Abstract
The organization of actin filaments into higher-ordered structures governs eukaryotic cell shape and movement. Global actin network size and architecture is maintained in a dynamic steady-state through regulated assembly and disassembly. Here, we used experimentally defined actin structures in vitro to investigate how the activity of myosin motors depends on network architecture. Direct visualization of filaments revealed myosin-induced actin network deformation. During this reorganization myosins selectively contracted and disassembled anti-parallel actin structures while parallel actin bundles remained unaffected. The local distribution of nucleation sites and the resulting orientation of actin filaments appeared to regulate the scalability of the contraction process. This “orientation selection” mechanism for selective contraction and disassembly suggests how the dynamics of the cellular actin cytoskeleton can be spatially controlled by actomyosin contractility.
Actin filament networks comprise a large variety of different structures. Their spatial organization supports complex cell shape regulation. The dynamics and mechanical properties of these structures result from the assembly of polarized actin filaments. Filopodia, retraction fibers and centripetal fibers, are built of parallel filaments (1, 2). Stress fibers and transverse arcs have filaments arranged in anti-parallel orientations (3, 4). The lamellipodium is a dense array of branched filaments (5).
The global architecture of the actin cytoskeleton is maintained through coordinated actions of a large number of regulatory proteins that modulate filament assembly and disassembly (6), as well as through contractility driven by myosin motor proteins (7). Myosin motor proteins can also promote filament disassembly (8). Collectively, these observations have supported a mechanism in which the coupling between myosin contractility and filament disassembly ensures a temporal synchrony between actin retrograde flow at the front and filament disassembly at the rear of migrating cells (9).
Central to this coupling mechanism is that filaments are selected for contraction or disassembly, but it is not known what factors determine the response to myosin contractile forces (10). Here we used micropatterning methods to assemble geometrically controlled and polarized actin filament networks (11) to evaluate how the overall polarity of actin filament architectures determines their response – reorganization and/or disassembly – to myosin contractile forces.
Actin filament growth on bar-shaped micropatterns covered with the Wiskott–Aldrich syndrome protein (WASp) pWA domain, an actin promoting factors, lead to the formation of a dense meshwork on the micropatterned region and parallel array of filaments with barbed ends oriented away from the nucleation site out of this region (11) (movie S1). Addition of myosins to the polymerization mix, including Arp2/3 complex, profilin and actin monomers, allowed us to investigate the contraction of this network (Fig. S1). We used double-headed (HMM ) myosin VI (12) a processive pointed-end directed motor that could sustain continuous force and motility without the need for self-assembly into minifilaments.
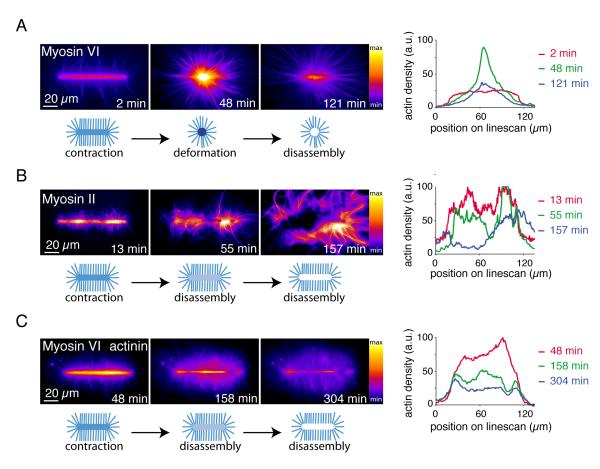
Green Fluorescent Protein (GFP)-tagged myosins and Alexa-568-labeled actin monomers allowed real-time tracking of actin growth and myosin-induced reorganization (Fig. 1). Myosins associated with the network and induced a clear two-phase process constituted by the deformation of actin networks followed by a massive filament disassembly of the condensed central meshwork (Fig. 1A and movie S2, short bars). Depending on the geometry of the pattern, this two-phase process could lead to the formation of a disassembly wave (Fig. S2, long bars). We then tested if a barbed-end directed myosin had a similar effect on network reorganization. Muscle myosin II bipolar filaments induced a two phase deformation-disassembly of the network similar to myosin VI, although the extent of deformation before disassembly was local and less pronounced (Fig. 1B and movie S3), presumably because of resistance from filament crosslinking (13). Consistent with this interpretation, the actin filament cross-linker, α-actinin, also minimized myosinVI-induced macroscopic deformation before network disassembly (Figs. 1C and S3, movie S4). Varying myosin concentration revealed that deformation and disassembly occurred above different concentration thresholds depending on the reticulated actin network (Fig. S3)
Fig. 1.
Myosin induced actin meshwork contraction and disassembly
(A) Time-series of myosin VI-induced network contraction on a bar-shaped micropattern. Actin filaments were visualized with fluorescent monomers. “Fire” look-up table color-coding reveals variations in actin network densities, quantified with a linescan along the bar at different time points. Actin density peaks because of network deformation after 48 minutes then falls off because of network disassembly. (B) Same as (A) with muscle myosin II-induced contraction. (C) Same as (A) with 100 nM alpha-actinin in the polymerization mix.
Parallel and polarized filaments emerging from the micropatterned regions with their barbed ends oriented outward (11) did not contract and disassemble with either myosin VI or II (Fig. 1A, B and movies S2, S3). Perhaps networks comprised of randomly oriented filaments can contract and disassemble, while parallel filament arrays can not. To understand the contribution of actin filaments polarity during actomyosin contraction, we used evanescent wave microscopy to follow in real time the effect of myosin on growing branched network (Fig. S4 and movie S5). Networks did not contract in the presence of myosin VI when they remained as individual patches of branched and parallel filaments. When individual sub-networks interacted in anti-parallel orientation, myosin rapidly induced a deformation of the network by its co-alignement into anti-parallel bundles (Fig. S4 and movie S5).
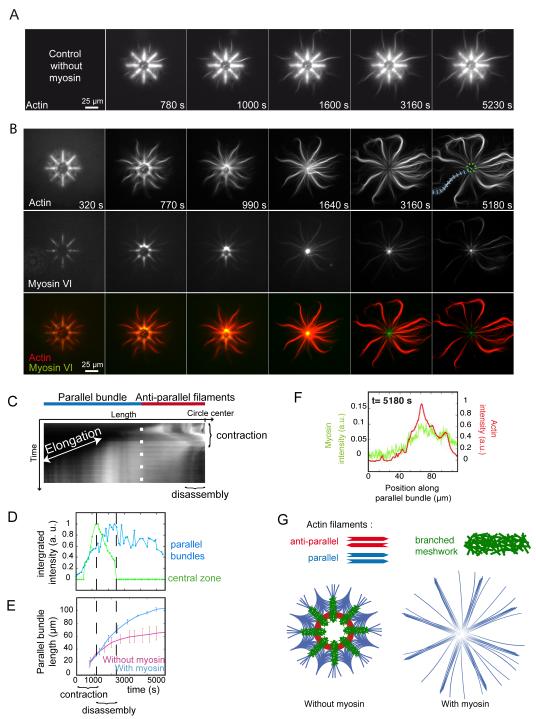
This “orientation selection” for selective contraction and disassembly of anti-parallel filaments by myosin was further tested on networks of controlled polarity and architecture. Filaments nucleated on eight branch radial array leads to the formation of all the diversity in actin organization found in a cell, meshwork of branched and randomly oriented actin filaments on the micro-pattern, bundles of aligned anti-parallel filaments in the most central part of the array and bundles of aligned parallel filaments in the distal part of the array (11) (Fig. 2A). This defined distinction between zones containing parallel, anti-parallel or branched filaments organization (Fig. 2G) enabled us to characterize the region-selectivity of myosin-induced reorganization. Myosin VI was chosen to induce contraction forces on these actin architectures because it is a pointed-end oriented motors and can pull on filaments with their barbed ends pointing out of the micropatterns (Fig. S5 and movie S6). The addition of myosin VI in solution lead to the rapid contraction of the anti-parallel bundles and branched meshwork, followed by their disassembly (Fig. 2B, central black hole after 1640 s, Fig. 2C, D and movies S7 and S8). The parallel bundles remained unperturbed and continue to elongate until the monomers freshly released by central disassembly were consumed (Fig. 2D, E and movie S8) although myosins were present on these bundles (Fig. 2F) on which they could move (Fig. S6). These processes could also be monitored on larger structures in which anti-parallel networks were easier to visualize (Fig. S7). Thus myosin-induced contraction is specific to bundles of anti-parallel filaments and branched meshwork and myosin-induced disassembly of these structures further supplies actin monomers for the growth of parallel filament bundles (Fig. 2G).
Fig. 2.
Regioselective action of myosins
(A) Time-series of network assembly on an eight-branch actin nucleating radial array. (B) Time-series of myosin VI-induced architecture selective contraction and disassembly (actin, myosin and an overlay are shown). (C) Kymograph of actin fluorescence along a parallel bundle (blue dashed line in B) 5180 s) and central region of actin filaments (dashed green circle in B) 5180 s), showing the different localization of elongation and contraction, disassembly. (D) Fluorescence intensity of a central zone (dashed green circle in B) and a parallel bundle (blue dashed line in B) over time. (E) Length variations of parallel bundles over time in absence or presence of myosins. (F) Linescan of fluorescence intensity along a parallel bundle confirming myosin presence all along. (G) Schematic representation of the final architecture on an eight-branch actin nucleating radial array in absence and in presence of myosins in solution.
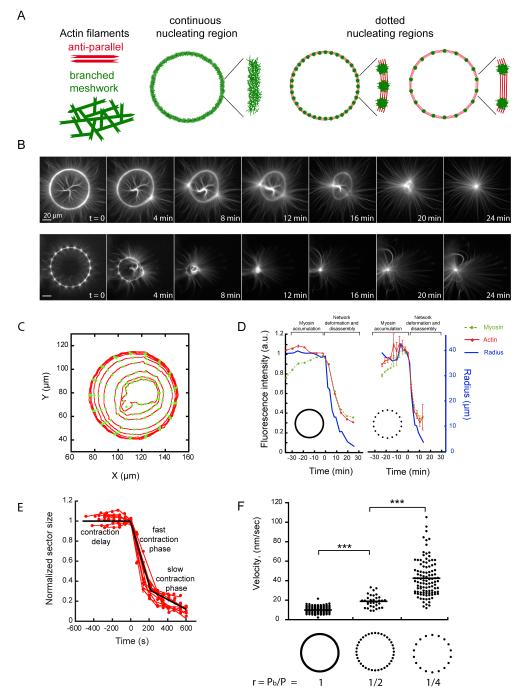
Next, we further characterized the contraction properties of bundles of anti-parallel filaments and branched meshworks. We compared the effect of myosins on actin rings in which the proportion of anti-parallel filaments zones were finely controlled (Fig. 3A). Filaments assemble into branched meshworks on full rings (Fig. 3A). On dotted rings, filaments formed branched meshworks on the dots, but specifically formed bundles of anti-parallel filaments between the dots (Fig. 3A). The proportion of bundles of anti-parallel filaments thus scales inversely with the number of dots in constant sized rings. We monitored actin network contraction and deformation upon the addition of myosin (Fig. 3B and movie S9). We measured the fluorescence intensity of actin and myosin in all angular sectors of the rings during contraction (Fig. 3C, D). Myosins first accumulated on the actin network without generating global deformation (Fig. 3D, green curve before time 0). Above a critical accumulation of myosins, deformation started (Fig. 3D, blue curve time 0). Network deformation was coupled to network disassembly (Fig. 3D, red curve). In addition, the total amounts of actin and myosin decreased following a similar decay as the radius of both full and dotted rings (Fig. 3D). As a consequence, the density of actin was constant during contraction (Fig. S8). Each sector of the rings followed three distinct phases during remodeling (Fig. 3E): First, a delay phase during which filaments were aligned; second, a fast contraction phase with a constant rate; and finally, a third phase during which the network was highly compacted at the ring center and the contraction slowed down. We measured the rate of the fast contraction phase, because it reflects the main amplitude of change in sector size. We compared the contraction rates of rings with continuous or dotted nucleating regions. Dot number and spacing were chosen to vary the ratio r between the total length of branched meshwork, Pb, and the rings perimeter, P. The contraction rate increased significantly as the ratio r decreased (Fig. 3F, movie S10). Thus for a given actin structure, the contraction rate is determined by the relative proportions of anti-parallel bundles and branched meshwork.
Fig. 3.
The proportion of anti-parallel filaments regulates network contraction rate.
(A) Schematic representation of actin networks nucleated on a full and dotted rings. (B) Time-series of myosin-induced contraction of actin networks nucleated from full (top) and dotted (bottom) rings. (C) Illustration of automated network contraction analysis (23). Each circle represents a time point. (D) The radius and total fluorescence intensities of both actin and myosin were recorded for all angular sectors over time. (E) Ring constriction kinetics. Time series of length values (red dots) could be fitted by three distinct phases (black line). (F) Fast contraction phase velocity measurements were compared between various ring compositions.
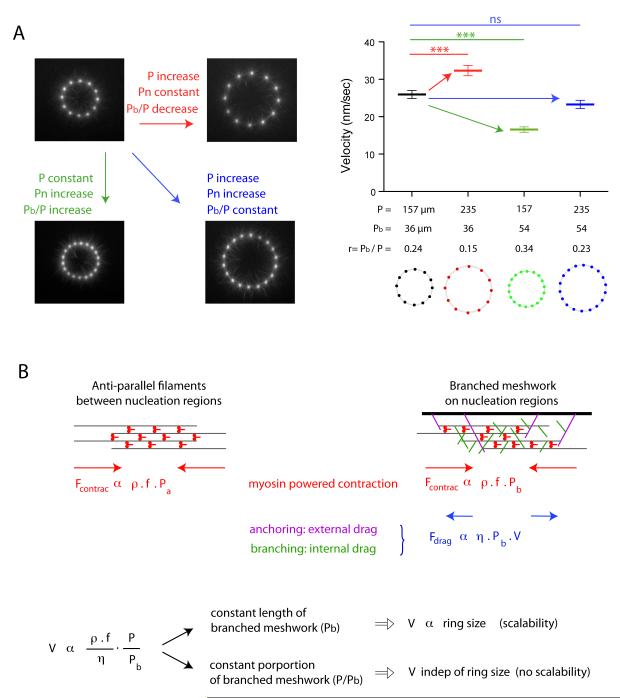
The contraction rate of in vivo structure such as the cytokinetic ring increases in proportion to their size, a process termed scalability, although no molecular determinants of the underlying mechanism have been established (14, 15). To evaluate the respective contributions of ring size and composition to the contraction rate, we varied the ring perimeter P and the portion of this perimeter that nucleates a branched meshwork Pb independently (Fig. 4A, movie S11). When P and Pb increased equally, the contraction rate was unaffected although the ring size increased (see black and blue rings in Fig. 4A). Thus, no scalability is observed when the proportion of anti-parallel bundles and branched meshwork is maintained constant during size increase. When P was increased and Pb kept constant, the contraction rate increased (see the pairs black, red rings and green, blue rings in Fig. 4A). Scalability is thus only observed when the size increase of the actin structure is coupled to an increase of the proportion of anti-parallel bundles.
Fig. 4.
The proportion of branched meshwork regulates the scalability of ring contraction.
(A) Respective effects of size and proportion of branched meshwork in contraction kinetics. We varied the ring perimeter P and the length of that perimeter nucleating a branched meshwork Pbranched (Pb) independently. Images show an early time point during actin network assembly on micropatterned dots. Fast contraction phase velocity measurements were compared between various ring configurations. (B) Model description. Filaments assemble into anti-parallel bundles between nucleation regions (left scheme). Nucleation regions (wide black bar, right scheme) generate branched actin meshwork. The contraction force is proportional to the density of myosins per unit length of filament, ρ, to the force per myosin head, f, and to the portion of the perimeter made of the relevant network, Pa for the antiparallel bundles and Pb for the branched meshwork. Myosin density is constant over the entire perimeter P=Pa+Pb. Anti-parallel bundles have a friction drag negligible compared to branched meshwork in which the effective friction coefficient η has two origins: an external drag due to network anchoring on the nucleation region and an internal drag due to entanglement of filament branches. The balance between the total contraction force and the frictional drag sets the contraction velocity V, which appeared to be proportional to the ratio P/Pb as observed in all our experiments.
These results demonstrate that contraction rate variations result from the proportion of anti-parallel filament bundle, which is controlled by the size of and distance between nucleation regions. In all conditions tested, the velocity V was proportional to the ratio P/Pb (Fig. S9). These observations could be captured by a simple physical model in which the contraction force was proportional to the amount of myosins per unit length of filament and the friction drag was proportional to the length of branched meshwork (Fig. 4B). In this model, network disassembly by myosins plays a passive role because it simply prevents the elastic reaction, which could arise from network compaction during contraction, but a more active role of network disassembly during contraction remains possible.
Thus myosins act on actin networks in a manner that depends on the actin filament orientation. Parallel filaments align and elongate whereas anti-parallel filaments contract and disassemble. We term such rules in myosin selectivity as an “orientation selection” mechanism that should not induce a global cell collapse but should instead support the overall spatial coordination of different actin structures by regulating their specific reorientation, deformation and disassembly.
Supplementary Material
Acknowledgments
22. We thank C. Sykes and J. Faix for muscle myosin II protein, F. Senger for image analysis and K. John for discussions regarding the computational model.This work was supported by grants from Human Frontier Science Program (RGP0004/2011 awarded to L.B. and E.M.D.L.C.), Agence Nationale de la Recherche (ANR-08-BLANC-0012 awarded to L.B.), INCA (INCA-2011-141 awarded to M.T.), the National Institutes of Health (GM097348 awarded to E.M.D.L.C.) and a PhD Fellowship from the IRTELIS program of the CEA (awarded to A.C.R.). E.M.D.L.C. is an American Heart Association Established Investigator, an NSF-CAREER Award recipient (MCB-0546353), and a Hellman Family Fellow.
Footnotes
Supplementary Materials Materials and Methods Figs. S1 to S9 Movies (S1 to S11) References (16-21)
References and Notes
- 1.Cramer LP, Mitchison TJ. Myosin is involved in post-mitotic cell spreading. J. Cell Biol. 1995;131:1. doi: 10.1083/jcb.131.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vignjevic D, et al. Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 2003;160:951. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cramer LP, Siebert M, Mitchison TJ. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 1997;136:1287. doi: 10.1083/jcb.136.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 1995;131:989. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 7.Veigel C, Schmidt CF. Moving into the cell: single-molecule studies of molecular motors in complex environments. Nat. Rev. Mol. Cell Biol. 2011;12:163. doi: 10.1038/nrm3062. [DOI] [PubMed] [Google Scholar]
- 8.Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A. A cytoskeletal demolition worker: myosin II acts as an actin depolymerization agent. J. Mol. Biol. 2008;375:325. doi: 10.1016/j.jmb.2007.09.066. [DOI] [PubMed] [Google Scholar]
- 9.Wilson CA, et al. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 2010;465:373. doi: 10.1038/nature08994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brawley CM, Rock RS. Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9685. doi: 10.1073/pnas.0810451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reymann A-C, et al. Nucleation geometry governs ordered actin networks structures. Nat. Mat. 2010;9:827. doi: 10.1038/nmat2855. [DOI] [PubMed] [Google Scholar]
- 12.De La Cruz EM, Ostap EM, Sweeney HL. Kinetic mechanism and regulation of myosin VI. J. Biol. Chem. 2001;276:32373. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 13.Koenderink GH, et al. An active biopolymer network controlled by molecular motors. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15192. doi: 10.1073/pnas.0903974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho A, Desai A, Oegema K. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell. 2009;137:926. doi: 10.1016/j.cell.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Calvert ME, et al. Myosin concentration underlies cell size-dependent scalability of actomyosin ring constriction. J. Cell Biol. 2011;195:799. doi: 10.1083/jcb.201101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isambert H, et al. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995;270:11437. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 17.Egile C, et al. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 1999;146:1319. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robblee JP, Olivares AO, de la Cruz EM. Mechanism of nucleotide binding to actomyosin VI: evidence for allosteric head-head communication. J. Biol. Chem. 2004;279:38608. doi: 10.1074/jbc.M403504200. [DOI] [PubMed] [Google Scholar]
- 19.Pollard TD. Wilson l., editor. Myosin purification and characterization. Methods and Perspectives in Cell Biology. 1982;24:333. doi: 10.1016/s0091-679x(08)60665-2. [DOI] [PubMed] [Google Scholar]
- 20.Vignaud T, et al. Reprogramming cell shape with laser nano-patterning. J. Cell Sci. 2012 doi: 10.1242/jcs.104901. DOI 10.1242/jcs.104901. [DOI] [PubMed] [Google Scholar]
- 21.Smith MB, et al. Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton (Hoboken) 2010;67:693. doi: 10.1002/cm.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.