Abstract
Oxidative stress has been identified as a common mechanism for cellular damage and dysfunction in a wide variety of disease states. Current understanding of the metabolic changes associated with obesity and the development of insulin resistance has focused on the role of oxidative stress and its interaction with inflammatory processes at both the tissue and organismal level. Obesity-related oxidative stress is an important contributing factor in the development of insulin resistance in the adipocyte as well as the myocyte. Moreover, oxidative stress has been linked to mitochondrial dysfunction, and this is thought to play a role in the metabolic defects associated with oxidative stress. Of the various effects of oxidative stress, protein carbonylation has been identified as a potential mechanism underlying mitochondrial dysfunction. As such, this review focuses on the relationship between protein carbonylation and mitochondrial biology and addresses those features that point to either the causal or casual relationship of lipid peroxidation–induced protein carbonylation as a determining factor in mitochondrial dysfunction.
Mitochondrial metabolism and generation of reactive oxygen species
Reactive oxygen species (ROS)4 are generated as a constitutive by-product of aerobic respiration. Pathologic states such as infection and disease are associated with considerably increased production of ROS in a variety of tissues (1–4). Although ROS play an important role in normal cellular signaling (5), increased levels or prolonged exposure to ROS can lead to pathologic changes to a variety of cellular components, including the modification of DNA, RNA, carbohydrates, proteins, and lipids (6). These changes have been associated with detrimental changes in cellular metabolism and induction of apoptosis or cell death (7).
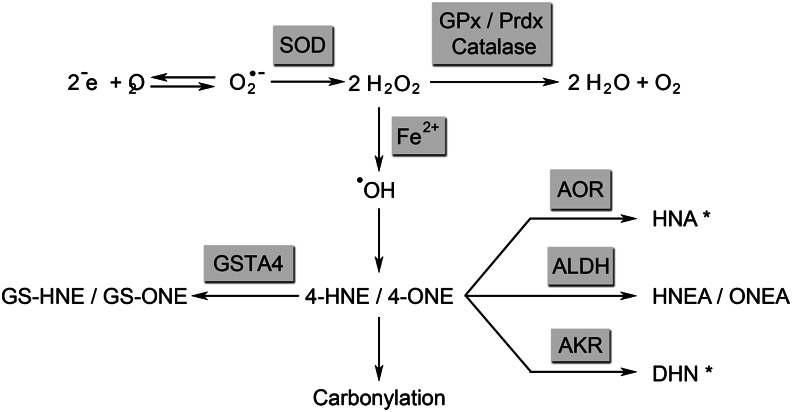
Free radicals such as the superoxide anion are chiefly generated by electron leakage from the mitochondrial electron transport chain, but are also produced via the action of enzymes including NAD(P)H oxidase, xanthine oxidase, lipoxygenases, and cyclooxygenases (1, 8). Electrons leaking from the electron-transport chain during mitochondrial respiration combine with surrounding oxygen to form the superoxide anion (Fig. 1). Superoxide is readily metabolized by superoxide dismutases, chief among them manganese superoxide dismutase (MnSOD), which acts on superoxide in the mitochondrial matrix to form hydrogen peroxide (9).
Figure 1.
Formation of lipid peroxidation products from superoxide (O2−). Asterisk denotes metabolites not yet characterized for 4-ONE. 4-HNE, 4-hydroxy trans-2,3-nonenal; 4-ONE, 4-oxo trans-2,3-nonenal; AKR, aldoketoreductase; AOR, alkenal/one oxidoreductase; DHN, 1,4-dihydroxynonene; GPx, glutathione peroxidase; GS-HNE, glutathionylated 4-HNE; GS-ONE, glutathionylated 4-ONE; GSTA4, glutathione S-transferase A4; HNA, 4-hydroxynonanal, HNEA: trans-4-hydroxy-2-nonenoic acid, ONEA: trans-4-oxo-2-nonenoic acid, Prdx: peroxyredoxin; SOD, superoxide dismutase.
Although mitochondrial ROS are generated from the superoxide anion, there is some debate as to which ROS is the key mediator of insulin resistance. MnSOD is an abundant and highly efficient enzyme, implying that superoxide is rapidly catalyzed to hydrogen peroxide under physiologic conditions (10, 11). Furthermore, the rate at which superoxide oxidizes thiols is relatively slow compared with its rate of catalysis by MnSOD (2), making it more likely a precursor rather than the primary second messenger connecting oxidative stress to insulin resistance. Hydrogen peroxide has been therefore proposed as the key signaling molecule (2).
Hydrogen peroxide may be metabolized by catalase, glutathione peroxidase, peroxiredoxins into water and oxygen (10, 12). Because catalase is largely, although not exclusively, localized to the peroxisome, its role in affecting mitochondrially-derived ROS is likely to be less important for insulin resistance. Alternately, hydrogen peroxide can react with iron or other metals in a Fenton reaction, generating the more potent hydroxyl radical (13). Hydroxyl radical acts on the polyunsaturated side chains of membrane phospholipids and triglycerides, leading to the addition of molecular oxygen, followed by chemical degradation (Hock cleavage). The most widely studied oxidized lipid degradation products are the α,β-unsaturated aldehydes, which include the (n-6) lipid peroxidation products 4-hydroxy trans-2,3-nonenal (4-HNE) and 4-oxo trans-2,3-nonenal (4-ONE) (6). 4-HNE can also be formed by an alternate pathway, whereby the 5-lipoxygenase product 5 S-hydroxyeicosatetraenoic acid can be oxidized by cyclooxygenase-2, yielding a bicyclic diendoperoxide. This diendoperoxide can further undergo hematin- and ferrous iron–catalyzed transformation to produce both 4-HNE and malondialdehyde (14).
Due to an electron-poor region at C3, 4-HNE and 4-ONE are highly susceptible to Michael addition, a nucleophilic attack by the side chains of histidine, cysteine, and lysine residues (7). These covalent modifications result in a free carbonyl attached to the protein, termed carbonylation. Carbonylation can target the proteins for selective degradation by the 26S proteasome (15). Further, as histidine, lysine, and cysteine residues are frequently components of enzyme-active sites or participate in protein–protein interactions, carbonylation typically leads to loss or alteration of protein function (7). In addition to carbonylation, reactive aldehydes can form a Schiff base with lysine side chains that may further react at the C3 producing inter- or intraprotein crosslinks. Schiff bases make up only a small percentage of lipid-modified proteins (16).
Although 4-HNE has been more widely studied, 4-ONE is more electrophilic (17) and reacts with model proteins at a faster rate than 4-HNE (18). This reactivity translates into higher toxicity of 4-ONE relative to 4-HNE with regard to mitochondrial function (17) and cell toxicity (18, 19). The relative importance of these 2 molecules as mediators of oxidative damage will require further investigation.
Antioxidant response
Lipid aldehydes can also be detoxified by a variety of processes. Phase I metabolism results in oxidation or reduction of carbonyl groups, decreasing reactivity. Phase I metabolism is carried out by aldehyde dehydrogenases, aldo-keto reductases, and alkenal/one oxidoreductase. 4-HNE is metabolized into its corresponding carboxylic acid, 4-hydroxy-2-nonenoic acid, and its corresponding alcohol, 1,4-dihydroxynonene by the enzymes aldehyde dehydrogenase and aldoketoreductase, respectively (20, 21) (Fig. 1). Additionally, saturation of the C2-C3 double by alkenal/one oxidoreductase forms 4-hydroxynonanal (22, 23) and carbonyl reductase saturates the C2-C3 double bond of 4-ONE (24).
Phase II metabolism of 4-HNE and 4-ONE results in glutathionylation at the reactive C3 (25). In the case of 4-HNE, this occurs chiefly via the action of glutathione S-transferases, such as glutathione S-transferase A4 (GSTA4), as well as nonenzymatically. Nonenzymatic glutathionylation of 4-ONE occurs very rapidly (26), whereas enzymatic glutathionylation of 4-ONE has not been characterized. The resultant glutathionylated metabolites are targeted for active transport out of the cell by RLIP76 or the multidrug-resistance protein 1 transporter (27, 28).
Although there are many factors that affect the production and detoxification of ROS, carbonylation itself directly influences cell-signaling pathways implicated in the antioxidant response and inflammation. Kelch-like–ECH-associated protein 1 (KEAP1) is bound to nuclear factor (erythroid-derived-2)-like 2 (Nrf2), sequestering it from entering the nucleus (29). Carbonylated KEAP1 is targeted for degradation, thereby releasing Nrf2, which heterodimerizes with other nuclear factors and binds to the antioxidant response element in a variety of antioxidant genes including glutathione-S-transferase (30). This effect is dampened by the carbonylation of the Rpt4 subunit of the proteasome by 4-HNE, leading to decreased proteasome activity and blocking of Nrf2 degradation (31). The modification of thioredoxin by 4-HNE releases apoptosis signal-regulating kinase 1, which allows for its autophosphorylation and activation. This in turn leads to a phosphorylation cascade involving SEK and c-Jun N-terminal kinase, ultimately leading to activation of the nuclear factor κ light-chain enhancer of activated B cells, a critical activator of multiple proinflammatory genes (30, 32).
Protein carbonylation in muscle
Causes
The normal function of skeletal muscle illustrates the importance of the fine balance between oxidant and antioxidant processes. Intense exercise promotes generation of ROS that at low levels serves an important role in normal force production (33) as well as glucose uptake during contraction (34). Exercise training is associated with improved antioxidant response over time (35). However, during the prolonged and extreme oxidative stress found in disease states, such as rats with a high tumor burden, poor antioxidant response and increased protein carbonylation are associated with muscle wasting (36). Additionally, more acute events such as tissue ischemia with subsequent reperfusion are associated with increased ROS production, protein carbonylation, and cellular injury (37).
High fat diet results in both increased intramyocellular lipid content (38) as well as increased levels of protein carbonylation (39). Further, insulin resistance in humans, as measured by hyperinsulinemic-euglycemic clamp, is related both to accumulation of intramyocellular lipid and increased levels of protein carbonylation in muscle cells (40). The effect of overnutrition on myocellular oxidative stress is not only seen in the obese. Long-term overfeeding of normal weight individuals >28 d resulted in both increased peripheral insulin resistance and increased carbonylation of cellular proteins, whereas no changes in markers of mitochondrial content were found (41).
Exercise is one of the most well-studied modulators of oxidative stress status in skeletal muscle. The effect of exercise on protein carbonylation in muscle depends on the duration, intensity, and type of exercise (42–44). After a single episode of intense exercise, the carbonylation of sarcoplasmic reticulum Ca2+-ATPase is increased 80%. This is paralleled by a 22% reduction in Ca2+-ATPase activity (42). The effect of acute exercise is transient, however, and within 1 h, levels of carbonylation are near normal (42). The effects of exercise on increasing carbonylation were measured in predominantly anaerobic “white” or fast-twitch muscles, but not in red muscles that have more oxidative muscle fibers and higher mitochondrial content (44). Supplementation with antioxidant vitamin E was able to partially reverse the level of carbonylation, both basally and post-exercise (44).
In contrast to the acute effects of exercise on muscle protein, long-term active mice show lower levels of carbonylation than both age-matched sedentary mice as well as younger mice that were not exercise trained (45). This effect is likely due in part to increased antioxidant activity in active mice, including increased superoxide dismutase and glutathione peroxidase activity (45, 46). In contrast, sedentary mice had increased expression of proteins involved in oxidative phosphorylation (aconitate hydratase, ADP/ATP translocase 1, and voltage-dependent anion-selective channel protein 1) (45).
In addition to exercise, there is a body of literature associating changes in respiratory dynamics with alterations in carbonylation of muscle proteins, both in intercostal and diaphragmatic muscles as well as in nonrespiratory skeletal muscle. These changes in pulmonary physiology represent alterations in the muscular forces required for breathing, but also affect overall exercise capacity. Obstructive sleep apnea is associated with both increased strain on respiratory muscles and repeated hypoxia-reoxygenation cycles. It is associated with increased markers of lipid peroxidation in the serum and increased carbonylation of intercostal muscle proteins (47). The severity and frequency of apnea events were positively correlated with increasing levels of carbonylation (47).
Chronic obstructive pulmonary disease is associated with muscular dysfunction as demonstrated by decreased peripheral muscle strength and exercise capacity. This is paralleled by increased production of ROS and reduced antioxidant capacity (48), and there is an inverse relationship between muscle carbonylation and both skeletal muscle exercise capacity and pulmonary function (48, 49). Whether the findings in chronic obstructive pulmonary disease relate to a direct increase in inflammatory state or an indirect effect of deconditioning and underuse of skeletal muscle is unclear.
On the opposite end of the spectrum from these disease states that require increased respiratory muscle activity, mechanical ventilation results in a prolonged underuse of respiratory muscles associated with muscle atrophy. Similar to the findings in the muscles of sedentary animals, underused respiratory muscles show 50% increased levels in protein carbonylation and 8-isoprostane, a marker of lipid peroxidation, after only 8 h of mechanical ventilation (50). These findings underline the importance of ongoing muscular function to minimize oxidative stress–related changes in skeletal muscle.
Another important cause of increased protein carbonylation is age. Studies in rats showed that fast-twitch glycolytic muscle fibers had overall higher levels of protein carbonylation relative to slow-twitch muscles, and this difference was increased with age. Furthermore, increased carbonylation in fast-twitch muscles was associated with decreased antioxidant capacity (51).
Finally, sepsis has been widely studied as a model of oxidative stress. In rats, sepsis is a strong trigger of oxidative stress in skeletal muscle, and protein carbonylation increases considerably within 12 h of induction of septic shock (52, 53). Pretreatment with the antioxidant N-acetylcysteine can reduce levels of oxidative stress, and this is reflected in the reversal of contractile dysfunction seen in sepsis (54). In such a system, it is not clear whether N-acetylcysteine functions to replenish glutathione pools or nonenzymatically alkylate 4-ONE and potentially 4-HNE, thereby reducing carbonylation.
Targets and effects
The constellation of carbonylated proteins resulting from exercise or other described stimuli of muscle oxidative stress varies somewhat between models; however, there are common themes in the classes of proteins affected (Table 1), including a large number of proteins that play a role in mitochondrial function.
Table 1.
Carbonylation targets identified in adipose and muscle tissue1
| Gene | Tissue | Ref. |
| Metabolism | ||
| Aspartate aminotransferase | M | (45, 69) |
| Aconitate hydratase (aconitase) | M | (45, 70) |
| Enolase 3β | A,M | (59,70, 71) |
| Aldolase | M | (70, 71) |
| Carbonic anyhdrase III | M | (70, 71) |
| Citrate synthase | M | (69) |
| Triose-phosphate isomerase | M | (70) |
| Hydroxyacyl-CoA dehydrogenase, mitochondrial | M | (45) |
| NADH-ubiquinone oxidoreductase | M | (69, 71) |
| 3-Hydroxyacyl-CoA dehydrogenase | M | (69) |
| Succinate dehydrogenase | M | (69) |
| Short-chain 3-hydroxyacyl-CoA dehydrogenase | M | (69) |
| Acetyl-CoA acetyltransferase | M | (69) |
| Fumarate hydratase 1 | A | (59) |
| Glyceraldehyde 3-phosphate dehydrogenase | A | (59) |
| Mitochondrial function | ||
| Electron transfer flavoprotein | M | (70) |
| Dihydrolipoamide dehydrogenase | M | (70) |
| NADH dehydrogenase 1 | M | (45, 69) |
| ATP synthase F0F1 complex | M | (45, 69, 71) |
| Voltage-dependent anion-selective channel protein 2 | M | (69) |
| Voltage-dependent anion-selective channel protein 1 | M | (45, 69) |
| Cytochrome C | M | (45, 69) |
| Antioxidant response | ||
| Superoxide dismutase [Mn] | M | (45) |
| Peroxiredoxin-1, -3, and -5 | A,M | (59, 69) |
| Glutathione peroxidase 1 | A | (59) |
| Glutathione S-transferase M1/ A4 | A | (59) |
| Aldehyde dehydrogenase | A,M | (59, 69, 71) |
| Signal transduction | ||
| Creatine kinase | M | (45,70,71) |
| Insulin receptor substrate 1 | A | (63) |
| Insulin receptor substrate 2 | A | (63) |
| Protein synthesis/degradation | ||
| Heat-shock protein beta-3 | M | (45) |
| Eukaryotic translation elongation factors 1β2 and 1α1 | A | (59) |
| Serine proteinase inhibitor, clade A, member 1a | A | (59) |
| Structure/contractile machinery | ||
| Actin | M | (71) |
| Myosin light and heavy chains | A,M | (59, 71) |
| Tropomysin | M | (71) |
| Lipid metabolism | ||
| Fatty acid binding protein 4 | A | (59,61) |
| Fatty acid binding protein 5 | A | (61,65) |
| Acyl-CoA dehydrogenases (short-, medium-, long-, and very long chain) | M | (45, 69) |
| Fatty acid synthetase | A | (59) |
| Triosephosphate isomerase 1 | A | (59) |
A, adipose; M, muscle.
In muscle, creatine kinase is a frequently identified target of carbonylation. This modification results in decreased activity, and carbonylation was associated with increased aggregation of creatine kinase into insoluble aggregates and high molecular weight oligomers (51). As a key signaling molecule in the homeostasis of creatine phosphate and creatine during increased muscle function, carbonylated creatine kinase may contribute to the loss of muscle function associated with age and disease.
In addition to muscle contractile function, there are several lines of evidence linking increased production of ROS in muscle with insulin resistance. Impaired insulin signaling resulting from hyperlipidemia, glucocorticoid therapy, and inflammation are all associated with increased myocellular ROS (55). Although increased ROS is associated with insulin resistance, decreasing ROS in rat skeletal muscle via increased expression of MnSOD can reverse the deleterious effect of a high-fat diet on insulin sensitivity (9). In sedentary humans, increased intramyocellular lipid content was associated with increased protein carbonylation and insulin resistance as measured by euglycemic-hyperinsulinemic clamp (40). Recent studies of 4-HNE treatment of both muscle tissue and L6 muscle cells showed decreased insulin signaling and decreased glucose uptake (56). These toxic effects on glucose metabolism in muscle are reversed by pretreatment with the antioxidants 3H-1,2-dithiole-3-thione, N-acetylcysteine, aminoguanidine, and S-adenosyl-methionine (56).
Protein carbonylation in fat
Causes
Inflammation has been linked to oxidative stress in adipose tissue, and the chronic low-grade inflammatory state associated with obesity is implicated as an important causal factor in the development of type 2 diabetes mellitus (reviewed in references 57 and 58). Increased protein carbonylation is observed in the adipose tissue of diet-induced obese mice (59, 60), and subcutaneous human adipose tissue similarly shows increased levels of protein carbonylation associated with increasing body weight (61). It is hypothesized that the increased carbonylation is due to tissue inflammation. This is supported by the observation that treatment of 3T3-L1 adipocytes with TNF-α leads to increased lipid peroxidation, increased protein carbonylation, and mitochondrial dysfunction (62). Obesity also is associated with decreased antioxidant capacity in adipose tissue, as demonstrated by decrease in adipose-specific GSTA4 expression (59). Other 4-HNE metabolizing enzymes such as aldehyde dehydrogenases (ALDHs) (ALDH1A1, ALDH1A7, ALDH2) and fatty ALDH (FALDH) were not significantly different (59).
Treatment of 3T3-L1 adipocytes with nontoxic levels of 4-HNE was shown to lead to increased carbonylation and decreased glutathione levels (59, 63). This effect was reversed by overexpression of FALDH and by treatment with rosiglitazone (63). Although rosiglitazone increases expression of FALDH, the effect of rosiglitazone could also be via modulation of other PPARγ-dependent genes or by a nontranscriptional mechanism. Indirect increase of 4-HNE levels via GSTA4 silencing in 3T3-L1 adipocytes was associated with increased carbonylation as well (60).
Targets and effects
Among the proteins carbonylated in adipocytes are enzymes involved in oxidative phosphorylation, branched chain amino acid catabolism, tricarboxylic acid cycle, fatty acid metabolism, insulin signaling, and the antioxidant response (Table 1) (59, 60).
Reduced antioxidant activity by knockout of GSTA4 in mice or knockdown of GSTA4 in 3T3-L1 adipocytes is associated with increased mitochondrial ROS, decreased mitochondrial membrane potential, and increased carbonylation (59, 60). These changes were associated with decreased mitochondrial respiration and inhibition of complex I, leading to increased superoxide anion production (62). As such, increased ROS leads to increased carbonylation, and, conversely, increased carbonylation leads to increased ROS, implying a feed-forward control loop amplifying small changes in cellular ROS or carbonylation level. Interestingly, overexpression of GSTA4 did not result in increased mitochondrial respiratory capacity over baseline (62). Although GSTA4 knockdown adipocytes had a generalized increase in protein carbonylation, the most prominent differentially carbonylated protein in knockdown versus GSTA4 overexpressing adipocytes was the mitochondrial inner membrane phosphate carrier that is thought to work in conjunction with F0F1 ATPase to facilitate ATP generation in response to electrochemical gradient across inner mitochondrial membrane (62). Directed knockdown of the phosphate carrier resulted in reduced basal, coupled, and maximal respiration (62). These changes in mitochondrial function were accompanied by a 75% reduction in mitochondrial DNA content and morphologic changes with irregularly shaped mitochondria and disorganized cristae, despite no change in mitochondrial number, area, or density (62). The mitochondrial dysfunction noted in GSTA4 knockdown adipocytes was accompanied by increased basal lipolysis, altered glucose transport, and decreased β-oxidation of fatty acids (62).
In both mouse and human, two of the major carbonylated proteins are adipocyte fatty acid binding protein (FABP4) and epidermal FABP (FABP5) (59, 61). The crystal structure of carbonylated FABP4 was solved, representing the first crystal structure of a 4-HNE–modified endogenous protein (59, 61). When covalently bound, 4-HNE does not reach as far into the binding cavity as a fatty acid; however, this modification of FABPs has been shown to inhibit their ability to bind fatty acids (64). In contrast, the noncovalently bound 4-HNE aldehyde extends as far into the binding cavity as a fatty acid carboxyl group. Because 4-HNE aligns in the binding cavity in a manner to similar that of fatty acids, this supports the hypothesis that bioactive aldehydes are a newly identified class of in vivo ligands for FABPs (59, 65). It remains to be determined whether either covalent or noncovalent binding of lipid peroxidation products by FABPs has an important biological function.
Two other targets of modification by products of lipid peroxidation include insulin receptor substrates 1 and 2. Treatment of 3T3-L1 adipocytes with 4-HNE results in carbonylation-mediated reduction in protein levels of insulin receptor substrates 1 and 2 by targeted degradation (63). Similar to the effect of 4-HNE treatment in muscle, adipocytes also exhibited impaired insulin signaling with reduction in both basal and insulin-stimulated phosphatidylinositol 3 kinase and protein kinase B activity (63). Treated adipocytes had impaired insulin-induced tyrosine phosphorylation of insulin receptor β, insulin receptor substrates 1 and 2, and increased serine 307 phosphorylation of insulin receptor β (63). These changes were associated with decreased basal and insulin-stimulated glucose transport and lipogenesis (63). Carbonylation may affect insulin signaling in other ways as well. In hepatocytes, the phosphatase and tensin homolog has been shown to be carbonylated, leading to inhibition of its lipid phosphatase activity (66, 67). Because phosphatase and tensin homolog activity inhibits insulin signaling, in this case carbonylation would have an activating effect (68).
Summary
Protein carbonylation is a product of elevated levels of ROS that occur when increased metabolic demand produces higher amounts of superoxide anion in the context of inadequate antioxidant mechanisms. Both myocytes and adipocytes are vulnerable to oxidative protein damage in pathologic states such as long-term underuse of muscle capacity or overnutrition resulting in obesity. Proteins important for mitochondrial function are targeted in both tissues, and oxidative stress has been associated with mitochondrial dysfunction.
The observation that carbonylation is transiently increased after exercise, a metabolically beneficial event, contrasted with increased carbonylation noted in long-term inactive muscle and obese adipose tissue implies that the timing and targets of carbonylation may play an important role in its effect on tissue function. Additionally, the relative abundance of carbonylated protein in healthy or dysfunctional tissue is likely to be important. Further study is necessary to understand the importance of the context of carbonylation in determining its relative beneficial or deleterious effects.
Although carbonylation of proteins plays a role in cellular signaling, modification of proteins by lipid aldehydes can have either an activating effect, as in the case of carbonylation of KEAP1 in antioxidant response and phosphatase and tensin homolog in insulin signaling, or an inactivating effect, as seen in the carbonylation of insulin receptor substrates 1 and 2, which decreases insulin signaling. In assessing the overall influence of carbonylation, there is little likelihood that alkylation will be a stoichiometric event. Although the activation of a small percentage of a given protein may be sufficient to exert demonstrable effect, inactivating modifications must typically affect a larger portion of the cellular protein pool to manifest changes in function. In this way, carbonylation may have opposing effects in times of low ROS production relative to higher ROS production. Future studies will need to further elucidate the importance of carbonylation in the function and abundance of key metabolic proteins.
Acknowledgments
The authors thank Dr. Eric Long for helpful discussions in preparing this manuscript. The authors have read and approved the final manuscript.
Footnotes
Abbreviations used: ALDH, aldehyde dehydrogenase; FABP, fatty acid binding protein; FALDH, fatty aldehyde dehydrogenase; GSTA4, glutathione S-transferase A4; 4-HNE, hydroxy trans-2,3-nonenal; KEAP, Kelch-like-ECH-associated protein, MnSOD, manganese superoxide dismutase; Nrf2, nuclear factor (erythroid-derived-2)-like 2; 4-ONE, 4-oxo trans-2,3-nonenal; ROS, reactive oxygen species.
Literature Cited
- 1.Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–8 [DOI] [PubMed] [Google Scholar]
- 2.Fisher-Wellman KH, Neufer PD. Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol Metab. 2012;23:142–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrades MÉ, Morina A, Spasić S, Spasojević I. Bench-to-bedside review: sepsis - from the redox point of view. Crit Care. 2011;15:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Ferranti S, Mozaffarian D. The perfect storm: obesity, adipocyte dysfunction, and metabolic consequences. Clin Chem. 2008;54:945–55 [DOI] [PubMed] [Google Scholar]
- 5.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42:318–43 [DOI] [PubMed] [Google Scholar]
- 7.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol Aspects Med. 2003;24:149–59 [DOI] [PubMed] [Google Scholar]
- 8.Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, Nakayama O, Makishima M, Matsuda M, Shimomura I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114:1752–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boden MJ, Brandon AE, Tid-Ang JD, Preston E, Wilks D, Stuart E, Cleasby M, Turner N, Cooney G, Kraegen E. Overexpression of manganese superoxide dismutase ameliorates high-fat diet-induced insulin resistance in rat skeletal muscle. Am J Physiol Endocrinol Metab. 2012;303:E798–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andriantsitohaina R, Duluc L, García-Rodríguez JC, Gil-del Valle L, Guevara-Garcia M, Simard G, Soleti R, Su D, Velázquez-Pérez L, Wilson J, et al. Systems biology of antioxidants. Clin Sci. 2012;123:173–92 [DOI] [PubMed] [Google Scholar]
- 11.Shi Q, Gibson GE. Oxidative stress and transcriptional regulation in Alzheimer disease. Alzheimer Dis Assoc Disord. 2007;21:276–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huh JY, Kim Y, Jeong J, Park J, Kim I, Huh KH, Kim YS, Woo HA, Rhee SG, Lee KJ, et al. Peroxiredoxin 3 is a key molecule regulating adipocyte oxidative stress, mitochondrial biogenesis, and adipokine expression. Antioxid Redox Signal. 2012;16:229–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandelker L. Introduction to oxidative stress and mitochondrial dysfunction. Vet Clin North Am Small Anim Pract. 2008;38:1–30 [DOI] [PubMed] [Google Scholar]
- 14.Griesser M, Boeglin WE, Suzuki T, Schneider C. Convergence of the 5-LOX and COX-2 pathways: heme-catalyzed cleavage of the 5S-HETE-derived di-endoperoxide into aldehyde fragments. J Lipid Res. 2009;50:2455–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kastle M, Grune T. Protein oxidative modification in the aging organism and the role of the ubiquitin proteasomal system. Curr Pharm Des. 2011;17:4007–22 [DOI] [PubMed] [Google Scholar]
- 16.Bruenner BA, Jones AD, German JB. Direct Characterization of Protein Adducts of the Lipid Peroxidation Product 4-Hydroxy-2-nonenal Using Electrospray Mass Spectrometry. Chem Res Toxicol. 1995;8:552–9 [DOI] [PubMed] [Google Scholar]
- 17.Picklo MJ, Azenkeng A, Hoffmann MR. Trans-4-oxo-2-nonenal potently alters mitochondrial function. Free Radic Biol Med. 2011;50:400–7 [DOI] [PubMed] [Google Scholar]
- 18.Lin D, Lee H, Liu Q, Perry G, Smith MA, Sayre LM. 4-Oxo-2-nonenal is both more neurotoxic and more protein reactive than 4-hydroxy-2-nonenal. Chem Res Toxicol. 2005;18:1219–31 [DOI] [PubMed] [Google Scholar]
- 19.West JD, Ji C, Duncan ST, Amarnath V, Schneider C, Rizzo CJ, Brash AR, Marnett LJ. Induction of apoptosis in colorectal carcinoma cells treated with 4-hydroxy-2-nonenal and structurally related aldehydic products of lipid peroxidation. Chem Res Toxicol. 2004;17:453–62 [DOI] [PubMed] [Google Scholar]
- 20.Alary J, Guéraud F, Cravedi J-P. Fate of 4-hydroxynonenal in vivo: disposition and metabolic pathways. Mol Aspects Med. 2003;24:177–87 [DOI] [PubMed] [Google Scholar]
- 21.Murphy TC, Amarnath V, Gibson KM, Picklo MJ., Sr Oxidation of 4-hydroxy-2-nonenal by succinic semialdehyde dehydrogenase (ALDH5A). J Neurochem. 2003;86:298–305 [DOI] [PubMed] [Google Scholar]
- 22.Dick RA, Kensler TW. The catalytic and kinetic mechanisms of NADPH-dependent alkenal/one oxidoreductase. J Biol Chem. 2004;279:17269–77 [DOI] [PubMed] [Google Scholar]
- 23.Kubátová A, Honzatko A, Brichac J, Long E, Picklo MJ. Analysis of HNE metabolism in CNS models. Redox Rep. 2007;12:16–9 [DOI] [PubMed] [Google Scholar]
- 24.Martin H-J, Ziemba M, Kisiela M, Botella JA, Schneuwly S, Maser E. The Drosophila carbonyl reductase sniffer is an efficient 4-oxonon-2-enal (4ONE) reductase. Chem Biol Interact. 2011;191:48–54 [DOI] [PubMed] [Google Scholar]
- 25.Curtis JM, Hahn WS, Long EK, Burrill JS, Arriaga EA, Bernlohr DA. Protein carbonylation and metabolic control systems. Trends Endocrinol Metab. 2012;23:399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doorn JA, Petersen DR. Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem Biol Interact. 2003;143–144:93–100 [DOI] [PubMed] [Google Scholar]
- 27.Singhal SS, Yadav S, Roth C, Singhal J. RLIP76: A novel glutathione-conjugate and multi-drug transporter. Biochem Pharmacol. 2009;77:761–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole SPC, Deeley RG. Transport of glutathione and glutathione conjugates by MRP1. Trends Pharmacol Sci. 2006;27:438–46 [DOI] [PubMed] [Google Scholar]
- 29.Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, Mann GE. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res. 2004;94:609–16 [DOI] [PubMed] [Google Scholar]
- 30.Jowsey IR, Smith SA, Hayes JD. Expression of the murine glutathione S-transferase alpha3 (GSTA3) subunit is markedly induced during adipocyte differentiation: activation of the GSTA3 gene promoter by the pro-adipogenic eicosanoid 15-deoxy-Delta12,14-prostaglandin J2. Biochem Biophys Res Commun. 2003;312:1226–35 [DOI] [PubMed] [Google Scholar]
- 31.Bardag-Gorce F, Li J, French BA, French SW. The effect of ethanol-induced CYP2E1 on proteasome activity: the role of 4-hydroxynonenal. Exp Mol Pathol. 2005;78:109–15 [DOI] [PubMed] [Google Scholar]
- 32.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent Proteasomal Degradation of Transcription Factor Nrf2 Contributes to the Negative Regulation of Antioxidant Response Element-driven Gene Expression. J Biol Chem. 2003;278:21592–600 [DOI] [PubMed] [Google Scholar]
- 33.Reid MB. Nitric oxide, reactive oxygen species, and skeletal muscle contraction. Med Sci Sports Exerc. 2001;33:371–6 [DOI] [PubMed] [Google Scholar]
- 34.Merry TL, Steinberg GR, Lynch GS, McConell GK. Skeletal muscle glucose uptake during contraction is regulated by nitric oxide and ROS independently of AMPK. Am J Physiol Endocrinol Metab. 2010;298:E577–85 [DOI] [PubMed] [Google Scholar]
- 35.Ji LL. Modulation of skeletal muscle antioxidant defense by exercise: Role of redox signaling. Free Radic Biol Med. 2008;44:142–52 [DOI] [PubMed] [Google Scholar]
- 36.Barreiro E, de la Puente B, Busquets S, López-Soriano FJ, Gea J, Argilés JM. Both oxidative and nitrosative stress are associated with muscle wasting in tumour-bearing rats. FEBS Lett. 2005;579:1646–52 [DOI] [PubMed] [Google Scholar]
- 37.Bulkley GB. Free radical-mediated reperfusion injury: a selective review. Br J Cancer Suppl. 1987;8:66–73 [PMC free article] [PubMed] [Google Scholar]
- 38.Hoppeler H, Billeter R, Horvath PJ, Leddy JJ, Pendergast DR. Muscle structure with low- and high-fat diets in well-trained male runners. Int J Sports Med. 1999;20:522–6 [DOI] [PubMed] [Google Scholar]
- 39.Paglialunga S, van Bree B, Bosma M, Valdecantos MP, Amengual-Cladera E, Jörgensen JA, van Beurden D, den Hartog GJM, Ouwens DM, Briedé JJ, et al. Targeting of mitochondrial reactive oxygen species production does not avert lipid-induced insulin resistance in muscle tissue from mice. Diabetologia. 2012;55:2759–68 [DOI] [PubMed] [Google Scholar]
- 40.Ingram KH, Hill H, Moellering DR, Hill BG, Lara-Castro C, Newcomer B, Brandon LJ, Ingalls CP, Penumetcha M, Rupp JC, et al. Skeletal muscle lipid peroxidation and insulin resistance in humans. J Clin Endocrinol Metab. 2012;97:E1182–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samocha-Bonet D, Campbell LV, Mori TA, Croft KD, Greenfield JR, Turner N, Heilbronn LK. Overfeeding reduces insulin sensitivity and increases oxidative stress, without altering markers of mitochondrial content and function in humans. PLoS ONE. 2012;7:e36320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsunaga S, Mishima T, Yamada T, Inashima S, Wada M. Alterations in in vitro function and protein oxidation of rat sarcoplasmic reticulum Ca2+-ATPase during recovery from high-intensity exercise. Exp Physiol. 2008;93:426–33 [DOI] [PubMed] [Google Scholar]
- 43.Veskoukis AS, Nikolaidis MG, Kyparos A, Kokkinos D, Nepka C, Barbanis S, Kouretas D. Effects of xanthine oxidase inhibition on oxidative stress and swimming performance in rats. Appl Physiol Nutr Metab. 2008;33:1140–54 [DOI] [PubMed] [Google Scholar]
- 44.Reznick AZ, Witt E, Matsumoto M, Packer L. Vitamin E inhibits protein oxidation in skeletal muscle of resting and exercised rats. Biochem Biophys Res Commun. 1992;189:801–6 [DOI] [PubMed] [Google Scholar]
- 45.Alves RMP, Vitorino R, Figueiredo P, Duarte JA, Ferreira R, Amado F. Lifelong physical activity modulation of the skeletal muscle mitochondrial proteome in mice. J Gerontol A Biol Sci Med Sci. 2010;65:832–42 [DOI] [PubMed] [Google Scholar]
- 46.Leeuwenburgh C, Fiebig R, Chandwaney R, Ji LL. Aging and exercise training in skeletal muscle: responses of glutathione and antioxidant enzyme systems. Am J Physiol. 1994;267:R439–45 [DOI] [PubMed] [Google Scholar]
- 47.Barreiro E, Nowinski A, Gea J, Sliwinski P. Oxidative stress in the external intercostal muscles of patients with obstructive sleep apnoea. Thorax. 2007;62:1095–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Couillard A, Maltais F, Saey D, Debigaré R, Michaud A, Koechlin C, LeBlanc P, Préfaut C. Exercise-induced quadriceps oxidative stress and peripheral muscle dysfunction in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2003;167:1664–9 [DOI] [PubMed] [Google Scholar]
- 49.Barreiro E, Gea J, Corominas JM, Hussain SNA. Nitric oxide synthases and protein oxidation in the quadriceps femoris of patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2003;29:771–8 [DOI] [PubMed] [Google Scholar]
- 50.Zergeroglu MA, McKenzie MJ, Shanely RA, Van Gammeren D, DeRuisseau KC, Powers SK. Mechanical ventilation-induced oxidative stress in the diaphragm. J Appl Physiol. 2003;95:1116–24 [DOI] [PubMed] [Google Scholar]
- 51.Feng J, Xie H, Meany DL, Thompson LV, Arriaga EA, Griffin TJ. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J Gerontol A Biol Sci Med Sci. 2008;63:1137–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barreiro E, Gea J, Di Falco M, Kriazhev L, James S, Hussain SNA. Protein carbonyl formation in the diaphragm. Am J Respir Cell Mol Biol. 2005;32:9–17 [DOI] [PubMed] [Google Scholar]
- 53.Peralta JG, Llesuy S, Evelson P, Carreras MC, Flecha BG, Poderoso JJ. Oxidative stress in skeletal muscle during sepsis in rats. Circ Shock. 1993;39:153–9 [PubMed] [Google Scholar]
- 54.Barreiro E, Sánchez D, Gáldiz JB, Hussain SNA, Gea J. N-acetylcysteine increases manganese superoxide dismutase activity in septic rat diaphragms. Eur Respir J. 2005;26:1032–9 [DOI] [PubMed] [Google Scholar]
- 55.Hoehn KL, Salmon AB, Hohnen-Behrens C, Turner N, Hoy AJ, Maghzal GJ, Stocker R, Van Remmen H, Kraegen EW, Cooney GJ, et al. Insulin resistance is a cellular antioxidant defense mechanism. Proc Natl Acad Sci U S A. 2009;106:17787–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillon NJ, Croze ML, Vella RE, Soulère L, Lagarde M, Soulage CO. The lipid peroxidation by-product 4-hydroxy-2-nonenal (4-HNE) induces insulin resistance in skeletal muscle through both carbonyl and oxidative stress. Endocrinology. 2012;153:2099–111 [DOI] [PubMed] [Google Scholar]
- 57.Hardy OT, Czech MP, Corvera S. What causes the insulin resistance underlying obesity? Curr Opin Endocrinol Diabetes Obes. 2012;19:81–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Styskal J, Van Remmen H, Richardson A, Salmon AB. Oxidative stress and diabetes: what can we learn about insulin resistance from antioxidant mutant mouse models? Free Radic Biol Med. 2012;52:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6:624–37 [DOI] [PubMed] [Google Scholar]
- 60.Curtis JM, Grimsrud PA, Wright WS, Xu X, Foncea RE, Graham DW, Brestoff JR, Wiczer BM, Ilkayeva O, Cianflone K, et al. Downregulation of adipose glutathione S-transferase A4 leads to increased protein carbonylation, oxidative stress, and mitochondrial dysfunction. Diabetes. 2010;59:1132–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frohnert BI, Sinaiko AR, Serrot FJ, Foncea RE, Moran A, Ikramuddin S, Choudry U, Bernlohr DA. Increased adipose protein carbonylation in human obesity. Obesity (Silver Spring). 2011;19:1735–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Curtis JM, Hahn WS, Stone MD, Inda JJ, Droullard DJ, Kuzmicic JP, Donoghue MA, Long EK, Armien AG, Lavandero S, et al. Protein carbonylation and adipocyte mitochondrial function. J Biol Chem. 2012;287:32967–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Demozay D, Mas J-C, Rocchi S, Van Obberghen E. FALDH reverses the deleterious action of oxidative stress induced by lipid peroxidation product 4-hydroxynonenal on insulin signaling in 3T3–L1 adipocytes. Diabetes. 2008;57:1216–26 [DOI] [PubMed] [Google Scholar]
- 64.Hellberg K, Grimsrud PA, Kruse AC, Banaszak LJ, Ohlendorf DH, Bernlohr DA. X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal. Protein Sci. 2010;19:1480–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bennaars-Eiden A, Higgins L, Hertzel AV, Kapphahn RJ, Ferrington DA, Bernlohr DA. Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J Biol Chem. 2002;277:50693–702 [DOI] [PubMed] [Google Scholar]
- 66.Shearn CT, Fritz KS, Reigan P, Petersen DR. Modification of Akt2 by 4-hydroxynonenal inhibits insulin-dependent Akt signaling in HepG2 cells. Biochemistry. 2011;50:3984–96 [DOI] [PubMed] [Google Scholar]
- 67.Shearn CT, Smathers RL, Stewart BJ, Fritz KS, Galligan JJ, Hail N, Petersen DR. Phosphatase and Tensin Homolog Deleted on Chromosome 10 (PTEN) Inhibition by 4-Hydroxynonenal Leads to Increased Akt Activation in Hepatocytes. Mol Pharmacol. 2011;79:941–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakashima N, Sharma PM, Imamura T, Bookstein R, Olefsky JM. The tumor suppressor PTEN negatively regulates insulin signaling in 3T3–L1 adipocytes. J Biol Chem. 2000;275:12889–95 [DOI] [PubMed] [Google Scholar]
- 69.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–63 [DOI] [PubMed] [Google Scholar]
- 70.Hussain SNA, Matar G, Barreiro E, Florian M, Divangahi M, Vassilakopoulos T. Modifications of proteins by 4-hydroxy-2-nonenal in the ventilatory muscles of rats. Am J Physiol Lung Cell Mol Physiol. 2006;290:L996–1003 [DOI] [PubMed] [Google Scholar]
- 71.Marin-Corral J, Fontes CC, Pascual-Guardia S, Sanchez F, Olivan M, Argilés JM, Busquets S, López-Soriano FJ, Barreiro E. Redox balance and carbonylated proteins in limb and heart muscles of cachectic rats. Antioxid Redox Signal. 2010;12:365–80 [DOI] [PubMed] [Google Scholar]