Abstract
Bacterial microcompartments (MCPs) are large proteinaceous structures comprised of a roughly icosahedral shell and a series of encapsulated enzymes. MCPs carrying out three different metabolic functions have been characterized in some detail, while gene expression and bioinformatics studies have implicated other types, including one believed to perform glycyl radical-based metabolism of 1,2-propanediol (Grp). Here we report the crystal structure of a protein (GrpN), which is presumed to be part of the shell of a Grp-type MCP in Rhodospirillum rubrum F11. GrpN is homologous to a family of proteins (EutN/PduN/CcmL/CsoS4) whose members have been implicated in forming the vertices of MCP shells. Consistent with that notion, the crystal structure of GrpN revealed a pentameric assembly. That observation revived an outstanding question about the oligomeric state of this protein family: pentameric forms (for CcmL and CsoS4A) and a hexameric form (for EutN) had both been observed in previous crystal structures. To clarify these confounding observations, we revisited the case of EutN. We developed a molecular biology-based method for accurately determining the number of subunits in homo-oligomeric proteins, and found unequivocally that EutN is a pentamer in solution. Based on these convergent findings, we propose the name bacterial microcompartment vertex for this special family of MCP shell proteins.
Keywords: OCAC, oligomeric state determination, EutN, bacterial microcompartments, protein assembly, capsid, glycyl radical, BMV, pentameric vertex protein, GrpN
Introduction
Microcompartments (MCPs) are polyhedrally shaped supramolecular protein assemblies that physically encapsulate select metabolic pathways in prokaryotes (reviewed in Refs.1–5). This encapsulation serves to concentrate and separate specific enzymes and their metabolic intermediates from the cytoplasm, thereby increasing reaction efficiency, retaining volatile intermediates, and/or protecting the cellular milieu from toxic intermediates.5 The carboxysome MCP increases carbon-fixation efficiency by encapsulating carbonic anhydrase and RuBisCO together, so the CO2 produced by the first enzyme can be delivered at high concentration to the second enzyme.4, 6 Cofactor B12-dependent MCPs for ethanolamine utilization (Eut) and 1,2-propandiol utilization (Pdu) function by retaining volatile or toxic aldehyde compounds—propionaldehyde and acetaldehyde—that occur as intermediates in those pathways.5, 7–11 Bioinformatic studies have suggested the existence of an MCP for glycyl radical-based 1,2-propanediol metabolism, referred to hereafter as Grp.12 Sequence analysis and gene expression data suggest that the Grp MCP, like the Pdu MCP, houses enzymes for metabolizing 1,2-propanediol, but that it uses a glycyl radical enzyme for the key dehydration reaction rather than B12-dependent enzymes.12,13 The Grp MCP has not yet been studied in detail.
Structural analyses have answered a number of questions regarding the geometry and mechanism of MCP assembly and function (reviewed in Ref.2, 3). Current models describe MCPs as being comprised of sheets of hexameric shell proteins (belonging to the BMC family of proteins) forming polyhedral facets,14–16 along with a much smaller number of special pentameric proteins (belonging to the EutN/PduN/CcmL/CsoS4 protein family) placed at the vertices of the polyhedral shell.17 This family of presumptive vertex proteins appears distinct in sequence and structure from the BMC family of proteins. Although crystallographic studies of (hexameric) BMC shell proteins have consistently supported this model of MCP organization, experiments on the vertex proteins have provided a less coherent picture. Two crystal structures of carboxysome vertex proteins, CcmL and CsoS4A, are indeed pentameric17 (PDB accession code 2QW7 and 2RCF). However, a third structure of a presumptive vertex protein, EutN from E. coli (PDB code 2Z9H), revealed a hexameric quaternary assembly with nearly hexagonal shape.18 Likewise, multiple genetic studies have shown that deleting the vertex proteins compromises the formation of closed polyhedral shells,19–21 yet closed shells can apparently be formed with lower efficiency in some cases.22 These equivocal results have complicated the interpretation of the architectural role of this protein family.
Electron microscopy studies indicate that carboxysomes tend to be more geometrically regular and more nearly icosahedral in shape compared to other types of MCPs that have been visualized23, 24; this has raised the possibility that pentameric units required for symmetric icosahedral architecture might be present only in carboxysome MCPs. Here we provide evidence that this family of proteins serves as the pentameric vertex element across multiple divergent types of MCPs. Specifically, we report the pentameric crystal structure of GrpN from the Grp MCP, and show also that EutN is a pentamer in solution, contrary to previous crystallographic findings.
Results and Discussion
An operon encoding enzymes and shell proteins for a presumptive Grp MCP has been described in R. rubrum.12 One of the encoded proteins, referred to hereafter as GrpN (Gene ID: 12642037), was identified as a representative of the EutN/PduN/CcmL/CsoS4 family of shell proteins (Pfam domain: PF03319), hereafter described as bacterial microcompartment vertex (or BMV) proteins. GrpN was overexpressed in E. coli and purified by metal affinity chromatography on the basis of a polyhistidine tail added to the protein. Single crystals were obtained in space group I23, and an X-ray structure was determined by molecular replacement and refined at a resolution of 3.2 Å. The final atomic model is 96% complete; residues 65–70 are in a disordered loop and could not be modeled accurately.
As expected, the tertiary fold follows closely that reported earlier for other members of this protein family (Fig. 1). The core structure of GrpN is comprised of five antiparallel beta strands that curve to create a small beta barrel. The C-termini and loop regions between beta strands extend away from the beta-barrels, making contacts with adjoining subunits. The atomic coordinates align well to CcmL, CsoS4A, and EutN, with RMSD values of 0.73 Å, 0.81 Å, and 0.80 Å, respectively, over C-alpha positions. Stabilization of the quaternary structure is largely mediated by contacts between loop regions from adjacent beta-barrels; minor contributions are made by barrel-to-barrel contacts. Structural comparisons between BMV structures show that the N-termini and the small beta barrels are highly conserved, while the C-termini and regions adjoining the core beta sheets differ slightly in length and secondary structure content. For example, GrpN and CsoS4A lack C-terminal beta hairpins present in both CcmL and EutN.
Figure 1.
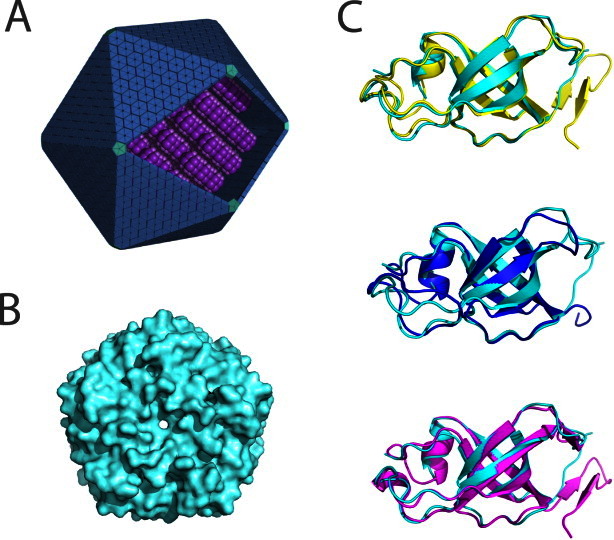
Comparison of the structure of GrpN with other bacterial microcompartment vertex (BMV) proteins. (A) An idealized model of an MCP showing pentameric units at the vertices of the polyhedral shell. (B) Space-filling model of the pentameric structure of GrpN. (C) Superposition of a GrpN monomer with CcmL (top, yellow), CsoS4A (middle, blue), and EutN (bottom, magenta). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
GrpN crystallized as a pentamer in the asymmetric unit of the crystal (Table I). The pentameric unit of GrpN matches those reported earlier for CcmL from the beta-type carboxysome and CsoS4A from the alpha-type carboxysome. One surprising aspect of the GrpN crystal structure is the arrangement of pentamers in the unit cell. Twelve crystallographically-related pentamers are arranged in a nearly icosahedral fashion. This 60-subunit icosahedral arrangement is unlikely to reflect the natural assembly of the much larger MCP; in native MCPs, pentameric units are presumably surrounded by BMC-type hexamers. In addition, the interactions between pentamers in the crystal involve relatively small sites of contact. Nonetheless, this spontaneous assembly could be an interesting starting point for engineering a novel icosahedral cage. The design of self-assembling protein cages by various methods has been discussed in a series of recent studies.27–31 The observation that GrpN is a pentamer establishes that other MCPs besides carboxysomes possess special pentameric shell proteins to explain the closure of an otherwise flat layer of hexameric units provided by the BMC-type proteins.
Table 1.
X-ray Data Collection and Model Refinement Statistics
| Statistics | Value |
|---|---|
| Wavelength (Å) | 0.9791 |
| Resolution range (Å) | 19.8–3.2 (3.314–3.2) |
| Space group | I23 |
| Unit cell (Å) | a = b = c = 150.7 |
| Total reflections recorded | 49,986 (3856) |
| Unique reflections | 9,248 (700) |
| Multiplicity | 5.4 (5.5) |
| Completeness (%) | 96.6 (99.6) |
| Mean I/sigma(I) | 21.0 (3.0) |
| Wilson B-factor (Å2) | 84.4 |
| R-meas25 | 7.3% (62%) |
| Model R-work | 0.267 (0.340) |
| Model R-free | 0.288 (0.381) |
| Number of non-hydrogen atoms | 2,930 |
| Macromolecules | 2,926 |
| Chloride | 3 |
| Water | 1 |
| Protein residues | 423 |
| Geometric deviations (rms) | |
| Bonds (Å) | 0.004 |
| Angles (°) | 0.69 |
| Ramachandran favored (%) | 95 |
| Ramachandran outliers (%) | 0 |
| Clashscore26 | 3813.35 |
| Average B-factor (Å2) | 87.6 |
| Protein atoms | 87.6 |
| Solvent (water) | 82.2 |
Statistics for the highest-resolution shell are shown in parentheses.
Our finding that GrpN forms a pentameric assembly left the earlier reported hexameric structure for E. coli EutN as an outlying observation. The hexameric state reported for EutN could either indicate an unusual structural role for this homolog, or it could represents a spurious or minor oligomeric form selected during crystallization. To distinguish between these two possibilities, we devised a novel method to determine the oligomeric state of EutN in solution (Fig. 2).
Figure 2.
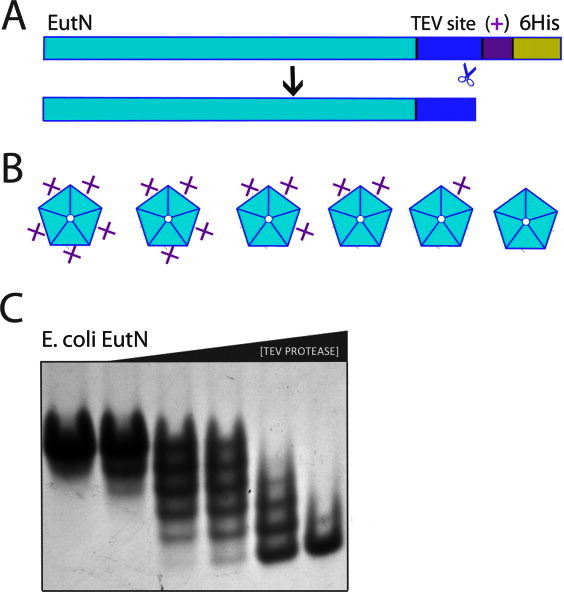
Application of the OCAC (oligomeric characterization by the addition of charge) method to the EutN shell protein. (A) Diagram of primary structure of EutN before and after cleavage with TEV protease. The plus symbol “ + ” denotes one additional positive charge. (B) For an n-oligomer, n + 1 possible charge states exist. Shown are the six possible charge-states of a pentamer. (C) EutN OCAC native gel. The engineered EutN(+) oligomer was incubated with TEV protease for 15 min, quenched with iodoacetamide, and then run on a native gel. From left to right, TEV protease concentrations are 0.0, 0.002, 0.01, 0.02, 0.1, and 0.5 mg/mL. The presence of six distinct bands shows that EutN is a pentamer. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The method we developed for determining oligomeric states is based on the change in charge-to-mass ratio that occurs after proteolytic cleavage of a small, charged N- or C-terminal sequence that is genetically appended at the end of the native protein subunit [Fig 2(A)]. Following different degrees of exposure to protease, a homo-oligomer composed of n subunits can be partially proteolyzed to generate a total of n + 1 distinct charge forms. The distinct charge forms can be enumerated by native gel electrophoresis. To be consistent with the nomenclature of established methods, we refer to our approach as OCAC, an acronym for oligomeric characterization by addition of charge [Fig. 2(B)]. A related method described earlier, OCAM (oligomeric characterization by addition of mass), was developed to determine the oligomeric state of membrane proteins.32 OCAM determines oligomeric state based on changes in the mobility of protein complexes on blue native gels, which arise from differential removal of relatively massive domains fused to the native protein subunits. In contrast, OCAC requires addition of relatively small terminal extensions to the native protein: essentially a protease cleavage site and as few as one or two directly adjacent charged amino acids. As with OCAM, the success of OCAC depends on a slow rate of exchange between subunits in different oligomeric complexes. Under conditions where we applied the method, native gels were capable of distinguishing between oligomers with just one unit charge difference. Another method based on counting distinct oligomeric forms was described several years ago, wherein the chemical modification of a genetically engineered cysteine residue by a charged reagent resulted in mobility shifts on SDS-PAGE.33 Unlike this earlier method, OCAC uses limited proteolysis to create alternative charge forms and does not require the complex to be stable in the presence of sodium dodecyl sulfate. The OCAC method also avoids the potential challenges of using chemical approaches when multiple cysteine residues are present in a protein.
We applied the OCAC method to examine the oligomeric state of EutN, which we had earlier reported to be a hexamer based on its crystal structure. EutN from E. coli was cloned to include a C-terminal TEV site followed by LEKK-6His. The resulting net charge difference between uncleaved and cleaved EutN is one charge unit (at pH 8.6 of the native gel running buffer). After recombinant expression and purification, this construct was subjected to a series of increasing concentrations of TEV protease, and then run on a native gel.
Surprisingly, the results clearly indicated that EutN is a stable pentamer in solution [Fig. 2(C)]. Six sharp bands are obtained by separation on a native gel. These arise from oligomers having 0, 1, 2, 3, 4, or 5 tails cleaved. This pentameric behavior contradicts two independent crystal structures in the PDB showing EutN in a hexameric arrangement18(PDB ID 2Z9H and 2HD3). A comparison of conditions used to obtain crystals of EutN did not suggest a clear explanation for the observed difference between crystalline and solution oligomer states. Although we see no evidence in the OCAC method for hexameric assembly, it is possible that a minor hexameric species of EutN exists in solution, which happens to crystallize preferentially. If the protein could equilibrate between pentameric and (minor) hexameric forms in solution, a low energy crystal form could drive the protein to a hexameric configuration by mass action. Such a scenario, however, would be difficult to reconcile with the conclusion that the pentameric form is stable at least over the time course of the OCAC experiment. Under one interpretation of the data, the hexameric form of EutN that is apparently populated under crystallization conditions represents a spurious form. Alternatively, the heterogeneous oligomerization behavior revealed by studies in solution and in crystals could reflect genuine complexity in the way these proteins are used to build microcompartment shells. A finer dissection of oligomeric states in intact MCPs must await higher resolution electron microscopy studies.
Notwithstanding the possibility of complex oligomerization phenomena, our combined results demonstrating the pentameric states of both GrpN and EutN help resolve an outstanding question about microcompartment architecture. Although the role of BMC type hexamers in forming the flat facets of MCPs has been apparent since the first structural studies,14, 15 the universal requirement for pentameric bacterial microcompartment vertex (BMV) proteins in MCPs has remained uncertain.18, 22 Here we confirm the first pentameric assemblies of BMV proteins from MCPs besides the carboxysome: GrpN from Rhodospirillum rubrum by X-ray crystallography, and EutN from E. coli in solution by the OCAC method. These results help reunify ideas for how different types of MCPs are constructed.
Materials and Methods
Cloning
A gene sequence was designed, using codons optimized for expression in E. coli, to encode GrpN with a C-terminal hexahistidine tag, using the online program DNAWorks.34 The resulting nucleotide sequence was synthesized by Biomatik, and then transferred to pET22b(+) vector using the NdeI restriction site via Isothermal Assembly (a.k.a. Gibson Assembly).35 We followed the isothermal protocol utilizing 20 nucleotide base pair complementary overhangs.
Full-length EutN was amplified from E. coli genomic DNA. Synthetic DNA oligomers, purchased from IDT, were used to add a TEV cleavage site and the positively charged tail onto the C-terminus of EutN with PCR. We refer to the resulting protein as EutN(+). Using Isothermal Assembly, EutN(+) was transferred into a pET22b(+) vector between NdeI and XhoI restriction sites to append a C-terminal 6xHis tag. The sequences of both GrpN and EutN(+) were verified by Laragen, Inc.
Expression and purification
The expression of GrpN-6His and EutN(+) were induced with 1mM IPTG in E. coli BL21 cells, shaking at 250 rpm, for 3 to 5 h at 37°C. Cells were spun down for 5 min at 6000 rpm and stored at –20°C. Cells were suspended in 50 mM Tris-HCl pH 7.6, 300 mM NaCl, 20 mM imidazole, with Protease Inhibitor Cocktail (Sigma, Cat # P8849) and lysed by sonication. Cells were spun down in rotor SS-34 at 16,500 rpm for 30 min, filtered through a 0.2 μm filter, and applied to a Hi-trap Nickel column by syringe at room temperature. Protein was eluted in one step with of 50 mM Tris–HCl pH 7.6, 300 mM NaCl, 400 mM Imidazole. GrpN was dialyzed into 2L of 10 mM Tris-HCl pH 7.6, 20 mM NaCl for 1 h at 4°C, and then again in 2L of fresh buffer overnight. GrpN-6His was highly soluble, but did show a tendency to precipitate over time. The protein was therefore centrifuged once or twice daily (20,800 rcf for 1–2 min at 4°C) and the pelleted precipitate was discarded.
Crystal structure determination
Initial crystallization screens were performed in 96-well, hanging drop trays, set up with the nanoliter liquid handling Mosquito from TTP LabTech. Upon optimization of condition G8 from Hampton Research screen HR2-110, cube-shaped crystals were obtained within the following condition ranges: 1.4–1.6 M ammonium sulfate, 0.1 M NaCl, 0.1 M HEPES pH 7.0–7.6, at protein concentrations between 20 and 40 mg/mL. Hanging drops were 1:1 well: protein with a total drop size between 2 and 4 μL. Crystals generally took between 1 and 10 days to grow.
Diffraction data extending to 3.2 Å resolution were collected at the Argonne National Laboratory, Advanced Photon Source (APS), beamline 24-ID-C. The structure of GrpN was phased by molecular replacement using the program PHASER.36 Coordinates for the CcmL pentamer (PDB accession code 2QW7) were used as the search model. The structure was built using the program COOT37 and refined using PHENIX38 and BUSTER39 with a final Rwork and Rfree of 0.2671 and 0.2885, respectively. 95% of the backbone dihedral angles are within the favored regions of a Ramachandran diagram. Coordinates and structure factors have been deposited with the PDB with ID code 4I7A.
OCAC assay
Aliquots of purified EutN(+) (in 50 mM Tris-HCl pH 7.6, 300 mM NaCl and 300 mM imidazole) were incubated on ice with fresh 1 mM DTT for 30 min. Because of weak UV absorbance at 280 nm, concentrations were adjusted based on band intensity as visualized by SDS-PAGE and Coomassie staining. TEV protease was added to EutN(+) aliquots in a series of dilutions, resulting in the following final concentrations: 0.0, 0.002, 0.01, 0.02, 0.1, and 0.5 mg/mL TEV protease. These TEV: EutN(+) samples were incubated at room temperature for 15 min. Protease reactions were stopped with addition of 5× native loading dye containing 10 mM iodoacetamide. Reactions were then run on a native gel (BioRad CAT# 456-1096) at 100 V for 2 h at room temperature.
Acknowledgments
We thank Michael Thompson, Julien Jorda, Thomas Bobik and members of the Yeates lab for helpful discussions. We thank Michael Sawaya and the staff of the UCLA-DOE X-ray Crystallography Core Facility. We thank M. Capel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov, and F. Murphy at NECAT beamlines 24-ID at APS.
In a new study published after this manuscript was submitted, Petit et al. demonstrate the production of an MCP that appears to metabolize 1,2-PD using a glycyl radical based reaction of the type discussed here and in refs (12, 13). See Petit E, Latouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. (2013). Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8(1):e54337.
Glossary
Abbreviations
- BMC
bacterial microcompartment (hexameric) shell protein
- BMV
bacterial microcompartment vertex protein (pentameric)
- Grp
glycyl radical-based propanediol utilizing MCP
- MCP
bacterial microcompartment
- OCAC
oligomeric characterization by addition of charge
- OCAM
oligomeric characterization by addition of mass
References
- 1.Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 2.Yeates TO, Thompson MC, Bobik TA. The protein shells of bacterial microcompartment organelles. Curr Opin Struct Biol. 2011;21:223–231. doi: 10.1016/j.sbi.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeates TO, Crowley CS, Tanaka S. Bacterial microcompartment organelles: protein shell structure and evolution. Annu Rev Biophys. 2010;39:185–205. doi: 10.1146/annurev.biophys.093008.131418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: their properties and paradoxes. Bioessays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price GD, Coleman JR, Badger MR. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stojiljkovic I, Bäumler A, Heffron F. Ethanolamine utilization in Salmonella typhimurium : nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bobik T, Xu Y, Jeter R, Otto K, Roth J. Propanediol utilization genes (pdu) of Salmonella typhimurium : three genes for the propanediol dehydratase. J Bacteriol. 1997;21:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorda J, Lopez D, Wheatley NM, Yeates TO. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 2012;22:179–195. doi: 10.1002/pro.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott KP, Martin JC, Campbell G, Mayer C, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium Roseburia inulinivorans. J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 15.Samborska B, Kimber MS. A dodecameric CcmK2 structure suggests β-carboxysomal shell facets have a double-layered organization. Structure. 2012;20:1353–1362. doi: 10.1016/j.str.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka S, Sawaya MR, Yeates TO. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science. 2010;327:81–84. doi: 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- 19.Price GD, Howitt SM, Harrison K, Badger MR. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol. 1993;175:2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J Bacteriol. 2011;193:1385–1392. doi: 10.1128/JB.01473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lünsdorf H, Rambach A, Gass MH, Bleloch A, McClean KJ, Munro AW, Rigby SE, Warren MJ, Prentice MB. Biochemical and structural insights into bacterial organelle form and biogenesis. J Bacteriol. 2008;283:14366–14375. doi: 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- 22.Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2leakage barrier. PLoS ONE. 2009;4:e7521. doi: 10.1371/journal.pone.0007521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 26.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 27.Lai Y-T, King NP, Yeates TO. Principles for designing ordered protein assemblies. Trends Cell Biol. 2012;22:653–661. doi: 10.1016/j.tcb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Lai Y-T, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336:1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Burkhard P. Encapsulation of gold nanoparticles into self-assembling protein nanoparticles. J Nanobiotech. 2012;10:42. doi: 10.1186/1477-3155-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, André I, Gonen T, Yeates TO, Baker D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wörsdörfer B, Woycechowsky KJ, Hilvert D. Directed evolution of a protein container. Science. 2011;331:589–592. doi: 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- 32.Gandhi CS, Walton TA, Rees DC. OCAM: a new tool for studying the oligomeric diversity of MscL channels. Protein Sci. 2011;20:313–326. doi: 10.1002/pro.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouaux J, Braha O, Hobaugh M, Song L, Cheley S, Shustak C, Bayley H. Subunit stoichiometry of staphylococcal alpha-hemolysin in crystals and on membranes: a heptameric transmembrane pore. Proc Natl Acad Sci USA. 1994;91:12828–12831. doi: 10.1073/pnas.91.26.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoover DM. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:43e–43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Headd JJ, Echols N, Afonine PV, Grosse-Kunstleve RW, Chen VB, Moriarty NW, Richardson DC, Richardson JS, Adams PD. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Cryst. 2012;D68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]


