lymphoma (NHL) (Amara et al., 2007; Butel, 2008; Butel and Lednicky, 1999; Gazdar et al., 2002; Shivapurkar et al., 2002; Vilchez et al., 2002b; Zekri et al., 2007); however, other studies have failed to detect an association (Butel, 2008; MacKenzie et al., 2003; Schüler et al., 2006; Sui et al., 2005). The pathogenesis of SV40 infections in humans has not been characterized, but lymphoid cells are thought to play an important role.
The Syrian golden hamster is the animal model for SV40-mediated oncogenesis because these rodents are highly susceptible to SV40-induced tumors (Butel et al., 1972; Butel and Lednicky, 1999; Carbone et al., 1989; Cicala et al., 1993; Diamandopoulos, 1972, 1973; Girardi et al., 1962). The types of neoplasms that develop in hamsters are influenced by the route of inoculation. The spectrum of tumors found to be SV40-positive in humans is the same as that observed in hamsters following SV40 inoculation (Gazdar et al., 2002; Vilchez and Butel, 2004).
It has been shown that biological differences exist among SV40 strains. The distinction of genetic strains is based on sequence differences in the C-terminal region of the large tumor antigen (T-ag) gene, which result in amino acid changes in T-ag, the viral replication protein and the major oncogenic protein (Butel and Lednicky, 1999; Forsman et al., 2004; Stewart et al., 1996, 1998). In addition, virus variants can be distinguished by rearrangements in the viral regulatory region (Lednicky and Butel, 2001). Recent results showed that SV40 isolates differ in oncogenic potential in hamsters following intraperitoneal (i.p.) inoculation (Sroller et al., 2008; Vilchez et al., 2004). The viral regulatory region was found to exert a major influence on tumorigenicity, as a significantly larger number of animals developed tumors following inoculation of viruses with simple (1E) regulatory regions than those exposed to viruses with complex (2E) regulatory regions (Sroller et al., 2008). The influence of T-ag strain differences was less pronounced, although the SVCPC T-ag was more oncogenic than either the 776 or VA45-54 T-ag domains. Together these studies suggest that virus-specific factors influence the oncogenic outcome in an SV40-infected host.
Intraperitoneal inoculation of weanling hamsters often results in mesotheliomas (Cicala et al., 1993). In contrast, intravenous (i.v.) inoculation of weanling hamsters reportedly induces leukemias, lymphomas, and osteosarcomas, as well as undifferentiated sarcomas (Diamandopoulos, 1972, 1973; Carbone et al., 1989; Cicala et al., 1992). The lymphomas have been characterized as being of B cell or histiocytic and macrophage origin (Carbone et al., 1989; Cicala et al., 1992; Coe and Green, 1975).
An evaluation of the oncogenicity of different SV40 strains administered via i.v. inoculation has not previously been performed. Characterization of SV40-mediated lymphomagenesis in hamsters may elucidate viral effects on lymphoid cells that will provide insights into a possible role of SV40 in human lymphoid tumors. Additionally, this model will allow for studies of a role that infected lymphoid cells may play in disseminating virus in a host. This report describes tumor induction in hamsters following i.v. inoculation of SV40, the development of monoclonal antibodies useful to characterize surface markers of hamster lymphoid cells, and the derivation and characterization of SV40 lymphoma tumor cell lines. We focused on SV40 strains SVCPC, highly oncogenic by the i.p. route of inoculation (Sroller et al., 2008; Vilchez et al., 2004), and VA45-54, used in the original studies of i.v. administration of SV40 (Diamandopoulos, 1972, 1973).
Results
Tumor induction in hamsters following i.v. inoculation of SV40
To study the process of SV40-induced lymphomagenesis in the hamster model, groups of 21-day-old Syrian golden hamsters were inoculated intravenously with 1 × 107 PFU of SV40. Viral strain-dependent tumorigenicity following this route of administration was evaluated by comparing two well-characterized strains, SVCPC and VA45-54. The latter virus had been found by Diamandopoulos (1972, 1973) to be lymphomagenic. To determine if unique T-ag sequences play a role in tumorigenesis following i.v. inoculation, recombinant viruses were included in which the T-ag C-terminal coding region (T-ag-C) from strains SVCPC and VA45-54 were present on the 776(2E) strain background, 776-CPC(2E) and 776-VA(2E). The role of the viral regulatory region on oncogenicity was examined by inclusion of single (1E) and double (2E) enhancer viruses, including both versions of strain VA45-54.
Inoculated animals were sacrificed at 22–34 weeks (5–8 months) postinoculation (p.i.) (Table 1). In each group, some animals died 2 months or later p.i. but were not necropsied. It is likely that these animals had tumors that caused the unexplained deaths, as such deaths were not observed in the control groups in this or previous studies (Sroller et al., 2008; Vilchez et al., 2004). Animal deaths less than 4 weeks p.i. were assumed to reflect complications of the surgical inoculation procedure and were removed from the animal counts. Tumor incidences have been calculated by both including and excluding the unnecropsied late (≥2 months p.i.) deaths (Table 1).
Table 1.
Tumor development in Syrian golden hamsters following intravenous inoculation with SV40 parental strains and recombinant virusesa
| Virus | No. tumors/No. necropsied animals (% tumors) | No. tumors + late deaths/No. animals (% tumors) | Median time to tumors, weeks (range)
|
Histologic types of tumorsb | |
|---|---|---|---|---|---|
| Necropsied animals | Necropsied + late deaths | ||||
| SVCPC | 3/6 (50) | 6/9 (67) | 24 (22–24) | 21 (8–24) | Mesothelioma , malignant fibrous histiocytoma (2), lymphoma, osteosarcoma, sarcomac |
| VA45-54(1E) | 2/12 (17) | 5/15 (33) | 34 (34) | 26 (13–34) | Not done |
| VA45-54(2E) | 9/12 (75) | 14/17 (82) | 28 (24–33) | 27 (20–33) | Osteosarcoma, malignant fibrous histiocytoma (2), sarcoma, lymphoma |
| 776-CPC(2E) | 2/13 (15) | 2/13 (15) | 30 (30) | 30 (30) | Mesothelioma |
| 776-VA(2E) | 3/9 (33) | 10/16 (62) | 30 (24–34) | 28 (23–34) | Mesothelioma |
| Totals: | 19/52 (36) | 38/71 (54) | |||
|
| |||||
| Controls (TC7 cell lysate) | 0/28 (0) | 0/28 (0) | |||
1 × 107 PFU of each virus was inoculated intravenously into 21-day-old weanling hamsters and animals were observed for 8 months. Animals that died less than 4 weeks p.i. were assumed to reflect complications of surgery and were removed from the group count. Both necropsy-proven tumors and animals that died more than 2 months p.i. (late deaths) without a necropsy are reported.
Tumors from 12 animals were examined histologically; tumors contained mixtures of types.
Sarcoma = poorly differentiated sarcoma.
Both SV40 strains SVCPC and VA45-54(2E) were highly oncogenic by the i.v. route of inoculation. SVCPC produced tumors in 50% of necropsied animals and that tumor incidence reached 67% (6/9) when late deaths were included. Strain VA45-54(2E) was most oncogenic, with tumors (confirmed at necropsy) in 75% (9/12) of the animals. Including late deaths, 14 of 17 (82%) animals of this group were affected. In contrast, the single-enhancer version of this virus [VA45-54(1E)] was much less oncogenic. Only 17% (2/12) of the necropsied animals in this group developed tumors, or 33% (5/15) when late deaths were also considered. This observation suggests that the rearranged regulatory region of the two-enhancer virus [VA45-54(2E)] may be an important factor in oncogenesis via the i.v. route of inoculation.
Results obtained with the recombinant constructs revealed a trend suggesting that tumor incidence may be influenced by the parental virus background. Only 33% (3/9) of necropsied animals had tumors following i.v. inoculation of 776-VA(2E) compared to 75% for VA45-54(2E), even though the inoculated viruses shared the same T-ag-C coding region and both had complex regulatory regions. When total numbers of animals with tumors were taken into account (necropsied + late deaths), the tumor incidence was 10 of 16 (62%) for 776-VA(2E), compared to 14 of 17 (82%) for VA45-54(2E). A comparison of tumor development by the matched recombinants 776-VA(2E) and 776-CPC(2E) revealed that the VA45-54 T-ag appeared to be more tumorigenic than the SVCPC T-ag [(3/9, 33% vs. 2/13, 15%; p=0.30) in necropsied animals, respectively, and (10/16, 62% vs. 2/13, 15%; p=0.01) when late deaths are included].
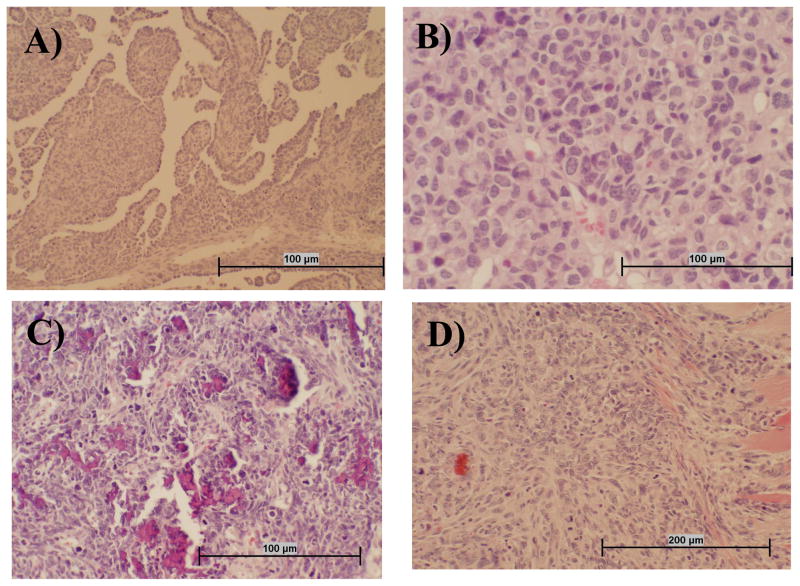
The morphologic types of tumors that developed following i.v. inoculation were identified by histologic examination of available preserved specimens (Table 1). Representative images of the different histologic types of tumors, including mesothelioma, lymphoma, osteosarcoa, and spindle cell sarcoma, are shown in Fig. 1. Multiple mesenchymal tumors including lymphomas, were often observed within a single malignancy. The morphologic diagnosis malignant fibrous histiocytoma was made on several of the tumors (not shown). This is a pleocellular neoplasm that presumably arises from primitive mesenchymal cells.
Fig. 1.
Representative tumor types that developed following i.v. inoculation of SV40. Sections of hamster tissue were collected at necropsy, formalin fixed, and stained with hematoxylin and eosin. Panel A: Mesothelioma tissue is characterized by infiltrative mass of atypical round to spindle cells often having papillary structures that are lined in one layer of round to cuboidal medium-sized cells with prominent nuclei. Panel B: Lymphoma tissue appears as highly cellular tumor tissue composed of homogeneous round cells that often have an indented nucleus and multiple nucleoli. Panel C: Osteosarcoma tissue is composed of atypical pleomorphic cells that are stellate, pyriform, and polyhedral, with tumor cells often oriented around small spicules of pale osteoid. There are also numerous fragments of bone in this section that are not decalcified. Panel D: The spindle cell carcinoma shown has an infiltrative mass into muscle composed of atypical enlongate to spindle cells with prominent nuclei and multiple nucleoli.
Immune response to SV40 T-ag
At the time of necropsy, blood was collected from each animal and serum samples were assayed by indirect immunofluorescence to measure antibody responses to SV40 T-ag (Table 2). The majority of tumor-bearing animals produced detectable levels of T-ag-specific antibody (93% for double-enhancer viruses and 100% for single-enhancer viruses). Median antibody titers for positive sera were similar: 100 (range 5–1000) and 100 (range 10–5000) for tumor-bearing animals exposed to 2E and 1E viruses, respectively. In contrast, viruses with complex or simple regulatory regions differed in their induction of T-ag-specific antibodies in non-tumor-bearing animals (20/20, 100% vs. 11/13, 85%, p=0.0001). In addition, the median antibody titers in the non-tumor-bearing animals were 100 (range 5–1000) for those inoculated with 2E viruses and 10 (range 5–1000) for those exposed to 1E viruses. None of the control animals tested (n=12) had detectable anti-T antibody levels. The antibody responses by the tumor-free animals following i.v. inoculation were similar to those observed in a previous study in which hamsters were inoculated i.p. (Sroller et al., 2008); 73% of non-tumor-bearing animals produced anti-T-ag antibody following i.p. inoculation of two-enhancer viruses, as compared to 41% for those exposed to single-enhancer viruses (p=0.0001).
Table 2.
Antibody responses to SV40 T-ag in Syrian golden hamsters inoculated intravenously with SV40
| Virus regulatory region | Presence of tumors | No. positivea/No. tested (%)
|
|
|---|---|---|---|
| Intravenous inoculationb | Intraperitoneal inoculationc | ||
| 2E (complex) | Yes | 13/14 (93) | 18/18 (100) |
| No | 20/20 (100)d | 99/136 (73)e | |
| 1E (simple) | Yes | 5/5 (100) | 57/64 (89) |
| No | 11/13 (85)d | 35/86 (41)e | |
Antibodies to SV40 T-ag were detected using an indirect immunofluorescence assay (Vilchez et al., 2004).
Sera were collected when tumor-bearing animals were sacrificed or when experiment was terminated 8 months after inoculation. Sera were not available from animals that died without necropsy.
From Sroller et al. (2008).
P = 0.0001.
P = 0.0001.
Hamster lymphoma cell lines
Enlarged spleens were observed at necropsy in several of the animals. Hamster 1087 had been inoculated with VA45-54(2E) and hamsters 1109 and 1113 with 776-VA(2E) (Table 3). The splenomegaly was greatest in hamster 1113 (Fig. 2). Splenomegaly may occur as part of the spleen’s normal functions in immunosurveillance and hematopoiesis, lymphoproliferation in response to viral infections, or infiltrations of leukemic or lymphoma cells. Histologic examination of the spleens from which the cell lines were derived revealed various types of tumor cells. The spleen sections from hamsters 1109 and 1113 that had been inoculated with SV40 strain 776-VA(2E) (Table 3) revealed fibrous sarcoma cells. The spleen from hamster 1087 that was inoculated with VA45-54(2E) contained a hematoma with extravasated erythrocytes and occasional foci of lymphocytes consistent with lymphoma. It is possible that the spleen sections available for microscopic examination did not contain the malignant cells that gave rise to the established lymphoma cell lines.
Table 3.
Characteristics of established hamster lymphoma cell lines
| Hamster No. | SV40 strain inoculated | Sacrificed (months p.i.) | Cell line designationa | Virus production (passage no.)b | Viral genomes per cell (passage no.)b | Ratio of early to late SV40 mRNA (passage no.)b | T-antigen expression |
|---|---|---|---|---|---|---|---|
| 1087 | VA45-54(2E) | 6 | S1087 | No (P2) | 4 (P6) | 55 (P31) | Yes |
| 1109 | 776-VA(2E) | 6 | S1109 | No (P2) | 7 (P4) | 43 (P31) | Yes |
| 1113 | 776-VA(2E) | 8 | S1113 | No (P2) | 6 (P1) | 15 (P33) | Yes |
Cell lines established from enlarged spleens of hamsters.
Passage no., passage level of the cell line tested.
Fig. 2.
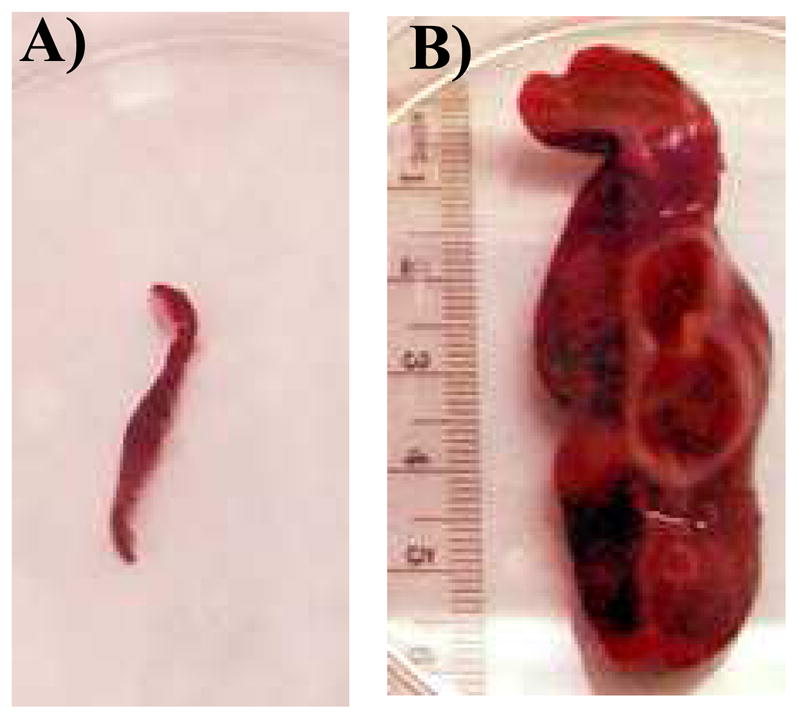
Spleens harvested from tumor-free hamsters (A) and 1113 (B), each at 8 months after i.v. inoculation with SV40 strain 776-VA(2E).
Single-cell suspensions were prepared from the enlarged spleens and cultured as described in Materials and methods. Cell lines were established and designated by the hamster number from which they were derived (S1087, S1109 and S1113). All three cell lines were immortalized in vitro as evidenced by long-term growth in tissue culture (>55 passages). Each cell line displayed a relatively heterogeneous morphology at all passage levels in which some cells were weakly adherent to the culture flask and others remained in suspension.
To rule out the possibility that the splenomegaly observed in the hamsters was related to an antiviral response to active replication of SV40, cells were analyzed for the production of infectious virus. Early passages (P2) of each of the three cell lines were subjected to three cycles of freezing and thawing and the lysates were tested for infectious virus by plaque assay. Infectious SV40 was not detected in any of the cell lines (Table 3).
SV40 DNA was quantified in the lymphoma cell lines using real-time quantitative PCR (RQ-PCR) to amplify T-ag DNA sequences. These values were normalized to cell number as determined by counting cells in a hemacytometer. A range of 4–7 copies per cell of SV40 DNA was detected in the cell lines (Table 3). Quantification of early (T-ag) and late (VP1) viral mRNAs revealed that both early and late transcripts were produced, with the ratio of early-to-late message ranging from 15- to 55-fold among the cell lines (Table 3). The low levels of late mRNA detected combined with the observations of low genome copy numbers and the absence of infectious virus production indicate that the cell lines were not supporting active virus replication (Ernoult-Lange and May, 1983).
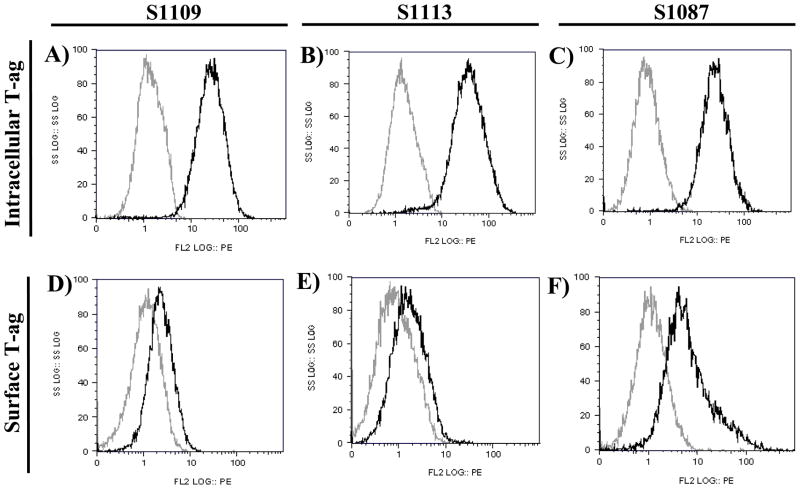
SV40 T-ag is the major transforming protein of the virus and its expression is likely involved in the immortalized phenotype of the hamster lymphomas. Expression of SV40 T-ag was confirmed in all three cell lines using monoclonal antibodies specific for T-ag in both flow cytometry and immunohistochemistry (IHC) assays. Flow cytometry revealed that all three cell lines uniformly expressed high levels of intracellular T-ag (Figs. 3A–C) and relatively low levels of surface T-ag (Figs. 3D–F). IHC staining confirmed that nearly all cells in the established cell lines expressed intranuclear T-ag (not shown). VP1 expression in the cell lines was not detected by immunofluorescence using anti-SV40 VP1 rabbit serum (data not shown).
Fig. 3.
Expression of SV40 T-ag detected by intracellular staining or surface staining and flow cytometry on hamster lymphoma cell lines S1109 (Panels A, D), S1113 (Panels B, E), and S1087 (Panels C, F). Intracellular expression (Panels A–C) was detected using PAb101 (black lines); negative control staining was done with mouse IgG2a (gray lines). Surface expression (Panels D–F) of SV40 T-ag was detected using PAb416 (black lines); negative control staining was done with IgG2a (gray lines).
Derivation of monoclonal antibodies specific to hamster lymphocytes
There are few commercially available immunological reagents that react with subsets of hamster lymphocytes. Monoclonal antibodies (mAbs) specific to hamster lymphocyte markers were developed as described in Materials and methods and were used to identify surface antigens of the lymphoma cell lines. Available antibodies displayed specificities directed to epitopes on the class II major histocompatibility complex (MHC) molecules and markers of B, T, and dendritic cells (Table 4). The optimal working dilution of hybridoma supernatants was determined by staining primary hamster splenocytes and peripheral blood lymphocytes followed by flow cytometric analysis (data not shown).
Table 4.
Derivation and characterization of monoclonal antibodies reactive to hamster lymphocytesa
| Clone name | Immunogenb | % Pos. splenocytes | IgG subclass | Specificity |
|---|---|---|---|---|
| H42A | PBMCs from multiple species | 50c | IgG2a | Class II MHC |
| HAL4A | Lymph node Mononuclear cells | 36 c | IgG3 | Class II MHC |
| HAL16A | Lymph node Mononuclear cells | 48 c | IgG1 | Class II MHC |
| HAT16A | Thymocytes | 67 | IgG2b | Class II MHC |
| HAT17A | Thymocytes | 40 | IgG1 | Class II MHC |
| HAB4A | Peripheral blood leukocytes | 18 c | IgG1 | Dendritic cell |
| HAT24A | Thymocytes | 12 c | IgG1 | T cell |
| HAL3A | Lymph node Mononuclear cells | 29 c | IgG2a | T cell |
| HAL9A | Lymph node Mononuclear cells | 1 | IgG1 | T cell |
| HAB1A | Peripheral blood leukocytes | 0.3 | IgG1 | T cell |
| HAT19A | Thymocytes | 14 | IgG2a | T cell |
| HASA7A | Mononuclear splenocytes | 33 | IgG1 | B cell |
| HAL24A | Lymph node Mononuclear cells | 6 | IgG2a | B cell |
| HAB6A | Peripheral blood leukocytes | 42 | IgG2a | Pan lymphocyte |
| HASA25A | Mononuclear splenocytes | 91 | IgG1 | CD45 |
| BAQ30A | PBMCs from multiple species | 75 | IgG1 | CD18 |
Washington State University Monoclonal Antibody Center.
Derived from normal tissues of Syrian golden hamsters.
Average of three or more experiments.
Identification of lymphoma cell lines of histiocytic origin
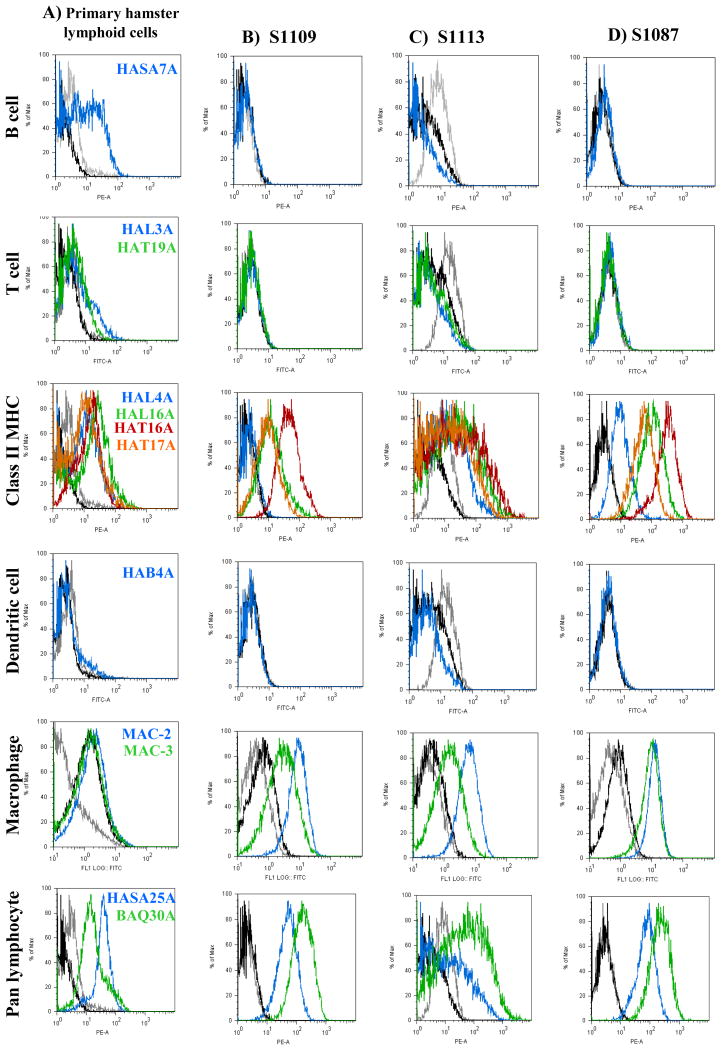
Hamster lymphoma cell lines S1109, S1113 and S1087 and control primary hamster splenocytes were stained with the anti-hamster monoclonal antibodies and were analyzed by flow cytometry (Fig. 4). All the described anti-hamster monoclonal antibodies recognized subpopulations in the primary hamster lymphoid cells (Fig. 4A). The percentage of positive cells for each antibody is shown in Table 4. All three cell lines were negative for expression of B-cell-specific epitopes recognized by HASA7A (Fig. 4B–D) and HAL24A (data not shown). None of the cell lines expressed T cell epitopes recognized by HAL3A, HAT19A (Fig. 4B–D), and HAT24A (data not shown). The cell lines expressed relatively high levels of class II MHC molecules, shown by reactivity with monoclonal antibodies HAL4A, HAL16A, HAT16A, HAT17A (Fig. 4B–D), and H42A (data not shown). Cell line S1113 showed the highest expression of a class II MHC epitope recognized by HAL4A, followed by S1087, whereas binding was undetected on S1109. A dendritic cell marker was not detected on any of the cell lines using antibody HAB4A. None of the cells lines reacted with the HAB6A monoclonal that is specific for a pan lymphocyte marker (data not shown). All three of the cell lines expressed relatively high levels of a CD45 homologue recognized by HASA25A, and a CD18 homologue recognized by BAQ30A (Fig. 4 B–D) (Saalmüller et al., 2005).
Fig. 4.
Surface expression of epitopes specific for lymphocyte subpopulations detected by flow cytometry. Panel A: primary hamster lymphoid cells; Panels B-D: established SV40-positive hamster lymphoma cell lines. For all histograms, negative controls are shown by gray lines representing unstained cells and black lines representing cells stained only with secondary antibody. Cells stained with the specific anti-hamster antibodies are color-coded to the histograms. Primary splenocytes from uninoculated hamsters were used as normal controls for assays with all monoclonal antibodies except MAC-2, for which primary hamster peripheral blood mononuclear cells were used as controls.
To further evaluate the possibility that the hamster lymphoma cell lines might be of B-cell origin, cells were stained with an anti-hamster Ig reagent that recognized the heavy chain of Ig that would be part of a hamster B-cell receptor. Surface Ig was not detected on any of the three lymphoma cell lines. This observation was in agreement with the negative results obtained with the B-cell-specific monoclonals, HASA7A (Fig. 4) and HAL24A. The cell lines were also stained with the MAC-2 and MAC-3 antibodies, commercially available reagents that are specific for a subpopulation of mouse mononuclear phagocytes, including macrophages, and reported to cross-react with hamster cells (Carbone et al., 1989). All three lymphoma cell lines were positive for MAC-2 and MAC-3 expression, although MAC-3 levels were relatively lower on cell lines S1109 and S1113 (Fig. 4B–D). Taken together, the detection of epitopes specific for macrophages and class II MHC molecules suggested that the SV40-transformed cell lines were derived from a histiocytic lineage.
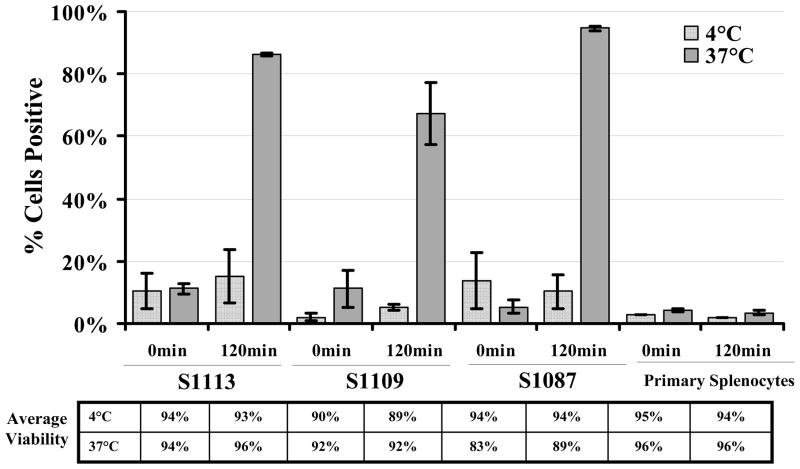
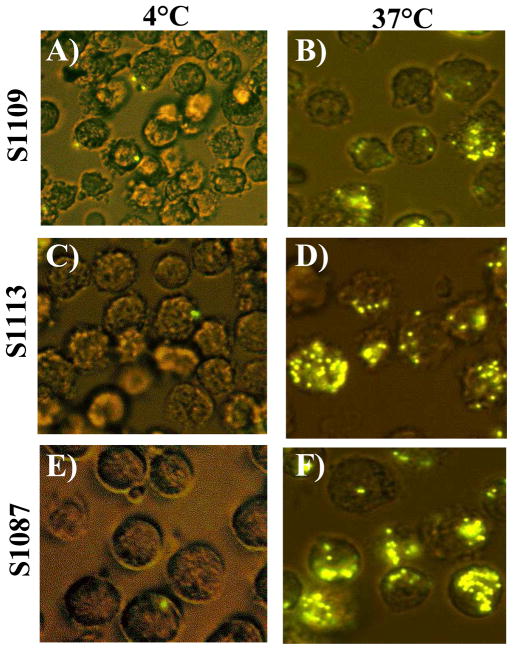
Functional assays were conducted to determine if the hamster lymphoma cell lines displayed phagocytic properties, characteristic of macrophage and dendritic cells. Primary hamster splenocytes and the S1113, S1109 and S1087 cell lines were incubated with fluorescent polystyrene beads at either 4 or 37 °C and analyzed by flow cytometry at 0 and 120 min after addition of the beads (Fig. 5). The human monocyte-macrophage cell line (MM6) was used as a positive control for phagocytic cells (data not shown) and primary hamster splenocytes served as nontransformed controls. All three hamster lymphoma cell lines were capable of taking up the beads at 37 °C, as greater than 70% of the cells were positive (Fig. 5). Cells incubated at 4 °C retained viability but failed to phagocytose beads, ruling out nonspecific surface sticking and showing that the positive results obtained at 37 °C were due to active uptake of beads. Primary hamster splenocytes failed to exhibit detectable levels of phagocytosis at either temperature. High levels of bead uptake were apparent by each of the hamster lymphoma cell lines without activation treatments. Confocal microscopy confirmed the intracellular localization of phagocytosed beads and showed that some cells ingested multiple beads (Fig. 6). Functional phagocytic activity supports the interpretation that the transformed lymphoma cell lines are of histiocytic origin.
Fig. 5.
Phagocytosis by hamster lymphoma cell lines measured by flow cytometry. The percentage of cells that internalized fluorescent-labeled beads, averaged from two independent experiments, are graphed with standard error of the mean. Control samples were incubated at 4 °C. The average viability for each sample from both experiments is shown below the graph.
Fig. 6.
Internalization of phagocytosed beads by hamster lymphoma cell lines measured by confocal microscopy. Cells are shown 120 min after the addition of 30 fluorescent-labeled beads per cell and incubation at either 4°C (Panels A, C, E) or 37 °C (Panels B, D, F). Shown are hamster cell lines S1109 (Panels A, B), S1113 (Panels C, D), and S1087 (Panels E, F). Original magnification, 20×.
Transplantability of SV40-positive hamster lymphoma cell lines
The ability of transformed cells to develop into tumors after being injected into animals is a conventional phenotypic characterization to indicate if malignant transformation has occurred. The tumor incidence and time required for tumor development following inoculation of transformed cells are often related to the cell dose of the inoculum (Lewis et al., 1999; Tevethia and Butel, 1973). To determine if the hamster lymphoma cell lines were tumorigenic, weanling hamsters were inoculated with various doses of lymphoma cells and observed for tumor growths. Each of the three cell lines was capable of inducing solid tumor growths near the subcutaneous site of injection following inoculation of ≥1 × 106 cells, demonstrating that the lymphoma cell lines are tumorigenic in intact animals.
Discussion
This study addressed tumor induction by different strains of SV40 in the Syrian golden hamster model following inoculation by the i.v. route. Previous studies have shown that the types of tumors induced by SV40 in hamsters are influenced by the route of inoculation (Butel and Lednicky, 1999; Cicala et al., 1993; Diamandopoulos, 1973). Recently, it has been demonstrated that viral strains differ in the frequency of tumor formation when virus is delivered intraperitoneally (Sroller et al., 2008). This study confirmed that SV40 is highly oncogenic by the i.v. route (Carbone et al., 1989; Cicala et al., 1993; Diamandopoulos, 1972, 1973), and it revealed that tumor formation is influenced by a combination of factors, including route of exposure and virus strain.
Our previous study demonstrated the significantly greater oncogenicity of viruses with simple regulatory regions (1E) compared to those with complex, rearranged regulatory regions (2E) when inoculated i.p. into weanling hamsters (Sroller et al., 2008). This was not observed in the present study with strain VA45-54 following i.v. inoculation. The viral dose (1 × 107 PFU) was the same in both studies, as was the age of the recipient hamsters (21-day-old weanlings), so the different patterns of oncogenicity must reflect the influence of the route of inoculation. Strain VA45-54(2E) was more tumorigenic than its one-enhancer counterpart (75% vs. 17% of necropsied animals). However, strain SVCPC, a single-enhancer virus that was highly oncogenic by the i.p. route, retained high oncogenicity by i.v. inoculation. To confirm that no mix-up had occurred with the VA45-54 viruses, several tumors were analyzed by PCR using SV40 regulatory region primers. Tumors were found to contain the appropriate regulatory region (1E or 2E) for the virus variants that were inoculated.
Analysis of the results obtained with strain 776-based recombinant viruses suggests that the 776 background lessens the oncogenicity of the VA45-54 strain. Whereas VA45-54(2E) induced tumors in 75% of hamsters, that incidence dropped to 33% when the T-ag region was transferred to a 776(2E) background. This approximate 2-fold reduction in tumor incidence with the same recombinant relative to parental VA45-54(2E) was also observed following i.p. inoculation (Sroller et al., 2008). These observations might reflect functioning of the viral regulatory regions in hamster cells. However, it is possible that SV40 small t-antigen (t-ag) may be functionally involved. The recombinant constructs utilized in this study contained the small t-ag coding region of the backbone virus (i.e., strain 776). There are two amino acid differences in small t-ag between SVCPC and 776 (residues 95 and 109) and three differences in small t-ag between VA45-54 and 776 (residues 95, 104 and 109) (Stewart et al., 1996). An effect of small t-ag on SV40 oncogenicity in hamsters has been reported (Carbone et al., 1989; Cicala et al., 1992; Matthews et al., 1987). Whereas wild-type virus induces a variety of tumor types, small t-ag deletion mutants preferentially induced lymphoid malignancies, suggesting that a small t-ag function is required for SV40 transformation of nondividing cells in vivo. Future studies are needed to address a potential role of the small t-ag in VA45-54 oncogenicity.
A broad spectrum of tumor types was obtained in this study following i.v. inoculation of SV40, as observed previously by others (Cicala et al., 1993; Diamandopoulos, 1972, 1973). A likely explanation is that the i.v. route efficiently delivers the virus into contact with a variety of susceptible cell types throughout the host. This is supported by the general trend of increased oncogenicity by SV40 strains following i.v. inoculation as compared to i.p. administration. For example, strain VA45-54(2E) was highly oncogenic when inoculated i.v. (75% of necropsied animals) but less so when inoculated i.p. (20–23%) (Sroller et al., 2008; Vilchez et al., 2004).
The majority of non-tumor-bearing animals produced durable antibodies to SV40 T-ag that were detectable at the termination of the experiments described here. We suggest that this high-frequency antibody response to T-ag in the absence of tumors reflects viral replication in some unidentified cells in the hamsters. Notably, the exposure to viruses with complex regulatory regions and viruses with single enhancers resulted in a statistically different frequency of T-antibody production (p=0.0001). This is in agreement with what had been observed following i.p. inoculation (Sroller et al., 2008). We speculate this difference in T-antibody production in non-tumor-bearing animals reflects dissimilar rates of SV40 replication and host response to infection between viruses with simple and complex regulatory regions. This hypothesis is supported by studies in cell culture which have shown that SV40 viruses with complex regulatory regions replicate faster and to higher titers than those with simple regulatory regions (Lednicky and Butel, 1997). In addition, studies with murine polyoma virus have shown that faster-growing viral variants are associated with high viral replication and a strong immune response, whereas slower-growing variants persist with low-level viral replication and no notable host response (Rochford et al., 1992).
This investigation confirmed that lymphoid cells can be malignantly transformed in vivo by SV40 under certain conditions (Carbone et al., 1989; Cicala et al., 1992; Diamandopoulos, 1972, 1973). Three spleen-derived lymphoma cell lines were established and found to be immortalized and tumorigenic. This in vivo transformation may predict a common outcome of SV40 infection of lymphocytes as the same types of tumors were induced in three out-bred animals, inoculated with two variants of SV40. It showed that cells of the monocyte lineage are susceptible to infection by wild-type SV40, as reported for a small t-ag deletion mutant of the virus (Carbone et al., 1989; Matthews et al., 1987). As surface immunophenotyping provides a direct method for characterizing populations of lymphoid cells, the newly developed anti-hamster antibodies described here were valuable tools for analyzing cells susceptible to infection and transformation in vivo by SV40. All three lymphoma cell lines appear to possess characteristics of histiocytic origin, including the ability to phagocytose particles, expression of high surface levels of MHC class II, and expression of the MAC-2 and MAC-3 antigens, markers of a subpopulation of mouse mononuclear phagocytes previously detected on other hamster lymphomas (Carbone et al., 1989; Cicala et al., 1992).
This study demonstrated the lymphomagenic property of SV40 strain VA45-54, the viral isolate utilized by Diamandopoulos (1972, 1973) in the original descriptions of lymphoma induction in hamsters by SV40. The virus is not exclusively lymphomagenic, as other types of malignancies, such as osteosarcomas, also developed. It remains to be determined what physiologic processes or viral characteristics contribute to lymphomagenesis by this strain when inoculated intravenously. Perhaps SV40 particles were internalized by circulating cells of histiocytic origin, such as monocytes, that trafficked to the spleen and differentiated into mature cells. Alternatively, other cells may have carried the virus to the spleen (and other organs) where target cells, such as the monocyte-derived cells recovered here, became infected. Expression of the VA45-54 early region presumably contributed to cell proliferation and transformation, and expression of the viral early proteins may have been influenced by the rearranged regulatory region of VA45-54(2E). This working model of SV40-induced lymphomagenesis will guide future studies into which additional hamster lymphocyte subsets are susceptible to SV40-induced infection and transformation and may provide insights into SV40 association with human lymphomas. The tumorigenic hamster lymphoma cell lines described here that retain phenotypic properties of cell origin will be valuable tools with which to study tumor progression and to evaluate immunotherapeutic drugs and treatment regimens for lymphomas.
Materials and methods
Viruses
SV40 strains to evaluate tumorigenicity following i.v. inoculation into hamsters included: SVCPC (GenBank, AF156108) and VA45-54, variants 1E and 2E (GenBank, AF156105). In addition, recombinant viruses were included: 776-CPC(2E) and 776-VA(2E). These recombinants were generated by cloning the large T-ag variable domain (T-ag-C) sequence from SVCPC and VA45-54, respectively, into the genome of SV40 strain 776 with a complex (2E) regulatory region (Sroller et al., 2008). Virus stocks were produced and quantified in TC-7 cells (Butel et al., 1999). Strain 776 was isolated from an adenovirus type 1 vaccine seed stock (Sweet and Hilleman, 1960). Strain VA45-54 was originally isolated from cultures of primary monkey kidney cells (Girardi et al., 1962) and has been detected in a human osteosarcoma (Lednicky et al., 1997). Strain SVCPC, with a simple (1E) regulatory region, has been isolated twice from human brain tumors (Krieg and Scherer, 1984; Lednicky et al., 1995) and has been detected in several human malignant specimens (Arrington et al., 2004; Vilchez et al., 2002a, 2002b). Both strains, VA45-54 and SVCPC, were found in the USSR poliovaccine seed stock (Cutrone et al., 2005).
Induction of tumors in hamsters
Weanling 21-day-old male and female Syrian golden hamsters were purchased from Harlan Sprague Dawley (Indianapolis, IN) and were housed in the biohazard facility at the Center for Comparative Medicine at Baylor College of Medicine. These studies were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine. Animals were maintained according to approved protocols and all care was in accordance with established national guidelines as outlined in DHEW Publication No. (NIH) 78-23, “Guide for the Care and Use of Laboratory Animals”. Using aseptic techniques, experimental animals under anesthesia were inoculated via the femoral vein with 1 × 107 PFU of virus in 0.2 ml. Control animals were inoculated similarly with lysates of uninfected TC7 cells. Animals were observed thrice weekly and were sacrificed when there was evidence of illness or tumor formation or at 8 months p.i. at the termination of the experiment. Necropsy was conducted following exsanguination by cardiac puncture with the animals anesthetized. Samples of normal and tumor tissue were collected, fixed in 10% buffered formalin, and paraffin embedded. Tissue sections were stained with hematoxylin and eosin and examined microscopically without knowledge of virus inoculum.
Establishment of splenocyte-derived cell lines
Enlarged spleens were observed in several animals. Single-cell suspensions were prepared by pushing fresh spleen tissue through a cell strainer (Falcon) into RPMI 1640 media (Gibco) supplemented with 10% fetal calf serum and insulin-transferrin-sodium selenite (Sigma). Cells were collected and incubated for 5 min at 25 °C in red cell lysis buffer (10 mM KHCO3, 150 mM NH4Cl, 0.1 mM EDTA pH 8.0). The recovered lymphocytes were washed and cultured in the same RPMI media. Cells were dissociated from flasks by pipetting or using cell scrapers (Sarstedt) and were subcultured once or twice per week for up to 60 passages.
Assays for SV40 DNA, RNA and infectious virus in hamster lymphoma cell lines
Cellular DNA was extracted following a proteinase K/phenol extraction protocol (Lednicky and Butel, 1998). RQ-PCR to detect and quantify SV40 DNA genome contents was performed as described (McNees et al., 2005). To quantify viral transcripts, total RNA was extracted from the lymphoma cell lines, using the RNAqueous-4PCR isolation kit (Ambion) followed by DNAse I treatment. RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System for RT-PCR kit (Invitrogen) with random hexamer primers. The SV40 early mRNAs and late mRNAs were detected by PCR using first-stranded cDNA and primer sets SVEARL1/2 and SVLATE1/2, designed to span SV40 introns so that PCR products from spliced mRNAs could be identified on gels. Quantification of SV40 early and late transcripts was made by RQ-PCR using the above cDNAs and targeting the large T-ag gene and the VP1 gene (McNees et al., 2005). An endogenous control was used for normalization by amplifying 18S ribosomal RNA using primers and probe targeting a sequence conserved across many species (Jelinek and Prchal, 2004). RNA processed without reverse transcriptase served as control for SV40 DNA contamination of RNA samples. Primer and probe sequences are shown in Table 5. Rescue of infectious virus from the hamster lymphoma cell lines was attempted as described (Sroller et al., 2008).
Table 5.
Sequences of primers and probes used in conventional PCR and RQ-PCR assays
| Oligonucleotides (5′ → 3′)a | Reference positionsb | Temperature | Size (bp) PCR product | |
|---|---|---|---|---|
| PCR, SV40 early region | ||||
| SVEARL1: | CAGCAGTAGCCTCATCATCAC | nt 4463–4483 | 52 | 669, viral DNA |
| SVEARL2: | GCAGCTAATGGACCTTCTAGGTCT | nt 5131–5108 | 602, small t-ag cDNA 322, large T-ag cDNA |
|
| PCR, SV40 late region | ||||
| SVLATE1: | CTCCGTTAAGGTTCGTAGGTC | nt 364–384 | 52 | 1283, viral DNA |
| SVLATE2: | CCTCAGTGAAGCTGTCTACTC | nt 1646–1626 | 346, 16S RNA cDNA | |
| RQ-PCR, SV40 early region, SVPENT primers and probe | ||||
| FP: | GATGGCATTTCTTCTGAGCAAA | nt 4486–4507 | N/A | N/A |
| RP: | GAATGGGAGCAGTGGTGGAA | nt 4550–4531 | ||
| Probe: | FAM-CAGGTTTTCCTCATTAAA-MGB | nt 4509–4526 | ||
| RQ-PCR, SV40 late region, SVVP1 primers and probe | ||||
| FP: | GGGCCCTTGTGCAAAGC | nt 2258–2274 | N/A | N/A |
| RP: | GTTGGTAAACAGCCCACAAATG | nt 2323–2302 | ||
| Probe: | VIC-ACAGCTTGTATGTTTCTG-MGB | nt 2277–2294 | ||
| RQ-PCR, human 18S ribosomal RNA | ||||
| FP: | TCGAGGCCCTGTAATTGGAA | N/A | N/A | N/A |
| RP: | CCCTCCAATGGATCCTCGTT | |||
| Probe: | FAM-AGTCCACTTTAAATCCTT-MGB | |||
FP = forward primer; RP = reverse primer.
Reference nucleotide positions in SV40 strain 776 (SV40-776).
PCR annealing temperature.
N/A = not applicable.
Detection of SV40 T-ag by flow cytometry
Intracellular and surface expression of SV40 T-ag was detected in the hamster lymphoma cell lines by flow cytometry. Intracellular staining was performed by fixing cells in 2% paraformaldehyde for 10 min at 37 °C, followed by 90% ice-cold ethanol overnight. Cells were washed with 1% bovine serum albumin (BSA) in phosphate buffered saline (PBS) and resuspended in the same buffer containing anti-T-ag monoclonal antibody PAb101 (Santa Cruz Biotechnology). Cells were incubated at 25 °C for 30 min, washed, and resuspended in buffer containing goat anti-mouse F(ab)′2-phycoerithrin (PE) (Southern Biotechnology Associates). For surface staining of T-ag, unfixed cells were stained on ice for 30 min with PAb416 (Oncogene Research Products) followed by goat anti-mouse F(ab)′2-PE. All samples were analyzed at the Flow Cytometry Core Facility at Baylor College of Medicine using a Beckman Coulter EPICS XL-MCL and FlowJo software (FlowJo, LLC, Ashland, OR).
Detection of SV40 VP1 by immunofluorescence
Staining for VP1 was performed by preparing Shandon Cytospin slides (Thermo Fisher Scientific) with each hamster lymphoma cell line for immunofluorescence staining per the manufacturer’s recommendations. Slides were processed as described previously and stained with anti-SV40 VP1 rabbit serum (Butel et al., 1984). A positive control of permissive SV40-infected TC7 cells was included, as well as two negative controls that were stained with either fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin (Ig) only, or with an anti-intact mouse mammary tumor virus rabbit serum followed by FITC-goat anti-rabbit.
Monoclonal antibodies against hamster lymphocytes
Mouse mAbs specific to hamster leukocyte differentiation molecules (hLDM) were produced by the Washington State University Monoclonal Antibody Center in the College of Veterinary Medicine, Washington State University, Pullman, WA. The mAbs were developed from 4 sets of BALB/c mice immunized with preparations of hamster peripheral blood leukocytes (HAB), thymocytes (HAT), lymph node mononuclear cells (HAL), or a mixture of nonadherent and adherent mononuclear splenocytes (HASA). Following 4 subcutaneous injections of 4 × 106 cells given every 2 weeks, the mice were given a final booster injection intravenously 3 days before spleen cells were harvested and fused with X63 Ag8.653 myeloma cells (Hamilton and Davis, 1995). Flow cytometry was used to screen the primary culture supernatants for hybridomas producing mAbs to hLDMs expressed on all or subsets of PBLs. Positive cultures were expanded and cryopreserved. Supernatants collected at the time of cryopreservation were screened again to select hybridomas for cloning and establishment of cell lines producing mAbs of interest. Two-color flow cytometry and a mAb (H42A, IgG2a) specific for a highly conserved epitope expressed on class II MHC molecules was used to identify mAbs specific for hamster MHC II molecules (Table 4) (Davis et al., 1987, 1995). The mAbs to class II MHC molecules were used in 2-color flow cytometry to identify mAbs specific for hLDMs expressed on T or B lymphocytes. Specificity was verified by 2-color flow cytometry that showed the predicted anti-T and anti-B mAbs labeled mutually exclusive populations of lymphocytes. Analysis revealed HAB1A recognized molecules expressed on a subset of HAL3A+ T lymphocytes and that HAB4A and HAT4A recognized a molecule expressed on a small subset (~3% of peripheral blood mononuclear cells) of cells predicted to be dendritic cells (data not shown). The HAB4A+ HAT4A+ cells were MHC II+.
Hamster lymphoma cell lines established from animals inoculated with SV40 were stained with the hybridoma supernatants at optimized dilutions, followed by anti-mouse isotype-specific fluorescent conjugates [anti-mouse IgG-PE, anti-mouse IgG1-FITC, anti-mouse IgG2a-PE (Southern Biotechnology Associates)]. Cell lines were stained also with FITC-conjugated rabbit anti-Syrian hamster IgG (H+L) (Jackson ImmunoResearch, MAC-2, and MAC-3, rat monoclonal antibodies specific for a subpopulation of mouse mononuclear phagocytes, including macrophages (Cedarlane, Ont., Canada), followed by Alexa Fluor 488 goat anti-rat IgG (H+L). Cells were analyzed at the Flow Cytometry Core Facility at Baylor College of Medicine.
Phagocytosis assay
Phagocytosis assays were performed by incubating 2 × 106 cells in suspension in 1 ml RPMI growth media with fluorescent polystyrene microspheres (Molecular Probes, Inc., Eugene, OR) at 30 beads per cell, a ratio determined to be optimal by dilution experiments (data not shown). After incubation at 4 or 37 °C for 0–120 min, cells were transferred to 15-ml conical tubes and washed 5 times with 5 ml of PBS containing 3% BSA. Cells were then resuspended in 0.5 ml of 1% BSA in PBS buffer and analyzed by flow cytometry. Additionally, cells were transferred to a 24-well plate, visualized and photographed using a Zeiss LSM 51D META confocal microscope (Carl Zeiss, Germany), and images were processed using LSM Image VisArt (Carl Zeiss, Inc., Thornwood, NY).
Tumorigenicity assays
Cell lines were harvested from culture, washed, and resuspended to the desired density in PBS. Cell suspensions in 0.1 ml were inoculated subcutaneously in the scapular region of weanling hamsters. Animals were observed twice weekly for tumor growth.
Antibody responses
Serum antibodies against SV40 T-ag were detected by an indirect immunofluorescence assay using SV40-transformed T-ag-positive cells as the target. Antibody titers were determined as the highest serum dilution that gave a detectable T-ag reaction (Sroller et al., 2008).
Statistical analysis
Group proportions among animal groups were tested with the Z test of proportions. Significance was defined as a p-value of <0.05.
Acknowledgments
This study was supported in part by grants R21 CA96951 and R01 CA104818 (J.S.B.) from the National Cancer Institute, T32 AI07471 from the National Institute of Allergy and Infectious Diseases (A.L.M.), and R25 GM56929 from the National Institute of General Medical Sciences (T.C.H.). Grants P30 AI36211 and P30 CA125123 from the National Institutes of Health provided core support. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, the National Institute of General Medical Sciences, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Adrienne L. McNees, Email: amcnees@bcm.edu.
Regis A. Vilchez, Email: regis.vilchez@spcorp.com.
Tiffany C. Heard, Email: heard@bcm.edu.
Vojtech Sroller, Email: vsroller@bcm.edu.
Connie Wong, Email: cwong@bcm.edu.
Alan J. Herron, Email: herron@bcm.edu.
Mary J. Hamilton, Email: mhamilto@vetmed.wsu.edu.
William C. Davis, Email: davisw@vetmed.wsu.edu.
Janet S. Butel, Email: jbutel@bcm.edu.
References
- Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M, Sriha B, Mokni M, Korbi S. Presence of simian virus 40 DNA sequences in diffuse large B-cell lymphomas in Tunisia correlates with aberrant promoter hypermethylation of multiple tumor suppressor genes. Int J Cancer. 2007;121:2693–2702. doi: 10.1002/ijc.23038. [DOI] [PubMed] [Google Scholar]
- Arrington AS, Moore MS, Butel JS. SV40-positive brain tumor in scientist with risk of laboratory exposure to the virus. Oncogene. 2004;23:2231–2235. doi: 10.1038/sj.onc.1207341. [DOI] [PubMed] [Google Scholar]
- Butel JS. SV40, human infections, and cancer: emerging concepts and causality considerations. In: Khalili K, Jeang KT, editors. Viral Oncology: Basic Science and Clinical Applications. Wiley-Blackwell; 2008. in press. [Google Scholar]
- Butel JS, Jafar S, Wong C, Arrington AS, Opekun AR, Finegold MJ, Adam E. Evidence of SV40 infections in hospitalized children. Hum Pathol. 1999;30:1496–1502. doi: 10.1016/s0046-8177(99)90173-9. [DOI] [PubMed] [Google Scholar]
- Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J Natl Cancer Inst. 1999;91:119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- Butel JS, Tevethia SS, Melnick JL. Oncogenicity and cell transformation by papovavirus SV40: The role of the viral genome. Adv Cancer Res. 1972;15:1–55. doi: 10.1016/s0065-230x(08)60371-1. [DOI] [PubMed] [Google Scholar]
- Butel JS, Wong C, Medina D. Transformation of mouse mammary epithelial cells by papovavirus SV40. Exp Mol Pathol. 1984;40:79–108. doi: 10.1016/0014-4800(84)90068-6. [DOI] [PubMed] [Google Scholar]
- Carbone M, Lewis AM, Jr, Matthews BJ, Levine AS, Dixon K. Characterization of hamster tumors induced by simian virus 40 small t deletion mutants as true histiocytic lymphomas. Cancer Res. 1989;49:1565–1571. [PubMed] [Google Scholar]
- Cicala C, Pompetti F, Carbone M. SV40 induces mesotheliomas in hamsters. Am J Pathol. 1993;142:1524–1533. [PMC free article] [PubMed] [Google Scholar]
- Cicala C, Pompetti F, Nguyen P, Dixon K, Levine AS, Carbone M. SV40 small t deletion mutants preferentially transform mononuclear phagocytes and B lymphocytes in vivo. Virology. 1992;190:475–479. doi: 10.1016/0042-6822(92)91237-o. [DOI] [PubMed] [Google Scholar]
- Coe JE, Green I. B-cell origin of hamster lymphoid tumors induced by simian virus 40. J Natl Cancer Inst. 1975;54:269–270. doi: 10.1093/jnci/54.1.269. [DOI] [PubMed] [Google Scholar]
- Cutrone R, Lednicky J, Dunn G, Rizzo P, Bocchetta M, Chumakov K, Minor P, Carbone M. Some oral poliovirus vaccines were contaminated with infectious SV40 after 1961. Cancer Res. 2005;65:10273–10279. doi: 10.1158/0008-5472.CAN-05-2028. [DOI] [PubMed] [Google Scholar]
- Davis WC, Davis JE, Hamilton MJ. Use of monoclonal antibodies and flow cytometry to cluster and analyze leukocyte differentiation molecules. In: Davis WC, editor. Methods in Molecular Biology, vol. 45: Monoclonal Antibody Protocols. Humana Press, Inc; Totowa, N.J: 1995. pp. 149–167. [DOI] [PubMed] [Google Scholar]
- Davis WC, Marusic S, Lewin HA, Splitter GA, Perryman LE, McGuire TC, Gorham JR. The development and analysis of species specific and cross reactive monoclonal antibodies to leukocyte differentiation antigens and antigens of the major histocompatibility complex for use in the study of the immune system in cattle and other species. Vet Immunol Immunopathol. 1987;15:337–376. doi: 10.1016/0165-2427(87)90005-5. [DOI] [PubMed] [Google Scholar]
- Diamandopoulos GT. Leukemia, lymphoma, and osteosarcoma induced in the Syrian golden hamster by simian virus 40. Science. 1972;176:173–175. doi: 10.1126/science.176.4031.173. [DOI] [PubMed] [Google Scholar]
- Diamandopoulos GT. Induction of lymphocytic leukemia, lymphosarcoma, reticulum cell sarcoma, and osteogenic sarcoma in the Syrian golden hamster by oncogenic DNA simian virus 40. J Natl Cancer Inst. 1973;50:1347–1365. doi: 10.1093/jnci/50.5.1347. [DOI] [PubMed] [Google Scholar]
- Ernoult-Lange M, May E. Evidence of transcription from the late region of the integrated simian virus 40 genome in transformed cells: location of the 5′ ends of late transcripts in cells abortively infected and in cells transformed by simian virus 40. J Virol. 1983;46:756–767. doi: 10.1128/jvi.46.3.756-767.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman ZH, Lednicky JA, Fox GE, Willson RC, White ZS, Halvorson SJ, Wong C, Lewis AM, Jr, Butel JS. Phylogenetic analysis of polyomavirus simian virus 40 from monkeys and humans reveals genetic variation. J Virol. 2004;78:9306–9316. doi: 10.1128/JVI.78.17.9306-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Butel JS, Carbone M. SV40 and human tumours: Myth, association or causality? Nat Rev Cancer. 2002;2:957–964. doi: 10.1038/nrc947. [DOI] [PubMed] [Google Scholar]
- Girardi AJ, Sweet BH, Slotnick VB, Hilleman MR. Development of tumors in hamsters inoculated in the neonatal period with vacuolating virus, SV40. Proc Soc Exp Biol Med. 1962;109:649–660. doi: 10.3181/00379727-109-27298. [DOI] [PubMed] [Google Scholar]
- Hamilton MJ, Davis WC. Culture conditions that optimize outgrowth of hybridomas. In: Davis WC, editor. Methods in Molecular Biology, vol. 45: Monoclonal Antibody Protocols. Humana Press Inc; Totowa, N.J: 1995. pp. 17–28. [DOI] [PubMed] [Google Scholar]
- Jelinek J, Prchal JT. Oxygen-dependent regulation of erythropoiesis. Methods Enzymol. 2004;381:201–210. doi: 10.1016/S0076-6879(04)81014-0. [DOI] [PubMed] [Google Scholar]
- Krieg P, Scherer G. Cloning of SV40 genomes from human brain tumors. Virology. 1984;138:336–340. doi: 10.1016/0042-6822(84)90357-x. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J Gen Virol. 1997;78:1697–1705. doi: 10.1099/0022-1317-78-7-1697. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Consideration of PCR methods for the detection of SV40 in tissue and DNA specimens. Dev Biol Stand. 1998;94:155–164. [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Simian virus 40 regulatory region structural diversity and the association of viral archetypal regulatory regions with human brain tumors. Semin Cancer Biol. 2001;11:39–47. doi: 10.1006/scbi.2000.0345. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Garcea RL, Bergsagel DJ, Butel JS. Natural simian virus 40 strains are present in human choroid plexus and ependymoma tumors. Virology. 1995;212:710–717. doi: 10.1006/viro.1995.1529. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Stewart AR, Jenkins JJ, III, Finegold MJ, Butel JS. SV40 DNA in human osteosarcomas shows sequence variation among T-antigen genes. Int J Cancer. 1997;72:791–800. doi: 10.1002/(sici)1097-0215(19970904)72:5<791::aid-ijc15>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Lewis AM, Jr, Alling DW, Banks SM, Soddu S, Cook JL. Evaluating virus-transformed cell tumorigenicity. J Virol Methods. 1999;79:41–50. doi: 10.1016/s0166-0934(98)00182-7. [DOI] [PubMed] [Google Scholar]
- MacKenzie J, Wilson KS, Perry J, Gallagher A, Jarrett RF. Association between simian virus 40 DNA and lymphoma in the United Kingdom. J Natl Cancer Inst. 2003;95:1001–1003. doi: 10.1093/jnci/95.13.1001. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Levine AS, Dixon K. Deletion mutations in the small t antigen gene alter the tissue specificity of tumors induced by simian virus 40. J Virol. 1987;61:1282–1285. doi: 10.1128/jvi.61.4.1282-1285.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees AL, White ZS, Zanwar P, Vilchez RA, Butel JS. Specific and quantitative detection of human polyomaviruses BKV, JCV, and SV40 by real time PCR. J Clin Virol. 2005;34:52–62. doi: 10.1016/j.jcv.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Rochford R, Moreno JP, Peake ML, Villarreal LP. Enhancer dependence of polyomavirus persistence in mouse kidneys. J Virol. 1992;66:3287–3297. doi: 10.1128/jvi.66.6.3287-3297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalmüller A, Lunney JK, Daubenberger C, Davis W, Fischer U, Göbel TW, Griebel P, Hollemweguer E, Lasco T, Meister R, Schuberth HJ, Sestak K, Sopp P, Steinbach F, Xiao-Wei W, Aasted B. Summary of the animal homologue section of HLDA8. Cell Immunol. 2005;236:51–58. doi: 10.1016/j.cellimm.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Schüler F, Dölken SC, Hirt C, Dölken MT, Mentel R, Gürtler LG, Dölken G. No evidence for simian virus 40 DNA sequences in malignant non-Hodgkin lymphomas. Int J Cancer. 2006;118:498–504. doi: 10.1002/ijc.21346. [DOI] [PubMed] [Google Scholar]
- Shivapurkar N, Harada K, Reddy J, Scheuermann RH, Xu Y, McKenna RW, Milchgrub S, Kroft SH, Feng Z, Gazdar AF. Presence of simian virus 40 DNA sequences in human lymphomas. Lancet. 2002;359:851–852. doi: 10.1016/S0140-6736(02)07921-7. [DOI] [PubMed] [Google Scholar]
- Sroller V, Vilchez RA, Stewart AR, Wong C, Butel JS. Influence of the viral regulatory region on tumor induction by simian virus 40 in hamsters. J Virol. 2008;82:871–879. doi: 10.1128/JVI.01626-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AR, Lednicky JA, Benzick US, Tevethia MJ, Butel JS. Identification of a variable region at the carboxy terminus of SV40 large T-antigen. Virology. 1996;221:355–361. doi: 10.1006/viro.1996.0386. [DOI] [PubMed] [Google Scholar]
- Stewart AR, Lednicky JA, Butel JS. Sequence analyses of human tumor-associated SV40 DNAs and SV40 viral isolates from monkeys and humans. J Neurovirol. 1998;4:182–193. doi: 10.3109/13550289809114518. [DOI] [PubMed] [Google Scholar]
- Stratton K, Almario DA, McCormick MC. Immunization Safety Review: SV40 Contamination of Polio Vaccine and Cancer. The National Academies Press; Washington, DC: 2003. [PubMed] [Google Scholar]
- Sui LF, Williamson J, Lowenthal RM, Parker AJC. Absence of simian virus 40 (SV40) DNA in lymphoma samples from Tasmania, Australia. Pathology. 2005;37:157–159. doi: 10.1080/00313020500058474. [DOI] [PubMed] [Google Scholar]
- Sweet BH, Hilleman MR. The vacuolating virus, S.V.40. Proc Soc Exp Biol Med. 1960;105:420–427. doi: 10.3181/00379727-105-26128. [DOI] [PubMed] [Google Scholar]
- Tevethia SS, Butel JS. Induction of common transplantation antigen by various isolates of papovavirus SV40 and by virus rescued from transformed cells. Intervirology. 1973;2:200–205. doi: 10.1159/000149424. [DOI] [PubMed] [Google Scholar]
- Vilchez RA, Brayton CF, Wong C, Zanwar P, Killen DE, Jorgensen JL, Butel JS. Differential ability of two simian virus 40 strains to induce malignancies in weanling hamsters. Virology. 2004;330:168–177. doi: 10.1016/j.virol.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Vilchez RA, Butel JS. Emergent human pathogen simian virus 40 and its role in cancer. Clin Microbiol Rev. 2004;17:495–508. doi: 10.1128/CMR.17.3.495-508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez RA, Lednicky JA, Halvorson SJ, White ZS, Kozinetz CA, Butel JS. Detection of polyomavirus simian virus 40 tumor antigen DNA in AIDS-related systemic non-Hodgkin lymphoma. J Acquir Immune Defic Syndr. 2002a;29:109–116. doi: 10.1097/00042560-200202010-00001. [DOI] [PubMed] [Google Scholar]
- Vilchez RA, Madden CR, Kozinetz CA, Halvorson SJ, White ZS, Jorgensen JL, Finch CJ, Butel JS. Association between simian virus 40 and non-Hodgkin lymphoma. Lancet. 2002b;359:817–823. doi: 10.1016/S0140-6736(02)07950-3. [DOI] [PubMed] [Google Scholar]
- Zekri AR, Mohamed W, Bahnassy A, Refat L, Khaled M, Shalaby S, Hafez M. Detection of simian virus 40 DNA sequences in Egyptian patients with different hematological malignancies. Leuk Lymphoma. 2007;48:1828–1834. doi: 10.1080/10428190701534408. [DOI] [PubMed] [Google Scholar]