Abstract
High throughput cellular studies require small sample volume to reduce costs and enhance sensitivity. Microfluidics-generated water-in-oil (W/O) single emulsion droplet systems, in particular, provide uniform, well defined and discrete microenvironment for cell culture, screening, and sorting. However, these single emulsion droplets are incapable of continuous supply of nutrient molecule and are not compatible with aqueous phase-based analysis. A solution is to entrap W/O droplets in another aqueous phase, forming water-in-oil-in-water (W/O/W) double emulsions. The external aqueous phase efficiently prevents desiccation and reduces the amount of organic component, and yet retaining the advantages of compartmentalization. The internal environment can also be programmed dynamically without the need of rupturing the droplets. In this study, we explore the potential application of W/O/W double emulsion droplets for cell cultivation, genetic activation and study of more complicated biological events such as bacteria quorum-sensing as an example. This study demonstrates the advantages and potential application of double emulsion for the study of complex biological processes.
Keywords: double emulsion, synthetic biology, gene circuits, microfluidics, cell analysis, quorum sensing
1. Introduction
Miniaturized bioreactor can facilitate biological studies: on one hand, it better mimics the micro-scale environment where typical biological events transpire; on the other, it improves the cost-effectiveness and sensitivity of high-throughput cellular studies by reducing sample consumption and enhancing signal response due to the small volume. For instance, confining cells in a defined array of microwells has enabled cultivation of both mammalian and microbial cells to facilitate study of complex biological activities such as cell growth and adhesion [1, 2], and quorum-sensitive bacterial growth [3–5]. However, such microwell strategy relies on continuous flow for perfusion of chemicals and may suffer from cross contamination among wells [6]. Recent advancement in nanofabrications has enabled more efficent manupulation of microscale volumes, improved the throughput and greatly expanded the options of miniaturization.
Compartmentalization of biological agents in discrete aqueous droplets dispersed in an oil phase is an attractive alternative to the microwell strategy. Typical water-in-oil (W/O) droplet has an aqueous volume from femtoliter to nanoliter, each representing an isolated micro-reactor or micro-incubator. The maximized surface-to-volume ratio of the micro-droplets enables highly efficient mass and heat transfer between the internal and external phases [7], resulting in precise and rapid perturbation of the microenvironment in the droplet. The application of W/O droplets for biological studies can be traced back to 1958 [8], when Nossal and Lederberg isolated single lymph-node cells and maintained them in individual microdroplets for a few hours. Antibody production was montinored and their ability to inhibit bacteria growth was evaluated. This study leads to the groundbreaking discovery that one lymphocyte cell could only produce one particular antibody, also known as the “one-cell-one-antibody” rule. Nonetheless, these experiments were extremely labourious, had slow throughput and thus was not adapted by the scientific community for general use. Moreover, the conventional W/O emulsions prepared in bulk also suffer from polydispersed size distribution, diminishing their appeal in many biological applications where reproducibility and quantification are paramount. Microfluidics, capable of generating homogeneous and uniform droplets, has emerged as an attractive configuration for cell cultivation [9, 10], screening [10], and sorting [11]. It has also been shown recently to improve the synthesis of DNA-nanocomplexes [12], detect single-cell enzyme activity [13] and protein expression levels [14]. However, for certain applications requiring sustained cell culture, the W/O scheme of an aqueous phase surrounded by an immiscible oil phase is undesirable. It suffers from lack of continuous supply of nutrients, desiccation due to evaporation of the oil phase, and incompatibility with aqueous phase-based analysis such as flow cytometry.
A solution is to entrap W/O droplets in another aqueous phase, forming water-in-oil-in-water (W/O/W) double emulsions. The external aqueous phase efficiently prevents desiccation and reduces the amount of organic component, and yet retaining the advantages of compartmentalization. The oil shell in the W/O/W droplet could function as a selective barrier to regulate molecule transport, allowing supply of nutrients or input of small inducer molecules from the external phase and retaining macromolecules in the internal phase. In this study, PDMS-based microfluidic chips were fabricated to generate stable and monodispersed double emulsions. Fluorinated oil was used as the organic phase over conventional hydrocarbon oil because of its improved oxygen permeability [15]. Fluoropolyether (PFPE)-polyethylene glycol (PEG) was synthesized as an organic phase surfactant for their low cellular toxicity and reduced protein adsorption at the interface [10]. We encapsulated bacteria carrying an inducible reporter gene to demonstrate that the microenvironment of the droplets could be easily controlled by chemical diffusion, which in turn triggered the reporter gene expression. Furthermore, we entrapped engineered bacteria in the droplets to observe oscillations of bacterial density programmed by a synthetic gene circuit. This study demonstrates the advantages and potential application of double emulsion for the study of complex biological processes.
2. Materials and Methods
2.1. Preparation of PDMS chips
Microfluidic chips were fabricated by conventional soft lithography techniques [16]. The chips were designed and drawn to-scale in AutoCAD, and printed to high resolution photo-masks on transparencies (CAD/Art Services, Inc., Bandon, OR). Patterned silicon mold was prepared from SU-8 3025 and SU-8 3035 (MicroChem, Newton, MA) negative photoresist following manufacturer’s protocol. PDMS pre-polymer and a cross-linker (Sylgard 184 Silicon Elastomer Kit, Dow Corning, Midland, MI) was used in recommended ratio of 10:1. The mixture was poured on top of patterned silicon wafer and degassed. After curing at 75°C for 1 hour, PDMS was cut and peeled off the wafers. Inlets and outlets were punched as through holes (Hole puncher, Technical Innovations, Brazoria, TX). A cover slide was bonded with the chip to close the channels after oxygen plasma treatment for 30 s at 20 W (Plasma Asher, Quorum Technologies, West Sussex, RH).
2.2. Coating PDMS chips
Microfluidic chips were fabricated by conventional soft lithography techniques [13]. To change the wettability of PDMS chips for double emulsion formation, the chips were coated following a two-step sol-gel coating procedure.[22] Briefly, tetraethylorthosilicate (TEOS), methyltriethoxysilane (MTES), (heptadecafluoro-1,1,2,2-tetrahydrodecyl)-triethoxysilane, trifluoroethanol and 3-(trimethoxysilyl )-propylmethacrylate were mixed at 2:1:4:1 volume ratio to form the sol-gel solution. 0.5 ml sol-gel was mixed with 0.9 ml methanol, 0.9 ml trifluoroethanol and 0.1 ml HCl solution (pH = 2). The mixture were heated at 85°C for 2 minutes to pre-convert various silane monomers to oligomers. The activated sol-gel solution was immediately filled to PDMS chips after plasma bonding and the device was placed on a hotplate set to 180°C for 1 minute. In the next step, sol-gel P DMS chips were coated with polyacrylic acid (PAA) via thermal initiated reaction. 500 μl de-ionized water, 200 μl acrylic acid, 100 μl ammonium persulfate (10 wt %), and 16 μl tetramethylethylenediamine (TEMED) were mixed and injected constantly to PDMS channels at 20 μl/min. The chip was placed on a hotplate set at 80°C for six minute s and then rinsed with de-ionized water.
2.3. Surfactant synthesis
PFPE-PEG surfactant was synthesized as previously described [17]. Briefly, 5 g Krytox 157FSH (DuPont, Wilmington, DE) was dissolved in 10 ml HFE7100 (3M, St. Paul, MN) and reacted with 0.5 ml SOCl2 under Argon purge overnight at 40°C. The product was concentrated on a rotary evaporator to remove solvent and excessive SOCl2. The product was then reacted with 0.3 g NH2-PEG-NH2 (Sigma, St. Louis, MO), after dissolving in 10ml HFE7100 and 6 ml benzotrifluoride (Sigma, St. Louis, MO), the reaction was purged by Argon and kept at 60°C overnight. After the product was cooled to RT, the mixture was concentrated on a rotary evaporator and then dried in vacuum to yield the surfactant.
2.4. Double emulsion droplets generation
Bacteria in M9 growth medium or fluorescence dye (50 μM for rhodamine B, 1 mg/ml for rhodamine B labeled BSA) in PBS were entrapped in the inner phase. 2 wt% PFPE-PEG surfactant in HFE7500 (3M, St. Paul, MN) was used as the organic phase, and 2 wt % pluoronic F-127 (Sigma, St. Louis, MO) was used as the outer continuous phase. Three syringe pumps (PHD2000, Harvard Apparatus, Holliston, MA) were used to control the flow rates of the three phases, which were 3 μl/min, 5 μl/min and 52 μl/min from the inside to the outside, respectively.
2.5. Partition coefficient determination
Water (inner aqueous phase) or water with 2% F127 (outer aquesou phase) containing 50 μM of rhodamine B is added gentally and dropwise on top of HFE7500 containing 2% PFPE-PEG surfactant. The solution was then left on an orbital shaker operating at 100 rpm for 24 hours to allow partition of chemical into the organic phase. Vigourous vortexing was avoided to prevent emulsion formation. After that, amount of Rhodamine B left in the aqueous phase was determined by intrapolating from a standard curve of rhodamine B. Rhodamine B concentration in the oil phase was thus determined by the difference before and after incubation. Partition coefficient was calculated by the following formula:
2.6. Plasmid preparation
The construction of the ePOP circuit is detailed in our prevous publication [18]. Briefly, the vector was constructed by inserting lux box fragment (140 bp upstream of luxI in V. fischeri) and E gene coding sequence from ΦX174 into host vector pLuxRI2. Each fragment was independently PCR-amplified and then combined by overlap PCR reaction. The fused fragment was inserted into host vector at AatII site. Due to a mutation in the circuit, the LuxR/LuxI quorum-sensing module is dysfunctional. Instead, the circuit generated population-level oscillations due to a cell density-mediated plasmid amplification.
2.7. Fluorescence microscopy
Droplets containing fluorescence-bearing bacteria were suspended in a 96-well plate and examined by Nikon Eclipse TE2000-U fluorescence inverted microscope. Fluorescence intensity was analyzed by Image J. For each sample, about 20 droplets were analysed to obtain average fluorescence intensity per droplet.
2.8. Flow cytometry analysis
E. coli constitutively expressing GFP were diluted in PBS buffer and encapsulated in the double emulsion droplets. Equal number of droplets were then suspended in PBS solution or M9 growth medium for comparison of cell growth over time by flow cytometry (FACSCanto II, BD Biosciences, Franklin Lakes, NJ). The FSC/SSC was gated with empty droplets and free bacteria to specifically determine the population of droplets with bacteria encapsulated. A 405 nm laser was chosen as the excitation and green fluorescence was recorded. More than 10000 droplets were measured each time to ensure reliable statistics. FlowJo (v.7.6, Tree Star, Ashland) was used to analyze the data. As the inner aqueous phase contained only PBS which offered little nutrients supply for bacterial growth, any observed difference between bacterial growth between these two groups was attribuited to nutrient transport across the oil layer.
3. Results and Discussion
3.1. Microchip design and double emulsion droplets generation
Microfluidic chips were fabricated with a standard soft lithographic protocol using poly(dimethyl siloxane) (PDMS). The dispersed and continuous phase converged on a narrow nozzle (Figure 1A) that was specially designed to create a focused shear, leading to discrete droplet generation. To minimize interaction of these discrete droplets with the channel wall, water-in-oil (W/O) droplets and oil-in-water (O/W) droplets were formed in hydrophobic and hydrophilic channels, respectively. Various materials, such as poly(acrylic acid) (PAA) [19], poly(vinyl alcohol) (PVA) [19] and poly(ethylene glycol) (PEG) [20] were previously reported to confer the naturally hydrophobic PDMS channels a hydrophilic surface. Differential wettability on an integrated single chip was previously achieved by either photo-masking [21] or flow-confinement [22] of reactive coating materials to specified regions in the microchip. To ease the optimization of surface treatment, double emulsions were generated using a setup of two stand-alone chips in our study, (Figure 1A). W/O droplets were first generated in chip 1, treated hydrophobic, then fed to chip 2, treated hydrophilic, to generate W/O/W double emulsions. Chip 2 was rendered hydrophilic by coating of acrylic acid following a two-step sol-gel method [22]. Surface hydrophilicity was confirmed by water contact angle measurement, which changed from 105° to 22° after coating. This two-chip modular design not only eases the pre-processing of PDMS but also enables combination of chips with different dimensions to form double emulsions of on-demand geometrical properties. Because of the simplicity and easier manipulation, this design is adopted by many other groups to generate double emulsion droplets with PDMS/Glass [23] or PMMA/PDMS [24] combinations for first and second chips.
Figure 1.
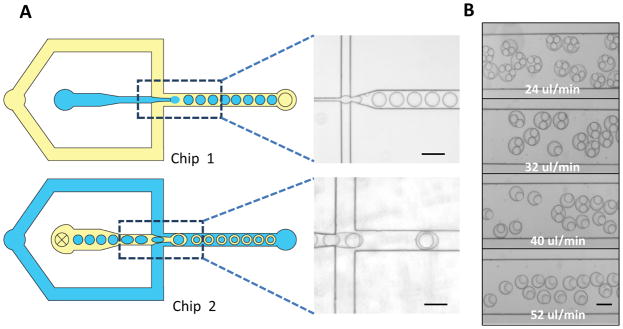
Chip design and double emulsion generation. A) A schematic of a two-chip setup for double emulsion formation: W/O droplets were firstly formed in an untreated PDMS channel (Chip 1) and then fed to a hydrophilic treated channel (Chip 2) to generate W/O/W double emulsion droplets; B) Number of W/O droplets encapsulated into double emulsion was modulated by the flow rate of continuous phase (water). (Scale bar: 100 μm)
Channel dimension is critical in determining droplet size. To achieve consistent and monodispersed double emulsion formation, chip 1 was fabricated to have an overall height of 25 μm, and a width of 15 μm at the narrowest point of the nozzle; chip 2 was made to be 50 μm in height and 50 μm in width at the narrowest point. This design ensured droplets formed from chip 1 were within appropriate size range to be encapsulated in chip 2. Relative flow ratio from different fluidic phases was another parameter to fine-tune droplet properties. By varying the continuous flow rate from chip 2 alone, we successfully produced double emulsions with a controllable number of W/O droplets engulfed in one organic oil shell (Figure 1B).
3.2. Selective chemical transport across oil layer
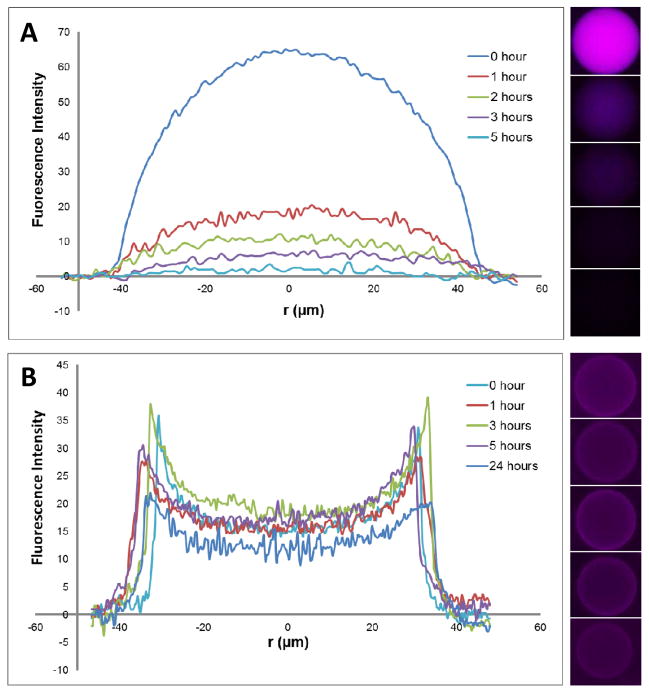
The oil layer in the W/O/W scheme serves as a selective membrane, through which small chemicals or molecules may diffuse in or out, rendering double emulsion an adjustable micro-incubator for cellular studies. For instance, growth of cells could be sustained via diffusion of small nutrient molecules and cell phenotype could be modulated by chemically induced gene expression. To show the selectivity of the transport across the oil layer, we encapusated a small fluorescent molecule, Rhodamine B, and BSA (67 Kda) labeled by Rhodamine B inside the droplets. Free dye molecules diffused out rapidly whereas the protein was trapped inside the droplets for more than 24 hours (Figure 2). Given the low viscosity (0.77cSt for HFE7500 and 1.01cSt for water) and thickness (~10 μm) of the organic layer, permeability of chemicals is primarily determined by the partition coefficient from the inner aqeous phase to the organic phase, and from the organic phase to the outer aqueous phase, which was 0.153 and 0.177 respectively for Rhodamine B according to experimental measurement. Partition coeffcient for Rhodamine B-labeled BSA could not be determined, as no measurable partition occured in the time course of the exeriment.
Figure 2.
Selective chemical transport across the oil phase of double emulsion. Diffusion of rhodamine B (A) and rhodamine B labeled BSA (B) from the core of double emulsion measured by change of fluorescence intensity per droplet. r is the distance from the center of emulsion.
3.3. Controlling droplet microenvironment via chemical diffusion
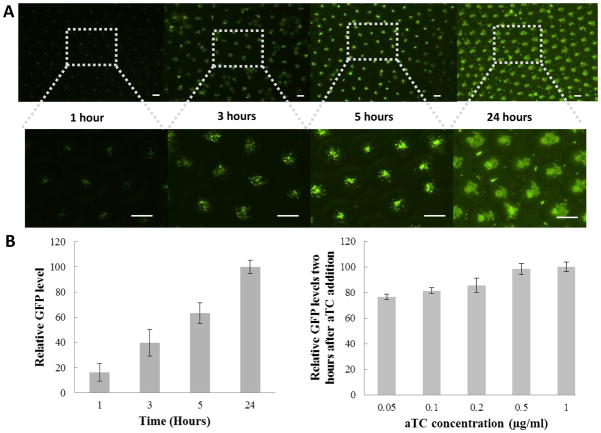
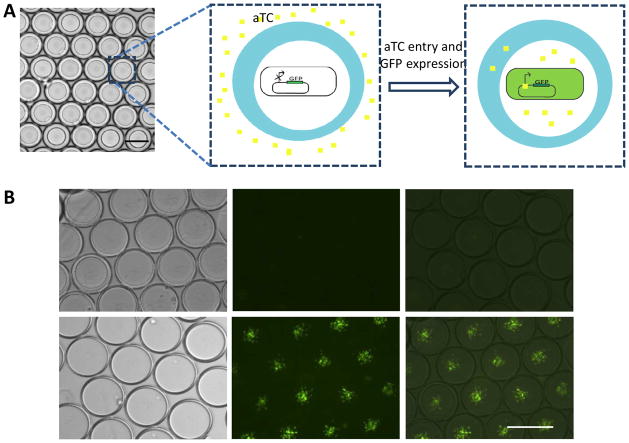
Inducible gene expression was chosen as one example to show the potential application of double emulsion as a pertubable microenvironment for cellular study. We encapsulated E. coli cells (MC4100Z1 cells) carrying a green fluorescence protein (GFP) gene controlled by a TetR-regulated promoter (Figure 2A). In MC4100Z1 cells, which constitutively expresses the TetR protein, GFP expression is inducible by a small chemical, anhydrotetracycline (aTC). Initially the inner aqueous phase consisted of bacteria in growth medium without aTC. Upon collection of the double emulsion droplets, aTC was added to the external aqueous phase. GFP expression became detectable one hour after aTC addition (Figure 2B), which was not detectable in the control. To observe GFP expression over time, a diluted population of bacteria bearing the same plamid was encapsulated in the double emulsion and subsequently suspended in medium solution containing aTC. Relative GFP expression level increased over time (Figure 4A and 4B), which was both contributed by increased GFP expression per cell (indicated by brighter bacteria cells), and bacteria growth over time. We also noted that the GFP expression was suppressed when the external medium was replaced with medium without aTC.
Figure 4.
aTC dependent GFP activation. A) Fluorescence microscope image showing GFP expression overtime after aTC addition; B) Relative GFP level per droplet after aTC addition as a function of time; C) Relative GFP level per droplet as a function of aTC concentration (Data were taken 2 hours after aTC addition). (Scale bar: 100 μm)
The rapid appearance of GFP signal in the bacterial cells suggests effective transport of aTC across the oil shell, which is consistent with our earlier note that the organic layer is permeable to small chemical dye. To further study the induction of GFP expression by aTC diffusion, bacteria-containing droplets were suspended in medium with various aTC concentrations. Fluorescence intensity per droplet was quantitated two hours after aTC addition. At low aTC concentrations, a concentration-dependent activation of GFP was seen (Figure 4D). At high aTC concentrations, however, GFP expression reached a plateau, indicating all bacterial cells were activated at their maximum gene expression limit. Noticeably, even at an extremely low aTC concentration, 0.05 μg/ml, the system was still able to achieve almost 80% of the maximum gene expression. This observation further confirms that aTC transport across the droplet was rather rapid.
3.4. Characterization of bacteria growth and nutrient transport by flow cytometry
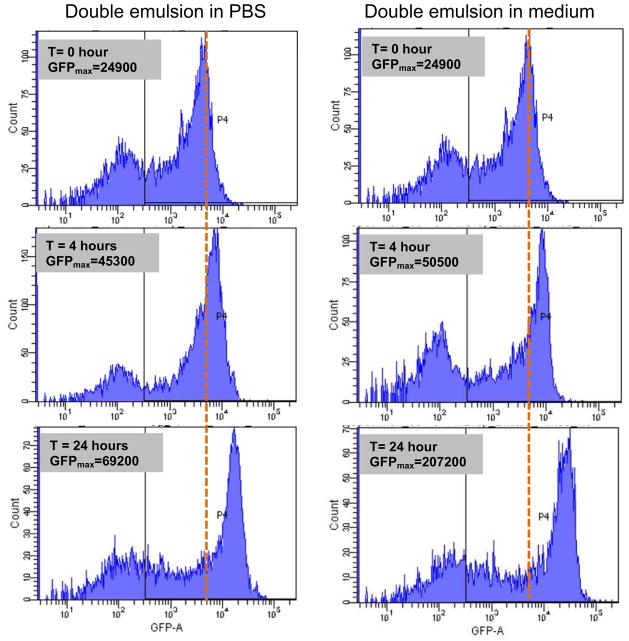
We also observed significant bacterial growth in the double emulsion (Figre 4A). To demonstrate ready transport of nutrient into the core through the oil layer, we suspended bacteria in PBS and enapsulated them in the droplets. Constitutive GFP-expressing bacteria, those with GFP constantly “ON”, were used so that relative GFP fluorescence intensity was only dependent on the total cell number per droplet. These emulsions were then transferred to bacteria growth medium or PBS solution. Because PBS provides little nutrients to the bacteria, the growth of bacteria in PBS would be slowed if not completely inhibited without additional nutrient supply. GFP signal change would correlate with bacteria growth. We assayed GFP intensity per droplet by flow cytometry. Droplets suspended in medium showed a much higher GFP level as compared to those in PBS at four hours and 24 hours (Figure 5). This again shows that double emulsion is a programmable micro-environment for biological studies by allowing transport of some chemical inducers and small nutrient molecules into the droplet. More importantly, the ability of assaying droplets in aqueous phase by flow cytometry opens up the possibility of high throughput screening and sorting.
Figure 5.
Bacteria growth monitored by flow cytometry. Constitutive GFP expressing bacteria was diluted to PBS solution, and encapsulated in double emulsion droplets. These droplets were then suspended to PBS solution (left) or medium solution (right) to compare bacteria growth over time.
3.5. Double emulsion as microcomparment to study cellular dynamics
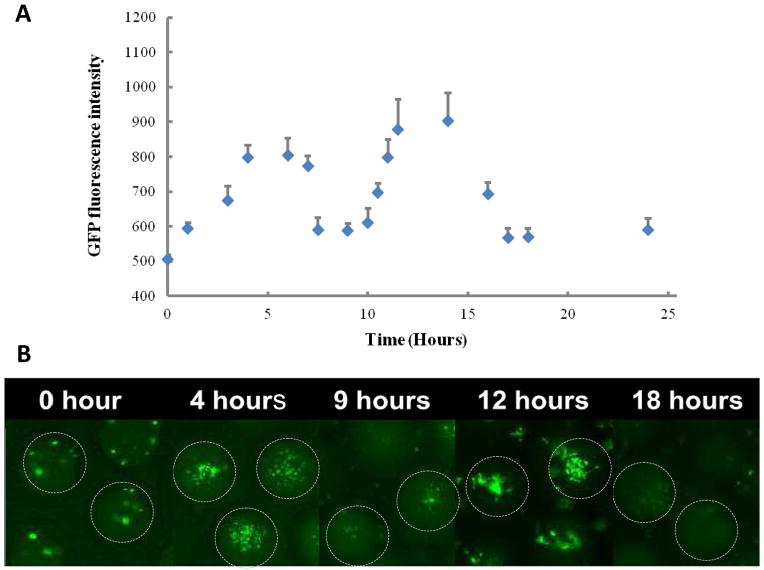
A major challenge in system biology and synthetic biology is to precisely control and quantitatively measure cellular processes. To this end, a small volume, uniformity, and tunability of a microfluidics-generated double emulsion droplet offers an attractive vehicle. To show this, we used engineered E. coli cells (MC4100 cells) programmed by a synthetic suicide circuit (the ePop circuit) [18]. The ePop circuit encodes a lysis protein, E protein, which is expressed at sufficiently high cell densities due to cell density-mediated plasmid amplification. Expression of E protein causes lysis of a subpopulation of cells and cell density reduction, which consequently turns off E protein expression. Transient silencing of E protein allows growth recovery, and the cycle repeats. To faciliate quantification of cell density inside droplets, we introduced a GFP reporter into these bacteria and use GFP intensity as a surrogate of the cell density.
When these bacteria were encapsulated in double emulsion droplets, two cycles of oscillations were observed during overnight culture (Figure 6). Oscillation was monitored via tracking changes in GFP expression over time using a fluorescence microscope. The number of bacteria and GFP expression decreased to a minimal level after 8 hours in the droplets, and grew back to the maximum at about 14 hours. It took another 3 hours for the bacterial population to revert to its minimal. A consistent pattern of relatively slow growth and fast decay was observed when compared to previous results conducted in conventional culture condition [18].
Figure 6.
Quorum-sensing activity in double emulsion microenvironment. A) GFP fluorescence intensity per droplet over time. B) Fluorescence microscope images showing change of GFP-expressing bacteria population over time. GFP-expressing bacteria were co-cultured with bacteria-expressing quorum sensing circuit. When the bacteria population reached a threshold density around 5 hours, the quorum-sensing bacteria released lytic protein to kill the bacteria in the droplets. The depletion caused the reduction of lytic protein expression and recovery of bacteria growth. (Scale bar: 50 μm)
In addition to characterization of synthetic gene circuits, enzymatic assays could be performed by supplying the substrate to external medium at any time and assay the enzyme level inside the droplets. Cellular response to various toxins and drugs could also be studied, and different chemicals could be applied sequentially to analyze cellular responses in a miniaturized environment. Moreover, retention of macromolecules within the aqueous core may also be advantageous in some studies where accumulation of a high concentration of macromolecules is required, for instance, cell-cell signaling involved in stem cell differentiation or antigen-dependent dendritic cell activation.
4. Conclusions
In conclusion, we have demonstrated efficient production of well-controlled double emulsions using a modular two-chip design. The selectively-permeable oil barrier creates a discrete microenvironment whose aqueous core could be well defined and precisely modulated for cell cultivation and study of biological activities that requires communication with an external aqueous environment.
Figure 3.
aTC diffusion and activation of GFP expression. A) Schematic illustration of the experiment design. GFP was expressed under aTC dependent promoter. Bacteria carry this expressing vector was encapsulated in the core without aTC. aTc was then added to the external medium, which diffuse into core of droplets to activate GFP expression; B) Fluorescence microscope images of droplets containing bacteria without aTC addition (top panel) and with aTC addition (bottom panel). Images were taken 1 hour after cell encapsulation. (Scale bar: 100 μm)
Acknowledgments
This work is funded by NSF EEC-0425626, NIH HL089764, HL109442. YP. Ho is grateful to the Danish Research Councils (11-116325/FTP) and Karen Elise Jensen Foundation for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6:1445–9. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 2.Hung PJ, Lee PJ, Sabounchi P, Aghdam N, Lin R, Lee LP. A novel high aspect ratio microfluidic design to provide a stable and uniform microenvironment for cell growth in a high throughput mammalian cell culture array. Lab Chip. 2005;5:44–8. doi: 10.1039/b410743h. [DOI] [PubMed] [Google Scholar]
- 3.Boedicker JQ, Vincent ME, Ismagilov RF. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chem Int Ed Engl. 2009;48:5908–11. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balagadde FK, You L, Hansen CL, Arnold FH, Quake SR. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–40. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 5.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–30. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, Tice JD, Ismagilov RF. A microfluidic system for controlling reaction networks in time. Angew Chem Int Ed Engl. 2003;42:768–72. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 7.Burns JR, Ramshaw C. The intensification of rapid reactions in multiphase systems using slug flow in capillaries. Lab Chip. 2001;1:10–5. doi: 10.1039/b102818a. [DOI] [PubMed] [Google Scholar]
- 8.Nossal GJ, Lederberg J. Antibody production by single cells. Nature. 1958;181:1419–20. doi: 10.1038/1811419a0. [DOI] [PubMed] [Google Scholar]
- 9.Koster S, Angile FE, Duan H, Agresti JJ, Wintner A, Schmitz C, et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip. 2008;8:1110–5. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 10.Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, Frenz L, et al. Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem Biol. 2008;15:427–37. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Baret JC, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9:1850–8. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 12.Ho YP, Grigsby CL, Zhao F, Leong KW. Tuning physical properties of nanocomplexes through microfluidics-assisted confinement. Nano Lett. 2011;11:2178–82. doi: 10.1021/nl200862n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juul S, Ho YP, Koch J, Andersen FF, Stougaard M, Leong KW, et al. Detection of single enzymatic events in rare or single cells using microfluidics. ACS Nano. 2011;5:8305–10. doi: 10.1021/nn203012q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huebner A, Srisa-Art M, Holt D, Abell C, Hollfelder F, Demello AJ, et al. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem Commun. 2007;12:1218–20. doi: 10.1039/b618570c. [DOI] [PubMed] [Google Scholar]
- 15.Riess JG, Krafft MP. Advanced fluorocarbon-based systems for oxygen and drug delivery, and diagnosis. Artif Cells Blood Substit Immobil Biotechnol. 1997;25:43–52. doi: 10.3109/10731199709118896. [DOI] [PubMed] [Google Scholar]
- 16.Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nat Protoc. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 17.Holtze C, Rowat AC, Agresti JJ, Hutchison JB, Angile FE, Schmitz CH, et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip. 2008;8:1632–9. doi: 10.1039/b806706f. [DOI] [PubMed] [Google Scholar]
- 18.Marguet P, Tanouchi Y, Spitz E, Smith C, You L. Oscillations by minimal bacterial suicide circuits reveal hidden facets of host-circuit physiology. PLoS One. 2010;5:e11909. doi: 10.1371/journal.pone.0011909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbier V, Tatoulian M, Li H, Arefi-Khonsari F, Ajdari A, Tabeling P. Stable modification of PDMS surface properties by plasma polymerization: application to the formation of double emulsions in microfluidic systems. Langmuir. 2006;22:5230–2. doi: 10.1021/la053289c. [DOI] [PubMed] [Google Scholar]
- 20.Wang AJ, Xu JJ, Chen HY. In-situ grafting hydrophilic polymer on chitosan modified poly(dimethylsiloxane) microchip for separation of biomolecules. J Chromatogr A. 2007;1147:120–6. doi: 10.1016/j.chroma.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 21.Schneider MH, Willaime H, Tran Y, Rezgui F, Tabeling P. Wettability Patterning by UV-Initiated Graft Polymerization of Poly(acrylic acid) in Closed Microfluidic Systems of Complex Geometry. Anal Chem. 2010;82 (21):8848–8855. doi: 10.1021/ac101345m. [DOI] [PubMed] [Google Scholar]
- 22.Abate AR, Thiele J, Weinhart M, Weitz DA. Patterning microfluidic device wettability using flow confinement. Lab Chip. 2010;10:1774–6. doi: 10.1039/c004124f. [DOI] [PubMed] [Google Scholar]
- 23.Okushima S, Nisisako T, Torii T, Higuchi T. Controlled production of monodisperse double emulsions by two-step droplet breakup in microfluidic devices. Langmuir. 2004;20:9905–8. doi: 10.1021/la0480336. [DOI] [PubMed] [Google Scholar]
- 24.Wu N, Oakeshott JG, Easton CJ, Peat TS, Surjadi R, Zhu Y. A double-emulsion microfluidic platform for in vitro green fluorescent protein expression. J Micromech Microeng. 2011;21(5):054032. [Google Scholar]