Abstract
A system to characterize normal liver, cirrhotic liver and hepatocellular carcinoma (HCC) evolved on cirrhotic liver is proposed in this paper. The study is performed with 56 real ultrasound images (15 normal, 16 cirrhotic and 25 HCC liver images) taken from 56 subjects. A total of 180 nonoverlapping regions of interest (ROIs), i.e. 60 from each image class, are extracted by an experienced participating radiologist. The multiresolution wavelet packet texture descriptors, i.e. mean, standard deviation and energy features, are computed from all 180 ROIs by using various compact support wavelet filters including Haar, Daubechies (db4 and db6), biorthogonal (bior3.1,bior3.3 and bior4.4), symlets (sym3 and sym5) and coiflets (coif1 and coif2). It is observed that a combined texture descriptor feature vector of length 48 consisting of 16 mean, 16 standard deviation and 16 energy features estimated from all 16 subband feature images (wavelet packets) obtained by second-level decomposition with two-dimensional wavelet packet transform by using Haar wavelet filter gives the best characterization performance of 86.6 %. Feature selection by genetic algorithm-support vector machine method increased the classification accuracy to 88.8 % with sensitivity of 90 % for detecting normal and cirrhotic cases and sensitivity of 86.6 % for HCC cases. Considering limited sensitivity of B-mode ultrasound for detecting HCCs evolved on cirrhotic liver, the sensitivity of 86.6 % for HCC lesions obtained by the proposed computer-aided diagnostic system is quite promising and suggests that the proposed system can be used in a clinical environment to support radiologists in lesion interpretation.
Keywords: Texture analysis, Liver ultrasound images, Cirrhosis, Hepatocellular carcinoma, Small hepatocellular carcinoma, Large hepatocellular carcinoma, Wavelet packet transform, Multiresolution analysis, Genetic algorithm, Support vector machines, Computer-aided diagnostic system
Introduction
Although biopsy is the “gold standard” for diagnosing liver diseases, ultrasonography is mostly preferred for screening, due to its noninvasive, nonradioactive and inexpensive nature. Echotexture of normal liver as it appears on ultrasound (US) is homogeneous with medium echogenicity, and it exhibits same or slightly increased echogenicity compared to the right kidney. Cirrhosis is considered to be the end stage of chronic hepatopathies which often leads to hepatocellular carcinoma (HCC). The diagnosis of cirrhosis is best achieved by looking at the granular structure of the liver parenchyma and the degree of nodularity present in the heterogeneous echotexture. HCC is viewed as most probable solid primary malignant liver lesion occurring on cirrhotic liver. Most small HCCs are diagnosed with a follow-up procedure for patients with cirrhosis. In few cases when HCC develops on normal liver parenchyma, it can be easily diagnosed by its sonographic appearance as it appears as a well-differentiated HCC or as fibrolamellar HCC (commonly appears with calcified areas). A lesion can be labeled as typical in appearance when its subjective diagnosis can be made with a good confidence level by looking at the US examination. The associated radiologists opined that no sonographic appearance is typical for HCCs as they exhibit a high degree of variability in terms of sonographic appearances even with in small HCCs (SHCCs) and large HCCs (LHCCs).
SHCCs frequently appear as hypoechoic nodule (solid tumor nodule without necrosis) or as hyperechoic nodule (solid tumor likely containing fat). In very few cases, SHCCs can also be isoechoic (same echogenicity as surrounding parenchyma). The HCC lesion may also exhibit hyperechoic echotexture with a hypoechoic halo (rim-like structure surrounding HCC lesion) or alternatively hypoechoic echotexture with hyperechoic halo sign. LHCCs appear much more complex and heterogeneous with mixed echogenicity (coarse irregular internal echoes) as a result of areas of necrosis, fibrosis as well as active growth areas [1].
There are few related researches in literature for characterization of liver tissue as normal, cirrhotic or HCC. The study in [2] reports characterization between normal, cirrhotic and HCC liver by using combination of roughness and granularity texture descriptors computed at various resolutions along with Fourier power spectrum-based features by using Bayes classifier. In another related research [3], multi-threshold dimension feature vector based on fractal geometry is proposed for characterization of normal, cirrhotic and HCC liver. The study in [4] used fractal dimensions of subimages obtained at various resolutions with M-band wavelet transform as fractal feature vector for characterization of normal, cirrhotic and HCC liver. The study in [5] used the same fractal feature vector and developed a system to characterize normal, cirrhotic and HCC by fusion of classifiers. The study in [4, 5] have used images scanned by high-resolution scanner with 32-pixel/cm and 8-bit/pixel resolution. Their study reports the use of region of interest (ROI) size of 64 × 64 pixels, i.e. 2 cm by 2 cm for their image resolution; however, in small HCC lesions (<2 cm), it is not possible to extract such a large ROI. The data description reported in studies [2–5] does not describe if only HCCs developed on cirrhotic liver are considered and number of SHCCs and LHCCs considered.
Keeping in view the research perspective in literature, the current study is different in the sense that only HCC lesions evolved on cirrhotic liver are considered and the representative data set of HCC images consisting of both small HCC images (SHCCIs) and large HCC images (LHCCIs) is used for classifier design.
Although detection of HCC in early stages has important clinical value, at the same time it is observed that in many cases HCCs are detected in advanced stages; therefore, the participating radiologists opined that isolation of a single case series, i.e. HCC lesions into incipient (small) and advanced (large) HCC, for characterization is not adequate as a ROI from HCC lesion representing primary malignancy of liver should be predicted as HCC irrespective of the fact whether the ROI belongs to SHCC or LHCC.
Given the fact that conventional gray scale B-mode US offers limited sensitivity for detection of lesions developed on cirrhotic liver, differentiating the texture patterns of HCCs on top of cirrhosis from its preceding stage of cirrhosis presents a daunting challenge even for experienced radiologists; therefore, a US tissue characterization system capable of providing adequate discrimination between cirrhotic changes in the liver and HCC is highly desirable. The work presented in this paper addresses this issue.
Materials and Methods
Data Collection and Description
Data Collection
For the present work, 56 B-mode liver US images comprising of 15 normal, 16 cirrhotic and 25 HCC liver images were collected from the Department of Radiodiagnosis and Imaging, Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh, India over the time period from March 2010 to May 2011. The consent of patients for using these images for research was taken prior to recording. The medical ethics committee of PGIMER, Chandigarh, granted the ethical clearance to carry out this study. The direct digital images recorded by using Philips ATL HDI 5000 US machine equipped with multifrequency transducer of 2–5 MHz range were used. The size of the images is 800 × 564 pixels with gray scale consisting of 256 tones and horizontal as well as vertical resolution is 96 dpi. The following protocols were followed for data collection:
(1) The judgment regarding the diagnostic quality (free from artifacts) and representativeness of each image class (normal liver, cirrhotic liver and HCC evolved on cirrhotic liver) was made by two domain experts (co-authors of this paper) with 13 and 23 years of experience in US imaging. (2) In case of HCCs, further categorization into SHCC and LHCC was made by observing the size of the lesion in transverse and longitudinal views (The HCC lesions less than 2 cm in size are considered as SHCCs). The acquired dataset had 25 HCC images with 25 solitary HCC lesions (14 SHCCs and 11 LHCCs). The distinction between SHCCs and LHCCs was made during data collection solely for the purpose of having representative data in training set for designing the classifier. (3) Only those images in which HCC lesion developed on top of cirrhosis were considered. (4) For cirrhosis, only those cases which have a clear indication of liver cirrhosis with no other hepatopathy were included.
Selection of Regions of Interest
The following protocols were followed for cropping the ROIs from the image database:
(1) The ROIs were cropped by an experienced participating radiologist by using a specially designed ROI manager software developed in biomedical instrumentation laboratory, Indian Institute of Technology, Roorkee, India. This ROI manager software provided the radiologist the flexibility to load the image, choose the ROI size and shape, move the ROI to any desired location over the image, freeze the ROI at any location and crop the ROIs together after the position of all the ROIs for a particular image is frozen. (2). For normal and cirrhotic liver images, ROIs were cropped from right lobe of the liver such that it contains only liver parenchyma with no inhomogeneous structures like blood vessels and hepatic ducts. (3) For HCC images, maximum non-overlapping ROIs were cropped from well within the boundary of the lesion. (4) Necrotic areas within the HCC lesions were avoided.
Interpretation by Radiologists
Experienced participating radiologists confirmed the presence of cirrhosis and HCC lesions evolved on cirrhotic liver by using liver image assessment criteria including: (1) visual interpretation of sonographic features based on their knowledge and expertise, (2) follow-up of clinical history of the patient and other associated findings, and (3) imaging appearance on dynamic helical computed tomography/magnetic resonance imaging/pathological examinations and biopsy, which is an invasive procedure.
Data Set Description
To design a robust classification system, it was ensured that the constituent HCC images in the data set offered a high degree of variability in terms of size and sonographic features. The size of SHCC varied from 1.5 to 1.9 cm, and the size of LHCC varied from 2.1 to 5.6 cm.
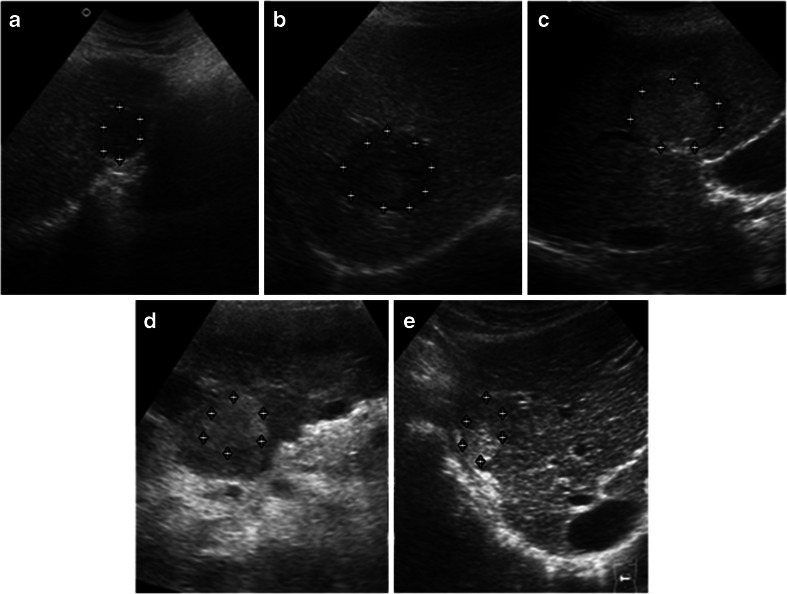
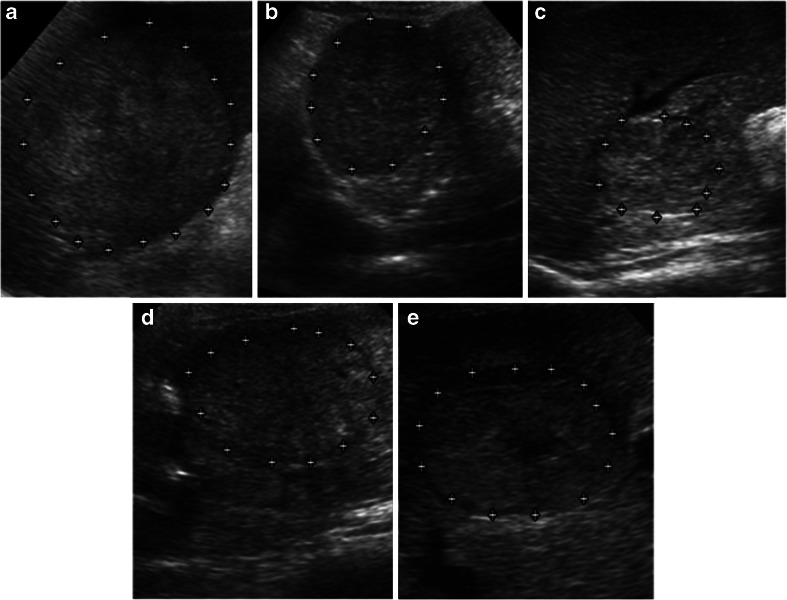
Figure 1a–e represents five SHCCI variants from the acquired image database. Figure 2a–e represents five LHCCI variants from the acquired image database.
Fig. 1.
a Hypoechoic SHCCI. b Hypoechoic SHCCI. c Hyperechoic SHCCI with hypoechoic halo. d Homogeneously hyperechoic SHCCI without halo. e Variant of SHCCI with mixed echogenicity (coexistence of hyperechoic and isoechoic areas)
Fig. 2.
a–e Heterogeneous echotexture represents complex and chaotic structure exhibited by LHCCI due to coexistence of areas of necrosis, fibrosis and active growth areas. Note: hypoechoic halo formation is visible in (d) and (e). Necrotic area is visible in the centre of LHHCI shown in (e)
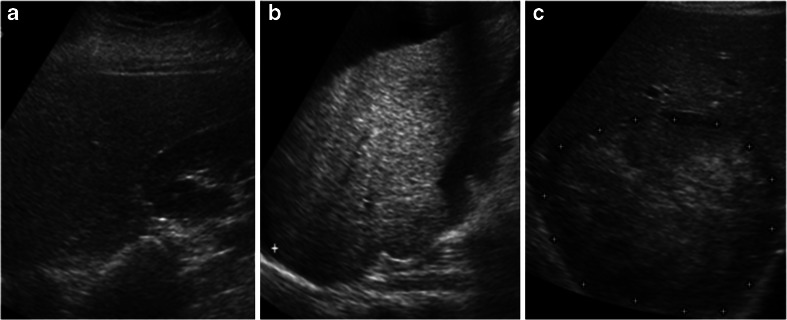
The highly experienced participating radiologists were of the view that the HCC image data set used in this study is a complete representative data offering a high degree of variability encountered during subjective analysis of these lesions in routine practice. Figure 3a–c represents samples of normal, cirrhotic and HCC liver images from the acquired image database.
Fig. 3.
a Normal liver image. b Cirrhotic liver image. c HCC liver image
Selection of ROI Size
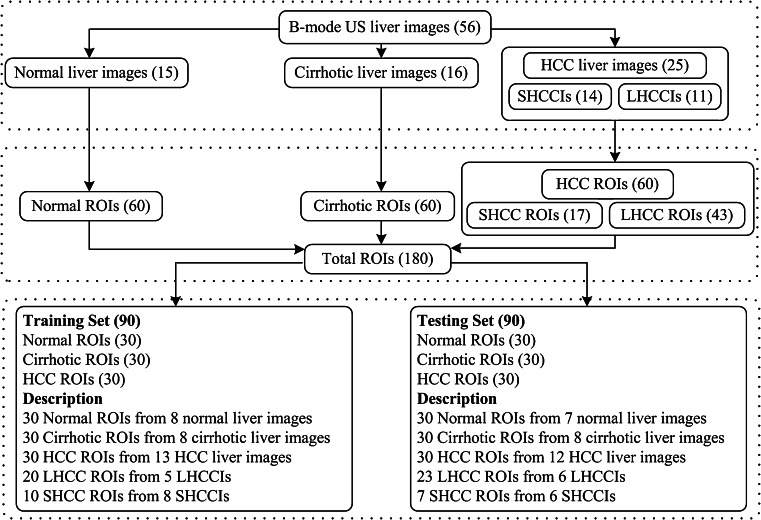
As the texture measurements are sensitive to the selection of ROI size, it should be chosen so as to provide a good statistical population. In other studies, it has been demonstrated that ROI size must be at least 800 pixels to provide good sampling distribution for estimating reliable statistics [6–8], whereas in few other related researches, a sample size of at least 1,000 pixels is suggested to estimate reliable statistics [9–11]. However, different ROI sizes ranging from 10 × 10 [12–15], 25 × 25 [16], 30 × 30 [3], 32 × 32 [2, 17–19], 40 × 40 [20–22], 50 × 50 [1, 17, 23, 24], 60 × 60 [6] and 64 × 64 pixels [1, 4, 5, 25] have been used for classification of liver diseases. After interaction with the participating radiologists, ROI size of 32 × 32 pixels was considered appropriate for the present study to estimate reliable statistics as well as to extract maximum ROIs from the acquired image database. The final data set consisting of total 180 ROIs with 60 normal ROIs (from 15 normal liver images), 60 cirrhotic ROIs (from 16 cirrhotic liver images) and 60 HCC ROIs (from 25 HCC liver images) was stored in a PC (Pentium Core-2-Duo, 2.67 GHz with 1.97 GB RAM). The distribution of acquired database among various liver image categories and its bifurcation into training and test set are summarized in Fig. 4.
Fig. 4.
Data set description (LHCCIs large HCC images, SHCCIs small HCC images, LHCC large HCC, SHCC small HCC)
Proposed Computer-Aided Diagnostic System
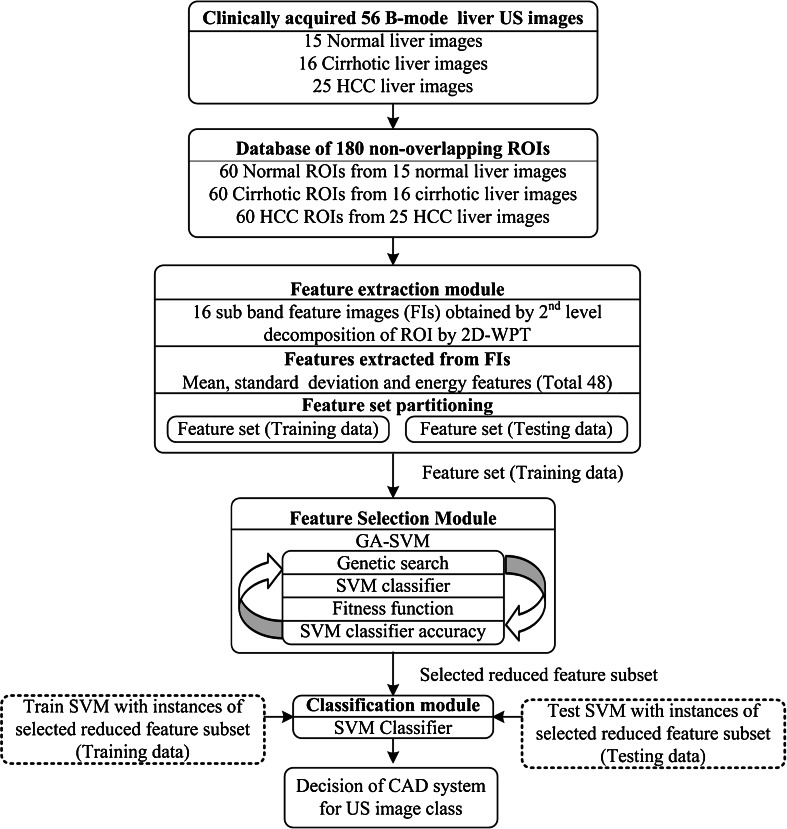
The block diagram of the proposed CAD system is shown in Fig. 5. For implementation of the proposed CAD system, database of 180 nonoverlapping ROIs was created from 56 clinically acquired US images. The CAD system consisted of three modules: (1) feature extraction module, (2) feature selection module and (3) classification module. In feature extraction module, each ROI in the database was decomposed up to second level of decomposition by two-dimensional WPT (2D-WPT), resulting in 16 subband feature images for each ROI. The texture descriptor feature vectors (TDFVs) of length 48 were extracted by estimating mean, standard deviation and energy features from all the 16 subband feature images for each ROI. In feature selection module, genetic algorithm–support vector machine (GA-SVM) feature selection method was used to find the optimal reduced TDFV which can significantly account for the textural variations exhibited by normal, cirrhotic and HCC liver. The instances of optimal reduced TDFV outputted by the feature selection module were passed to the classification module. In classification module, a multiclass SVM classifier is implemented using LibSVM library [26].
Fig. 5.
Proposed CAD system
Feature Extraction Module
Multiresolution Analysis
Feature extraction can be carried on a single scale by considering the spatial interactions which exist over small neighbourhoods for example by using gray-level co-occurrence matrix, gray level run length matrix, gray level difference statistics, neighbourhood gray tone dependence matrix, statistical feature matrix, etc. Feature extraction in transform domain is carried out over various scales by using multiresolution schemes such as discrete wavelet transform (DWT), stationary wavelet transform (SWT) and wavelet packet transform (WPT). Computing texture descriptors in transform domain is much more logical in the sense that human visual system processes images in a multiscale way and scale is a dominant aspect for analysis of texture [27]. In case of two-dimensional discrete wavelet transform (2D-DWT) as only the low-frequency subimage is recursively decomposed; it may not be efficient for texture characterization as most significant texture information usually appears in the middle- and high-frequency channels [28].
A speckled image of liver tissue is produced on US. It is a well-known fact that speckle in US images carries useful information and therefore cannot be treated as a typical random noise [29, 30]. As speckle represents high-frequency components of the US image, the 2D-WPT which is considered as richer space-frequency multiresolution analysis scheme may offer appropriate texture descriptors with reduced or no effect of speckle noise. Other researches where multiresolution wavelet packet texture descriptors have shown remarkable performances are [25, 31].
Wavelet Packet Transform
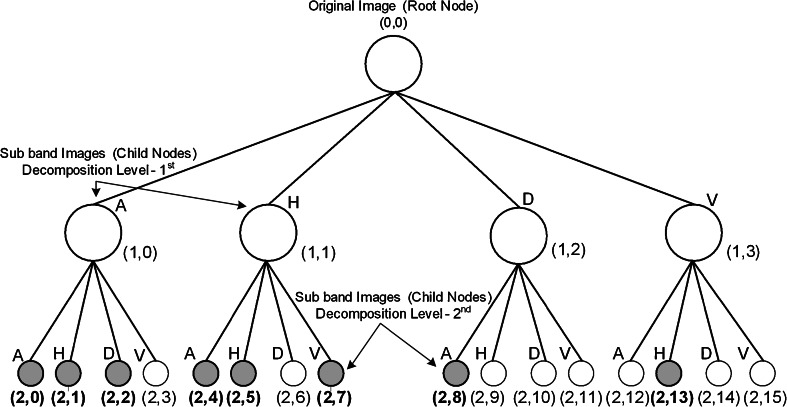
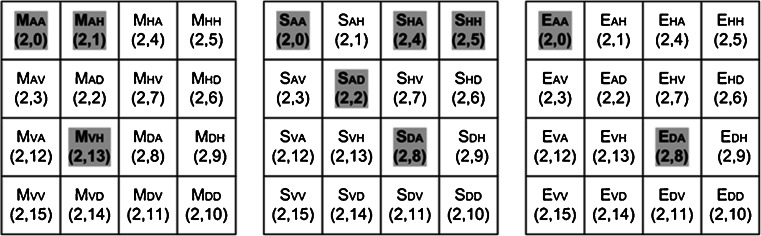
As a result of 2D-WPT decomposition, the complete frequency plane is subdivided into equal size bands. The 2D-WPT tree up to second level of decomposition results in 16 subband feature images (wavelet packets) each representing a band in the frequency plane as depicted in Fig. 6. Mean, standard deviation and energy features estimated from all 16 subband feature images result in a TDFV of length 48. Eight subband feature images (wavelet packets) are shaded and indicated in bold in Fig. 6. Ten texture descriptors (three mean features, five standard deviation features and two energy features) estimated from these eight subband feature images are selected by GA-SVM feature selection method shown as shaded and indicated in bold in Fig. 7.
Fig. 6.
2D-WPT tree up to second level of decomposition [from (2,0) to (2,15)] represents 16 subband feature images (wavelet packets). A approximate subband, H horizontal subband, D diagonal subband and V vertical subband
Fig. 7.
Features extracted from 2D-WPT tree at second level of decomposition (M mean, S standard deviation and E energy features). Ten features (three mean features, five standard deviation features and two energy features) estimated from eight subbands [(2,0), (2,1), (2,2), (2,4), (2,5), (2,7), (2,8) and (2,13)] feature images selected by GA-SVM feature selection method are shaded and indicated in bold
Selection of Wavelet Filter
The review of literature for texture characterization in transform domain using multiresolution features indicates that the choice of wavelet filter is important as the properties of these decomposition filters play a significant role in description of texture; specifically with 2D-DWT and 2D-WPT schemes, the choice of an appropriate wavelet filter affects the characterization performance. Studies in literature [25, 31–34] have shown empirical success by using different wavelet filters on specific classification tasks. The criteria like support width, shift invariance, orthogonality or biorthogonality and symmetry are important and must be considered for selecting an appropriate wavelet filter. Usually compact support wavelet filters are desired for ease of implementation. Orthogonality is another important property for conservation of energy at each decomposition level. Symmetry is required to avoid dephasing in processing images. For information on guidelines for selecting an appropriate wavelet filter, the readers may also look [32, 35]. In the present work, ten compact support wavelet filters including Haar, Daubechies (db 4 and db6), biorthogonal (bior3.1, bior3.3 and bior4.4), symlets (sym3 and sym5) and coiflets (coif1 and coif2) are considered for analysis with 2D-WPT. Comparison of important properties of wavelet filters used in this work is summarized in Table 1.
Table 1.
Comparison of properties of wavelet filters used
| Wavelet | Biorthogonal | Orthogonal | Symmetry | Asymmetry | Near symmetry | Compact support |
|---|---|---|---|---|---|---|
| Db | No | Yes | No | Yes | No | Yes |
| Haar | No | Yes | Yes | No | No | Yes |
| Bior | Yes | No | Yes | No | No | Yes |
| Coif | No | Yes | No | No | Yes | Yes |
| Sym | No | Yes | No | No | Yes | Yes |
Selection of Wavelet Packet Texture Descriptors
Extensive literature survey on texture classification in transform domain using multiresolution approaches like DWT, SWT and WPT reveals that mean, standard deviation and energy features are frequently used not only for classification of natural texture (i.e. Brodatz image database) [28, 33, 35, 36] but also for medical images [25, 31, 32]. For the present study, mean, standard deviation and energy features are computed for each subband feature image using the Eqs. (1), (2) and (3).
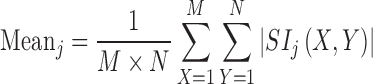 |
1 |
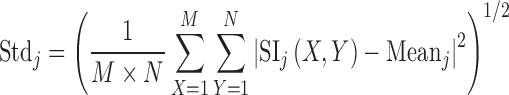 |
2 |
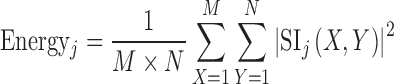 |
3 |
Here, SIj are subimages of size M × N at level j = 1, 2…5. As the size of the ROI is 32 by 32 pixels, decomposition up to fifth level is possible.
The study in [32] determined the best level of decomposition for 2D-WPT with entropy criterion-based best level algorithm according to which decomposition is carried out until the entropy of the subband is less than the sum of entropy of all its child subbands. However, in another study [37], it is reported that such a criterion for obtaining the best level of decomposition may not be suitable for texture classification tasks as small entropy value obtained from a particular subband may not necessarily indicate that the subband will separate the texture classes effectively. By second-level decomposition of a ROI with 2D-WPT, 16 subimages are obtained; computing mean, standard deviation and energy features from these 16 subimages yields a TDFV of length 48 (16 × 3). By subsequent third-, fourth- and fifth-level decomposition of ROI with 2D-WPT 64, 256 and 1,024 subimages are obtained; computing mean, standard deviation and energy features from these subimages results in large TDFVs of lengths 192 (64 × 3), 768 (256 × 3) and 3,072 (1,024 × 3), respectively. These large TDFVs were not considered for analysis due to computational efficiency constraints.
Wavelet packet decomposition tree up to second level of decomposition yields 16 subimages (wavelet packets) as shown in Fig. 6. Only mean feature estimated from these 16 subimages results in mean TDFV of length 16; similarly, the standard deviation TDFV and energy TDFV of length 16 are obtained. The combined TDFVs like mean Std TDFV consisting of mean and standard deviation features are of length 32; similarly, mean energy TDFV and Std energy TDFV of length 32 are obtained. The mean Std energy TDFV consisting of mean, standard deviation and energy features is of length 48.
Feature Selection
Designing CAD systems with smallest number of features is always desired as interference of irrelevant features can lead to reduced learning performance of the classifier which further increases the time taken to perform classification task and reduces the classification accuracy. GA-SVM feature selection method is used in this work to remove irrelevant features. For applying genetic algorithm (GA) to any problem, two steps are extremely important, adequate representation and appropriate fitness function. In the present work, binary representation is used for representing all possible feature subspaces of a given feature set, and the training accuracy obtained by the SVM classifier is used as fitness function. The main steps of GA-SVM feature selection method are:
Initialization: an initial population of possible candidate solutions (individuals or chromosomes) is created randomly.
Representation: each chromosome is a 48-bit binary mask where each bit corresponds to a single feature; 0 at any location in the bit string indicates that the corresponding feature is excluded and 1 indicates that it is included.
Fitness evaluation: the performance of each individual or chromosome is gauged by appropriate fitness function.
Selection: roulette wheel selection—the individuals or chromosomes which are deemed fit have high probability to enter the mating pool than those deemed unfit.
Crossover: the selected individuals in the mating pool are recombined with the probability Pc using crossover operator to produce next-generation offspring.
Mutation: a mutation operator is applied to these offspring with a low probability Pm to ensure that there is a always a variability added to the pool of solutions.
Fitness check: the offspring are then again evaluated using the fitness function, and those which have higher fitness values than the earlier individuals are considered for forming a new population.
At the end of one iteration, the new population formed after selection, crossover, mutation and fitness check contains old individuals which are fitter than the new individuals and those which are comparatively fitter amongst the new individuals. In this way, in subsequent generations, the chromosomes which are fittest mate more often and propagate their genetic material to the offspring which form potential candidate solutions, thus biasing the search space towards promising candidate subspaces. The GA search procedure terminates when either there is no improvement in the fitness value after a fixed number of successive iterations or after a predefined number of generations. In this study, single-point crossover was used, and the other GA run parameters are set as crossover rate equal to 0.6, mutation rate equal to 0.033, population size equal to 20 by manual optimization after a series of trails.
Classification
A generalization capability of the classifier is tested with instances of the feature vector which are not used in classifier design. The data set used in the present work consists of total 180 ROIs (60 normal ROIs taken from 15 normal images, 60 cirrhotic ROIs taken from 16 cirrhotic images and 60 HCC ROIs taken from 25 HCC images). The training set and testing set consist of 90 ROIs (30 ROIs from each image class). A complete description of dataset used in this study is summarized in Fig. 4.
SVM Classifier
It has been argued in [38, 39] that classifier designs which use regularization like support vector machines are less prone to overfitting and obtain good generalization performance to a certain extent even without feature space dimensionality reduction. For the present work, SVM classifier is chosen for the classification task. The SVM classifier attempts to construct an optimum hyper plane in the higher dimensional feature space to separate the training data with minimum expected risk. Kernel functions that satisfy Mercer’s theorem are used for nonlinear mapping of the training data from input space to higher dimensional feature space. In the present work, the performance of Gaussian radial basis function kernel is investigated. To avoid the bias caused by unbalanced feature values, all the extracted features were normalized in the range of [0, 1] by using min–max normalization procedure. For a detailed description of SVM approach, additional information can be found in [38, 39].
For multiclass classification, LibSVM library [26] uses one-against-one technique by constructing M(M − 1)/2 binary sub-classifiers where M is the number of classes. Each binary sub-classifier is trained to separate a pair of classes, and prediction is made by majority voting technique. In present three-class problem, the prediction of the class for an instance of TDFV of the test data set is made by majority voting mechanism on the predictions of three binary sub-classifiers, i.e. SVM (normal/cirrhosis), SVM (normal/HCC) and SVM (cirrhosis/HCC).
A crucial step for obtaining good generalization performance with SVM classifier is the correct choice of the regularization parameter C and kernel parameter γ. The regularization parameter C attempts to maximize the margin while keeping low value for training error. In the present work, extensive search is carried out in the parameter space for the values of C є {2−4, 2−3,……., 215} and γ є {2−12, 2−11,……., 25} using tenfold cross-validation to obtain optimal values of C and γ for training the SVM model.
Classification Performance
In addition to the classification accuracy, the sensitivity, i.e. true positive rate, for each class is also estimated. Sensitivity for a particular class say for HCC ROIs abbreviated as sensitivityH is the ratio of the number of correctly classified HCC ROIs over the total number of actual HCC ROIs.
Results
Initially, all the seven TDFVs, i.e. mean TDFV, standard deviation TDFV, energy TDFV, mean Std TDFV, mean energy TDFV, Std energy TDFV and mean Std energy TDFV estimated from all 16 subband feature images obtained from second-level decomposition of a ROI with 2D-WPT by using ten compact support wavelet filters including Haar, Daubechies (db 4 and db6), biorthogonal (bior3.1, bior3.3 and bior4.4), symlets (sym3 and sym5) and coiflets (coif1 and coif2) were used for classification with SVM classifier to compare the capability of different wavelet filters to characterize textural variations for normal, cirrhotic and HCC liver. The maximum and minimum classification accuracies obtained by all the seven TDFVs with the corresponding wavelet filters are reported in Table 2.
Table 2.
Comparison of maximum and minimum accuracy obtained by all seven TDFVs with the corresponding wavelet filter
| Classification performance SVM | |||||
|---|---|---|---|---|---|
| TDFVs | L | Max. acc. | Wavelet filter | Min. acc. | Wavelet filter |
| Mean | 16 | 84.4 | Haar | 73.3 | sym3 |
| Std | 16 | 84.4 | Haar | 70.0 | sym5 |
| Energy | 16 | 81.1 | Haar | 72.2 | coif2, db6 |
| Mean Std | 32 | 84.4 | Haar | 76.6 | bior3.3 |
| Mean energy | 32 | 85.5 | Haar | 71.1 | sym3 |
| Std energy | 32 | 84.4 | Haar | 74.4 | coif1 |
| Mean Std energy | 48 | 86.6 | Haar | 74.4 | bior3.1 |
Note that for all seven TDFVs maximum accuracy is obtained by using Haar wavelet filter. However, the highest accuracy of 86.6 % (indicated in bold) is obtained by using Haar wavelet filter with Mean Std energy TDFV of length 48.
Accuracy values are expressed in percentage
TDFVs texture descriptor feature vectors, L length of TDFV
From Table 2, it is interesting to note that for all seven TDFVs, maximum classification accuracy is obtained by using Haar wavelet filter. The highest accuracy of 86.6 % is obtained by using Haar wavelet filter with mean Std energy TDFV (seventh row of Table 2).
The classification results (overall accuracy and sensitivity of each image class) obtained by all seven TDFVs using Haar wavelet filter are reported in Table 3. From Table 3, it can be visualized that the second highest accuracy of 85.5 % is obtained by using mean energy TDFV (5th row of Table 3). By including standard deviation features with mean and energy features, the sensitivity for cirrhosis has increased from 80.0 to 83.3 % (compare fifth and seventh rows of Table 3). It can be concluded that mean, standard deviation and energy features all contribute to capture the textural variations of normal, cirrhotic and HCC ROIs with highest accuracy of 86.6 % obtained by mean Std energy TDFV.
Table 3.
The classification results (overall accuracy and sensitivity of each image class) obtained by all seven TDFVs using Haar wavelet filter
| Classification performance SVM | |||||
|---|---|---|---|---|---|
| TDFVs | L | Accuracy | SensitivityN | SensitivityC | SensitivityH |
| Mean | 16 | 84.4 | 93.3 | 76.6 | 83.3 |
| Std | 16 | 84.4 | 90.0 | 83.3 | 80.0 |
| Energy | 16 | 81.1 | 86.6 | 70.0 | 76.6 |
| Mean Std | 32 | 84.4 | 93.3 | 80.0 | 80.0 |
| Mean energy | 32 | 85.5 | 93.3 | 80.0 | 83.3 |
| Std energy | 32 | 84.4 | 90.0 | 80.0 | 83.3 |
| Mean Std energy | 48 | 86.6 | 93.3 | 83.3 | 83.3 |
Accuracy and sensitivity values are expressed in percentage
TDFVs texture descriptor feature vectors, L length of TDFV, SensitivityN sensitivity for normal, SensitivityC sensitivity for cirrhosis, SensitivityH sensitivity for HCC
The classification results (overall accuracy and sensitivity of each image class) obtained by using mean Std energy TDFVs with all ten wavelet filters are reported in Table 4. It can be observed that the highest classification accuracy of 86.6 % is obtained by mean Std energy TDFV with Haar wavelet filter (fourth row of Table 4). It can also be noted that the highest sensitivity of 83.3 % for cirrhosis and HCC cases is obtained in this case.
Table 4.
The classification results (overall accuracy and sensitivity of each image class) obtained by using mean Std energy TDFVs with all ten compact support wavelet filters
| Classification performance SVM | ||||
|---|---|---|---|---|
| Wavelet filter | Accuracy | SensitivityN | SensitivityC | SensitivityH |
| bior3.1 | 74.4 | 93.3 | 73.3 | 56.6 |
| bior3.3 | 77.7 | 100.0 | 66.6 | 66.6 |
| bior4.4 | 76.6 | 83.3 | 70.0 | 76.6 |
| Haar | 86.6 | 93.3 | 83.3 | 83.3 |
| db4 | 80.0 | 86.6 | 80.0 | 73.3 |
| db6 | 80.0 | 93.3 | 76.6 | 70.0 |
| sym3 | 78.8 | 90.0 | 73.3 | 73.3 |
| sym5 | 76.6 | 83.3 | 73.3 | 73.3 |
| coif1 | 81.1 | 90.0 | 83.3 | 70.0 |
| coif2 | 78.8 | 86.6 | 73.3 | 76.6 |
Accuracy and sensitivity values are expressed in percentage
SensitivityN sensitivity for normal, SensitivityC sensitivity for cirrhosis, SensitivityH sensitivity for HCC
It can be concluded that compactly supported, orthogonal and symmetric Haar wavelet filter is suitable for use with 2D-WPT multiresolution scheme along with SVM classifier for characterizing the normal, cirrhotic and HCC liver. However, since the length of mean Std energy TDFV (16 mean, 16 standard deviation and 16 energy features) is 48 and the total number of training instances is 90, feature selection with GA as search procedure and classification accuracy of the SVM classifier as fitness function is used for removing noisy, non-informative and redundant features. The GA-SVM feature selection method selected a subset of 10 features, i.e. 3 mean, 5 standard deviation and 2 energy features (highlighted in Fig. 7) out of total 48 features, i.e. 16 mean, 16 standard deviation and 16 energy features (shown in Fig. 7). It can also be noted that the ten features of optimal reduced TDFV (highlighted in Fig. 7) are estimated from eight subband feature images (highlighted in Fig. 6).
The classification results obtained by using mean Std energy TDFV with Haar wavelet filter and optimal reduced TDFV selected by GA-SVM method by using SVM classifier are reported in Table 5. It can be observed that optimal reduced TDFV of length 10 selected by GA-SVM feature selection method gives the overall accuracy of 88.8 %; however, the accuracy achieved by using mean Std energy TDFV of length 48 is 86.6 %. The other interesting fact is that by use of optimal reduced TDFV, the sensitivity for detecting abnormal cases, i.e. cirrhotic as well as HCC cases, has increased. The sensitivity for HCC cases has increased from 83.3 to 86.6 %, and sensitivity for cirrhosis cases has increased from 83.3 to 90.0 %. The results obtained are promising as the sensitivity of conventional gray scale B-mode US for detecting HCC lesions evolved on cirrhotic liver is limited, and it is reported that contrast enhanced US improves the sensitivity to around 85 % [40]. The sensitivity of the proposed CAD system for detecting HCCs is 86.6 % with conventional B-mode US images. The results of the study indicate that optimal reduced TDFV consisting of ten features (three mean and five standard deviation and two energy features) estimated from eight subband feature images (wavelet packets) obtained by 2D-WPT using Haar wavelet filter can significantly account for textural variations exhibited by a variety of HCCs evolved on cirrhotic liver as well as cirrhotic and normal liver.
Table 5.
The classification results obtained by using mean Std energy TDFV with Haar wavelet filter and optimal reduced TDFV selected by GA-SVM method
| Classification performance SVM | |||||||
|---|---|---|---|---|---|---|---|
| TDFVs | L | Confusion matrix | Sensitivity | Accuracy | |||
| Mean Std energy TDFV | 48 | N | C | H | 86.6 | ||
| N | 28 | 0 | 2 | 93.3 | |||
| C | 0 | 25 | 5 | 83.3 | |||
| H | 1 | 4 | 25 | 83.3 | |||
| Optimal reduced TDFV | 10 | N | C | H | 88.8 | ||
| N | 27 | 0 | 3 | 90.0 | |||
| C | 0 | 27 | 3 | 90.0 | |||
| H | 2 | 2 | 26 | 86.6 | |||
Mean Std energy features, 16 mean features, 16 standard deviation features and 16 energy features; optimal reduced TDFV, three mean, five standard deviation and two energy features selected by GA-SVM method. Accuracy and sensitivity values are expressed in percentage
TDFVs texture descriptor feature vectors, L length of TDFV, N normal, C cirrhosis, H HCC
Discussion
Brief details of CAD systems proposed in literature for characterization of normal, cirrhotic and HCC liver from B-mode US images are given in Table 6. The study in [4] and [5] has used images scanned by a high-resolution scanner with 32-pixel/cm and 8-bit/pixel resolution. They have used the ROI size of 64 × 64 pixels, i.e. 2 × 2 cm for their images; however, in small HCC lesions (<2 cm), it is not possible to extract such a large ROI. The data description reported in studies [2–5] does not describe if only HCCs developed on cirrhotic liver are considered and the number of SHCCs and LHCCs considered. The direct comparison of the present study with other related researches is not possible because image databases and image acquisition methods are different. However, it can be stated that the proposed approach for characterization between normal, cirrhotic and HCC liver yields comparable results with use of comprehensive and representative training data for classifier design.
Table 6.
Brief details of CAD systems proposed in literature for characterization of normal, cirrhotic and HCC liver
| Dataset description | Classification performance SVM | |||||||
|---|---|---|---|---|---|---|---|---|
| Authors | Patients | Images per class | No. of ROIs | ROI Size | SensitivityN | SensitivityC | SensitivityH | Accuracy |
| Wu et al. [2](1992) | 45 | Normal–15 | 90 | 32 × 32 | 86.6 | 100 | 83.3 | 90 |
| Cirrhotic–15 | ||||||||
| HCC–15 | ||||||||
| Wu et al. [3](1993) | – | – | 90 | 30 × 30 | 80 | 90 | 93.3 | 87.8 |
| Le et al. [4](2004) | – | – | 150 | 64 × 64 | 92 | 100 | 96 | 96 |
| Le et al. [5](2007) | – | – | 432 | 64 × 64 | 100 | 91.5 | 94.5 | 95.3 |
| Present study (2012) | 56 | Normal–15 | 180 | 32 × 32 | 90 | 90 | 86.7 | 88.8 |
| Cirrhotic–16 | ||||||||
| HCC–25 | ||||||||
Twenty-five HCC images consisting of 25 solitary HCC lesions (14 small HCC lesions and 11 large HCC lesions) developed on top of cirrhotic liver are used in this study. Accuracy and sensitivity values are expressed in percentage
Misclassification Analysis
The 12 HCC images of the test data were reviewed by an experienced participating radiologist, and the remarks are summarized in Table 7. From Table 7, it can be observed that HCC ROIs predicted as normal belong to two different LHCC images (image nos. 3 and 12 in Table 7) and the remaining two misclassified HCC ROIs predicted as cirrhotic belong to a single SHCC image (image no. 7 in Table 7). Experienced participating radiologists opined that ROI with identification number 3* might have been misclassified due to close proximity with the halo, although it was confirmed that the ROIs with identification numbers 3, 10, 11 and 29 are actual misclassifications of the proposed CAD system. ROIs with identification numbers 3 and 29 are patches inside HCC lesion which are predicted as normal by the proposed CAD system. As US is commonly used to facilitate liver lesion biopsy, the participating radiologist were of the view that any ROI inside the HCC lesion if predicted as normal should be avoided for taking the sample for biopsy.
Table 7.
Review remarks from experienced radiologists for 12 HCC images of the test set
| Image no. in test data | LHCCI/SHCCI | ROI identification no. | Prediction of proposed CAD | Remarks |
|---|---|---|---|---|
| 1 | SHCCI | 1 | Predicted as HCC | Correct |
| 2 | SHCCI | 2 | Predicted as HCC | Correct |
| 3 | LHCCI | 3a, 4, 5, 6 | 3 misclassified as normal | Incorrect |
| Remaining 4, 5, 6 Predicted as HCC | Correct | |||
| 4 | SHCCI | 7 | Predicted as HCC | Correct |
| 5 | SHCCI | 8 | Predicted as HCC | Correct |
| 6 | SHCCI | 9 | Predicted as HCC | Correct |
| 7 | SHCCI | 10, 11 | 10, 11 both misclassified as cirrhotic | Incorrect |
| 8 | LHCCI | 12,13,14 | All predicted as HCC | Correct |
| 9 | LHCCI | 15,16,17 | All predicted as HCC | Correct |
| 10 | LHCCI | 18,19,20 | All predicted as HCC | Correct |
| 11 | LHCCI | 21, 22, 23, 24, 25, 26, 27, 28 | All predicted as HCC | Correct |
| 12 | LHCCI | 29, 30 | 29 misclassified as normal | Incorrect |
| 30 predicted as HCC | Correct |
ROIs misclassified by the proposed CAD system are indicated in bold.
aMisclassified as normal and remarked as incorrect
Conclusion
In the present work, a CAD system for characterizing normal, cirrhotic and HCC liver has been developed by multiresolution texture analysis of B-mode liver US images. The proposed CAD system achieved overall classification accuracy of 88.8 % with the sensitivity of 90.0 % for normal and cirrhotic liver and 86.6 % for HCC liver with optimal reduced TDFV obtained by GA-SVM feature selection method and SVM classifier. Considering limited sensitivity of conventional B-mode gray scale US for detecting HCCs evolved on cirrhotic liver, the sensitivity of 86.6 % for HCC lesions obtained by the proposed CAD system is quite promising and suggests that the proposed system can be used in a clinical environment to support radiologists in lesion interpretation thereby improving diagnostic accuracy which can avoid unnecessary biopsies.
Acknowledgments
Author Jitendra Virmani would like to acknowledge Ministry of Human Resource Development (MHRD), India for financial support. The authors wish to acknowledge the Department of Electrical Engineering, Indian Institute of Technology, Roorkee, India and Department of Radiodiagnosis and Imaging, Postgraduate Institute of Medical Education and Research, Chandigarh, India for their constant patronage and support in carrying out this research work. The authors would like to thank the anonymous reviewers for their substantive and informed review, which led to significant improvements in the manuscript.
Contributor Information
Jitendra Virmani, Phone: +91-9876-053170, FAX: +91-1332-273560, Email: jitendra.virmani@gmail.com.
Vinod Kumar, Email: vinodfee@gmail.com.
Naveen Kalra, Email: navkal2004@yahoo.com.
Niranjan Khandelwal, Email: khandelwaln@hotmail.com.
References
- 1.Mitrea D, Nedevschi S, Lupsor M, Badea R: Exploring texture-based parameters for noninvasive detection of diffuse liver diseases and liver cancer from ultrasound Images. In: 8th WSEAS International Conference on Mathematical Methods and Computational Techniques in Electrical Engineering, MMACTEE'06. IEEE, Wisconsin, USA, 2006, pp 259–265
- 2.Wu CM, Chen YC, Hsieh KS. Texture features for classification of ultrasonic liver images. IEEE Trans Med Imag. 1992;11(2):141–152. doi: 10.1109/42.141636. [DOI] [PubMed] [Google Scholar]
- 3.Wu CM, Chen YC. Multi-threshold dimension vector for texture analysis and its application to liver tissue classification. Pattern Recogn. 1993;26(1):137–144. doi: 10.1016/0031-3203(93)90095-E. [DOI] [Google Scholar]
- 4.Lee WL, Hsieh KS, Chen YC. A study of ultrasonic liver images classification with artificial neural networks based on fractal geometry and multiresolution analysis. Biomed Eng Appl Basis Commun. 2004;16(2):59–67. doi: 10.4015/S1016237204000104. [DOI] [Google Scholar]
- 5.Lee WL, Hsieh KS: Computer assisted characterization for ultrasonic liver tissue by fusion of classifiers. In: Proceedings of Second IEEE International Conference on Innovative Computing, Information and Control, ICICIC'07. IEEE, Kumamoto, Japan, 2007, pp 100
- 6.Kadah YM, Farag AA, Zurada JM, Badawi AM, Youssef AM. Classification algorithms for quantitative tissue characterization of diffuse liver disease from ultrasound images. IEEE Trans Med Imaging. 1996;15(4):466–478. doi: 10.1109/42.511750. [DOI] [PubMed] [Google Scholar]
- 7.Singh M, Singh S, Gupta S: A new measure of echogenicity of ultrasound images for liver classification. In: Proceedings of 24th IEEE Canadian Conference, CCECE-2011. IEEE, Niagara Falls, Canada, 2011, pp 317–320
- 8.Fukunaga K. Introduction to Statistical Pattern Recognition. New York: Academic; 1990. [Google Scholar]
- 9.Badawi AM, Derbala AS, Youssef ABM. Fuzzy logic algorithm for quantitative tissue characterization of diffuse liver diseases from ultrasound images. Int J Med Inf. 1999;55:135–147. doi: 10.1016/S1386-5056(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 10.Badawi AM, Emara SM, Youssef ABM: Ultrasound tissue characterization of diffuse liver diseases using fuzzy rules. In: Proceedings of Third International Conference on Fuzzy Systems Systems and Intelligent Control, (IFSIC’94). Louisville, KY, 1994. pp 288–298
- 11.Emara SM, Badawi AM, Youssef ABM: Fuzzy similarity measures for ultrasound tissue characterization. In: Proceedings of Nonlinear Image Processing SPIE, 1995, (2424), pp 556–566
- 12.Balasubramanian D, Srinivasan P, Gurupatham R: Automatic classification of focal lesions in ultrasound liver images using principal component analysis and neural networks. In: Proceedings of 29th IEEE EMBS Annual International Conference. IEEE, Lyon, France, pp. 23–26, 2007 [DOI] [PubMed]
- 13.Poonguzhali S, Deepalakshmi, Ravindran G: Optimal feature selection and automatic classification of abnormal masses in ultrasound liver images. In: Proceedings of IEEE International Conference, ICSCN’07. IEEE, Chennai, India, 2007, pp 503–506
- 14.Poonguzhali S, Ravindran G. Automatic classification of focal lesions in ultrasound liver images using combined texture features. Inf Tech J. 2008;7(1):205–209. doi: 10.3923/itj.2008.205.209. [DOI] [Google Scholar]
- 15.Sujana S, Swarnamani S, Suresh S. Application of artificial neural networks for the classification of liver lesions by image texture parameters. Ultrasound Med Biol. 1996;22(9):1177–1181. doi: 10.1016/S0301-5629(96)00144-5. [DOI] [PubMed] [Google Scholar]
- 16.Mittal D, Kumar V, Saxena SC, Khandelwal N, Kalra N. Neural network based focal liver lesion diagnosis using ultrasound images. Int J Comput Med Imaging Graph. 2011;35(4):315–323. doi: 10.1016/j.compmedimag.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Mitrea D, Nedevschi S, Lupsor M, Badea R. Textural models based on ultrasound images for the detection of hepatocellular carcinoma in early and advanced stages. Int J Comput. 2008;2(1):66–73. [Google Scholar]
- 18.Kyriacou E, Pavlopoulos S, Koutsoris D, Zoumpoulis P, Theotokas I: Computer assisted characterization of liver tissue using image texture analysis techniques on B-scan images. In: Proceedings of 19th IEEE EMBS International Conference. IEEE, Chicago, IL, 1997, pp 806–809
- 19.Gangeh MJ, Duin RPW, Eswaran C, Romeny BMH: Scale space texture classification using combined classifiers with application to ultrasound tissue characterization. In: IFMBE Proceedings Biomed ’06, 2006, 15: pp 287–290
- 20.Virmani J, Kumar V, Kalra N, Khandelwal N: A rapid approach for prediction of liver cirrhosis based on first order statistics. In: Proceedings of IEEE International Conference on Multimedia, Signal Processing and Communication Technologies, IMPACT-2011, 212–215, 2011
- 21.Virmani J, Kumar V, Kalra N, Khandelwal N: Prediction of cirrhosis based on singular value decomposition of gray level co-occurrence matrix and a neural network classifier. In: Proceedings of IEEE International Conference on Developments in E-systems Engineering, DeSE-2011. IEEE, Dubai, UAE 2011, pp 146–151
- 22.Virmani J, Kumar V, Kalra N, Khandelwal N: Prediction of cirrhosis from liver ultrasound B-mode images based on Laws’ masks analysis. In: Proceedings of IEEE International Conference on Image Information Processing, ICIIP-2011. Waknaghat, HP, India, 2011, pp 1–5
- 23.Mitrea D, Nedevschi S, Lupsor M, Socaciu M, Badea R: Experimenting various classification techniques for improving the automatic diagnosis of the malignant liver tumors based on ultrasound images. In: Proceedings of International Congress on Image and Signal Processing, CISP-2010. IEEE, Yantai, China, 2010, pp 1853–1858
- 24.Mitrea D, Nedevschi S, Lupsor M, Socaciu M, Badea R: Improving the textural model of the hepatocellular carcinoma using dimensionality reduction methods. In: Proceedings of International Congress on Image and Signal Processing, CISP-2009. IEEE, Tianjin, China 2009, pp 1–5
- 25.Yoshida H, Casalino DD, Keserci B, Coskun A, Ozturk O, Savranlar A. Wavelet packet based texture analysis for differentiation between benign and malignant liver tumors in ultrasound images. Phys Med Biol. 2003;48:3735–3753. doi: 10.1088/0031-9155/48/22/008. [DOI] [PubMed] [Google Scholar]
- 26.Chang CC, Lin CJ: LIBSVM, A library of support vector machines, Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm. Accessed 15 March 2012.
- 27.Daugman J. An information-theoretic view of analog representations in the striate cortex. In: Schwartz EL, editor. Computational Neuroscience. Cambridge: MIT Press; 1990. pp. 403–424. [Google Scholar]
- 28.Chang T, Kuo CCJ. Texture analysis and classification with tree structured wavelet transform. IEEE Trans Image Proc. 1993;2(4):429–441. doi: 10.1109/83.242353. [DOI] [PubMed] [Google Scholar]
- 29.Sudha S, Suresh GR, Sukanesh R. Speckle noise reduction in ultrasound images using context-based adaptive wavelet thresholding. IETE J Res. 2009;55:135–143. doi: 10.4103/0377-2063.54905. [DOI] [Google Scholar]
- 30.Hiremath PS, Akkasaligar PT, Badiger S. Speckle reducing contourlet transform for medical ultrasound images. Int J Compt Inf Engg. 2010;4(4):284–291. [Google Scholar]
- 31.Wan J, Zhou S: Features extraction based on wavelet packet transform for B-mode ultrasound images. In: Proceedings 3rd International Congress on Image and Signal Processing, CISP-2010. IEEE, Yantai, China, 2010, pp 949–955
- 32.Tsiaparas NN, Golemati S, Andreadis I, Stoitsis JS, Valavanis I, Nikita KS. Comparison of multiresolution features for texture classification of carotid atherosclerosis from B-mode ultrasound. IEEE Trans Inf Tech Biomed. 2011;15(1):130–137. doi: 10.1109/TITB.2010.2091511. [DOI] [PubMed] [Google Scholar]
- 33.Avci E. Comparison of wavelet families for texture classification by using wavelet packet entropy adaptive network based fuzzy inference system. Appl Soft Comput. 2008;8:225–231. doi: 10.1016/j.asoc.2007.01.003. [DOI] [Google Scholar]
- 34.Rydgren E, Thomas EA, Amiel F, Rossant F, Amara A: Iris features extraction using wavelet packets. In: Proceedings of International Conference on Image Processing, ICIP-04. IEEE, Singapore, 2004, 2:861–864
- 35.Mojsilovic A, Popovic MV, Rackov DM. On the selection of an optimal wavelet basis for texture characterization. IEEE Trans Imag Proc. 2000;9(12):2043–2050. doi: 10.1109/83.887972. [DOI] [PubMed] [Google Scholar]
- 36.Xiaobin L, Zheng T: Neural Information Processing. Berlin: Springer Berlin, 2006, LNCS-4233: pp 474–480
- 37.Rajpoot N: Local discriminant wavelet packet basis for texture classification. In: Proceedings of SPIE Wavelets X. SPIE, San Diego, California, USA, 2003, 5207:774–783
- 38.Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min Knowl Disc. 1998;2(2):1–43. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- 39.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. J Machine Learn. 2002;46(1–3):1–39. [Google Scholar]
- 40.Harding J, Callaway M: Ultrasound of focal liver lesions. RAD Magazine, 2010, 36(424): pp 33–34