Abstract
The development of normal lung tissue toxicity after radiation exposure results from multiple changes in cell signaling and communication initiated at the time of the ionizing event. The onset of gross pulmonary injury is preceded by tissue hypoxia and chronic oxidative stress. We have previously shown development of debilitating lung injury can be mitigated or prevented by administration of AEOL10150, a potent catalytic antioxidant, 24 hours after radiation. This suggests that hypoxia-mediated signaling pathways may play a role in late radiation injury, but the exact mechanism remains unclear. The purpose of this study was to evaluate changes in the temporal expression of hypoxia-associated genes in irradiated mouse lung and determine whether AEOL10150 alters expression of these genes. A focused oligo array was used to establish a hypoxia-associated gene expression signature for lung tissue from sham-irradiated or irradiated mice treated with or without AEOL10150. Results were further verified by RT-PCR. 44 genes associated with metabolism, cell growth, apoptosis, inflammation, oxidative stress and extracellular matrix synthesis were upregulated after radiation. Elevated expression of 31 of these genes was attenuated in animals treated with AEOL10150, suggesting that expression of a number of hypoxia-associated genes are regulated by early development of oxidative stress after radiation. Genes identified herein could provide insight into the role of hypoxic signaling in radiation lung injury, suggesting novel therapeutic targets, as well as clues to the mechanism by which AEOL10150 confers pulmonary radioprotection.
Keywords: radiation-induced lung injury, oxidative stress, hypoxia-inducible factor, inflammation
INTRODUCTION
The risk of radiation-induced normal tissue toxicity limits the therapeutic dose of radiation that can be used in treating human cancers. This limitation is a significant barrier to achieving successful tumor control in all tumor types, but it is particularly confounding in treating tumors of the thoracic region because of the extreme radiosensitivity of the lungs. Although the mechanisms underlying radiation-induced normal tissue injury have been partially elucidated, the way in which radiation affects specific inter- and intracellular signaling pathways and inflammatory mediators is not completely understood.
It is well known that the initial ionizing event not only directly damages DNA causing apoptosis or mitotic catastrophe, but also affects cellular macromolecules, and lipid membranes. The initial cellular damage begins a series of reactions that ultimately result in chronic oxidative stress, tissue hypoxia, inflammation, and fibroproliferation. If left unresolved, these conditions may result in tissue damage, diminished quality of life, and complete organ failure.
The development of chronic oxidative stress within irradiated tissue begins during the initial exposure and is perpetuated by molecules activated within minutes or hours post irradiation (1). Transforming growth factor-β (TGF-β1), a pro-inflammatory and pro-fibrogenic cytokine, is a prominent effector in the response of tissue to radiation. Within seconds of radiation exposure, TGF-β1 is activated by cleavage of the active protein from its latency-associated complex by hydroxyl radical (2). Once activated, TGF-β1 can induce synthesis of NADPH oxidase, an oxidant-generating enzyme (3, 4). The increase in active NADPH oxidase causes chronic ROS/RNS overproduction, creating an imbalance in free radical production and the antioxidant capabilities of the cell. This self-perpetuating increase in production of radical species in the cell causes continuous activation of pro-apoptotic, hypoxia-mediated, pro-inflammatory, and pro-fibrogenic pathways long after the initial radiation exposure.
Continuous activity of these pathways contributes significantly to vascular dysfunction and tissue hypoxia (1, 5). As areas of hypoxia develop, inflammatory cells, most notably macrophages, are recruited to the site of injury where they undergo the respiratory burst further increasing tissue levels of ROS/RNS(6). These high levels of ROS/RNS are responsible for activation of molecules involved in radiation-induced normal tissue injury, including TGF- β1, NF-κB, and HIF-1α (7, 8). The cyclic nature of these events creates a self-perpetuating pathologic environment characterized by chronic oxidative stress, tissue hypoxia, and continual activation of redox-regulated signaling molecules and pathways involved in inflammation and fibroproliferation.
In our previous studies, we have shown the potent catalytic antioxidant, AEOL10150 (9) can mitigate or prevent radiation-induced lung injury in a rodent model when given prior to or following radiation exposure(10, 11). In these studies, treatment with AEOL10150 significantly reduced oxidative damage to DNA (8-OHdG), TGF-β mediated fibrosis, and expression of hypoxia-inducible factor-1alpha (HIF-1α) and its downstream products CAIX and VEGF (11).
In this study we hypothesized the development of radiation-induced lung injury is facilitated by genes upregulated by hypoxia and that expression of these genes is affected by redox-mediated signaling. To investigate this hypothesis, we used a hypoxia signaling pathway specific Oligo DNA microarray to examine hypoxia-associated gene expression in C57BL/6J mouse lungs 1 day, 3 days, 1 week, 3 weeks, 6 weeks, and 6 months after a single dose of 15 Gy to the whole thorax. Next, we evaluated whether the potent catalytic antioxidant AEOL10150 could influence expression of select hypoxia-associated genes when administered for four weeks following radiation exposure.
The transcriptome profile for hypoxia-associated genes identified in this study provides valuable insight into the sequence of events associated with development of normal tissue injury following radiation exposure. We hypothesize that the protective effect of AEOL10150, shown in our previous studies (10, 12), is due to its effects on redox-regulated cell signaling, which may include, but is not limited to, hypoxia induced gene expression. Additional evaluation of hypoxia-associated genes not affected by AEOL10150 may lead to a better understanding of the pathological mechanisms underlying the development of injury and may identify new targets for therapeutic intervention.
MATERIALS AND METHODS
Animal Irradiation
Female C57BL/6J mice (8–10 weeks) received a single 15 Gy dose of x-ray irradiation (Therapax 320, Pantak Inc., East Haven, CT) at a dose rate of 67cGy/min using a beam energy of 320 kV (75 SSD, HVL≈ 2.00 mm Al). Radiation was delivered to the whole thorax through adjustable apertures (1.25–1.5 cm) with 8 mm lead shielding of the rest of the body. Sham-irradiated animals were subjected to the same scenario, but the radiation source was not turned on. All mice were anesthetized before irradiation with an intraperitoneal (i.p.) injection of a ketamine (100 mg/kg)/xylazine (10 mg/kg) mixture. Radiation dosimetry was performed using a calibrated ionization chamber and single-use MOSFET radiation detectors placed in tissue equivalent material or within the chest cavity of non-study related C57BL/6J mice (13, 14). The variation in dose rate across the field profile was less than 6%. Animals were euthanized and tissue harvested at pre-determined time points of 1 day, 3 days, 1 week, 3 weeks, 6 weeks, and 6 months. At the time of euthanasia (>250mg/kg sodium pentobarbital, i.p.), lungs were snap frozen in liquid nitrogen and stored at −80°C (n = 7/group). All studies were performed and approved by the Institutional Animal Care and Use Committee at Duke University Medical Center.
AEOL10150 regimen
To determine whether AEOL10150 affects the transcriptome profile of hypoxia-associated genes, a separate cohort of animals was randomized into the following groups: a) sham-irradiation, b) AEOL10150 alone, c) 15 Gy whole thorax irradiation (WTI), and d) 15 Gy WTI + AEOL10150. Each group contained 6 animals. AEOL10150 was supplied by Aeolus Pharmaceuticals (San Diego, CA). Animals receiving AEOL10150 were given a loading dose of 40 mg/kg/day by subcutaneous injection 2 hrs after radiation. For the remainder of the study animals were treated with a maintenance dose of 20 mg/kg every other day for 4 weeks.
Histopathology and Immunohistochemistry
To visualize histopathologic damage in sham-irradiated and irradiated tissue at each of the predetermined time points, hematoxylin and eason (H&E) staining was carried out as previously described(15). Immunostaining for carbonic-anhydrase was performed as previously described by Gauter-Fleckenstein et al (16). Briefly, 5 micron thick paraffin embedded tissue sections were deparaffinized and rehydrated using xylene and graded alcohol (100-80%) concentrations. Hydrogen peroxide (10%) was used to block endogenous peroxidase activity and antigen retrieval performed with citrate buffer (Biogenex, San Ramon, CA). Slides were incubated at 4°C overnight with a rabbit polyclonal antibody to carbonic anhydrase IX (1:200; Abcam, Cambridge, MA) followed by washing in phosphate buffered saline and incubation for one hour at room temperature with a donkey anti-rabbit secondary antibody (1:200, Jackson ImmunoResearch, West Grove, PA). Slides were counterstained with Harris hematoxylin and visualized under light microscopy using a 40x objective.
Pimonidazole
Pimonidazole hydrochloride (70 mg/kg i.p, Chemicon International Inc, Temecular, CA) was injected three hours prior to euthanasia and immunostaining performed as previously described(17).
RNA isolation
Snap frozen whole lung tissue was homogenized in 5 ml TRIzol reagent (Invitrogen, Carlsbad, CA, USA) with the PowerGen 125 homogenizer (Fisher Scientific, Pittsburgh, PA, USA). The total RNA was isolated using TRIzol reagent following the manufacturer’s instruction (Invitrogen, Carlsbad, CA, USA). The RNA was repeatedly purified until the A260:A280 was greater than 2.0 and A260:A230 was greater than 1.7.
Probe preparation
cDNA synthesis, cRNA synthesis, and cRNA labeling were performed using the TrueLabeling-AMP 2.0 kit (SA Bioscience, Frederick, MD, USA) according to the manufacturer’s manual. Briefly, 10 µg of pooled total RNA was used for the cDNA synthesis. The RNA pool was produced by mixing equal amount of total RNA from each sample in the same group. The cDNA was then subjected to cRNA synthesis and labeling with Biotinylated-UTP overnight at 37°C. After purification with SA Biosciences ArrayGrade cRNA cleanup kit (SA Bioscience, Frederick, MD, USA), 15 µg of biotin-labeled cRNA was used for hybridization.
Hybridization
A mouse hypoxia signaling pathway specific Oligo GE Array DNA microarray (SA Bioscience, Frederick, MD, USA, Cat#: OMM-032) was used to determine temporal changes in the expression of 113 genes following thoracic irradiation. Prior to hybridization, array membranes were pre-wetted with 5 ml of deionized water for 5 minutes and then pre-hybridized with 2 ml pre-warmed GEAhyb hybridization solution (SA Bioscience, Frederick, MD, USA, Cat#: H-01) for 4 hrs at 60°C. The array membrane was hybridized with 15 µg of purified biotin-labeled cRNA probes overnight at 60°C with continuous agitation. The membranes were then washed with Wash Solution 1 (2x SSC, 1% SDS) for 20 min at 60°C and followed by Wash Solution 2 (0.1x SSC, 0.5% SDS) for 15 min at 60°C with continuous agitation.
Chemiluminescent detection and data analysis
The signal was visualized using the Chemiluminescent Detection kit (SA Bioscience, Frederick, MD, USA, Cat#: D-01). Briefly, after blocking, the membranes were bound with alkaline phosphatase-conjugated streptavidin. After washing, the membranes were incubated with Chemiluminescent substrate for 5 min and exposed to x-ray film. Gene spots with signal intensity higher than the blank on the same membrane were considered detectable. The densitometry of GAPDH in the radiation group was normalized with that in control group. Densitometry for each gene spot was then normalized with the adjusted GAPDH. For the time point experiment, the average increased densitometry from two repeated experiments was graded in 5 expression degrees. One represents a weak increase and five represents the highest increase. If the gene spot was undetectable in both the control and irradiated mice in same time point, it was designated as 0. For the AEOL10150 regimen, the densitometry of each gene in 4 groups was directly compared. The percentage of reversal by AEOL10150 was calculated.
RT-PCR
Total RNA isolation was performed as described above. Reverse transcription was performed by incubating 3 µg of pooled total RNA, 2.5 µM of random primer, and 200 units of Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA), for a total reaction volume of 20 µL, for 10 min at 25°C followed by 60 min at 50°C. PCR amplification was carried out using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) and two pairs of primers in one reaction, one amplifying the target gene fragment and another amplifying a 1 kb GAPDH fragment to serve as an internal control. PCR products were visualized on 1.5% agarose gel containing 0.5 µg/ml ethidium bromide. The ratio of PCR product of target gene to GAPDH better reflect the RNA level in the samples. Primer sequences and the size of PCR products are listed in Table 1.
Table 1.
RT-PCR primers and product size
| Gene Symbol | Primer sequence | Fragment size (bp) |
|---|---|---|
| Adm-F | 5’-AGCTGGTTTCCATCACCCTG-3’ | 502 |
| Adm-R | 5’-CCTGGGAGGACTCCACAGTC-3’ | |
| Angpt14-F | 5’-GTGAGGACACAGCCTACAGC-3’ | 333 |
| Angpt14-R | 5’-AGGCTGCTGTAGCCTCCATG-3’ | |
| CTGF-F | 5’-CTGCCTACCGACTGGAAGAC-3’ | 495 |
| CTGF-R | 5’-ACATCTTCCTGTAGTACAGG-3’ | |
| Eno-1α-F | 5’-GCCAGAGAGATCTTTGACTC-3’ | 443 |
| Eno-1α-R | 5’-CCGTTGATCACATTGAAAGC-3’ | |
| Gna11-F | 5’-CAGAGCGCAGGAAGTGGATC-3’ | 427 |
| Gna11-R | 5’-ACTCCTTCAGGTTCAGCTGC-3’ | |
| HIF-1α-F | 5’-CTCAGTTTGAACTAACTGGAC-3’ | 680 |
| HIF-1α-R | 5’-GTCGTGCTGAATAATACCAC-3’ | |
| HIF-Fα-F | 5’-CGTGGTGACCCAAGACGGTGA-3 | 630 |
| HIF-2α-R | 5’-AGCATCCGGTACTGGCCAGA-3’ | |
| HIF-2β-F | 5’-CAGAACATATCCCAGATCTC-3’ | 439 |
| HIF-2β-R | 5’-GGTCAGCAAAGTCCTCGATG-3’ | |
| Ppara-F | 5’-TGAAGCCATCTTCACGATGC-3’ | 446 |
| Ppara-R | 5’-ATCTCTTGCAACAGTGGGTG-3’ | |
| Rora-F | 5’-TCATTACGTGTGAAGGCTGC-3’ | 495 |
| Rora-R | 5’-CCAGATGCTGGTGTGTAGTC-3’ | |
| Sord-F | 5’-GAACAAGGTCCTTGTGTGTG-3’ | 427 |
| Sord-R | 5’-GTCTTCGATGCAAGCATGGA-3’ | |
| GAPDH-F | 5’-GGTGAAGGTCGGTGTGAACG-3’ | 990 |
| GAPDH-R | 5’-TGGAGGCCATGTAGGCCATG-3’ |
Western blotting
Snap frozen left lungs were prepared for western blot analysis by immersing the lung in homogenization buffer, containing protease and phosphatase inhibitors (Roche Applied Sciences, Indianapolis, IN), and sonicating samples to disrupt cell membranes. Samples were centrifuged to remove undigested tissue. The supernatants were diluted 1:1 in Laemmli Sample Buffer (95% LSB, 5% β-Mercaptoethanol) (BioRad, Berkeley, CA). The prepared samples were then boiled and centrifuged prior to loading (10 µL per well) onto 10% SDS-PAGE gels. Following electrophoresis, protein was transferred onto polyvinylidene fluoride (PVDF) membranes and probed with a specific antibody to HIF-1α (Novus Biologicals, Littleton, CO), HIF-2α (Abcam, Cambridge, MA), or HIF-3α (Novus Biologicals, Littleton, CO) diluted in 5% non-fat milk in TBS+0.1% Tween20. Following secondary antibody incubation (Cell Signaling, Beverly, MA), signals were visualized using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL). Films were scanned using an HP ScanJet 3110. Band intensity was quantified using ImageJ Software (NIH, Bethesda, MD). Measured intensities were normalized using α-tubulin expression and then averaged within groups. Comparisons between irradiated and non-irradiated controls at each time point were made using a student’s t-test.
RESULTS
Temporal changes in tissue oxygenation
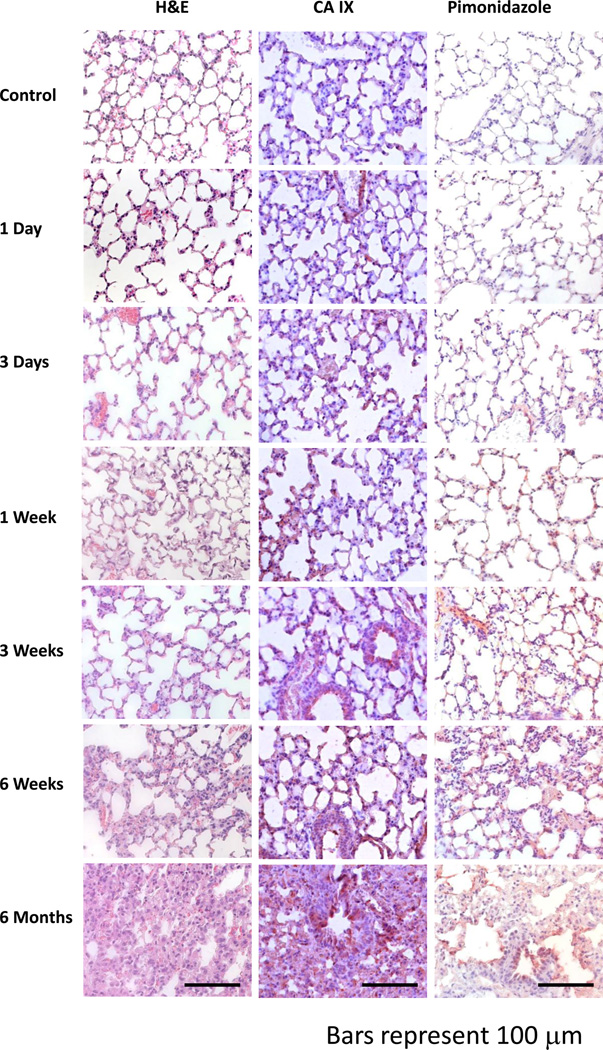
Positive staining for carbonic-anhydrase IX, an endogenous marker of hypoxia, and the exogenous hypoxia marker, pimonidazole, was evident three days after radiation and was strongly increased at one week (Figure 1). The development of tissue hypoxia occurred prior to the overt onset of histopathologic damage (H&E stain, Figure 1). By six months, architectural distortion and tissue fibrosis can be seen.
Figure 1. The temporal progression of tissue hypoxia in lung after radiation.
Tissue hypoxia is first noticeable around three days (CAIX, Pimonidazole) post-radiation and progressively increases throughout the follow-up period (6 months). At six weeks, the first histopathologic lesions are seen (H&E). These are generally focal in nature and are characterized by thickening of the alveoli wall and increased inflammatory cell infiltrate. By six months, tissue damage had considerably worsened and a greater number of focal lesions were observed. Error bars represent 100 µm.
Changes in HIF-1α, 2α, 2β, 3α mRNA expression after radiation
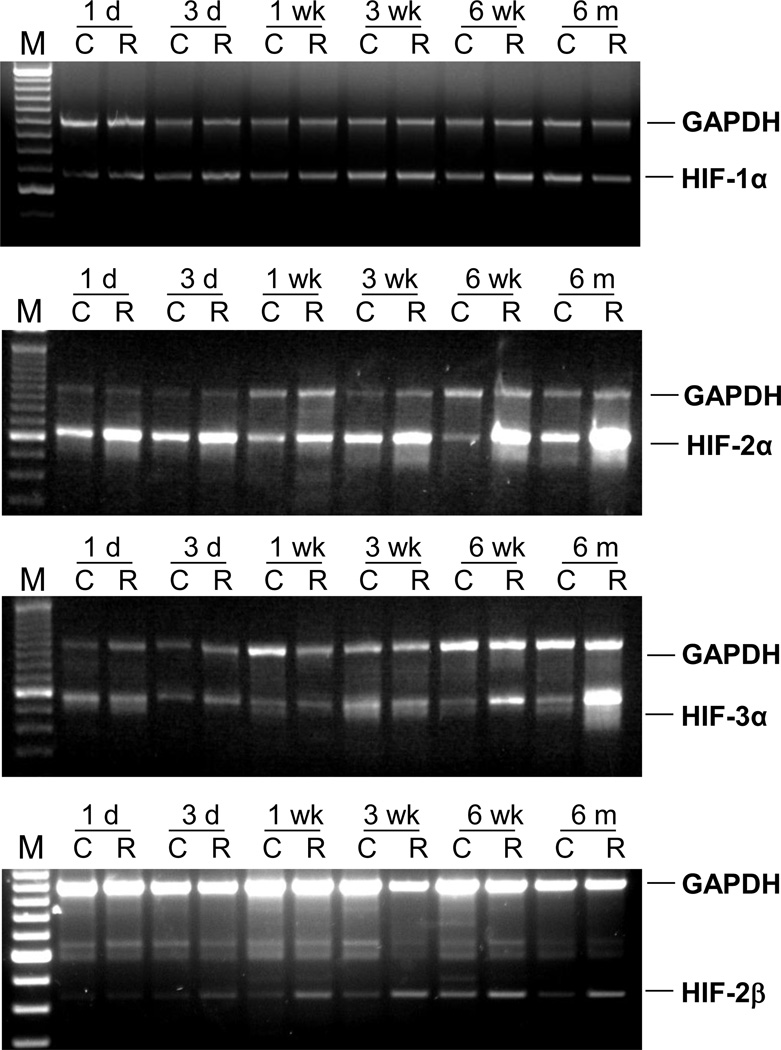
Differences in expression of HIF-1α, 2α, 2α, and 3α were examined by comparing RT-PCR of pooled total RNA from the lungs of the control and irradiated mice sacrificed at each time point (n= 7). HIF-1α mRNA expression was constant across all of the time points examined, whereas expression of HIF-2α was upregulated in irradiated mice at multiple time points. HIF-2β expression was upregulated in irradiated animals beginning 1 week after radiation whereas expression of HIF-3α was not upregulated until 6 months post-radiation (Figure 2).
Figure 2. Temporal expression of HIF mRNA.
The amplified PCR fragments were visualized on 1.5% agarose gel containing 0.5 µg/ml ethidium bromide. The GAPDH and HIF genes were amplified in the same reaction. The top band shows a 1 kb GAPDH fragment and the bottom band shows the HIF gene fragment.
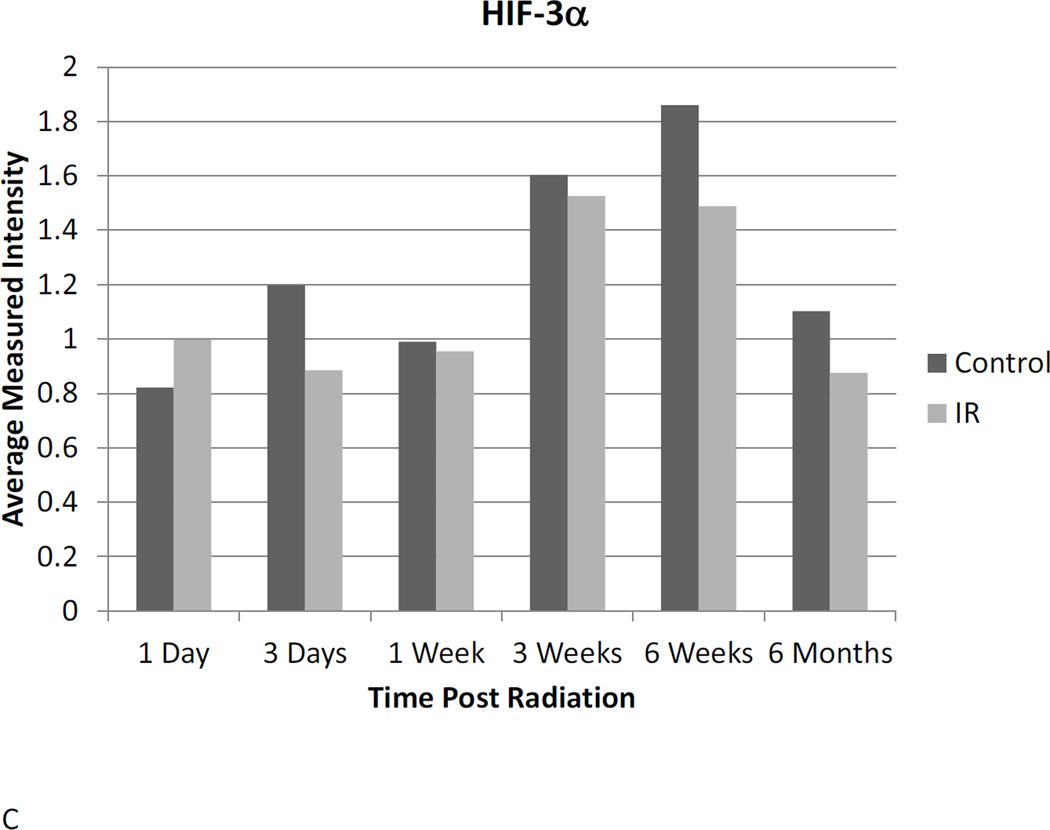
HIF-1α, 2α, 2β, 3α protein expression
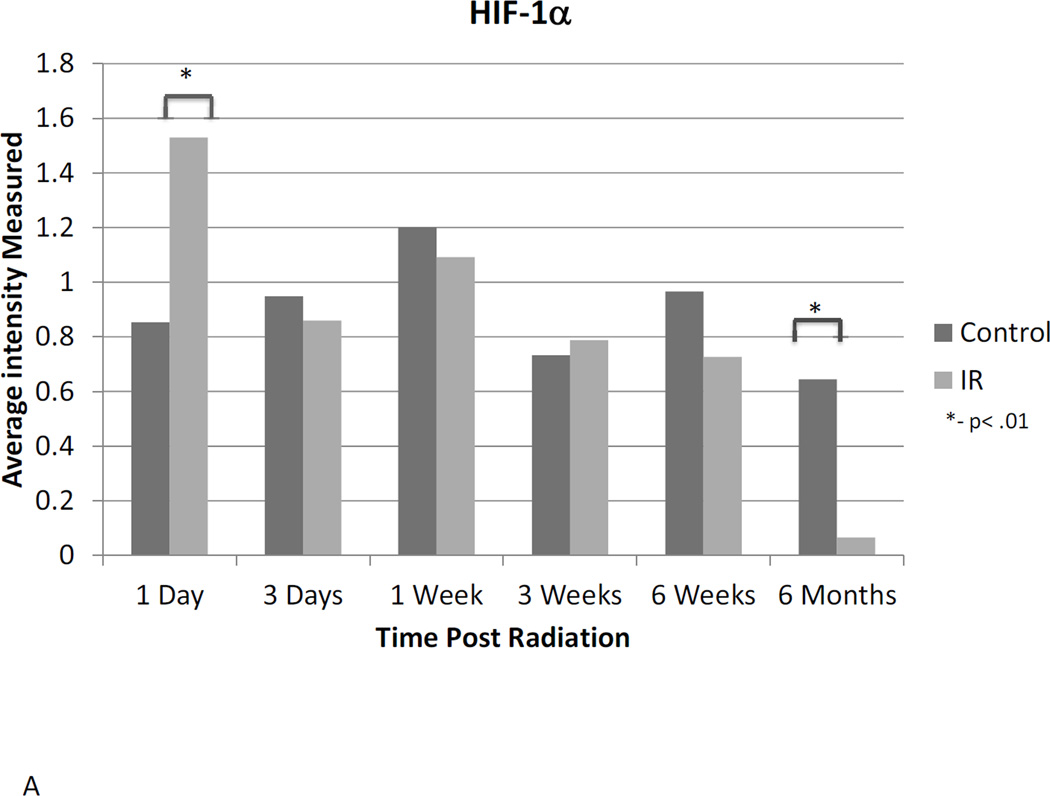
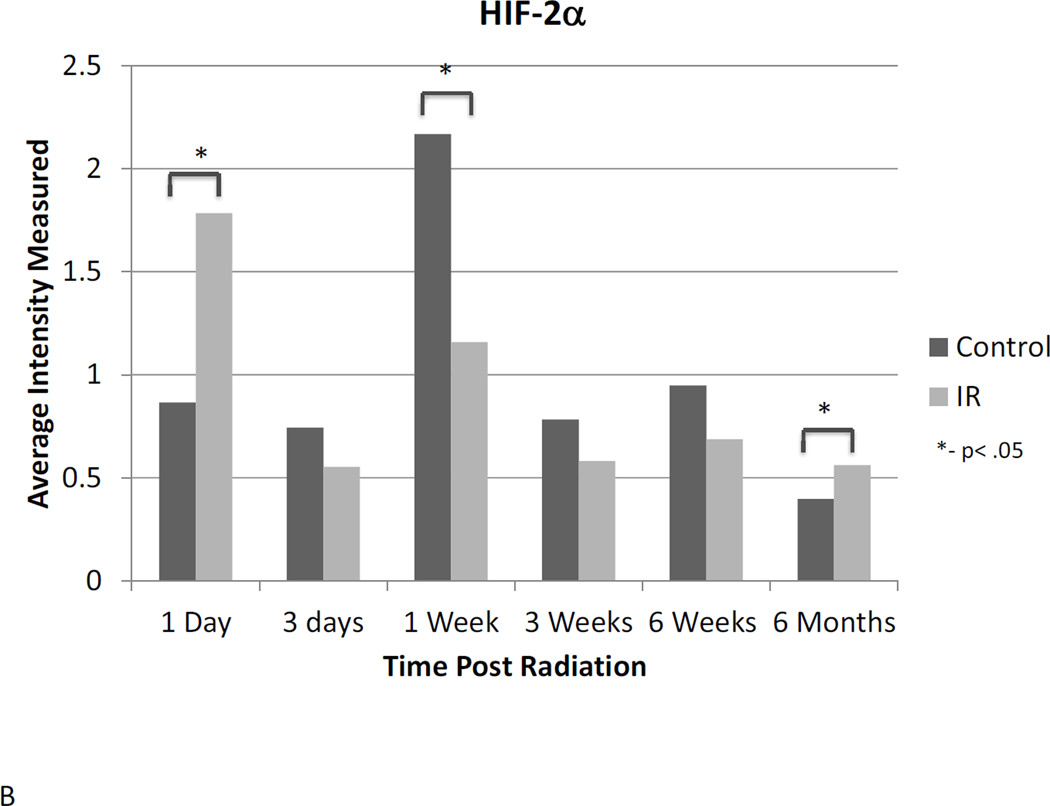
Western blot analysis suggests a dynamic role for HIF-1α, -2α, and -3α. Assessment of protein expression showed that HIF-1α protein expression is elevated in irradiated animals 24 hours after radiation, but does not differ significantly at any other time point until 6 months post-irradiation, when HIF-1α expression is reduced relative to untreated controls (Figure 3A). HIF-2α expression was elevated 24 hours post-irradiation and again 6 months following radiation. Differential expression was also observed at the one-week time point, but was not different between irradiated and control animals at any intervening time points (Figure 3B). No differential expression of HIF-3α protein was observed at any point following radiation (Figure 3C).
Figure 3. Western blot analysis illustrates the dynamic changes in stabilization of HIF-1α (A) and HIF-2α (B) proteins changes over time following irradiation, while HIF-3α (C) remains constant.
Western blot bands were visualized and normalized based on α-tubulin expression. The relative intensities were measured and averaged within groups and irradiated animals were compared to time matched controls. 1d: 1 day, 3d: 3 days, 1wk: I week, 3wk: 3 weeks; 6wk: 6 weeks, 6m: 6 months, C: control, R: radiation.
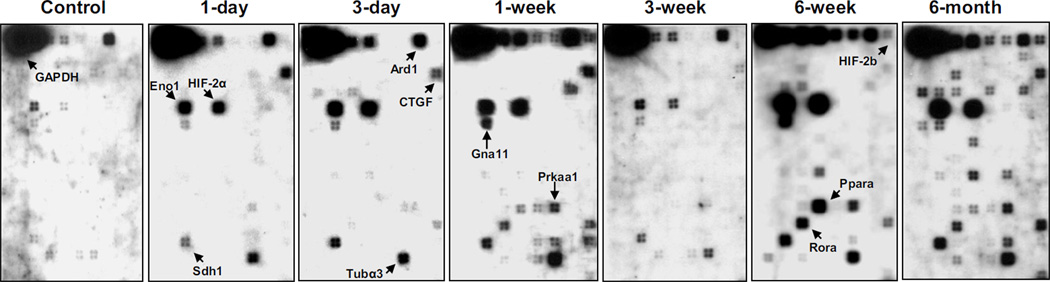
Genes with elevated post-radiation expression at all time points
Oligo array assay revealed that beginning one day after radiation, messenger RNA expression of Adm, Agpat2, Ard1, CTGF, Eno1, HIF-2α, Gna11, Prkaa1, Sdh1, and Tubα3 was elevated in irradiated animals relative to control. This increase continued throughout the six-month follow-up period (Table 2). One week after radiation increased expression of Agtpbp1, Angptl4, HIF-2β, Nmyc1, Ppar-α, and Th was observed. Expression remained elevated in all later time points (Figure 4).
Table 2.
Increased ratio of gene expression
| Gene Symbol | Description | Increased fold | |||||
|---|---|---|---|---|---|---|---|
| 1d | 3d | 1w | 3w | 6w | 6m | ||
| Adm | Adrenomedulin | 1 | 3 | 4 | 1 | 5 | 5 |
| Agpat2 | 1-acylglycerol-3-phosphate O-acyltransferase 2 | 2 | 3 | 4 | 1 | 5 | 5 |
| Agtpbp1 | RIKEN cDNA A230056J06 gene | 0 | 0 | 1 | 2 | 5 | 2 |
| Angptl4 | Angiopoietin-like 4 | 0 | 0 | 1 | 1 | 4 | 2 |
| Ard1 | N-acetyltransferase ARD1 homolog | 1 | 3 | 5 | 1 | 3 | 2 |
| HIF-2β | Aryl hydrocarbon receptor nuclear translocator 2 | 0 | 0 | 2 | 1 | 2 | 3 |
| Car12 | Carbonic anyhydrase 12 | 0 | 0 | 0 | 0 | 1 | 2 |
| Cdc42 | Cell division cycle 42 homolog | 0 | 0 | 0 | 0 | 1 | 2 |
| CTGF | Connective tissue growth factor | 4 | 3 | 5 | 2 | 2 | 4 |
| Cygb | Cytoglobin | 1 | 1 | 0 | 0 | 2 | 4 |
| Dapk3 | Death-associated kinase 3 | 1 | 1 | 0 | 0 | 2 | 3 |
| Dctn2 | Dynactin 2 | 0 | 1 | 0 | 0 | 2 | 3 |
| Dr1 | Down-regulator of transcription 1 | 0 | 0 | 0 | 0 | 0 | 3 |
| Eef1a1 | Eukaryotic translation elonation factor 1 alpha 1 | 0 | 0 | 0 | 0 | 1 | 3 |
| EGFR | Epidermal growth factor receptor | 0 | 0 | 1 | 1 | 0 | 0 |
| Eno1 | Enolase 1, alpha non-neuron | 4 | 5 | 5 | 2 | 5 | 5 |
| HIF-2α | Endothelial PAS domain protein 1 | 5 | 5 | 5 | 3 | 5 | 5 |
| Fabp4 | Fatty acid binding protein 4, adipocyte | 0 | 0 | 0 | 0 | 0 | 1 |
| Fnbp3 | Formin binding protein 3 | 0 | 0 | 0 | 0 | 0 | 1 |
| Gap43 | Growth associated protein 43 | 0 | 1 | 0 | 0 | 1 | 3 |
| Gna11 | Guanine nucleotide binding protein, alpha 11 | 2 | 3 | 4 | 2 | 5 | 3 |
| Htatip | HIV-1 tat interactive protein, homolog | 0 | 0 | 0 | 0 | 1 | 3 |
| IL1p | Interleukin 1 beta | 0 | 0 | 0 | 0 | 1 | 1 |
| HIF-3α | Hypoxia inducible factor 3, alpha subunit | 0 | 0 | 0 | 0 | 0 | 1 |
| Lep | Leptin | 0 | 0 | 0 | 0 | 1 | 2 |
| Lipe | Lipase, hormone sensitive | 0 | 0 | 0 | 0 | 1 | 1 |
| Man2b1 | Mannosidase 2, alpha B1 | 0 | 1 | 1 | 0 | 3 | 4 |
| Mmp14 | Matrix metalloproteinase 14 | 0 | 0 | 0 | 0 | 1 | 3 |
| Nmyc1 | Neuroblastoma myc-related oncogene 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| Plod3 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | 0 | 0 | 0 | 0 | 1 | 2 |
| Ppara | Peroxisome proliferator activated receptor alpha | 0 | 0 | 2 | 1 | 5 | 3 |
| Ppp2cb | Protein phosphatase 2a, catalytic subunit, beta isoform | 0 | 0 | 2 | 0 | 1 | 2 |
| Prkaa1 | Protein kinase, AMP-activated, alpha 1 catalytic subunit | 1 | 1 | 4 | 1 | 5 | 4 |
| Rora | RAR-related orphan receptor alpha | 0 | 1 | 3 | 0 | 5 | 4 |
| Rps7 | Ribosomal protein S7 | 0 | 2 | 3 | 0 | 3 | 4 |
| Sdh1 | Sorbitol dehydrogenase 1 | 3 | 4 | 4 | 2 | 5 | 5 |
| Slc2a1 | Solute carrier family 2, member 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Slc2a8 | Solute carrier family 2, member 8 | 0 | 0 | 2 | 0 | 1 | 1 |
| Snrp70 | U1 small nuclear ribonucleoprotein polypeptide A | 0 | 0 | 3 | 0 | 2 | 4 |
| Sod2 | Superoxide dismutase 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| TGF-β1 | Transforming growth factor, beta 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| Th | Tyrosine hydroxylase | 0 | 0 | 1 | 2 | 1 | 2 |
| Tuba1 | Tubulin, alpha 1 | 0 | 0 | 2 | 1 | 0 | 1 |
| Tuba3 | Tubulin, alpha 3 | 3 | 4 | 5 | 2 | 4 | 5 |
d: day, w: week, m; Month. 0 = undetectable, 1 = weak increase, 5 = highest increase.
Figure 4. Representative Oligo GEArray hybridization images.
The control for all of these time points shows a similar pattern. Arrows indicate gene spots with significant increases.
Genes exhibiting a bi-phasic increase after radiation
As shown in Table 2, mRNA expression of Cygb and Dapk3 is briefly increased at two time points, the first beginning one day after radiation and lasting no more than a few days, and the second beginning 6 weeks after radiation and continuing to the 6 month time point. Dctn2, Gap43, Man2b1, Rora, and Rps7 mRNA expression was elevated for periods of differing duration 3 days after radiation and again 6 weeks post-radiation.
Genes expressed during the development of symptomatic injury
Six weeks after radiation, an increase in mRNA expression of Car12, Cdc42, Eef1a1, Htatip, IL-1β, Lep, Lipe, Mmp14, Plod3, and TGF-β1 was observed in all irradiated animals. Expression of Dr1, Fabp4, Fnbp3 and Slc2α1 was elevated only at 6 months after post-radiation (Table 2).
AEOL10150 normalizes hypoxia-associated gene expression 6 weeks after radiation
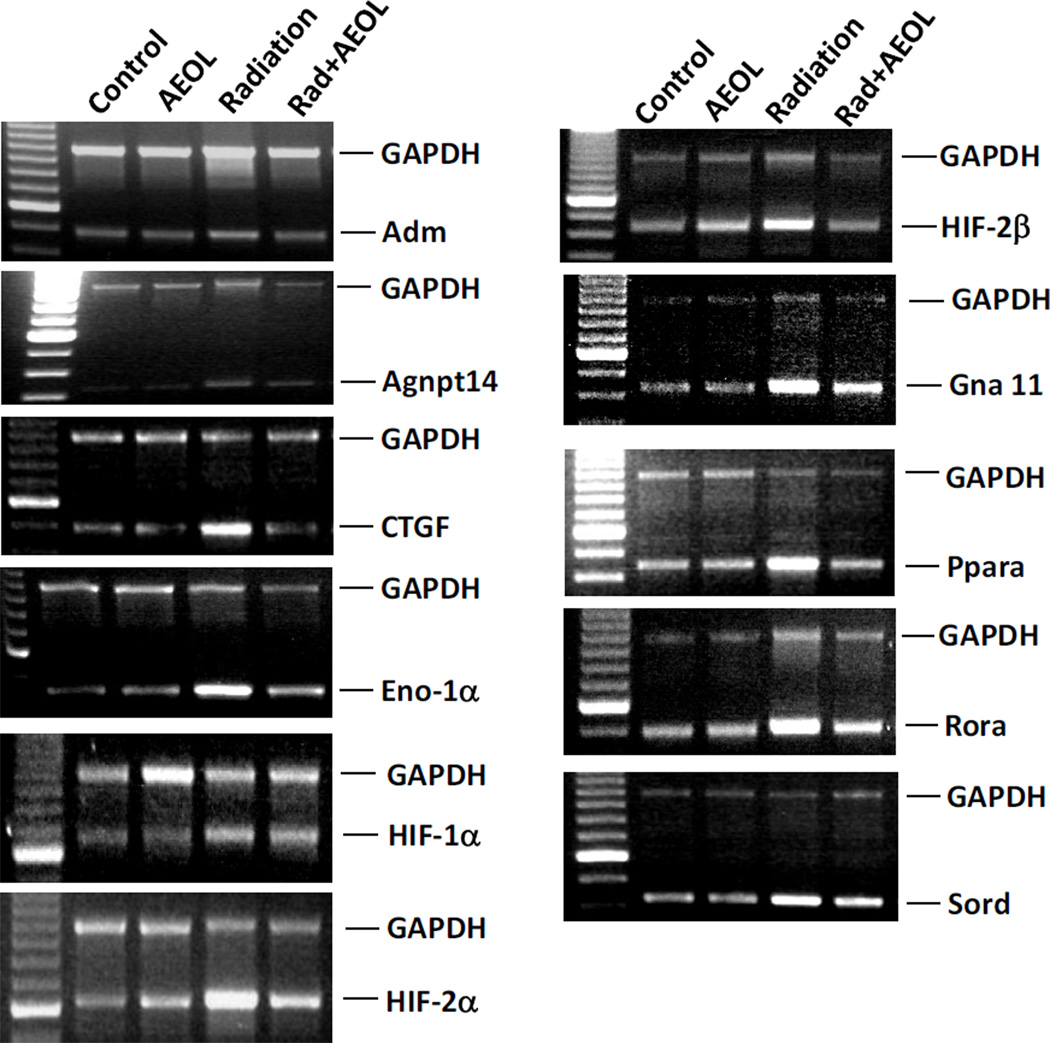
To further investigate changes in the hypoxia-associated transcriptome, we analyzed expression of the same 113 genes, 6 weeks after radiation and treatment with AEOL10150, using a hypoxia signaling pathway specific Oligo DNA microarray. Thirty of the 31 genes observed to be upregulated in mice following radiation alone showed no detectable increase in expression following treatment with AEOL10150 (Table 3). The only hypoxia gene that remained upregulated even when AEOL10150 was administered following radiation was Add1, the gene encoding adducin-α (Table 3). Messenger RNA expression was further confirmed by RT-PCR for 11 genes (Figure 5). Both RT-PCR and microarray showed no changes in HIF-1α mRNA.
Table 3.
The increased ratios of gene expression and reversal by AEOL10150
| Gene Symbol | Description | Increased folds with radiation |
% reversal with AEOL |
|---|---|---|---|
| Add1α | Adducin 1 (alpha) | 10.1 | 0 |
| Adm | Adrenomedulin | 11.6 | 84.1 |
| Agpat2 | 1-acylglycerol-3-phosphate O-acyltransferase 2 | 10.8 | 85.2 |
| Agtpbp1 | RIKEN cDNA A230056J06 gene | 8.6 | 94.2 |
| Angptl4 | Angiopoietin-like 4 | 8.8 | >99 |
| Ard1 | N-acetyltransferase ARD1 homolog | 9 | 91 |
| HIF-2β | Aryl hydrocarbon receptor nuclear translocator 2 | 5.7 | >99 |
| Car12 | Carbonic anyhydrase 12 | 1.3 | >99 |
| Cdc42 | Cell division cycle 42 homolog | 1.6 | >99 |
| CTGF | Connective tissue growth factor | 1.7 | >99 |
| Dapk3 | Death-associated kinase 3 | 1.6 | >99 |
| Dctn2 | Dynactin 2 | 2.9 | >99 |
| Eef1α1 | Eukaryotic translation elonation factor 1 alpha 1 | 1.2 | >99 |
| Eno1 | Enolase 1, alpha non-neuron | 11.2 | 76.3 |
| Hif-2α | Endothelial PAS domain protein 1 | 11.4 | 80.8 |
| Gna11 | Guanine nucleotide binding protein, alpha 11 | 8.1 | 95.2 |
| IL1b | Interleukin 1 beta | 1.7 | >99 |
| Man2αb1 | Mannosidase 2, alpha B1 | 2.8 | >99 |
| Mmp14 | Matrix metalloproteinase 14 | 1.5 | >99 |
| Nmyc1 | Neuroblastoma myc-related oncogene 1 | 1.3 | >99 |
| Pparα | Peroxisome proliferator activated receptor alpha | 7.1 | >99 |
| Ppp2cβ | Protein phosphatase 2a, catalytic subunit, beta isoform | 1.7 | >99 |
| Prka1 | Protein kinase, AMP-activated, alpha 1 catalytic subunit | 3.7 | >99 |
| Rorα | RAR-related orphan receptor alpha | 5.1 | 96.3 |
| Rps7 | Ribosomal protein S7 | 1.9 | >99 |
| Sdh1 | Sorbitol dehydrogenase 1 | 5.9 | 95.7 |
| Snrp70 | U1 small nuclear ribonucleoprotein polypeptide A | 1.5 | >99 |
| Sumo2 | SMT3 suppressor of mif two 3 homolog 2 | 1.7 | >99 |
| TGF-β1 | Transforming growth factor, beta 1 | 1.4 | >99 |
| Th | Tyrosine hydroxylase | 1.6 | >99 |
| Tubα3 | Tubulin, alpha 3 | 5.5 | 89.4 |
Figure 5. Expression of genes after AEOL10150 treatment.
The amplified PCR fragments were visualized on 1.5% agarose gel containing 0.5 µg/ml ethidium bromide. The GAPDH and target gene were amplified in the same reaction. The top band shows a 1 kb GAPDH fragment and the bottom band shows the targeted gene fragment.
DISCUSSION
The development of tissue hypoxia is a key step in the progression of radiation-induced lung injury (1, 17, 18). In the current study, we aimed to identify genes induced by tissue hypoxia between the time of irradiation and development of lung injury. We furthermore sought to determine whether the introduction of a potent antioxidant, AEOL10150, during the first four weeks post-irradiation could mitigate expression of those genes six weeks into the course of disease development. This study found dynamic changes in hypoxia-inducible gene expression in lung tissue during the six months between the time of thoracic irradiation and development of pulmonary injury. Furthermore, we found hypoxia-inducible gene expression could be modulated by changes in the cellular redox environment resulting in normalization of 30 of 31 genes upregulated after radiation. These findings are consistent with previous studies by other groups, who have suggested oxidative stress is a potent inducer of hypoxia-associated genes(19).
It is believed that the first signs of tissue hypoxia can occur within days following thoracic irradiation due to endothelial cell swelling leading to capillary occlusion (20). One of the most prominent gene families regulated by hypoxia are the hypoxia-inducible factors (HIF). HIF proteins are transcriptional complexes composed of alpha and beta subunits that can activate a wide number of downstream genes involved in cellular response to stress. The alpha subunit of the HIF complex is the main hypoxia sensor. Under normoxic conditions, the alpha subunit is rapidly degraded by the proteasome(21). However, under hypoxic conditions or oxidative/nitroxidative stress, the alpha subunit is stabilized and associates with its binding partner, HIF-1β to form an activated transcriptional complex (21, 22). Three alpha subunit isoforms have recently been identified in mammalian tissues: HIF-1α, HIF-2α and HIF-3α, all of which were evaluated in this study.
HIF-1α and HIF-2α are closely homologous and recognize hypoxia-responsive elements (HREs) in the promoter regions of a vast array of genes, including many of those found to be upregulated in this study (23–26). One of the most interesting findings in the current study was the contrast in mRNA expression between HIF-1α and HIF-2α following radiation. Although no increase in HIF-1α mRNA was observed in irradiated lung tissue at any of the time points evaluated, HIF-2α mRNA was strongly increased as early as 24 hours post-radiation. However, an increase in protein stabilization of both HIF-1α and HIF-2α subunits was observed. The stabilization of the alpha subunits of both HIF proteins was coupled with a mild increase in tissue hypoxia measured by CA-IX and pimonidazole staining. This data provides evidence that hypoxia develops in irradiated mouse lung within the first few days after exposure.
Recent studies using HIF-1α or HIF-2α knockout mice revealed different characteristics and functions of the two isoforms(27, 28). Unlike HIF-2α-knockout mice, which develop multiple-organ deficiency and biochemical abnormalities (29), HIF-1α -knockout mice mainly show abnormal vascular development and embryonic lethality(30). Furthermore, distinct transcriptional targets for HIF-1α and HIF-2α have also been proposed(31). Thus, the regulation of HIF exhibits a complex pattern in irradiated lung. Further clarification of the mechanisms and consequences of HIF activation in lung post-radiation is warranted.
One of the primary mechanisms by which the alpha subunit of HIF is stabilized under both normoxic and hypoxic conditions is through s-nitrosylation of a cysteine residue (Cys533) in the oxygen dependent domain(32). It is thought that oxidative stress is a more potent stabilizer of HIF than the prolyl hydroxylase pathway(33). Chronic oxidative stress is an important feature of radiation-induced lung injury and can be exacerbated by tissue hypoxia(1). Thus, it is not surprising that many of the genes induced after radiation in this study were those coding for oxidoreductases such as cytoglobin (Cygb), sorbitol dehydrogenase (Sdh1), procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 (Plod3), and tyrosine hydrolase (Th). Treatment with AEOL10150 resulted in reduced expression of both Sdh1 and Th (Table 3). Increased expression of oxidoreductases is consistent with the known increase in oxidative/nitroxidative stress in the lung following radiation exposure and attenuation of this increase in oxidative stress with antioxidant therapy has been shown to mitigate lung injury(1, 11, 16, 34, 35).
The induction of the gene encoding for cytoglobin at one to three days and again at six weeks to six months was particularly interesting. Cytoglobin is an oxygen-binding globin predominantly expressed in activated fibroblasts under hypoxic conditions (36). The primary role of cytoglobin is to maintain the redox status of the cell during periods of oxidative stress by scavenging hydrogen peroxide, nitric oxide, and peroxynitrite (37–39). Several recent studies have evaluated cytoglobin as a therapeutic target in fibrotic disease (38–41). Xu et al. (39) found viral-mediated overexpression of cytoglobin in primary hepatic stellate cells prevented H2O2 mediated fibroblast differentiation, TGF-β1 expression, and collagen synthesis. Thus, cytoglobin may prove to be a potential therapeutic target in the lung and warrants further exploration.
Our prior studies have shown a continuous and persistent increase in cell death between twenty four hours and six months post-radiation (42, 43). In this study, increased expression of the gene encoding for death-associated protein kinase (Dapk), a protein involved in programmed cell death, autophagy, and inflammation (44), was observed one and three days post-radiation and again at six weeks and six months. Death-inducing signals, such as TGF-β, IFN-γ, and Myc, as well as DNA damaging agents, can influence Dapk gene expression and increase Dapk-mediated apoptosis and autophagy of nutrient-starved cells (44). Dapk expression was reduced by more than 99% in animals treated with AEOL10150 after radiation. This observation is consistent with our previous studies, which demonstrated a significant decrease in cell death six weeks post-radiation in animals treated with AEOL10150 (42, 43).
Other interesting genes upregulated after radiation were those encoding dynactin subunit 2 (Dctn2) and growth associated protein-43 (Gap43), both of which are involved in tissue regeneration and cell proliferation/differentiation and genes involved in cell cycle progression (CdC42), and DNA damage repair (Dr1). AEOL10150 mitigated expression of both Dctn2 and Cdc42, genes primarily involved in cytoskeletal reorganization and biogenesis, by more than 99%.
It is well known that hypoxia can stimulate both resident lung cells and inflammatory cells, such as macrophages, to secret pro-inflammatory cytokines, chemokines, and growth factors (6, 45, 46). Interleukin-1β (IL-1β), a cytokine produced by activated macrophages, is an important mediator of cell proliferation, differentiation, and apoptosis as part of the inflammatory response (47–49). The gene encoding IL-1β, as well as those encoding fatty acid binding protein 4 (Fabp4) and Leptin, both of which are involved the inflammatory response, were all upregulated at six weeks after radiation. TGF-β1 and matrix metalloproteinase-14 (MMP-14), which are involved in collagen deposition and fibrogenesis were also upregulated in this time frame. These later changes are indicative of initiation of late injury disease processes. Expression of peroxisome proliferator activated receptor-α (Pparα), which plays a role in the anti-inflammatory response by negatively regulating macrophage foamy cell differentiation (50), is upregulated at all time points after radiation. Increased Pparα expression is likely a compensatory response to the increase in foamy macrophages observed in C57BL/6J mice following radiation (51). Expression of IL-1β, TGF-β1, and Pparα was reduced in irradiated lungs of mice treated with AEOL10150 suggesting a reduction in overall pulmonary inflammation, which is consistent with the reduced lung weights and decreased macrophage infiltrate seen in rodents treated with AEOL10150 (11).
In conclusion, upregulation of hypoxia-associated genes begins following radiation and continues at least throughout the first six-month post-exposure. These genes participate in a variety of physiological and pathological functions, including hypoxia response, inflammation, oxidative stress, transcriptional regulation, signal transduction, cell metabolism, proliferation and differentiation, and apoptosis. Radiation-induced overexpression of 31 hypoxia related genes was suppressed six weeks after radiation following four weeks of post-radiation administration of AEOL10150, a potent catalytic antioxidant. It has been previously demonstrated that AEOL10150 can attenuate development of late radiation-induced pulmonary injury in rats following single dose(10) or fractionated radiation(12). A relationship between early signaling changes and late injury would support the initiation of therapeutic interventions in the first few weeks post-exposure in order to protect the lungs from developing acute and chronic lung damage. This may not be the only therapeutic option, however, as the biphasic wave of gene expression observed in this study and in previous studies (52) suggests that there may be a larger window of opportunity for intervening in the progression of disease processes. More importantly to this study, however, was that the observed reversal of radiation-induced gene expression following treatment with AEOL10150 suggests that these genes may be involved in the cellular response to oxidative stress or the generation of ROS/RNS. Identifying the roles of these genes in pulmonary radiation injury could provide insight into the role of hypoxia-associated genes in radiation lung injury, widen the temporal window for therapeutic interventions, and introduce novel therapeutic targets.
Highlights.
Oxidative stress and tissue hypoxia underlie the pathogenesis of radiation injury in lung tissue
Hypoxia-associated gene expression is dynamically altered in lung tissue after radiation
mRNA expression and protein stabilization of HIF-1 alpha and HIF-2 alpha differ with respect to time
AEOL10150 reduces expression of 31 of 32 hypoxia-associated genes elevated in response to radiation
ACKNOWLEDGEMENTS
We would like to thank Ross McGurk and Greta Toncheva for performing radiation dosimetry, validation, and quality assurance. This study was supported by the National Institutes of Health Grant U19-AI067798-06 and by AEOLUS Pharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
Isabel L. Jackson is a consultant for AEOLUS Pharmaceuticals, Inc. All other authors declare no conflict of interest.
REFERENCES
- 1.Fleckenstein K, Zgonjanin L, Chen L, Rabbani Z, Jackson IL, Thrasher B, et al. Temporal onset of hypoxia and oxidative stress after pulmonary irradiation. International journal of radiation oncology, biology, physics. 2007;68(1):196–204. doi: 10.1016/j.ijrobp.2006.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barcellos-Hoff MH, Dix TA. Redox-mediated activation of latent transforming growth factor-beta 1. Molecular endocrinology (Baltimore, Md. 1996;10(9):1077–1083. doi: 10.1210/mend.10.9.8885242. [DOI] [PubMed] [Google Scholar]
- 3.Cucoranu I, Clempus R, Dikalova A, Phelan PJ, Ariyan S, Dikalov S, et al. NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ Res. 2005;97(9):900–907. doi: 10.1161/01.RES.0000187457.24338.3D. [DOI] [PubMed] [Google Scholar]
- 4.Sturrock A, Cahill B, Norman K, Huecksteadt TP, Hill K, Sanders K, et al. Transforming growth factor-beta1 induces Nox4 NAD(P)H oxidase and reactive oxygen species-dependent proliferation in human pulmonary artery smooth muscle cells. American journal of physiology. 2006;290(4):L661–L673. doi: 10.1152/ajplung.00269.2005. [DOI] [PubMed] [Google Scholar]
- 5.Ward WF, Solliday NH, Molteni A, Port CD. Radiation injury in rat lung. II. Angiotensin-converting enzyme activity. Radiation research. 1983;96(2):294–300. [PubMed] [Google Scholar]
- 6.Bosco MC, Puppo M, Blengio F, Fraone T, Cappello P, Giovarelli M, et al. Monocytes and dendritic cells in a hypoxic environment: Spotlights on chemotaxis and migration. Immunobiology. 2008;213(9–10):733–749. doi: 10.1016/j.imbio.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22(37):5734–5754. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- 8.Yakovlev VA, Rabender CS, Sankala H, Gauter-Fleckenstein B, Fleckenstein K, Batinic-Haberle I, et al. Proteomic analysis of radiation-induced changes in rat lung: Modulation by the superoxide dismutase mimetic MnTE-2-PyP(5+) International journal of radiation oncology, biology, physics. 2010;78(2):547–554. doi: 10.1016/j.ijrobp.2010.03.037. PMCID: 2939237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batinic-Haberle I, Spasojevic I, Tse HM, Tovmasyan A, Rajic Z, St Clair DK, et al. Design of Mn porphyrins for treating oxidative stress injuries and their redox-based regulation of cellular transcriptional activities. Amino Acids. 2010 doi: 10.1007/s00726-010-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabbani ZN, Batinic-Haberle I, Anscher MS, Huang J, Day BJ, Alexander E, et al. Long-term administration of a small molecular weight catalytic metalloporphyrin antioxidant, AEOL 10150, protects lungs from radiation-induced injury. International journal of radiation oncology, biology, physics. 2007;67(2):573–580. doi: 10.1016/j.ijrobp.2006.09.053. PMCID: 1819401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabbani ZN, Mi J, Zhang Y, Delong M, Jackson IL, Fleckenstein K, et al. Hypoxia Inducible Factor 1 alpha Signaling in Fractionated Radiation-Induced Lung Injury: Role of Oxidative Stress and Tissue Hypoxia. Radiation research. 2010;173(2):165–174. doi: 10.1667/RR1816.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabbani ZN, Salahuddin FK, Yarmolenko P, Batinic-Haberle I, Thrasher BA, Gauter-Fleckenstein B, et al. Low molecular weight catalytic metalloporphyrin antioxidant AEOL 10150 protects lungs from fractionated radiation. Free radical research. 2007;41(11):1273–1282. doi: 10.1080/10715760701689550. [DOI] [PubMed] [Google Scholar]
- 13.Briere TM, Lii J, Prado K, Gillin MT, Sam Beddar A. Single-use MOSFET radiation dosimeters for the quality assurance of megavoltage photon beams. Physics in medicine and biology. 2006;51(5):1139–1144. doi: 10.1088/0031-9155/51/5/006. [DOI] [PubMed] [Google Scholar]
- 14.Beddar AS, Salehpour M, Briere TM, Hamidian H, Gillin MT. Preliminary evaluation of implantable MOSFET radiation dosimeters. Physics in medicine and biology. 2005;50(1):141–149. doi: 10.1088/0031-9155/50/1/011. [DOI] [PubMed] [Google Scholar]
- 15.Jackson IL, Vujaskovic Z, Down JD. A Further Comparison of Pathologies after Thoracic Irradiation among Different Mouse Strains: Finding the Best Preclinical Model for Evaluating Therapies Directed Against Radiation-Induced Lung Damage. Radiation research. 2011 doi: 10.1667/RR2421.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauter-Fleckenstein B, Fleckenstein K, Owzar K, Jiang C, Reboucas JS, Batinic-Haberle I, et al. Early and late administration of MnTE-2-PyP5+ in mitigation and treatment of radiation-induced lung damage. Free radical biology & medicine. 2010;48(8):1034–1043. doi: 10.1016/j.freeradbiomed.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vujaskovic Z, Anscher MS, Feng QF, Rabbani ZN, Amin K, Samulski TS, et al. Radiation-induced hypoxia may perpetuate late normal tissue injury. International journal of radiation oncology, biology, physics. 2001;50(4):851–855. doi: 10.1016/s0360-3016(01)01593-0. [DOI] [PubMed] [Google Scholar]
- 18.Jackson IL, Chen L, Batinic-Haberle I, Vujaskovic Z. Superoxide dismutase mimetic reduces hypoxia-induced O2*-, TGF-beta, and VEGF production by macrophages. Free radical research. 2007;41(1):8–14. doi: 10.1080/10715760600913150. [DOI] [PubMed] [Google Scholar]
- 19.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nature reviews. 2008;8(6):425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maisin JR. The influence of radiation on blood vessels, circulation. Chapter 3. Ultrastructure of the vessel wall. Current topics in radiation research quarterly. 1974;10(1):29–57. [PubMed] [Google Scholar]
- 21.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(14):7987–7992. doi: 10.1073/pnas.95.14.7987. PMCID: 20916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, et al. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26(1):63–74. doi: 10.1016/j.molcel.2007.02.024. PMCID: 2905600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284(25):16767–16775. doi: 10.1074/jbc.M901790200. PMCID: 2719312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9(6):677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 25.Pugh CW, Ratcliffe PJ. The von Hippel-Lindau tumor suppressor, hypoxia-inducible factor-1 (HIF-1) degradation, and cancer pathogenesis. Seminars in cancer biology. 2003;13(1):83–89. doi: 10.1016/s1044-579x(02)00103-7. [DOI] [PubMed] [Google Scholar]
- 26.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11(1):72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberger C, Mandriota S, Jurgensen JS, Wiesener MS, Horstrup JH, Frei U, et al. Expression of hypoxia-inducible factor-1alpha and -2alpha in hypoxic and ischemic rat kidneys. J Am Soc Nephrol. 2002;13(7):1721–1732. doi: 10.1097/01.asn.0000017223.49823.2a. [DOI] [PubMed] [Google Scholar]
- 28.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10(5):413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 29.Scortegagna M, Ding K, Oktay Y, Gaur A, Thurmond F, Yan LJ, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat Genet. 2003;35(4):331–340. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- 30.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17(11):3005–3015. doi: 10.1093/emboj/17.11.3005. PMCID: 1170640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25(13):5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. PMCID: 1157001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, et al. Regulation of HIF-1alpha stability through S-nitrosylation. Molecular cell. 2007;26(1):63–74. doi: 10.1016/j.molcel.2007.02.024. PMCID: 2905600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dewhirst MW. Relationships between cycling hypoxia, HIF-1, angiogenesis and oxidative stress. Radiation research. 2009;172(6):653–665. doi: 10.1667/RR1926.1. PMCID: 2790140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epperly MW, Sikora CA, DeFilippi SJ, Gretton JE, Bar-Sagi D, Archer H, et al. Pulmonary irradiation-induced expression of VCAM-I and ICAM-I is decreased by manganese superoxide dismutase-plasmid/liposome (MnSOD-PL) gene therapy. Biol Blood Marrow Transplant. 2002;8(4):175–187. doi: 10.1053/bbmt.2002.v8.pm12014807. [DOI] [PubMed] [Google Scholar]
- 35.Moulder JE, Cohen EP. Future strategies for mitigation and treatment of chronic radiation-induced normal tissue injury. Seminars in radiation oncology. 2007;17(2):141–148. doi: 10.1016/j.semradonc.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt M, Gerlach F, Avivi A, Laufs T, Wystub S, Simpson JC, et al. Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J Biol Chem. 2004;279(9):8063–8069. doi: 10.1074/jbc.M310540200. [DOI] [PubMed] [Google Scholar]
- 37.Burmester T, Gerlach F, Hankeln T. Regulation and role of neuroglobin and cytoglobin under hypoxia. Advances in experimental medicine and biology. 2007;618:169–180. doi: 10.1007/978-0-387-75434-5_13. [DOI] [PubMed] [Google Scholar]
- 38.Lv Y, Wang Q, Diao Y, Xu R. Cytoglobin: a novel potential gene medicine for fibrosis and cancer therapy. Current gene therapy. 2008;8(4):287–294. doi: 10.2174/156652308785160656. [DOI] [PubMed] [Google Scholar]
- 39.Xu R, Harrison PM, Chen M, Li L, Tsui TY, Fung PC, et al. Cytoglobin overexpression protects against damage-induced fibrosis. Mol Ther. 2006;13(6):1093–1100. doi: 10.1016/j.ymthe.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 40.Nakatani K, Okuyama H, Shimahara Y, Saeki S, Kim DH, Nakajima Y, et al. Cytoglobin/STAP, its unique localization in splanchnic fibroblast-like cells and function in organ fibrogenesis. Lab Invest. 2004;84(1):91–101. doi: 10.1038/labinvest.3700013. [DOI] [PubMed] [Google Scholar]
- 41.Mimura I, Nangaku M, Nishi H, Inagi R, Tanaka T, Fujita T. Cytoglobin, a novel globin, plays an antifibrotic role in the kidney. Am J Physiol Renal Physiol. 2010;299(5):F1120–F1133. doi: 10.1152/ajprenal.00145.2010. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Jackson IL, Zhang Y, Rabbani Z, Xu P, Hadley C, et al. Hypo-CpG methylation controls PTEN expression and cell apoptosis in irradiated lung. 2011 doi: 10.1080/10715762.2016.1189078. Unpublished Data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Zhang X, Rabbani ZN, Jackson IL, Vujaskovic Z. Oxidative stress mediates lung injury by inducing apoptosis Int. J Radiat Oncol. doi: 10.1016/j.ijrobp.2011.08.005. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y, Hupp TR, Stevens C. Death-associated protein kinase (DAPK) and signal transduction: additional roles beyond cell death. FEBS J. 2010;277(1):48–57. doi: 10.1111/j.1742-4658.2009.07411.x. [DOI] [PubMed] [Google Scholar]
- 45.Drakopanagiotakis F, Xifteri A, Polychronopoulos V, Bouros D. Apoptosis in lung injury and fibrosis. Eur Respir J. 2008;32(6):1631–1638. doi: 10.1183/09031936.00176807. [DOI] [PubMed] [Google Scholar]
- 46.Lewis JS, Lee JA, Underwood JC, Harris AL, Lewis CE. Macrophage responses to hypoxia: relevance to disease mechanisms. J Leukoc Biol. 1999;66(6):889–900. doi: 10.1002/jlb.66.6.889. [DOI] [PubMed] [Google Scholar]
- 47.Hempel SL, Monick MM, Hunninghake GW. Effect of hypoxia on release of IL-1 and TNF by human alveolar macrophages. American journal of respiratory cell and molecular biology. 1996;14(2):170–176. doi: 10.1165/ajrcmb.14.2.8630267. [DOI] [PubMed] [Google Scholar]
- 48.Carmi Y, Voronov E, Dotan S, Lahat N, Rahat MA, Fogel M, et al. The role of macrophage-derived IL-1 in induction and maintenance of angiogenesis. J Immunol. 2009;183(7):4705–4714. doi: 10.4049/jimmunol.0901511. [DOI] [PubMed] [Google Scholar]
- 49.McCourtie AS, Farivar AS, Woolley SM, Merry HE, Wolf PS, Mackinnon-Patterson B, et al. Alveolar macrophage secretory products effect type 2 pneumocytes undergoing hypoxia-reoxygenation. Ann Thorac Surg. 2008;86(6):1774–1779. doi: 10.1016/j.athoracsur.2008.07.071. PMCID: 2659526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384(6604):39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 51.Chiang CS, Liu WC, Jung SM, Chen FH, Wu CR, McBride WH, et al. Compartmental responses after thoracic irradiation of mice: strain differences. International journal of radiation oncology, biology, physics. 2005;62(3):862–871. doi: 10.1016/j.ijrobp.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 52.Rubin P, Johnston CJ, Williams JP, McDonald S, Finkelstein JN. A perpetual cascade of cytokines postirradiation leads to pulmonary fibrosis. International journal of radiation oncology, biology, physics. 1995;33(1):99–109. doi: 10.1016/0360-3016(95)00095-G. [DOI] [PubMed] [Google Scholar]