Abstract
The Bacillus subtilis KinD signal-transducing histidine kinase is a part of the sporulation phosphorelay known to regulate important developmental decisions such as sporulation and biofilm formation. We have determined crystal structures of the extracytoplasmic sensing domain of KinD, which was copurified and crystallized with a pyruvate ligand. The structure of a ligand-binding site mutant was also determined; it was copurified and crystallized with an acetate ligand. The structure of the KinD extracytoplasmic segment is similar to that of several other sensing domains of signal transduction proteins and is composed of tandem Per-Arnt-Sim (PAS)-like domains. The KinD ligand-binding site is located on the membrane distal PAS-like domain and appears to be highly selective; a single mutation, R131A, abolishes pyruvate binding and the mutant binds acetate instead. Differential scanning fluorimetry, using a variety of monocarboxylic and dicarboxylic acids, identified pyruvate, propionate, and butyrate but not lactate, acetate, or malate as KinD ligands. A recent report found that malate induces biofilm formation in a KinD-dependent manner. It was suggested that malate might induce a metabolic shift and increased secretion of the KinD ligand of unknown identity. The structure and binding assays now suggests that this ligand is pyruvate and/or other small monocarboxylic acids. In summary, this study gives a first insight into the identity of a molecular ligand for one of the five phosphorelay kinases of B. subtilis.
Keywords: signal transduction, sporulation, ligand-binding, Bacillus subtilis, bacterial development, two-component system, histidine kinase, KinD
Introduction
Bacteria must function in continuously changing environments. To cope with these changes, they dedicate a significant fraction of their genome to adaptive response systems. Bacilli and Clostridia offer an example of an ultimate survival strategy. Under adverse conditions, these bacteria differentiate into a spore, a dormant life form resistant to harsh environmental conditions and possibly able to survive for millions of years.1
Many of the bacterial adaptive responses are mediated by so-called two-component systems.2 These are signal transduction systems that connect a signal and its response via two proteins, a sensor histidine kinase and a response regulator. Communication between these proteins occurs via phosphoryl group transfer. The response regulator acts most commonly as a transcription factor that modulates the expression of target genes in response to its phosphorylation status.3 The histidine kinase serves as the signal receptor; it often spans the cytoplasmic membrane and may transduce extracytoplasmic and cytoplasmic signals to initiate the appropriate response in the cytoplasm.4
Bacilli sporulation is controlled by the phosphorelay, a more elaborate variation of the two-component system theme involving numerous proteins (see Fig. 1).5 Here, five kinases (KinA-KinE) with different regulatory roles merge to phosphorylate a single-domain response regulator, Spo0F, which in turn determines the phosphorylation levels of the terminal response regulator/transcription factor, Spo0A, via a phosphotransfer protein, Spo0B.6 Structurally, Spo0B is related to the sensor histidine kinase, but it lacks the capability to autophosphorylate.7 The phosphorelay can thus be thought of as two two-component systems arranged in tandem. This complex architecture provides additional checkpoints for initiating sporulation and indeed a number of phosphatases and inhibitor proteins have been identified that control sporulation at various levels of the phosphorelay.8–10
Figure 1.
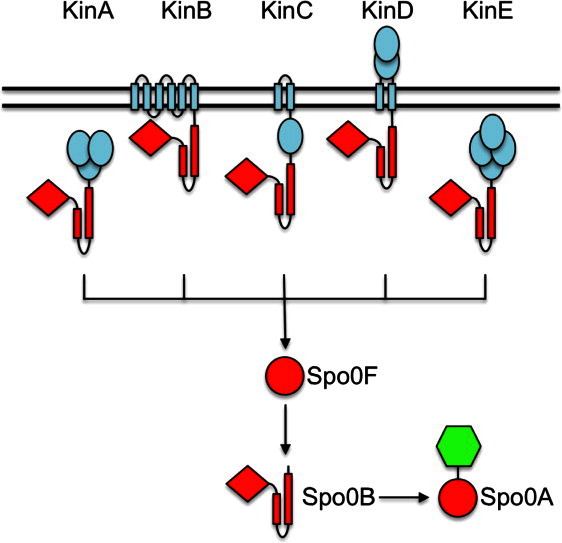
Topology and transmembrane organization of the sporulation kinases and the phosphorelay signal transduction pathway of Bacillus subtilis. Of the five kinases feeding into the phosphorelay, only KinD has an extensive extracytoplasmic sensor domain. Phosphoryl groups are transferred from the kinases to the response regulator, Spo0A, via two phosphotransfer proteins, the response regulator Spo0F and the kinase-like protein Spo0B. Phosphorylated Spo0A regulates the transcription of a large number of genes whose products are important for the transition to stationary phase and for the initiation of important developmental pathways such as sporulation. Potential signal input domains and transmembrane helices of the kinases are colored in blue. The enzymatic and regulatory domains that are subject to phosphorylation and phosphoryl group transfer are in red. The DNA-binding output domain is depicted in green. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In B. subtilis, the five kinases, KinA-KinE, are known to increase the flux of phosphoryl groups through the phosphorelay.6 Low levels of phosphorylated Spo0A (Spo0A∼ P) contribute to the transition from exponential growth to stationary phase, whereas higher levels of Spo0A∼ P are required to initiate sporulation.6, 11 In a laboratory culture, only KinA and KinB kinases are capable of generating sufficiently high Spo0A phosphorylation levels to initiate sporulation. KinA contains three cytosolic Per-Arnt-Sim (PAS) domains that apparently sense starvation signals, but the identity of this signal is unknown.12 In contrast, KinC and KinD appear to have a role in initiating the transition from exponential growth to stationary phase by generating low levels of Spo0A∼ P.6, 13 KinC is activated in response to potassium membrane leakage and this activation depends on a single cytosolic PAS sensory domain.14 KinD has been shown to delay the onset of sporulation15 and has been proposed to act as a phosphatase under certain conditions, which regulates the level of Spo0A∼ P. Its signal has been unidentified as of yet. KinE is a close sequence homolog of the other four kinases and has a domain architecture reminiscent of KinA but it fails to autophosphorylate in vitro.6 The only evidence that it too is a phosphorelay kinase comes from a back-transfer reaction; KinE is capable of receiving phosphate from Spo0F.6
Despite these advances and decades of detailed studies, the exact identities of the molecular stimuli that activate any of the phosphorelay kinases still remain unknown. This is a problem common to many signaling systems. As is the case for KinC,14 sometimes conditions under which a given protein is activated have been identified but even then the identity of the signaling ligand remains elusive.
One approach to gain insights on the identity of a molecular ligand is to solve the structure of the signal detection domain of a sensor histidine kinase. This may reveal a ligand-binding site and provide clues or direct evidence for the signaling molecule. When examining the molecular architecture of the five phosphorelay kinases, KinA and KinE are found to be entirely cytoplasmic and KinB and KinC are found to span the membrane but have no significant extracytoplasmic domains. KinD however has a predicted extracytoplasmic segment flanked by two transmembrane helices in addition to its cytoplasmic catalytic domains (Fig. 1). The N-terminal sensory domain (Pfam02743) shows sequence similarity to domains found in animal Ca2+ channel subunits and prokaryotic chemotaxis receptors,16 and is predicted to have structural similarity to PAS-like domains.17 The KinD topology suggests that it is the most likely one of the five phosphorelay kinases capable of responding to ligands that accumulate or are being depleted in the extracellular milieu; the small periplasmic loops connecting transmembrane helices of KinB and KinC might in principle also be responsive to such signals.
To obtain insight into the ligand-binding properties of the KinD extracellular segment sensor domain (KinD-SD), we have determined its structure. The structure revealed, as predicted from its amino acid sequence, two PAS-like domains, and it contained an electron density for a small molecule that was most consistent with pyruvate bound in a pocket at the membrane distal domain. This small molecule was copurified and cocrystallized with the protein. Subsequent cocrystallization trials with added pyruvate confirmed this assignment. Residue R131 was identified as one that may play a crucial role in pyruvate binding; therefore, the R131A mutant was made and investigated further. Surprisingly, crystal structures of the KinD-SD R131A mutant showed that it complexed only with acetate, even in the presence of excess pyruvate. Ligand binding was confirmed by differential scanning fluorimetry (DSF). Our structures and binding studies detail the ligand recognition framework and implicate pyruvate, propionate, and/or butyrate as possible ligands that may modulate KinD autophosphorylation.
Results
The sensor domain of KinD
Similar to other histidine kinases, the KinD-SD is sandwiched between two transmembrane helices near the N-terminus of the 506 amino acid protein. To determine the boundaries of the domains, the program TMHMM18 was used, which predicted residues 19–36 and 251–273 as transmembrane segments. The expressed sensor domain of KinD contained residues Glu37-Leu250 and included a Ser-Asn-Ala sequence at the N-terminus as a result of cloning into the ligation-independent cloning (LIC) site derived from the vector pMCSG7 and cleavage of the His6 tag with TEV protease.19
We have determined four structures designated here as follows: (1) KinD_W_PYR1 is a wild-type sensor domain with an apparent pyruvate moiety bound in the ligand-binding site. The pyruvate occupancy is partial and the ligand must have been acquired during protein expression in Escherichia coli; (2) KinD_W_PYR2 is a wild-type sensor domain, which was cocrystallized with 30 mM pyruvate to confirm the ligand assignment for the partially occupied structure. Clear electron density for the ligand at near 100% occupancy is found in the binding site; (3) KinD_R131A_ACY is the KinD-SD R131A mutant that was crystallized in the presence of an acetate-containing solution and an acetate was found in the binding site; (4) KinD_R131A_“PYR” is the KinD-SD R131A mutant, where 100 mM pyruvate was added to the protein prior to crystallization and incubated for 30 min at room temperature and the protein was crystallized in a solution without acetate. Surprisingly, acetate but not pyruvate was found in the binding site of the two monomers of the R131A mutant in the crystal asymmetric unit even though it was preincubated with pyruvate. The two wild-type crystals that contain pyruvate, KinD_W_PYR2 and KinD_W_PYR1, diffracted X-rays to the highest resolutions of 2.03 and 2.30 Å, respectively; their cell dimensions are also very similar. The acetate-containing R131A mutant, KinD_R131A_ACY, diffracted to 2.40 Å. The R131A mutant that was cocrystallized with pyruvate but found to have an acetate in its binding site, KinD_R131A_“PYR,” diffracted to 2.64 Å. The cell dimensions of the R131A mutant crystals differ slightly from the wild-type ones.
Structure of the KinD-SD monomer
The structure of the KinD-SD consists of two tandem PAS-like domains, reminiscent of periplasmic ligand-binding domains of other signal transducers.20 Its N-terminal α helix (α1) comprising residues 38–79 is part of both domains of the KinD-SD structure. The α1 helix has a slight 15° bend at residue 55. The two PAS-like domains are aligned along α1: the membrane distal domain consists of residues 66–181, and the membrane proximal domain consists of residues 38–65 and 182–242. Each domain is characterized by a central β-sheet, one face of which interacts with a portion of the long N-terminal α1 helix. The other face is presented to the solvent and can potentially form a ligand-binding site (Fig. 2).
Figure 2.
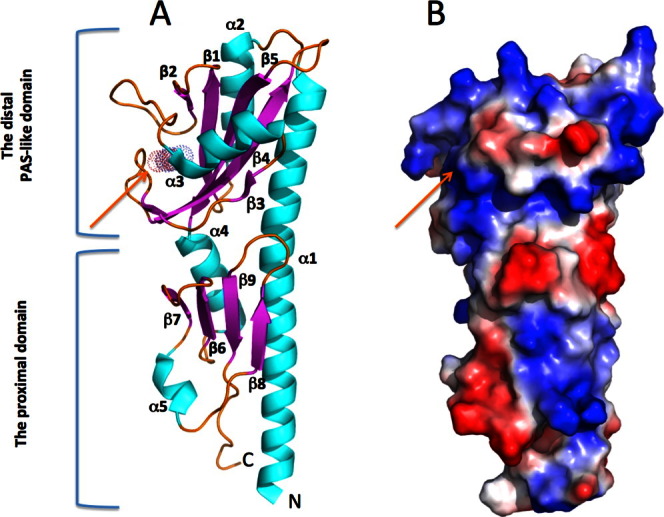
A. Ribbon presentation and surface charge of the KinD structure. A KinD monomer consists of two tandem PAS-like domains. Helices are colored in cyan, the β-sheets are in magenta and loops are in yellow. The N- and C-termini are labeled. A small ligand, pyruvate, is presented as a colored atom dot surface. B. Surface charge of the KinD monomer. Blue and red colors show positive and negative potential, respectively. The pyruvate molecule is buried in the positively charged pocket pointed by the orange arrow. An interactive view is available in the electronic version of the article. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The membrane distal domain consists of the C-terminal segment of helix α1, residues (66–79), and helix α2 (86–97) that are antiparallel to each other. These are followed by the central β-sheet that is composed of five antiparallel β-strands: β1 (103–110), β2 (115–118), β3,1 (144–145), β4 (157–166), and β5 (172–181). The beta segment, β3,2 (149–150), is part of the loop between β3,1 and β4. The helix, α3 (132–140), is located between β-strands 2 and 3 on the opposite side of the N-terminal helices. The arrangement of the secondary structure elements in the distal domain creates a concave pocket decorated with several positively charged residues (Fig. 2).
The helix α4 (182–193) connects the distal to the proximal domain and it is antiparallel to the N-terminal end of helix α1 (38–65). The central β-sheet of the proximal domain is composed of only four antiparallel β-strands: β6 (197–202), β7 (207–211), β8 (223–226), and β9 (232–237). Helix α5 (214–217), a 310 helix, connects the β7 and β8 segments (Fig. 2). The solvent-presented surface of the proximal domain is flat, with no obvious cavity or binding pocket.
The buried surface area between the distal and proximal domains is 1239 Å2. Residues from the β-strands and regions between them from both domains as well as helical segments contribute to this largely hydrophobic core between domains.
Dimerization of KinD sensor domains
In all four crystal structures presented here, two monomers, designated as A and B, form the asymmetric unit (Fig. 3). The monomer structures vary slightly due to crystal packing; the root mean square (RMS) deviation between α-carbons for residues 43–242 is 0.53 Å. The two monomers are related by a noncrystallographic twofold axis of about 174° which is roughly parallel to the long α helix (α1). The N-terminal helix in KinD-SD forms a 15° bend at residue 55. The angle between the two long helices is 15°, and the closest distance between the α carbons is 6.5 Å at residue Tyr64. The interacting surfaces within the dimer of the KinD-SD monomers are mainly formed from the α1 and α2 helices of the distal domains. Within the monomers, the long α1 and short α2 helices are antiparallel to each other and in the dimer they form a slightly distorted four-helix bundle. The Aα1 helix interacts with Aα2 and Bα1, and the Bα1 helix interacts with Bα2 and Aα1. The bend in α1 results in an arrangement where the C-terminal regions of Aα1 and Bα1 wrap around each other. The dimer interface including the four-helix bundle is predominantly hydrophobic with a total buried surface area between the monomers of 1890 Å2. This arrangement creates a well-defined structural unit composed of tandem PAS-like domains. The N- and C-terminals of both chains point in the same direction and approach each other very closely (∼ 5 Å). In the full-length protein, these regions would extend into the transmembrane helices, which therefore are likely to interact in the membrane (Fig. 3). It is probable that a four-helix bundle is formed in the membrane by the four transmembrane helices, consistent with the knowledge that sensor histidine kinases are stable homodimers that modulate autokinase activity in response to ligand binding by a conformational change within the dimer, rather than by altering their oligomeric state.21, 22 Recent NMR structural data and the results from molecular dynamics simulations have essentially confirmed a four-helix bundle arrangement for histidine kinase transmembrane regions.23, 24 The ligand binding may control the relative orientation of these transmembrane helices.20
Figure 3.
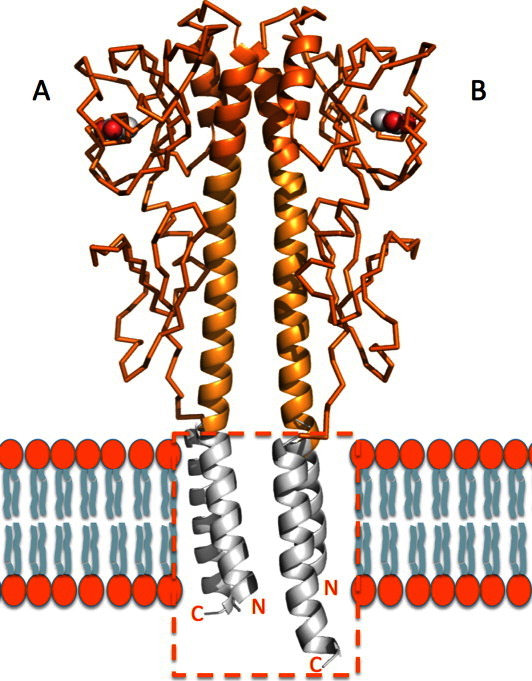
KinD-SD dimer. Two monomers are related with a noncrystallographic twofold axis of about 174°. The two long N-terminal helices are presumed to be closely perpendicular to the cytoplasmic membrane and would be connected to the transmembrane helices that could form a four-helix bundle in the cell membrane with helices connected to the C-terminals. The four-helix bundle through the putative lipid bilayer (marked with orange box) was modeled based on the transmembrane helical structure of rhodopsin II–transducer (PDB entry 1H2S).
Another noncrystallographic dimer in the crystal is formed by monomers with their proximal domains related by a local twofold axis. However, in this pair, the N- and C-terminals of the two monomers point in opposite directions. Therefore, this dimer arrangement is not likely to be of physiological relevance. The interface contains mostly polar residues and the buried surface area between them is 1590 Å2.
DSF identifies pyruvate, propionate, and butyrate as KinD ligands
Pyruvate was copurified and cocrystallized with the KinD-SD occupying a ligand-binding pocket in the membrane distal PAS domain. This suggested that pyruvate might be a physiological KinD ligand; however, it was also possible that of those molecules present during overexpression in E. coli with the propensity to bind to the KinD ligand pocket, pyruvate was the most abundant. To test whether pyruvate binding was specific or whether other small carboxylic acids of similar size were capable of binding to KinD-SD, we performed DSF in the presence of various different compounds (Fig. 4). This assay detects receptor ligands by measuring an increase in the thermal denaturation temperature of the receptor, a consequence of ligand binding. We found that pyruvate, butyrate, and propionate were all able to bind to and stabilize the KinD-SD. On the contrary, lactate failed to bind and stabilize KinD. As lactate and pyruvate are only distinguished by their C-α substituents (hydroxyl- versus keto-group), binding of ligands appears to be highly specific. Notably, the dicarboxylic acids, malate (Fig. 4), fumarate, and succinate (not shown), also failed to bind KinD. The binding of acetate was weaker than pyruvate.
Figure 4.
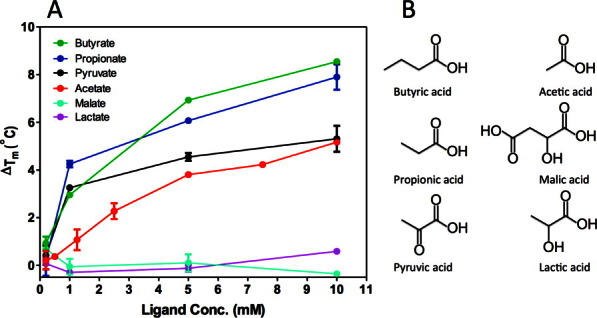
Binding of different small organic acids by KinD-SD. A. Concentration-dependent stabilization of KinD-SD by the ligands: sodium butyrate (green), sodium propionate (blue), sodium pyruvate (black), sodium acetate (red), sodium malate (cyan), and sodium lactate (violet). Tm values were measured by DSF, and the ΔTm values were calculated relative to the Tm value in the absence of ligand. Each point represents the average of two measurements. For some data, the error bars are smaller than the size of the dot. B. Representations of ligands used in the DSF assay.
The ligand-binding site
In our structures, the membrane distal PAS-like domain was found to be occupied by the ligands, whereas the proximal domain was unoccupied. Pyruvate was found in the ligand-binding site of the wild-type KinD-SD and an acetate was found in the KinD-SD R131A mutant (see Fig. 5). The ligand-binding site is a concave pocket composed of one face of the central β-sheet, the loop between β2 and α3 segments (residues 120–130), the helix α3 (131–139), and the loop between β3 and β4 segments (residues 152–156). The ligand-binding site is composed of residues Ser104, Tyr107, Leu128, Arg131, Phe133, Tyr149, Ser151, Arg152, Val177, and Ala179.
Figure 5.
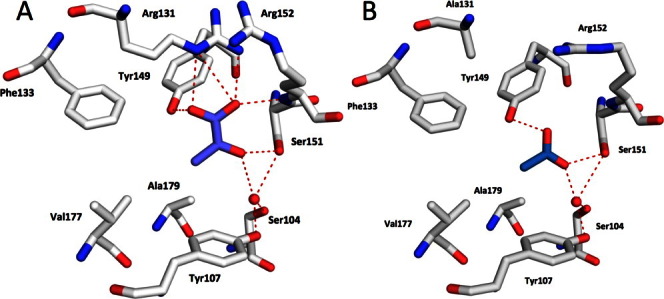
Representations of ligands in the binding pockets of the wild-type KinD and the R131A mutant. A. In wild-type KinD-SD, the pyruvate molecule (in slate/red) forms a salt bridge with Arg131, it also forms hydrogen bonds with side chains of Ser151 and with the nitrogen atom of the main chain of Arg152. Pyruvate also interacts with side chains of Ser104 and Tyr107 through a common water molecule. The methyl group of pyruvate points toward the hydrophobic residues, Phe133, Val177, Ala179, and Leu128 (not shown here). B. In the R131A mutant, the acetate ion (in sky blue/red) interacts with side chains of Ser151, and Tyr149; it also interacts with side chains of Ser104 and Tyr107 through a common water molecule. The methyl group of acetate points toward the hydrophobic residues, Phe133, Val177, Ala179, and Leu128 (not shown here). Key residues are labeled. Hydrogen bonds and salt bridges are indicated by dashed lines. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In the KinD_W_PYR2 structure, the carboxylate moiety of pyruvate forms a salt bridge with Arg131 (3.0 Å), and it hydrogen bonds with the main chain nitrogen atom of Arg152 (2.8 Å) and Tyr149 (2.6 Å) (Fig. 5A). Its keto-group forms a hydrogen bond with Ser151 (2.8 Å), and through a water molecule it interacts with Ser104 and Tyr107. These interactions are likely important to distinguish pyruvate from lactate, which failed to bind KinD. In addition to these polar interactions, the methyl group of pyruvate is placed in a hydrophobic pocket formed by side chains of Leu128, Phe133, Val177, and Ala179 in the binding site. As pyruvate is an asymmetric ligand, its hydrophobic segment plays an important role in determining pyruvate orientation and binding in the binding pocket. Interestingly, the KinD-SD is stabilized better by the more hydrophobic propionate and butyrate than by pyruvate in the DSF assays (see Fig. 4), suggesting that the hydrophobic component of the pyruvate molecule is important to its binding to KinD-SD. This hydrophobic pocket is likely responsible for distinguishing pyruvate from the dicarboxylic acids, fumarate, succinate, and malate, all of which failed to bind KinD.
In the R131A mutant structure (KinD_R131A_ACY), the acetate ion is shifted with respect to pyruvate and its carboxylate maps into the 2-oxo position of pyruvate. It now forms hydrogen bonds with Ser151 (2.8 Å) and Tyr149 (2.8 Å) (Fig. 5B). The carboxylate group of acetate interacts with Ser104 and Tyr107 through a water molecule on one side and interacts through another water molecule with the main chain nitrogen of Arg152 on the other side. The methyl group of acetate assumes the same position as in pyruvate and points toward the hydrophobic residues, Leu128, Phe133, Val177, and Ala175. The fact that acetate is bound preferentially to the mutated binding pocket in the presence of 100 mM pyruvate underscores the importance of the salt bridge with Arg131 in providing a significant fraction of binding energy. Thus, specificity seems to be achieved by a combination of polar and hydrophobic interactions.
Discussion
KinD sensor domain
B. subtilis has 35 signal-transducing sensor histidine kinases. Thirteen of these proteins are predicted to have a periplasmic sensor domain positioned between two transmembrane segments following a cytoplasmic N-terminus, and these kinases might therefore respond to extracellular signals. Molecular ligands are thus far only known for three of these kinases, CitS, DctS, and YufL, which respond to the citric acid cycle intermediates, citrate, fumarate/succinate, and malate, respectively.25 Conversely, molecular ligands for all other B. subtilis histidine kinases are unknown. Out of a total of 13 extracytoplasmic sensor domains, 11 are predicted to have PAS-like structures and two of these have predicted tandem PAS-like structures, namely YesM and KinD, the subject of this study. KinD consists of 506 amino acids with transmembrane helices predicted to be between 19–36 and 251–273 followed by a catalytic segment of the HisKA and HATPase domains in the cytoplasm. Residues 37–250 form the periplasmic sensor domains. Our structures confirmed the structural prediction for KinD-SD, displaying the expected tandem PAS-like structure and adding to an emerging family of receptor domains for signal transduction proteins common to the bacterial phylum20 but also found in plant cytokinin receptors.26
The DALI server27 was used to identify similar structures in the Protein Data Bank. The membrane distal PAS-like domain of the KinD-SD is similar to the structures of the distal domains of other two-domain sensors but the membrane proximal domain of KinD-SD is smaller than those of the others with the exception of the one from Arabidopsis thaliana.26 Of the proteins that have a single PAS-like sensor domain, the CitA sensor domain (PDB ID 1P0Z)28 is structurally closest to the membrane distal domain of KinD-SD. Interestingly, the PhoR sensor domain (PDB ID 3CWF)17 is more closely related to the membrane proximal domain of KinD-SD than any of the proximal PAS-like domains from other two-domain sensors.
The membrane distal PAS-like domain is likely to be the ligand-binding domain for most tandem PAS-like sensors, as small molecule ligands are bound in many of the known crystal structures. However, the function of the membrane proximal domain of these two-domain sensors is presently unknown; small molecules have not been observed to bind there. We offer the following speculations on the function for this domain: (1) it serves as a spacer to position the membrane distal domain appropriately at optimal concentration gradients for its ligand; (2) it serves as an input domain for a second signal; (3) it is an evolutionary rudiment that conserved its structure to allow transmission of a conformational change from the ligand-binding domain to the catalytic domains; (4) it serves to interact with other factors (lipids and proteins) perhaps to position the transducer at an appropriate subcellular compartment. An example in support of (2) has emerged for the McpC chemotaxis receptor29; McpC also features a predicted tandem PAS-like domain.30 Recent evidence suggests that McpC mediates chemotaxis toward 11 proteinogenic amino acid ligands by directly binding to the membrane distal PAS-like domain. Six additional proteinogenic amino acids are detected indirectly via several ligand-binding lipo-proteins that were shown to interact with the membrane proximal PAS-like domain.29 It remains to be seen whether such a dual sensing function can be confirmed for other tandem-PAS sensors. In this context, it is interesting to note that KinD signaling has been linked to the lipoprotein, Med.31
Ligand binding
In the initial KinD sensor structure (KinD_W_PYR1), an unexplained electron density was observed in the concave pocket of the distal domain. The shape of the density and the nature of the surrounding residues suggested that it was a pyruvate moiety. To further prove the nature of this ligand, the KinD-SD sample was preincubated with pyruvate prior to crystallization. The resulting structure (KinD_W_PYR2) showed excellent density for pyruvate in exactly the same position as the unexplained density in KinD_W_PYR1. In KinD_W_PYR1, pyruvate was very likely acquired by the protein during protein expression in E. coli as it was not present in any of the buffers during protein purification or crystallization. The DSF assay confirmed the ability of the KinD-SD to bind pyruvate, propionate, and butyrate.
Comparison of the KinD-SD structures with bound pyruvate and acetate
In the wild-type protein, residue R131 forms a salt bridge with pyruvate. To determine the importance of this interaction, a mutant of KinD-SD was made in which R131 was replaced by an alanine. In the mutant (KinD_R131A_ACY) structure, acetate was found in the membrane distal domain at a position very similar but not identical to where the pyruvate was bound in the native structure. The mutant was crystallized from a solution that contained acetate. Surprisingly, R131, which plays a key role in pyruvate binding, is not required for the interaction between the protein and acetate. To verify KinD-SD R131A's preference for acetate, the mutant protein was preincubated with a high concentration (100 mM) of pyruvate and crystallized under conditions not containing acetate (KinD_R131A_“PYR”). This structure also showed acetate in the binding site. Again, in this later case, the acetate is very likely acquired by the KinD-SD R131A mutant during protein expression in E. coli because acetate was not added during protein purification or crystallization. Inadvertently, by a single mutation in the binding site, we have switched the specificity of KinD-SD from pyruvate to acetate. The KinD ligand-binding site appears to be specific for carboxylic acids and can accept pyruvate, propionate, and butyrate but rejects lactate and malate. These acids differ in the length of the aliphatic chain and stereochemistry/substitution of the alpha carbon. Our structures predict that this site would also accept 2-oxo-butyrate but would reject 2-hydroxy butyrate, alanine, or 2-amino butyrate, and it binds weakly acetate as well.
A question arose whether a comparison of wild-type and mutant structures would reveal differences that might give insights on the conformational changes induced by ligand binding. However, the wild-type and the mutant structures are essentially identical; the RMS deviation of the A chain alpha carbons is 0.30 Å and that of the B chain is 0.31 Å (residues 43–242 were overlapped for both chains). The angle of rotation that relates the two monomers is 174.4° for the wild type and 173.8° for the R131A mutant. When the alpha carbons of the two dimers (43–242 from both monomers) are overlapped, the RMS deviation is 0.42 Å. The slightly different mode of dimerization observed between the wild-type and mutant KinD-SD might be caused by the fact that the two crystal unit cells are slightly different; the crystals were obtained from different precipitants.
Is pyruvate the physiological ligand of KinD?
Pyruvate is a key metabolite feeding the Krebs cycle and is involved in several other metabolic pathways. In the presence of carbohydrates, B. subtilis produces and secretes large quantities of pyruvate into the extracellular medium where its concentration peaks toward the end of exponential growth.32 Upon exhaustion of glucose, pyruvate is reimported and further metabolized by lactate dehydrogenase.33 From all available data, it seems plausible that the external concentration of pyruvate can reflect B. subtilis growth conditions and its metabolic status.
This data show that KinD-SD interacts tightly with pyruvate, but it cannot unambiguously identify pyruvate as the true physiological ligand. We attempted a number of experiments to identify whether pyruvate might indeed activate KinD in vivo. A B. subtilis strain deleted for all other sporulation kinases does not sporulate and we hoped to find that the addition of pyruvate to the media might induce sporulation. This however was not the case (data not shown), perhaps because sufficient pyruvate for KinD activation was already secreted by this strain, and perhaps KinD alone is not capable of producing sufficient levels of Spo0A-P to initiate sporulation. In 1993, a report described that a mutation in the alanine dehydrogenase, the enzyme that converts alanine to pyruvate, resulted in a sporulation defect that could be partially rescued by the addition of pyruvate to the media.34 We hypothesized that perhaps KinD serves as a phosphatase in the absence of pyruvate and thus drains phosphoryl groups from the system. If this were true, we would have expected that an aldkinD double mutant sporulates better than the ald single mutant. However, this was not the case and in addition, when placing the ald mutation in B. subtilis strains having only the gene for either KinA, KinB, or KinD, all strains were affected by the ald mutation (data not shown), suggesting that the effect on sporulation occurs downstream of the kinases and perhaps downstream of the entire phosphorelay. Of course, none of these experiments rule out that pyruvate is the physiological ligand for KinD but they also do not confirm it. Apparently more detailed experiments than those described are required to confirm the relevance of the pyruvate ligand to KinD signaling. An in vitro approach with the full-length kinase would perhaps be most straight-forward. Thus far, the expression of the full-length transmembrane histidine kinase has eluded us.
While this manuscript was in preparation, a study was published that explored the developmental response of B. subtilis to the presence of tomato roots.35 The authors found that a developmental phenomenon regulated by the sporulation phosphorelay, namely biofilm formation, could be induced by the presence of tomato roots and that this phenomenon was dependent on KinD. They identified malate to be at least partially responsible for induction of biofilm formation mediated by KinD. As large concentrations of malate were required to observe this effect, the authors were careful in their interpretation of the data, speculating that it might not be malate directly that serves as a signal for KinD, but perhaps that the presence of a large amount of malate triggers a metabolic shift in B. subtilis that then itself secretes increasing concentrations of the KinD signal. Alternatively, they argued that malate along with other unidentified compounds might serve as the KinD signal directly. We concur with the former conclusion, as the KinD-SD structure is unlikely to accommodate a malic acid molecule in the binding pocket that is currently occupied by pyruvate. Consistent with this notion, neither lactate, malate, fumarate, nor succinate showed any binding to the KinD-SD in our DFS assays.
In this context, it is interesting to compare the structures of the KinD-SD in complex with pyruvic acid to that of the dicarboxylic acid sensor, DctB-SD, in complex with succinic acid.36 The sensor histidine kinase, DctB, is known to activate in response to several dicarboxylic acids, including succinate and malate.36 Overall, the ligand-binding domains of DctB and KinD are remarkably similar and the respective ligands occupy similar binding pockets. Strikingly, the carboxy group of pyruvate and succinate in the respective proteins is coordinated by a conserved arginine (R131 in KinD). The remainder of the residues lining the binding pockets however is different. Most notably, the second carboxy group of succinate forms a salt bridge with Lys197 in DctB. This residue is an alanine in KinD and forms a hydrophobic pocket together with the surrounding residues. It thus seems unlikely that malic acid is directly sensed by KinD. Pyruvate and/or perhaps some other monocarboxylic acids seem to be the more likely physiological ligands. The DctB example also suggests that there might not be just a single physiological ligand, but instead an array of related molecules including propionic acid and butyrate that could also serve as KinD activators. As mentioned above, the chemotaxis sensor, McpC, binds and responds to 11 amino acids directly, adding to the dual PAS-like sensors that respond to multiple ligands.29
Comparison of periplasmic sensors composed of tandem PAS-like domains
The quorum sensor, LuxQ, represented the first structural example of a tandem PAS-like sensor domain.37 In recent years, the tandem PAS-like signal detection domain has been recognized as a common feature among bacterial signal transduction proteins, including histidine kinases, chemotaxis receptors, and diguanylate cyclases, and has also been found in the plant histidine kinase where it binds and responds to cytokinins.
To date, structures of at least eight sensors with tandem PAS-like domains from bacteria have been determined (PDB IDs 3LI9, 3LIA, 3LIC, 3E4P, 3E4Q, 3C8C, 3T4J, and 4JGP [this work]). Although at first glance the various tandem PAS domain sensors look very similar, on closer examination they all display individual characteristics. Each protein features a characteristic bend at specific but differing locations in the long N-terminal alpha helices (as determined by the program HELANAL38); furthermore, the distance and angle between the N-terminal helices of the dimer differ for each protein (see Table I). Two ortholog sensor domains were determined from different bacteria.36, 39 Furthermore, several forms of the sensors DctB (3BY936) and LuxQ (3C3037) were determined in more than one crystal form with different ligands bound to them. The structures are not altered significantly by crystal packing in different unit cells when crystallized from different precipitating agents.
Table I.
Characteristics and Interactions of the N-terminal Long Helices
| Bend of N-terminal helix | Dimer | ||||||
|---|---|---|---|---|---|---|---|
| Distance between helices | Rotation angle between monomers (°) | ||||||
| PDB code | Protein | Location | Angle (°) | Angle between helices (°) | Location | Distance (Å) | |
| 4JGP | KinD_pyr | Leu19 | 9 (12)a | 14.9 | Tyr28 | 6.5 | 174.4 |
| 3LI9 | Z2 | Asp59 | 29 | 7.5 | Ala60 | 4.0 | 180b |
| 3LIA | Z2 | Asp59 | 26 (30)a | 8.9 | Ala60 | 3.8 | 180 |
| 3LIC | Z6 | Leu38 | 20 | 28.6 | Phe61 | 11.8 | 180b |
| 3E4P | DctB/MLA | Leu82 | 21 (23)a | 17.6 | Ala74 | 5.6 | 171.6 |
| 3E4Q | DctB/Apo | 84 (86)a | 29 (35)a | 15.1 | Ala74 | 5.5 | 172.4 |
| 3C8C | Mcp_N | Glu72 | 17 | 8.5 | Gln83 | 11.9 | 176.2 |
| 3T4J | A. thaliana | 132 (169)a | 11 (8)a | 30.2 | Val155 | 8.0 | 179.2 |
Value in parentheses is for monomer 2.
Monomers related by a crystallographic twofold axis.
The tandem PAS sensors are generally monomers in solution (see Ref.20 and KinD-SD). In the crystal structures, monomers, “head to tail” and “parallel” dimers are all observed. The “parallel” dimers are believed to be of physiological relevance consistent with the fact that sensor histidine kinases form stable dimers and because the N- and C-termini in this dimer form are placed such that they connect to the adjacent N- and C-terminal transmembrane helices.
The rotation angles that relate the monomers in the “parallel” dimers vary from 180° to 140°. The following rotation angles were observed: 171.6 and 172.4° in 3E4P and 3E4Q (DctB29, 30), 174.4° in KinD-SD (this work), 176.2° in 3C8C,32 179.2° in 3T4J34 and 180° in 3LIA, 3LI8, and 3LI9.31 In 3LI8 and 3LI9, the two monomers are related by a crystallographic twofold axis; in 3LIA, the local twofold is close to the b-axis of the unit cell. The rotation angles between the two monomers that vary from 180° are probably the result of accommodating the molecules in the crystals, to optimize their packing. A 140° angle was observed only in the LuxQP-AI-2 complex 2HJ9,37 and is likely due to the optimization of the molecular interactions within the complex. Different relative orientations of monomers in the “parallel” dimers and failure to detect dimers in solution suggest that interactions between monomers are weak and can be influenced by crystal packing forces. We thus caution that shifts in the dimer interface of these domains in response to ligand binding might be artificial in some cases and might not always be of physiological relevance. Regardless, a wide body of literature suggests that signal detection is accompanied by quaternary structural changes (e.g., Refs.37,40, and41), and the weak associations detected here and elsewhere might be necessary to allow for such changes.
In summary, the KinD structures add additional observations on how a sensor domain detects small molecule ligands. The structures add to the growing number of tandem PAS-like sensor domains used by many signal transducers and implicate pyruvate and other small monocarboxylic acids as ligands for this developmentally important kinase.
Materials and Methods
Protein cloning, expression, purification, and production of the R131A mutant
The PCR-amplified KinD-SD gene fragment from B. subtilis (gi: 16078430; residues 37–250) was cloned into the vector pMCSG7 using a modified LIC protocol as described earlier.19 KinD-SD was produced as an N-terminal His6-tagged protein in E. coli BL21 (DE3) carrying plasmids pMAGIC that encodes one rare E. coli tRNA (covering Arg codons [AGG/AGA]) (Scientific Reagents). Cells were grown with ampicillin and kanamycin at 100 and 30 μg/mL, respectively. A seleno-methionine (SeMet) derivative of the expressed protein was prepared as described earlier.42 The harvested cells, containing SeMet-labeled protein, were resuspended in lysis buffer (500 mM NaCl, 5% [v/v] glycerol, 50 mM HEPES pH 8.0, 10 mM imidazole, and 10 mM 2-mercaptoethanol) and stored at −80°C. The protein was purified using Ni-affinity chromatography as described previously.43 Briefly, the harvested cells were resuspended in lysis buffer supplemented with 1 mg/mL lysozyme and 100 mL of protease inhibitor (Sigma, P8849) per 2 g of wet cells. This mixture was kept on ice for 20 min and then sonicated. The lysate was clarified by centrifugation at 36,000g for 1 h and filtered through a 0.44 μm membrane. The lysate was applied to a 5 mL HiTrap Ni-NTA column (GE Health Systems) on an ÄKTAxpress system (GE Health Systems). The His6-tagged protein was released with elution buffer (500 mM NaCl, 5% glycerol, 50 mM HEPES pH 8.0, 250 mM imidazole, and 10 mM 2-mercaptoethanol) and the fusion tag was removed by treatment with recombinant His7-tagged TEV protease (a gift from Dr. D. Waugh, NCI). Subtractive Ni-NTA affinity chromatography was used to remove the His6-tag, uncut protein, and His7-tagged TEV protease. The KinD-SD was dialyzed against crystallization buffer containing 250 mM NaCl, 20 mM HEPES pH 8.0, and 2 mM dithiothreitol and then concentrated to 25 mg/mL for crystallization using an Amicon Ultra centrifugal filter device with a 10,000 MW cutoff (Millipore), flash-cooled, and stored in liquid nitrogen.
The Polymerase Incomplete Primer Extension cloning method44 was used to create an Arg131 to Ala131 (R131A) site-directed mutant. The forward primer, CGAAAGTAAACCTGGCCGACGCATCTTTTTTTATAAAAGCAAAGG, and reverse primer, CCTTTGCTTTTATAAAAAAAGATGCGTCGGCCAGGTTTACTTTCG, were designed and the aforementioned pMCSG7 construct of the KinD sensor domain was used as the template. Using PfuUltra Hotstart PCR Master Mix (Stratagene, La Jolla, CA), the mutagenesis was performed in a final volume of 50 μL. The produced mutant clone was sequenced for confirmation. The R131A mutant was produced using the same procedure as wild-type KinD-SD.
Protein crystallization
The KinD-SD proteins, the wild-type and the R131A mutant were crystallized using sitting drop vapor diffusion at 297 K in a CrystalQuick® 96-well round-bottomed plate (Greiner Bio-One North America). Protein solution (400 nL) was mixed with 400 nL of crystallization reagent using the Mosquito® nanoliter liquid workstation (TTP LabTech) and allowed to equilibrate against 135 μL of crystallization reagent. The plate was then incubated at 24°C in a RoboIncubator® plate storage system (Rigaku). Automated crystal visualization (CrystalTrak, Rigaku) was used for droplet imaging using Minstrel III® (Rigaku). Five different crystallization screens were used: Index (Hampton Research), SaltRx (Hampton Research), PEGs II Suite (Qiagen—NeXtal), and MCSG-1 and MCSG-2 (Microlytic). The best crystals of KinD-SD were obtained in PEGs II Suite condition B3, which contains 0.1M HEPES pH 7.0 and 25% (w/v) PEG 1000. The R131A mutant was crystallized in MCSG-1 condition B3 consisting of 0.2M ammonium acetate, 0.1M Bis-Tris pH 5.5, and 25% (w/v) PEG3350. Cocrystals of the wild-type protein or the R131A mutant protein and pyruvate were prepared by preincubating 1 mM protein with 30 mM or 100 mM pyruvate. The best crystals of the wild-type protein–pyruvate complex were obtained with 30 mM pyruvate in the MCSG-2 B5 condition (0.2M ammonium sulfate, 0.1M sodium cacodylate:HCl pH 6.5 and 20% [w/v] PEG 8000). The R131A mutant with pyruvate was crystallized in the MCSG-1 D2 condition (0.2M sodium chloride, 0.1M Bis-Tris pH 6.5, and 25% [w/v] PEG3350) in the presence of 100 mM pyruvate. All crystals were cryoprotected using the solution prepared by adding 10–15% (v/v) glycerol to the crystallization condition and flash-cooled in liquid nitrogen. All crystals belong to orthorhombic space group, P212121, with similar unit cell dimensions of a = 75.8–76.7, b = 82.2–83.4, c = 90.4–91.2 Å (Table II) and diffracted X-rays to 2.12–2.76 Å with crystals of the wild-type protein diffracting to higher resolutions than the R131A mutant.
Table II.
Summary of Data Collection
| Data collection | ||||
|---|---|---|---|---|
| KinD_PYR1 | KinD_PYR2 | KinD_R131A_ACY | KinD_R131A_“PYR” | |
| Crystal | Cocrystallized with pyruvate | Cocrystallized with pyruvate | ||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell (a,b,c, Å) | 76.1, 82.5, 90.4 | 76.4, 82.2, 90.7 | 76.7, 83.4, 91.0 | 75.8, 83.0, 91.2 |
| Highest resolution bin (Å) | 2.34–2.30 | 2.07–2.03 | 2.44–2.40 | 2.69–2.64 |
| Number of observed reflectionsa | 24,870 (1075) | 36,578 (1704) | 22,228 (999) | 17,054 (787) |
| Rmerge (%)a,b | 0.133 (0.524) | 0.063 (0.638) | 0.088 (0.610) | 0.051 (0.648) |
| Completeness (%)a | 99.3 (94.1) | 99.8 (100) | 99.2 (99.7) | 99.7 (100) |
| I/σIa | 5.1 (1.62) | 15.7 (4.9) | 10.1 (4.61) | 16.1 (4.28) |
Numbers in parentheses are values for the highest resolution bin.
Rmerge = 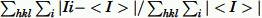 ,, where Ii is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l.
,, where Ii is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l.
X-ray diffraction and structure determination
Diffraction data were collected at 100 K at the 19-BM beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory. For all crystals, single-wavelength anomalous dispersion (SAD) data at 0.97924 Å (12.6605 keV) near the absorption peak for selenium were collected from a single SeMet-labeled protein crystal (0.2 × 0.05 × 0.05 mm3). The absorption peak energy was determined using the X-ray fluorescence scan. Data collection was done using Omega rotation by exposing a crystal to the X-ray beam for 5 s per 1.0° rotation at a crystal to detector distance of 200 mm. The data were recorded on a CCD detector ADSC Q210r scanning a full 180° on omega. All the data were collected using SBCCOLLECT and processed and scaled by the HKL3000 suite.45 Crystal data statistics are listed in Table II.
The structures were determined by SAD phasing using HKL300045 with the following steps: the 10 selenium (Se) sites were located by SHELXC and SHELXD,46 the handedness of the substructure Se sites was checked by SHELXE,46 phasing was done by MLPHARE,47 density modification was done as published,48 and the first model was built by ARP/wARP.49 For each structure, the initial model from HKL3000 was then completed manually using COOT.50 All ligand molecules, either pyruvate or acetate, in the structures were evident from the electron density calculated using the SAD-density modified phases. Multiple iterative cycles of manual adjustment using COOT together with refinement using PHENIX51 led to the final models with good R and R-free values and stereochemistry evaluated by MolProbity and Ramachandran plot. The final refined models of the structures contain two chains of the KinD-SD and well-defined ligands, either pyruvate or acetate and between 38 and 298 solvent molecules (Table III). In some of the structures, there was no interpretable electron density for 1–3 N- and 5–7 C-terminal residues as well as the loop region near Cys99, and these parts of the protein are missing in the final models. The combined model covers residues 37–245 of the extracytoplasmic domain. Main chain torsion angles of all residues of the four structures fell within acceptable regions of Ramachandran plots. The details of the structure determination, refinement, and model quality are shown in Table III.
Table III.
Summary of Structure Determination and Refinement
| Phasing (SAD) | ||||
|---|---|---|---|---|
| KinD_PYR1 | KinD_PYR2 | KinD_R131A_ACY | KinD_R131A_“PYR” | |
| Phasing power/FOM | 1.31/0.21 | 1.78/0.32 | 1.43/0.27 | 1.7/0.29 |
| Resolution range (Å) | 36.28–2.30 | 37.4–2.03 | 37.9–2.40 | 37.9–2.64 |
| Number of SeMet | 10 | 10 | 10 | 10 |
| Refinement | ||||
| Resolution range (Å) | 36.28–2.30 | 37.4–2.03 | 37.9–2.40 | 37.9–2.64 |
| Rcryst (%) | 19.3 | 18.0 | 20.1 | 19.3 |
| Rfree (%) | 24.1 | 21.6 | 25.2 | 24.8 |
| Number of protein residues | 416 | 408 | 407 | 404 |
| Solvent molecules | 153 | 298 | 175 | 38 |
| RMS bond (Å) | 0.008 | 0.007 | 0.008 | 0.009 |
| RMS angle (°) | 1.144 | 1.043 | 1.097 | 1.21 |
| RMS dihedral (°) | 15.6 | 14.9 | 16.91 | 17.4 |
| Avg. B (Å2) | 44.3 | 34.8 | 47.7 | 33.7 |
| Wilson B (Å2) | 34.0 | 22.3 | 40.8 | 25.8 |
| Ramachandran Plot | ||||
| Preferred (%)a | 98.8 | 99.8 | 98.8 | 99.3 |
| Gen. allowed (%)a | 1.2 | 0.2 | 1.2 | 0.7 |
| Disallowed (%)a | 0 | 0 | 0 | 0 |
| PDB ID | 4JGO | 4JGP | 4JGR | 4JGQ |
As defined in phenix.refine (PHENIX).
Characterization of the α helices
The N-terminal helices were characterized by the program HELANAL.38 The bend in the helix and its direction cosine were determined by the program, and the angle between the helices was determined from the direction cosines.
Binding of small acid to KinD-SD
The stability of KinD-SD in the presence of different acid molecules was measured by DSF as described.52, 53 DSF experiments were performed using an MX4000 RT-PCR instrument (Stratagene) with excitation at 492 nm and emission at 610 nm. KinD-SD was diluted to 10 μM in a solution containing 100 mM HEPES pH 7.5, 150 mM NaCl, and 5× SYPRO orange. Ligand concentrations ranged from 0.2 to 10 mM. SYPRO orange was obtained as a 5000× stock solution in dimethyl sufoxide from Invitrogen. Samples (40 μL) were arrayed in white quantitative PCR plates with optical caps (ABgene). Samples were heated from 10°C to 80°C at 1°C increments. For each data point, the sample was equilibrated at the indicated temperature for 1 min prior to data measurement. Each sample was analyzed in duplicate. Tm values were obtained by fitting the data in the thermal transition region to the Boltzman sigmoidal equation using the program Prism (GraphPad Software).
Acknowledgments
The authors would like to thank members of the Structural Biology Center at Argonne National Laboratory for their help in conducting these experiments. The authors wish to thank Gekleng Chhor for proofreading of this manuscript.
References
- 1.Cano RJ, Borucki MK. Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science. 1995;268:1060–1064. doi: 10.1126/science.7538699. [DOI] [PubMed] [Google Scholar]
- 2.Bourret RB, Silversmith RE. Two-component signal transduction. Curr Opin Microbiol. 2010;13:113–115. doi: 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szurmant H, White RA, Hoch JA. Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol. 2007;17:706–715. doi: 10.1016/j.sbi.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 7.Zapf J, Sen U, Madhusudan, Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8:851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 8.Perego M, Brannigan JA. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides. 2001;22:1541–1547. doi: 10.1016/s0196-9781(01)00490-9. [DOI] [PubMed] [Google Scholar]
- 9.Bick MJ, Lamour V, Rajashankar KR, Gordiyenko Y, Robinson CV, Darst SA. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J Mol Biol. 2009;386:163–177. doi: 10.1016/j.jmb.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Grau R, Perego M, Hoch JA. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Tomchick DR, Brautigam CA, Machius M, Kort R, Hellingwerf KJ, Gardner KH. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry. 2008;47:4051–4064. doi: 10.1021/bi7021156. [DOI] [PubMed] [Google Scholar]
- 13.McLoon AL, Kolodkin-Gal I, Rubinstein SM, Kolter R, Losick R. Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J Bacteriol. 2011;193:679–685. doi: 10.1128/JB.01186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci USA. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis Biofilms. MBio. 2010;1:1–7. doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anantharaman V, Aravind L. Cache—a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem Sci. 2000;25:535–537. doi: 10.1016/s0968-0004(00)01672-8. [DOI] [PubMed] [Google Scholar]
- 17.Chang C, Tesar C, Gu M, Babnigg G, Joachimiak A, Pokkuluri PR, Szurmant H, Schiffer M. Extracytoplasmic PAS-like domains are common in signal transduction proteins. J Bacteriol. 2010;192:1156–1159. doi: 10.1128/JB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 19.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dago AE, Schug A, Procaccini A, Hoch JA, Weigt M, Szurmant H. Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc Natl Acad Sci USA. 2012;109:E1733–1742. doi: 10.1073/pnas.1201301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szurmant H, Bu L, Brooks CL, 3rd, Hoch JA. An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxiliary proteins. Proc Natl Acad Sci USA. 2008;105:5891–5896. doi: 10.1073/pnas.0800247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maslennikov I, Klammt C, Hwang E, Kefala G, Okamura M, Esquivies L, Mors K, Glaubitz C, Kwiatkowski W, Jeon YH, Choe S. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. Proc Natl Acad Sci USA. 2010;107:10902–10907. doi: 10.1073/pnas.1001656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka K, Kobayashi K, Ogasawara N. The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium. Microbiology. 2003;149:2317–2329. doi: 10.1099/mic.0.26257-0. [DOI] [PubMed] [Google Scholar]
- 26.Hothorn M, Dabi T, Chory J. Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat Chem Biol. 2011;7:766–768. doi: 10.1038/nchembio.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 29.Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol Chem. 2012;287:39412–39418. doi: 10.1074/jbc.M112.413518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glekas GD, Foster RM, Cates JR, Estrella JA, Wawrzyniak MJ, Rao CV, Ordal GW. A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J Biol Chem. 2009;285:1870–1878. doi: 10.1074/jbc.M109.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banse AV, Hobbs EC, Losick R. Phosphorylation of Spo0A by the histidine kinase KinD requires the lipoprotein med in Bacillus subtilis. J Bacteriol. 2011;193:3949–3955. doi: 10.1128/JB.05199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speck EL, Freese E. Control of metabolite secretion in Bacillus subtilis. J Gen Microbiol. 1973;78:261–275. doi: 10.1099/00221287-78-2-261. [DOI] [PubMed] [Google Scholar]
- 33.Yashphe J, Hoch JA, Kaplan NO. Regulation of lactate dehydrogenase synthesis in Bacillus subtilis. Biochim Biophys Acta. 1978;544:1–7. doi: 10.1016/0304-4165(78)90203-9. [DOI] [PubMed] [Google Scholar]
- 34.Siranosian KJ, Ireton K, Grossman AD. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J Bacteriol. 1993;175:6789–6796. doi: 10.1128/jb.175.21.6789-6796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo JH, Losick R. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol. 2012;85:418–430. doi: 10.1111/j.1365-2958.2012.08109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou YF, Nan B, Nan J, Ma Q, Panjikar S, Liang YH, Wang Y, Su XD. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J Mol Biol. 2008;383:49–61. doi: 10.1016/j.jmb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bansal M, Kumar S, Velavan R. HELANAL: a program to characterize helix geometry in proteins. J Biomol Struct Dyn. 2000;17:811–819. doi: 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- 39.Cheung J, Hendrickson WA. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J Biol Chem. 2008;283:30256–30265. doi: 10.1074/jbc.M805253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 41.Ayers RA, Moffat K. Changes in quaternary structure in the signaling mechanisms of PAS domains. Biochemistry. 2008;47:12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultrafast protein structure determination. Acta Crystallogr D Biol Crystallogr. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 43.Kim Y, Dementieva I, Zhou M, Wu R, Lezondra L, Quartey P, Joachimiak G, Korolev O, Li H, Joachimiak A. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klock HE, Lesley SA. The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009;498:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 45.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 46.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z. Daresbury study weekend proceedings. In: Wolf W, Evans PR, Leshe AGW, editors. Warrington, UK: SERC Daresbury Laboratory; 1991. pp. 80–85. [Google Scholar]
- 48.Cowtan K. DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 49.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 53.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Jr, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, Hui R, Marsden BD, Nordlund P, Sundstrom M, Weigelt J, Edwards AM. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]


