Abstract
Mammals exhibit multiple adaptive mechanisms that sense and respond to fluctuations in dietary nutrients. Consumption of reduced total dietary protein or a protein diet that is deficient in 1 or more of the essential amino acids triggers wide-ranging changes in feeding behavior and gene expression. At the level of individual cells, dietary protein deficiency is manifested as amino acid (AA) deprivation, which activates the AA response (AAR). The AAR is composed of a collection of signal transduction pathways that terminate in specific transcriptional programs designed to catalyze adaptation to the nutrient stress or, ultimately, undergo apoptosis. Independently of the AAR, endoplasmic reticulum stress activates 3 signaling pathways, collectively referred to as the unfolded protein response. The transcription factor activating transcription factor 4 is one of the terminal transcriptional mediators for both the AAR and the unfolded protein response, leading to a significant degree of overlap with regard to the target genes for these stress pathways. Over the past 5 y, research has revealed that the basic leucine zipper superfamily of transcription factors plays the central role in the AAR. Formation of both homo- and heterodimers among the activating transcription factor, CCAAT enhancer-binding protein, and FOS/JUN families of basic leucine zipper proteins forms the nucleus of a highly integrated transcription factor network that determines the initiation, magnitude, and duration of the cellular response to dietary protein or AA limitation.
Current status of knowledge
Amino acid response
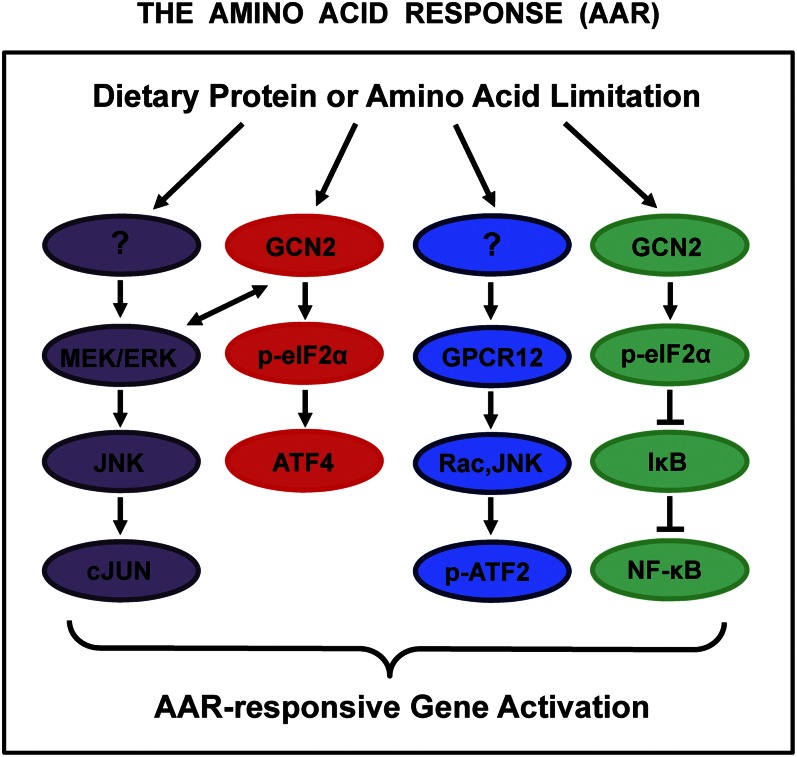
A broad spectrum of adaptive mechanisms has evolved in mammals to sense and respond to fluctuations in dietary nutrients. For example, it has been demonstrated that mammals can detect the quality of dietary protein. A diet deficient in even a single essential amino acid (AA)3 will be avoided (1, 2), a response mediated at least in part by general control nonderepressible 2 (GCN2), which serves as an AA sensor (3, 4). Beyond changes in feeding pattern, a protein-/AA-deficient diet also leads to changes in metabolism beyond protein/AA, such as lipid metabolism (5). mRNA (messenger RNA) microarray analysis has documented that a diet deficient in total protein results in significant changes in gene expression (6) and, in the case of pregnancy, results in fetal epigenetic changes, including DNA methylation (7). Likewise, consumption of dietary protein that is naturally deficient in 1 or more of the indispensible AA, as many of the grains are, also triggers an altered gene expression profile (6). At the level of organs and individual cells, a reduction in total dietary protein or a protein source with an imbalanced AA composition is manifested as AA deprivation, which activates an AA response (AAR) that is composed of multiple signal transduction pathways (Fig. 1). As reviewed previously (8), activation of the AAR regulates gene expression at many steps including chromatin structure, transcription start site, transcription rates, mRNA splicing, RNA export, RNA turnover, and translation initiation.
Figure 1.
Signal transduction pathways of the amino acid (AA) response (AAR). The AAR represents a collection of signal transduction pathways that are activated by AA deprivation of mammalian cells. There are differences between cell types, such as which mitogen-activated protein kinases are activated, but the central pathway of general control nonderepressible 2 (GCN2)–eukaryotic initiation factor 2–activating transcription factor 4 (ATF4) appears to be ubiquitously expressed and regulated in all tissues. Likewise, the phosphorylation of ATF2 may well be a universal response, but the tissue distribution of the nuclear factor κB (NF-κB) activation has not been established. The autoregulatory induction of cJUN by the AAR occurs in cultured transformed cells to a much greater extent than in nontransformed counterparts, although the response has not yet been studied in vivo. The initial AA sensor for the cJUN and phosphorylated ATF2 (p-ATF2) pathways has not been determined conclusively. Interestingly, the 4 pathways shown rely on a variety of molecular mechanisms. The synthesis of ATF4 protein is translationally controlled, the phosphorylation of preexisting cJUN and ATF2 protein is mediated by mitogen-activated protein kinase signaling, and NF-κB activation occurs as a consequence of disassociation, but not degradation, of IκB. See the text for additional details. GPCR, G protein–coupled receptor; IκB, inhibitory kappa beta; JNK, c-Jun N-terminal kinase; MEK/ERK, mitogen-activated protein kinase/extracellular-regulated kinase.
Although the initial AA sensor for some of the AAR-associated pathways has not been identified definitively, the GCN2 kinase is well established as the AA monitoring mechanism for the activating transcription factor (ATF) 4 pathway. GCN2 has the ability to sense the level of each of the AA because the GCN2 kinase activity is activated when the protein binds any one of the uncharged transfer RNA (tRNA) molecules. GCN2 phosphorylates the translation eukaryotic initiation factor 2 (eIF2)α (4, 9–13). Phosphorylation of eIF2α (phospho-eIF2α) functions as an inhibitor of eIF2B (14), which catalyzes the exchange of GDP for GTP during activation of the eIF2 complex, a necessary step for the assembly of the 43S ribosomal subunit. Consequently, phospho-eIF2α suppresses general protein synthesis, but promotes a paradoxical increase in translation of selected mRNA species. Among these are ATF4 and ATF5, discussed in detail in the following, as well as growth arrest and DNA damage–inducible 34 (GADD34) (15). GADD34 is a component of the feedback loop that permits reactivation of general translation by targeting protein phosphatase 1 to phospho-eIF2α, which is then dephosphorylated, thereby permitting translation of the up-regulated stress-responsive mRNA species (16–18).
GCN2 is only 1 of 4 eIF2α kinases that respond to a spectrum of cellular stresses (reviewed in Reference 19). Another eIF2α kinase is the double-stranded RNA–activated protein kinase-like endoplasmic reticulum (ER) kinase (PERK). PERK, activated by ER stress, is 1 of 3 ER stress-signaling pathways collectively termed the unfolded protein response (UPR). Activation of the PERK eIF2α kinase also increases ATF4 synthesis and an associated downstream transcriptional program that often overlaps with the AAR and oxidative stress, leading Harding et al. (20) to propose the integrated stress response with ATF4 as the central protein mediator. One can, in many cases, use either AA deprivation or ER stress to investigate common ATF4-responsive target genes for these pathways. However, there are also differences in the gene profile activated by GCN2 and PERK signaling (21), indicating that factors other than ATF4 add specificity to each pathway. The mechanism underlying this specificity represents one of the many interesting questions yet to be answered. Some insight has been gained from 1 ATF4 target gene, the system A sodium-dependent neutral AA transporter 2 (SNAT2). SNAT2 contains an ATF4-responsive site that binds ATF4 and triggers increased transcription during the AAR (22). However, despite increased ATF4 binding to this same SNAT2 genomic site during UPR activation, transcription activity is not enhanced. Simultaneous activation of the AAR and the UPR revealed that the UPR generates a signal that acts downstream of ATF4 binding to repress the AAR action on SNAT2.
AAR in health and disease
There are circumstances in which protein/AA deprivation is associated with positive health responses. For example, there are data illustrating extension of life expectancy after protein restriction and even after dietary restriction for a single essential AA (23–25). However, more often protein malnutrition or poor AA assimilation is a negative factor associated with a wide range of disease states, and AA-dependent gene expression has been recognized as a component in some cases. For example, protein malnourishment of hospitalized patients is a common occurrence that is characterized by decreased expression of insulin-like growth factor-1 (IGF-1) and other genes, which can contribute to morbidity in adults and can lead to slowed growth in children (26). Similar reductions in IGF-1 expression are observed in nonhospitalized people with disease-related protein malnutrition. In healthy adults, consumption of a low-protein diet results in a decrease in IGF-1 expression and an increase in the counterregulatory action of IGF binding protein 1 (IGFBP-1) (27). The AA-dependent control of the IGF-1 and IGFBP-1 genes has been reported in both in vivo and cell culture models (28–32).
In the United States, ∼10 to 15% of all babies are born small for gestational age, and a significant number of those are associated with intrauterine growth retardation (IUGR) (reviewed in Reference 33). More than 50% of stillbirths have preceding IUGR, and worldwide, ∼30 million newborns have IUGR (reviewed in Reference 34). Although IUGR is typically the result of placental deficiencies that limit nutrient delivery, it can also be caused by poor maternal nutrition during the pregnancy. One of the hallmarks of IUGR in both humans and animal models is a reduction of maternal to fetal transfer of essential AA, and there is a positive correlation between the 2 with regard to the degree of IUGR (35). Intracellular AA signaling within the placenta has not been extensively investigated, but it has been documented that human placentas from IUGR pregnancies have increased eIF2α phosphorylation compared with controls (36). Consistent with these observations, a balanced protein-energy supplement regimen is one of the promising approaches to prevent IUGR (34). The long-term consequences of fetal AA deprivation can be far-reaching. Dietary protein deprivation not only has immediate effects on gene expression, but also has long-term effects through epigenetic mechanisms. Protein/AA restriction in utero causes genomewide changes in fetal hepatic DNA methylation and changes in gene regulation during adulthood of the offspring (7), possibly leading to metabolic syndrome as predicted by the fetal origins of adult disease (37). Interestingly, a low-protein diet during pregnancy induces hepatic Igf2 and H19 gene expression in the newborn offspring, possibly by altering DNA methylation (38). The activation of these genes in the offspring was reversed when the low-protein diet was supplemented with folate by an unknown mechanism.
A critical consequence of the protein malnutrition associated with many disease states is global tissue wasting. For example, protein-energy wasting is a common feature of chronic kidney disease, in particular in those who undergo maintenance dialysis (39). Most dialysis patients consume ∼25% less protein than the recommended dietary protein intake values, contributing to the tissue wasting and, in children, to growth retardation. Another example of tissue wasting is cancer-associated cachexia, which is characterized by decreased food intake (anorexia) and loss of muscle mass, as well as increased morbidity and mortality (40, 41). An aspect of cancer that is not fully understood is the relationship between tumor growth and dietary protein/AA availability. Tumors may be subjected to AA limitation in several circumstances: 1) during protein malnutrition of cancer patients; 2) in tumor regions for which sufficient vascularization has yet to develop; or 3) when tumor vessels are compromised, which occurs often. Consequently, hypoxic areas of tumors, which are likely to be deprived of other nutrients, including AA, exhibit increased ATF4 expression (42). In fact, Ye et al. (43) showed that the GCN2-eIF2α-ATF4 pathway is activated in tumor tissue, and knockdown of either GCN2 or ATF4 expression in human xenograft tumors caused reduced proliferation. The researchers concluded that the GCN2-eIF2α-ATF4 pathway confers prosurvival and proliferative capabilities to tumor cells undergoing nutrient limitation (43). Ye et al. (43) also revealed a correlation between reduced asparagine synthetase (ASNS) expression and blocked tumor growth, an observation supported by the demonstration that exogenous expression of just ASNS alone in the ATF4 knockdown cells restored proliferation of the tumor xenografts. These in vivo tumor studies complement observations by our laboratory showing that increased ectopic expression of ASNS alone replicates the asparaginase (ASNase) drug–resistant, highly proliferative phenotype that can develop during therapy of children with acute lymphoblastic leukemia (ALL) (44, 45). Children with ALL are treated with l-ASNase to deplete the leukemia cells of asparagine (46), which have low expression of ASNS and consequently undergo cell cycle arrest and eventually apoptosis (44, 47). ASNase sensitivity of some ovarian cancer cell lines suggests that asparagine deprivation may represent an exciting new approach to treating ovarian tumors (48).
In vivo observations in model organisms
A majority of the work focused on the actions of transcription factors during the AAR has been performed in cell cultures as a model system for a diet that is protein deficient or is unbalanced with regard to essential AA. Some protein sources are deficient in selected AA, including legumes, which can be deficient in methionine, grains that are low in lysine content, or corn, which does not supply sufficient amounts of tryptophan or lysine. Several studies have demonstrated that feeding mice a leucine-free diet can induce the AAR (49, 50). Indeed, within 1 h after initiating a leucine-free diet, increased hepatic transcription of AA-responsive genes can be detected, and within 2 h after replacing the leucine in the diet, the transcriptional changes are reversed (50). Analysis of genomic structure, epigenetic changes to DNA and histones, and measurement of transcription activity in vivo can be complicated because most organs represent more than 1 cell type, which can compromise the interpretation of mechanistic data given the possibility of cell-specific responses. Indeed, we know from in vivo studies after dietary leucine deprivation that cell-specific responses play a critical role in modulating the AAR, as shown for the liver and muscle (49). Also, whole animal studies do not permit a direct test of the regulatory effect of AA alone because circulating AA levels can influence the release and action of hormones and growth factors. How the AAR pathway is differentially regulated and affects interorgan AA metabolism must await direct comparisons and molecular analysis because a systematic and thorough survey of tissues for AA-responsive signaling, transcription factor expression/activation, and control of target genes has not been reported after dietary protein/AA deprivation.
Activating ATF family
Although activation of the AAR increases transcription from the ATF4 gene by ∼2-fold (51), the primary regulation of ATF4 protein content is through translational control of preexisting mRNA. The ATF4 mRNA contains 2 upstream open reading frames (uORF), uORF1 and uORF2, that are located 5′ to the ATF4 coding sequence, and both are translated in the nonstressed condition to the exclusion of ATF4 itself because uORF2 overlaps with the ATF4 coding sequence, but is out of frame (52, 53). During AA limitation, the uORF1 is translated but as a consequence of phospho-eIF2α inhibition of eIF2B and a corresponding decrease in functional eIF2 complex, ribosome scanning bypasses the uORF2 start site and translation reinitiation occurs at the ATF4 coding region. Thus, synthesis of ATF4 protein is selectively increased in response to AA deficiency (or after PERK activation by ER stress). ATF4 triggers increased transcription by binding to CCAAT enhancer-binding protein (C/EBP)-ATF response elements (CARE), so named because they are composed of a half-site for the C/EBP family and a half-site for the ATF family of the basic leucine zipper (bZIP) transcription factor superfamily (54, 55). The products of these CARE-containing genes modulate a wide spectrum of cellular events designed to adapt to dietary stress. Under continued stress of sufficient magnitude, ATF4-induced apoptosis can also occur. Given that ATF4 is induced by both the AAR and the UPR, these CARE sequences are usually, but not always, functional in response to both stress pathways. In the context of protein or AA deprivation, the CARE sites are often referred to as AA response elements (AARE). Consistent with ATF4’s role as a critical transcription factor within the AAR, the ATF half-site is highly conserved, whereas the C/EBP half-site is quite divergent (5′-TGATGXAAX-3′) (8). It is possible that different ATF4 dimerization partners bind to the C/EBP half-site to provide transcriptional specificity to the ATF4 signal that is generated by the 4 independent eIF2α kinases, although the identity and properties of these proposed heterodimers have not been studied extensively. If ATF4 requires a dimerization partner, the abundance of that protein is not limiting, even in the basal or “fed” state. Support for this hypothesis comes from the use of HEK293 cells stably expressing a tetracycline (Tet)-inducible ATF4 construct (56). Tet-induced ATF4 expression in these cells maintained in control culture medium, to prevent activation of other aspects of the AAR, still resulted in transcriptional induction of all 5 ATF4-responsive genes that were tested. In association with the Tet-dependent enhancement of ATF4 binding to the CARE site on an AAR target gene, recruitment of the general transcription machinery, including RNA polymerase II, was observed at a level comparable to that induced by activation of the AAR. These results indicate that increased ATF4 expression alone is sufficient to trigger enhanced transcription of CARE-containing genes. Ameri and Harris (57) published a comprehensive review of ATF4.
Among the ATF4-activated genes are the transcription factors C/EBPβ (58), ATF3 (59, 60), and CHOP (61), which act as feedback repressors of the ATF4 signal. Using the ASNS gene as a model, a self-limiting mechanism has been characterized in which prolonged AA deprivation leads to feedback suppression due to ATF4 activation of C/EBPβ and ATF3 expression (62). More recent studies have added C/EBP homologous protein (CHOP) as a negative regulator of ATF4 action during the AAR (63). Chromatin immunoprecipitation (ChIP) analysis of the ASNS gene was used to illustrate that ATF4 binding to the CARE site occurs as early as 30 to 45 min after AA deprivation, and elevated ATF4 binding continues for ∼4 h. After 4 to 6 h of AA limitation, there is a gradual decrease in ATF4 binding to the CARE site, an increase in C/EBPβ, ATF3, and CHOP recruitment, and a parallel decrease in ASNS transcription activity (62, 63). This self-limiting cycle of ATF4 action has now been demonstrated for a number of CARE-containing genes (60, 64). The mechanistic details at the molecular level of how these factors interact with the general transcription machinery to control the rate of initiation or elongation have not been established.
For some ATF4-regulated genes, increased transcription also requires phosphorylated ATF2 (p-ATF2), which encodes an active histone acetyltransferase (65–67). As illustrated in Figure 1, it has been demonstrated that AA deprivation triggers a signaling cascade that terminates with the c-Jun N-terminal kinase (JNK) 2–dependent phosphorylation of ATF2 at Thr-69 and Thr-71, which then leads to recruitment of p-ATF2 to the CARE sequence of selected AAR target genes (67). Bruhat et al. (68) first demonstrated the importance of ATF2 in mediating the transcriptional induction of the CHOP gene after leucine starvation and subsequently showed that leucine deprivation led to an increase in the abundance of the active p-ATF2, whereas there was no detectable change in the total ATF2 protein levels (65). Activation of the AAR in ATF2 knockout fibroblasts failed to induce ATF3 and CHOP mRNA, further demonstrating the essential role of ATF2 for those genes. Conversely, other AA-responsive genes, such as ASNS (65) and TRB3 (50), are largely unaffected by the lack of ATF2, suggesting that it does not contribute to transcriptional control of all the AAR targets equally. However, small interfering RNA (siRNA) knockdown of ATF2 in HepG2 hepatoma cells did result in a partial suppression of ASNS induction (69), so it appears that both gene specificity and tissue specificity may influence ATF2 action. Activation of the AAR leads to enhanced recruitment of p-ATF2 to the CARE of the CHOP gene and stress-induced acetylation of histones H2B and H4 (66), a result also shown for the ATF3 gene (67).
As discussed in more detail in the following, autoactivation of the cJUN gene by the AAR is dependent on the formation and binding of ATF2-cJUN dimers to the 2 AP-1 sequences in the proximal promoter of cJUN, although 1 of the sites appeared to show a preference for cJUN homodimers (69). Co-immunoprecipitation revealed an increase in p-ATF2/p-cJUN heterodimer formation in response to AAR activation. Interestingly, similar to the observation for cJUN, overexpression of ATF2 stabilized ATF4 protein, and, conversely, a dominant negative form of ATF2 caused a complete block of the AAR-induced increase in ATF4 protein abundance (69). It has been reported that ATF4 can be acetylated by p300, but not by p300/CBP-associated factor (70). However, p300/CBP-associated factor binds to ATF4 and can act as a cofactor in the ATF4-dependent induction of CHOP during the AAR (61). Conversely, Lassot et al. (71) reported that the p300-dependent stabilization of ATF4 protein was due to inhibition of ubiquitination and independent of p300 acetyltransferase activity. Direct tests for ATF4 acetylation by ATF2 have not been reported, but it is possible that ATF2-mediated acetylation also stabilizes the ATF4 protein.
As reviewed elsewhere, ATF5 has been implicated in regulation of cell growth and development, especially in the nervous system (72). ATF5 is a member of the ATF family of the bZIP transcription factors and is highly homologous to ATF4 (73). Like ATF4, the mRNA for ATF5 also contains upstream uORFs that control translation in an AA-dependent manner (74, 75). As expected, activation of the GNC2 kinase and eIF2 phosphorylation are both required for the increase in ATF5 expression. Interestingly, ATF4 is necessary both to maintain ATF5 mRNA levels and to support the ATF5-dependent transcription from the ASNS promoter (73). Al Sarraj et al. (73) documented that the bZIP transcription factor CHOP acts to counterregulate the induction of the ASNS promoter by ATF5 acting at the CARE site, similar to the CHOP antagonisms of ATF4 (63). The stimulatory action of ATF5 via a CARE sequence was also reported for the CHOP gene itself (76). How ATF4 and ATF5 both act at a CARE site when they are simultaneously expressed in response to AA deprivation has not been investigated. ChIP sequencing to survey the genome for ATF4-ATF5 binding sites and quantitative re-ChIP analysis for their binding at specific genes are necessary to reveal mechanistic insight into their possible interaction. In contrast to studies showing action of ATF5 at CARE sequences, Li et al. (77) used an unbiased selection approach to identify an ATF5 binding sequence, 5′-YTCTYCCTT-3′. The authors showed that the EGR1 gene contains 2 of these ATF5 binding sequences and that they were necessary for basal EGR1 promoter activity. Whether these elements mediate induction of ATF5 target genes in response to AA limitation or other stimuli is unknown.
ATF5 polymorphisms have been linked to an altered response to therapy for children with ALL. As a key component of the multidrug treatment for childhood ALL, patients are given ASNase, which depletes the body of asparagine (and glutamine to a lesser extent). Due to a suppressed ability of ALL cells to rapidly up-regulate ASNS protein content, relative to most other tissues in the body, the leukemia cells are preferentially sensitive to ASNase (44, 45). Rousseau et al. (78) discovered that patients with a T1562C polymorphism within the ATF5 gene exhibit lower event-free survival times when treated with ASNase. The T1562C polymorphism leads to higher ATF5 promoter activity, and the authors speculate that ATF5-driven expression of ASNS causes decreased sensitivity to ASNase therapy. Analysis of the relationships among ATF5 action, ASNS protein levels, and ASNase sensitivity is an interesting avenue for further research in the ALL field.
ATF3 expression is increased in response to a broad spectrum of stress signals and contributes to the control of many cellular activities (reviewed in References 79 and 80). Expression of ATF3 is induced in response to activation of the AAR and the UPR pathways, and these inductions require the eIF2α kinases GCN2 and PERK, respectively (59, 81), as well as ATF4 (20, 60). In response to AAR activation, both increased transcription (60) and mRNA stabilization (82) contribute to induction of ATF3 expression. Yaman et al. (83) showed that during the AAR, HuR is shifted from the nucleus to the cytoplasm after which it binds to and stabilizes the CAT-1 cationic AA transporter mRNA. Subsequently, Pan et al. (82) reported similar results for the ATF3 mRNA. The AA-dependent shift to cytoplasmic accumulation of HuR paralleled the kinetics of ATF3 mRNA accumulation and suppression of HuR protein using siRNA partially blocked the increase in ATF3 mRNA in histidine-deprived cells. The human ATF3 promoter has a consensus CARE site that was first reported to be responsible for autoregulation of the ATF3 gene (84). Transient overexpression experiments and studies on knockout fibroblasts showed that ATF4, acting via the CARE site, increased transcription from the ATF3 gene and that C/EBPβ and ATF3 functioned as feedback repressors (60). However, there are reports that the AA-dependent induction of ATF3 mRNA is observed after short hairpin RNA knockdown of ATF4 (85). Interestingly, the ATF3 gene exhibits extensive alternative splicing, and AA availability regulates exon selection such that several different ATF3 isoforms are synthesized (81). Two of these isoforms exhibit opposite transcriptional activities when overexpressed, full-length ATF3 strongly antagonizes ATF4 action, whereas ATF3∆ZIP3, an isoform with a truncated leucine zipper domain, further enhances ATF4 activation at CARE sites. Although the physiologic consequences of these opposing activities have yet to be explored fully, differential expression of these isoforms may provide further modulation of the ATF3 signal, add gene specificity to the ATF4 signal, or serve to provide cell-specific stress responses from the ATF3 gene.
C/EBP family
The C/EBPβ mRNA contains 3 methionine residues that are used as translational start sites, which leads to 3 separate isoforms, liver-enriched transcriptional activator protein* (LAP*, 345 AA in humans), a protein of 23 fewer N-terminal AA called LAP, and the liver-enriched transcriptional inhibitory protein (LIP, 147 AA in humans), which corresponds to the C-terminal half of the LAP form (86). LIP lacks the transactivation domain of LAP*/LAP, but contains the bZIP dimerization region and, therefore, through dimerization with other C/EBP members, can serve as a dominant negative repressor. The ratio of the C/EBPβ isoforms can be controlled by translation-associated regulatory mechanisms mediated by the mammalian target of rapamycin (mTOR) (87). Decreased translation rates, as would occur during AA deprivation, results in a preference for LAP synthesis. Conversely, increased mTOR activity, which is enhanced by AA sufficiency and suppressed by AA limitation (88, 89), promotes enhanced translation rates and synthesis of the LIP isoform.
As mentioned previously, the expression of C/EBPβ is increased by ATF4 action and subsequently binds to the CARE sequence and, along with ATF3, acts to feedback repress the ATF4 signal (62). Studies in C/EBPβ knockout MEF cells or HepG2 hepatoma cells treated with an siRNA against C/EBPβ revealed that AAR-induced transcription from the endogenous ASNS gene or luciferase driven by the ASNS promoter was enhanced in the absence of C/EBPβ expression (58). All 3 isoforms appear to function as repressors of the AAR-dependent induction, with LIP also required for basal ASNS transcription. Li et al. (90) used UPR activation to demonstrate that LIP levels follow a biphasic response such that during the early phase, LIP levels decrease, but then subsequently increase significantly. The large change in the LIP/LAP ratios was linked to LIP-dependent inhibition of ATF4-driven gene expression for genes known to respond to ATF4 via CARE sequences. Similar changes were observed after AA deprivation, but the magnitude was considerably less. Induction of CARE-containing genes such as ASNS, SNAT2, and CAT-1 is enhanced in C/EBPβ-deficient fibroblasts, as reported by Thiaville et al. (58) for AA deprivation. In contrast, Li et al. (90) identified the CARE-containing gene CHOP and 2 of its downstream targets, GADD34 and TRB3, as genes for which UPR-triggered induction was suppressed in C/EBPβ-deficient cells, and Carraro et al. (50) concluded that C/EBPβ does not contribute to AA-dependent TRB3 transcription. Interestingly, LIP protein stabilization during the latter phase of the UPR is dependent on the formation of a LIP-CHOP dimer, and, conversely, nuclear import of CHOP is enhanced by this dimerization, a process that Chiribau et al. termed molecular symbiosis (91).
Expression of CHOP, also known as growth arrest and DNA damage protein 153 (GADD153), is activated by a wide range of stress stimuli. Deleting the CHOP gene documented a link between CHOP expression and apoptosis induced by cellular stress (92), an observation that has now been reproduced for many cells and tissues. AA limitation increases CHOP expression by both transcriptional (93, 94) and posttranscriptional mechanisms (95, 96). Jousse et al. (94) used deletion analysis to show that the CHOP CARE sequence (5′-TGATGCAAT-3′), originally identified as an arsenic-responsive element (54, 55), also functions as an AARE. The induction of the CHOP gene following AA limitation requires both ATF2 phosphorylation and increased ATF4 expression (65).
An intriguing question is why CHOP cooperates with ATF4 to activate the TRB3 gene, whereas CHOP inhibits the ATF4-dependent transcription of others, such as ASNS (63). Ohoka et al. (97) demonstrated that the flanking sequence of the TRB3 AARE element might contain 1 or more binding sites for additional factors that contribute to the distinct function of the ATF4/CHOP heterodimer. The stress response unit of the TRB3 gene contains 3 identical tandem repeats each consisting of 33 bp. Each of the repeats contains a CCAAT-like element (5′-CTAAT-3′), a CARE sequence (5′-TGATGCAAA-3′), a CHOP binding site (5′-TGCAAAC-3′) that overlaps with the CARE sequence, and a cytosine/guanine-rich palindrome (5′-CCCGGTCCGGG-3′) of unknown function. Mutagenesis analysis showed that the CCAAT-like element, the CHOP binding site, and the CARE site were each essential for TRB3 induction (50, 97). The ASNS promoter contains a sequence similar to the overlapping CARE and CHOP sequences in the TRB3 gene, but it does not have a TRB3 CCAAT-like element. ASNS promoter activity is repressed by CHOP overexpression, and, conversely, siRNA-mediated knockdown of CHOP further enhances the induction of ASNS by either AA deprivation or ER stress (63). These contrasts suggest the possibility that the CCAAT-like element may be involved in the discrimination between activation and repression by the ATF4/CHOP heterodimer. The CHOP-dependent repression of the ASNS gene required the entire CHOP protein, arguing against the possibility of simple sequestration of ATF4 by the CHOP leucine zipper domain, and ChIP analysis showed an association of CHOP with the ASNS and TRB3 promoters (63). Collectively, these results document that CHOP is a member of the AAR transcription factor network that can either activate or repress the stress-induced regulation of specific CARE-containing genes, perhaps based on specificity provided by flanking elements.
FOS/JUN family
There have been several isolated reports suggesting a link between the AA availability and expression of members of FOS/JUN family. For example, methionine limitation of Chinese hamster ovary cells increased cJUN, cFOS, and Jun-B mRNA levels (98), and total AA deprivation of a pancreatic tumor cell line led to an increase in both total cJUN and phosphorylated cJUN protein levels (99). Gietzen et al. (100) discovered that regions of the mouse brain that exhibit increased eIF2α and extracellular regulated kinase activation in response to a threonine-free diet also contained elevated levels of cJUN. After expression mRNA microarray analysis of HepG2 cells, Shan et al. (101) observed that both cFOS and cJUN were increased in their expression by the AAR. Those results led Fu et al. (69) to complete the first systematic analysis of AA-responsive expression for the FOS/JUN family of transcription factors. The authors discovered a novel ATF4-independent AAR pathway that centers on transcriptional induction of selected FOS/JUN members. In human hepatocellular carcinoma cells, expression of cJUN, JUN-B, cFOS, and FOS-B was induced by the AAR, whereas that for JUN-D, FRA-1, and FRA-2 was not. For several human liver, prostate, and ovarian cell lines, the AAR-induced increase in cJUN expression was considerably greater in transformed cells compared with nontransformed counterparts, an effect independent of cell growth rate. Of the 4 AA-responsive FOS/JUN members, cJUN made the largest contribution to the induction of several known AAR downstream target genes, including ASNS, CHOP, and ATF3. Rather than direct cJUN transcriptional action on these genes, the effect appeared to be indirect, a result of cJUN-dependent stabilization of ATF4 protein by an unknown mechanism (69).
The AAR-induced transcription from the cJUN gene was autoregulatory and dependent on mitogen-activated protein kinase–mediated phosphorylation of preexisting cJUN protein. A primary dimerization partner for cJUN is ATF2 (102), and it has been demonstrated that for a number of stimuli, transcription from the cJUN gene is induced by cJUN-cJUN homodimers or cJUN-ATF2 heterodimers binding to the 2 AP1 sites within the cJUN proximal promoter (103–105). Fu et al. (69) showed that the AAR activates a cascade that involves activation of the mitogen-activated protein kinase/extracellular-regulated kinase and JNK arms of the mitogen-activated protein kinase signaling pathways, followed by phosphorylation of preexisting ATF2 and cJUN protein, and then formation of cJUN-ATF2 activated heterodimers. In response to AA deprivation, the cJUN-ATF2 dimers are recruited to the 2 AP-1 sites within the cJUN proximal promoter, which leads to autoactivation of the cJUN gene. These results are the first to document that some AP-1 sequences can function as a transcriptional AARE. The mechanism by which the mitogen-activated protein kinase/extracellular-regulated kinase and JNK pathways are activated is unknown, but the ability to induce cJUN transcription through the use of amino alcohols to block tRNA charging suggests that GCN2 or another tRNA-monitoring mechanism is likely to be the initial sensor.
As the name indicates, JUN dimerization protein 2 (JDP2) was initially identified as a cJUN binding partner (106), but it can homodimerize or heterodimerize with several members of the JUN family and with ATF2 (107). JDP2 inhibits ATF2-dependent transcription by recruiting an HDAC3-containing complex to target genes (108). Chérasse et al. (109) documented that JDP2 is bound to the CHOP CARE site in the basal state. The amount of JDP2 associated with the gene decreases after activation of the AAR, leading the authors to suggest that JDP2 functions as a repressor of regulated CHOP transcription. Support for this proposal came from additional experiments showing that knockdown of endogenous JDP2 by siRNA caused an increase in basal and AAR-induced levels of CHOP mRNA, whereas JDP2 overexpression blocked the increase in CHOP promoter-driven luciferase reporter activity. Consistent with the data of Jin et al. (108), Chérasse et al. also showed that HDAC3 association with the CHOP CARE region decreased as JDP2 decreased and ATF4 association increased (109). Although the possible direct regulation of JDP2 expression by the AAR has not been reported, expression mRNA microarray data revealed a 10-fold increase in JDP2 mRNA after AAR activation (101). JDP2 has been reported to inhibit transcription from the ATF3 gene by an unknown mechanism (110). Given that the full-length ATF3 protein is the primary isoform produced during the AAR and that it inhibits ATF4 function (81), suppression of ATF3 expression by JDP2 may indirectly enhance ATF4 action. In contrast to this and many other reports of transcriptional repression by JDP2, its dimerization with CHOP activates transcription of certain genes via AP-1 sites (111). If JDP2 functions in the “fed” state to suppress transcription from AA-responsive genes such as CHOP, activation of JDP2 expression by the AAR presents a paradox, unless it is also part of a feedback repression mechanism. It would be interesting to further investigate the primary role of JDP2 within the AAR network of transcription factors.
Transcription factors outside of the bZIP superfamily
As mentioned previously, Igfbp-1 expression is up-regulated in rats fed a protein-free diet and in cells cultured with medium deprived of AA (32). An E-box like AA responsive element was identified at −77 to −112 bp upstream of the transcriptional start site within the Igfbp-1 gene, and electrophoretic mobility shift analysis using nuclear extracts of liver tissue from rats fed a control or protein-free diet revealed increased binding of both USF-1 and USF-2 (112). Consistent with this observation, the abundance of nuclear USF-1 and USF-2 proteins was significantly increased in livers of the rats receiving the protein-free diet. Averous et al. (28) demonstrated that AA deprivation of HepG2 hepatoma cells induced IGFBP-1 through both transcriptional and posttranscriptional mechanisms. The authors also showed that the transcriptional induction was independent of GCN2 and ATF4. Given that USF-1 and USF-2 are expressed in a wide variety of tissues and that E-box elements are common to many genes, the possible role of USF-1 and USF-2 within the AAR requires further investigation.
Nuclear factor κB (NF-κB) family members are transcription factors that regulate a wide range of processes such as immunity, stress responses, apoptosis, and differentiation. Microarray analysis of several central nervous system tumor cell lines after methionine deprivation identified NF-κB as a gene up-regulated during this AA stress condition (113). In some cell lines, the NF-κB protein was retained in the cytoplasm, whereas in others, nuclear transfer was observed. Likewise, by microarray analysis of HepG2 cells after activation of the AAR, Shan et al. (101) observed that the mRNA expression levels for many of the NF-κB family members were increased. Jiang et al. (114) used wild-type and knockout mouse embryonic fibroblasts to establish that during AA deprivation, NF-κB is activated in a GCN2-dependent manner, and during ER stress, NF-κB activation is PERK dependent. In wild-type cells, NF-κB was activated within 1 h after leucine deprivation and remained increased as long as 6 h. In contrast, only a modest increase in NF-κB binding activity was observed in GCN2-deficient fibroblasts. Consistent with the role of the GCN2 and PERK, in fibroblasts expressing eIF2α with a Ser-51-Ala (A/A) mutation that cannot be phosphorylated by GCN2 or PERK, no activation of NF-κB was detected (114). The authors showed that the phosphorylated eIF2α-dependent activation of NF-κB was the result of a disassociation from its inhibitor IκB (inhibitory kappa beta). In many instances, NF-κB is activated by signals that lead to the degradation of IκB, but for the phosphorylated eIF2α–triggered response, IκB total abundance was unchanged. The mechanism by which the AAR/UPR pathways cause NF-κB/IκB disassociation in the absence of IκB turnover has not been established. Interestingly, GCN2 also mediates the induction of NF-κB activity by UV irradiation (115), but how UV activates the GCN2 kinase is unknown.
As an important tissue for nutrient homeostasis, the liver expresses many metabolic pathways controlled by liver-specific transcription factors, such as the FOXA1, FOXA2, and FOXA3 proteins (116). Imae et al. (117) showed that hepatic FOXA3 expression is up-regulated in mice fed a protein-free diet or a diet in which casein was replaced by an equal amount of gluten (deficient in lysine and threonine). Su et al. (118) extended that observation to show that of the 3 family members, FOXA2 and FOXA3 are induced by the AAR, both in intact animals fed a low-protein diet (20% vs. 8%) and in hepatoma cells in culture treated with histidinol to increase uncharged His-tRNA. The expression of FOXA1 was unaffected. The mechanism by which FOXA2/3 expression was induced was primarily transcriptional in nature, but not through the ATF4-dependent pathway. The FOXA2/3 genes do not appear to have a functional ATF4-responsive CARE sequence within their proximal gene locus, and the AAR induction was not blocked by knockdown of ATF4. Perhaps consistent with the ATF4-independence, triggering the UPR pathway has little or no effect on FOXA2/3 expression (118), reminiscent of the SNAT2 gene described earlier. Thus, FOXA2/3 join a small list of genes that are induced by the AAR, but not by the UPR. The FOXA2/3 proteins do not appear to play a major role in the control of currently recognized AAR target genes. Further studies are necessary to establish the functions of the FOXA transcription factors during the AAR.
Conclusions
Investigation of the signaling pathways and gene expression programs triggered by the AAR has identified several transcriptional mediators of this response. ER stress exhibits a significant degree of overlap with regard to these mediators as a consequence of ATF4 induction. What these investigations show is that the bZIP superfamily of transcription factors plays the central role during these cellular stress responses (Fig. 2). Formation of homo- and heterodimers among the ATF, C/EBP, and FOS/JUN families of proteins forms an integrated transcription factor network that determines the initiation, magnitude, and length of the cellular response to AA limitation. Despite the recent progress in characterizing the transcriptional programs associated with the AAR, many questions remain. For example, how does ATF4 signaling, triggered by 4 different eIF2α kinases, become selective for the appropriate subset of genes within a given tissue or cell type? A related question is what role do the ATF4 dimerization partners play in determining the specificity of the response? Microarray analysis of GCN2-deficient cells indicates that there are GCN2-independent, AA-responsive genes (119), suggesting that there are additional AA sensors to be discovered. What is the identity of these sensors and how do they function? One possibility that has been put forth that deserves more investigation is that plasma membrane AA transporters serve as sensors (120). Another interesting possibility comes from the observations that 1 or more members of the class 3 G protein–coupled receptors have AA-sensing capabilities (reviewed in Reference 121). Further support for this hypothesis comes from the data of Chaveroux et al. (67) who showed that Gα12 was necessary for increased ATF2 phosphorylation after leucine deprivation. They concluded that this phosphorylation was triggered independently of GCN2 signaling because it occurred in GCN2-deficient fibroblasts, but it did not occur in wild-type cells incubated with leucinol. However, Fu et al. (69) showed that a robust increase in ATF2 phosphorylation is observed after activation of the AAR by treatment of HepG2 cells with histidinol. It is possible that signaling, metabolism, or other cell-specific differences accounts for this apparent discrepancy.
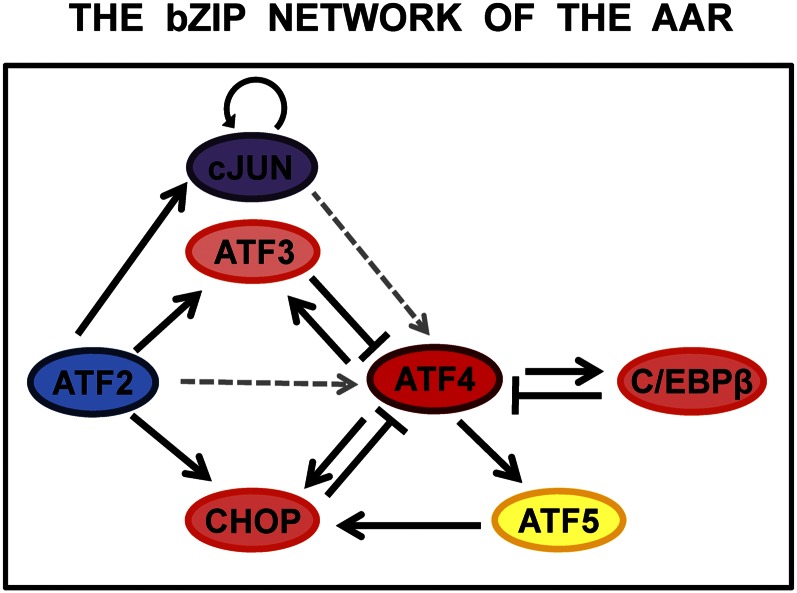
Figure 2.
The basic leucine zipper (bZIP) transcription factor network of the amino acid response (AAR). The core of the transcription factor network that has been identified thus far to mediate the AAR consists of primarily bZIP family members. Several are transcriptionally activated by activating transcription factor (ATF) 4 [ATF3, CCAAT enhancer-binding protein homologous protein (CHOP), and CCAAT enhancer-binding protein (C/EBP) β] and then subsequently act to feedback inhibit the ATF4 signal as part of a self-limiting ATF4 program described in the text. C/EBPβ and cJUN are illustrated in the figure, but other members of the C/EBP and FOS/JUN families are known to be induced in expression by the AAR and/or contribute to regulation of AAR target genes. The solid arrows represent direct activation of the indicated gene, whereas the solid bars represent transcriptional antagonism of ATF4 action on AAR target genes. The dashed gray arrows indicate that ATF2 and cJUN stabilize the ATF4 protein by an unknown mechanism.
Thus, although considerable progress in understanding AA-dependent control of gene expression has been achieved, innumerable important mechanistic questions remain.
Acknowledgments
All authors have read and approved the final manuscript.
Footnotes
Supported by the National Institutes of Health (DK-92062, DK-94729) to M.S.K.
Author disclosures: M. S. Kilberg, M. Balasubramanian, L. Fu, and J. Shan, no conflicts of interest.
Abbreviations used: AA, amino acid; AAR, amino acid response; AARE, amino acid response element; ALL, acute lymphoblastic leukemia; ASNase, asparaginase; ATF, activating transcription factor; ASNS, asparagine synthetase; bZIP, basic leucine zipper; CARE, CCAAT enhancer-binding protein–activating transcription factor response element; C/EBP, CCAAT enhancer-binding protein; ChIP, chromatin immunoprecipitation; CHOP, CCAAT enhancer-binding protein homologous protein; eIF2, eukaryotic initiation factor 2; ER, endoplasmic reticulum; GADD, growth arrest and DNA damage inducible; GCN2, general control nonderepressible 2; IGF-1, insulin-like growth factor-1; IGFBP-1, insulin-like growth factor binding protein 1; IκB, inhibitory kappa beta; IUGR, intrauterine growth retardation; JDP2, JUN dimerization protein 2; JNK, c-Jun N-terminal kinase; LAP, liver-enriched transcriptional activator protein; LIP, liver-enriched transcriptional inhibitory protein; mRNA, messenger RNA; NF-κB, nuclear factor κB; p-ATF, phosphorylated activating transcription factor; PERK, PKR-like endoplasmic reticulum kinase; phospho-eIF2α, phosphorylation of eukaryotic initiation factor 2α siRNA, small interfering RNA; SNAT2, sodium-dependent neutral amino acid transporter 2; Tet, tetracycline; tRNA, transfer RNA; UPR, unfolded protein response.
Literature Cited
- 1.Gietzen DW, Rogers QR. Nutritional homeostasis and indispensable amino acid sensing: a new solution to an old puzzle. Trends Neurosci. 2006;29:91–9 [DOI] [PubMed] [Google Scholar]
- 2.Koehnle TJ, Russell MC, Morin AS, Erecius LF, Gietzen DW. Diets deficient in indispensable amino acids rapidly decrease the concentration of the limiting amino acid in the anterior piriform cortex of rats. J Nutr. 2004;134:2365–71 [DOI] [PubMed] [Google Scholar]
- 3.Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, Koehnle TJ, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–8 [DOI] [PubMed] [Google Scholar]
- 4.Maurin AC, Jousse C, Averous J, Parry L, Bruhat A, Cherasse Y, Zeng H, Zhang Y, Harding HP, Ron D, et al. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 2005;1:273–7 [DOI] [PubMed] [Google Scholar]
- 5.Guo F, Cavener DR. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5:103–14 [DOI] [PubMed] [Google Scholar]
- 6.Endo Y, Fu Z, Abe K, Arai S, Kato H. Dietary protein quantity and quality affect rat hepatic gene expression. J Nutr. 2002;132:3632–7 [DOI] [PubMed] [Google Scholar]
- 7.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–6 [DOI] [PubMed] [Google Scholar]
- 8.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–62 [DOI] [PubMed] [Google Scholar]
- 10.Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2. Genetics. 2000;154:787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimball SR, Anthony TG, Cavener DR, Jefferson LS, Nutrient signaling through mammalian GCN2. New York: Springer-Verlag; 2005 [Google Scholar]
- 13.Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. The GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent, L-asparaginase. J Biol Chem. 2009;284:32742–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimball SR, Jefferson LS. Role of amino acids in the translational control of protein synthesis in mammals. Semin Cell Dev Biol. 2005;16:21–7 [DOI] [PubMed] [Google Scholar]
- 15.Lee YY, Cevallos RC, Jan E. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J Biol Chem. 2009;284:6661–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2. J Cell Biol. 2001;153:1011–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22:1180–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11 [DOI] [PubMed] [Google Scholar]
- 20.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33 [DOI] [PubMed] [Google Scholar]
- 21.Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics. 2009;38:328–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gjymishka A, Palii SS, Shan J, Kilberg MS. Despite increased ATF4 binding at the C/EBP-ATF composite site following activation of the unfolded protein response, system A transporter 2 (SNAT2) transcription activity is repressed in HepG2 cells. J Biol Chem. 2008;283:27736–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmerman JA, Malloy V, Krajcik R, Orentreich N. Nutritional control of aging. Exp Gerontol. 2003;38:47–52 [DOI] [PubMed] [Google Scholar]
- 24.Ooka H, Segall PE, Timiras PS. Histology and survival in age-delayed low-tryptophan-fed rats. Mech Ageing Dev. 1988;43:79–98 [DOI] [PubMed] [Google Scholar]
- 25.Segall PE, Timiras PS. Patho-physiologic findings after chronic tryptophan deficiency in rats: a model for delayed growth and aging. Mech Ageing Dev. 1976;5:109–24 [DOI] [PubMed] [Google Scholar]
- 26.Thissen JP, Ketelslegers JM, Underwood LE. Nutritional regulation of the insulin-like growth factors. Endocr Rev. 1994;15:80–101 [DOI] [PubMed] [Google Scholar]
- 27.Smith WJ, Underwood LE, Clemmons DR. Effects of caloric or protein restriction on insulin-like growth factor-I (IGF-I) and IGF-binding proteins in children and adults. J Clin Endocrinol Metab. 1995;80:443–9 [DOI] [PubMed] [Google Scholar]
- 28.Averous J, Maurin AC, Bruhat A, Jousse C, Arliguie C, Fafournoux P. Induction of IGFBP-1 expression by amino acid deprivation of HepG2 human hepatoma cells involves both a transcriptional activation and an mRNA stabilization due to its 3′UTR. FEBS Lett. 2005;579:2609–14 [DOI] [PubMed] [Google Scholar]
- 29.Jousse C, Bruhat A, Ferrara M, Fafournoux P. Physiological concentration of amino acids regulates insulin-like-growth-factor-binding protein 1 expression. Biochem J. 1998;334:147–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanamoto R, Yokota T, Hayashi SI. Expressions of c-myc and insulin-like growth factor-1 mRNA in the liver of growing rats vary reciprocally in response to changes in dietary protein. J Nutr. 1994;124:2329–34 [DOI] [PubMed] [Google Scholar]
- 31.Straus DS, Burke EJ, Marten NW. Induction of insulin-like growth factor binding protein-1 gene expression in liver of protein-restricted rats and in rat hepatoma cells limited for a single amino acid. Endocrinology. 1993;132:1090–100 [DOI] [PubMed] [Google Scholar]
- 32.Takenaka A, Komori K, Morishita T, Takahashi SI, Hidaka T, Noguchi T. Amino acid regulation of gene transcription of rat insulin-like growth factor-binding protein-1. J Endocrinol. 2000;164:R11–6 [DOI] [PubMed] [Google Scholar]
- 33.Figueras F, Gardosi J. Intrauterine growth restriction: new concepts in antenatal surveillance, diagnosis, and management. Am J Obstet Gynecol. 2011;204:288–300 [DOI] [PubMed] [Google Scholar]
- 34.Imdad A, Bhutta ZA. Effect of balanced protein energy supplementation during pregnancy on birth outcomes. BMC Public Health. 2011;11: Suppl 3:S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown LD, Green AS, Limesand SW, Rozance PJ. Maternal amino acid supplementation for intrauterine growth restriction. Front Biosci (Schol Ed). 2011;3:428–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yung HW, Calabrese S, Hynx D, Hemmings BA, Cetin I, Charnock-Jones DS, Burton GJ. Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction. Am J Pathol. 2008;173:451–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker DJ. Adult consequences of fetal growth restriction. Clin Obstet Gynecol. 2006;49:270–83 [DOI] [PubMed] [Google Scholar]
- 38.Gong L, Pan YX, Chen H. Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics. 2010;5:619–26 [DOI] [PubMed] [Google Scholar]
- 39.Kalantar-Zadeh K, Cano NJ, Budde K, Chazot C, Kovesdy CP, Mak RH, Mehrotra R, Raj DS, Sehgal AR, Stenvinkel P, et al. Diets and enteral supplements for improving outcomes in chronic kidney disease. Nat Rev Nephrol. 2011;7:369–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May PE, Barber A, D'Olimpio JT, Hourihane A, Abumrad NN. Reversal of cancer-related wasting using oral supplementation with a combination of beta-hydroxy-beta-methylbutyrate, arginine, and glutamine. Am J Surg. 2002;183:471–9 [DOI] [PubMed] [Google Scholar]
- 41.Morley JE. Calories and cachexia. Curr Opin Clin Nutr Metab Care. 2009;12:607–10 [DOI] [PubMed] [Google Scholar]
- 42.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, DePanis DN, Bobrovnikova-Marjon E, Diehl A, Ron D, Koumenis C. The GCN2–ATF4 pathway is critical for tumor cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aslanian AM, Fletcher BS, Kilberg MS. Asparagine synthetase expression alone is sufficient to induce L-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem J. 2001;357:321–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su N, Pan YX, Zhou M, Harvey RC, Hunger SP, Kilberg MS. Correlation between asparaginase sensitivity and asparagine synthetase protein content, but not mRNA, in acute lymphoblastic leukemia cell lines. Pediatr Blood Cancer. 2008;50:274–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–48 [DOI] [PubMed] [Google Scholar]
- 47.Aslanian AM, Kilberg MS. Multiple adaptive mechanisms affect asparagine synthetase substrate availability in asparaginase-resistant MOLT-4 human leukaemia cells. Biochem J. 2001;358:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorenzi PL, Weinstein JN. Asparagine synthetase: a new potential biomarker in ovarian cancer. Drug News Perspect. 2009;22:61–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, McNurlan MA, Wek RC. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem. 2004;279:36553–61 [DOI] [PubMed] [Google Scholar]
- 50.Carraro V, Maurin AC, Lambert-Langlais S, Averous J, Chaveroux C, Parry L, Jousse C, Ord D, Ord T, Fafournoux P, et al. Amino acid availability controls TRB3 transcription in liver through the GCN2/eIF2alpha/ATF4 pathway. PLoS ONE. 2010;5:e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem. 2002;277:24120–7 [DOI] [PubMed] [Google Scholar]
- 52.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfgang CD, Chen BP, Martindale JL, Holbrook NJ, Hai T. gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol Cell Biol. 1997;17:6700–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J. 1999;339:135–41 [PMC free article] [PubMed] [Google Scholar]
- 56.Shan J, Ord D, Ord T, Kilberg MS. Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptional induction via CCAAT enhancer-binding protein-activating transcription factor response elements. J Biol Chem. 2009;284:21241–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21 [DOI] [PubMed] [Google Scholar]
- 58.Thiaville MM, Dudenhausen EE, Zhong C, Pan YX, Kilberg MS. Deprivation of protein or amino acid induces C/EBP beta synthesis and binding to amino acid response elements, but its action is not an absolute requirement for enhanced transcription. Biochem J. 2008;410:473–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang HY, Wek SA, McGrath BC, Lu D, Hai TW, Harding HP, Wang XZ, Ron D, Cavener DR, Wek RC. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol Cell Biol. 2004;24:1365–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem J. 2007;401:299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chérasse Y, Maurin AC, Chaveroux C, Jousse C, Carraro V, Parry L, Deval C, Chambon C, Fafournoux P, Bruhat A. The p300/CBP-associated factor (PCAF) is a cofactor of ATF4 for amino acid-regulated transcription of CHOP. Nucleic Acids Res. 2007;35:5954–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation. J Biol Chem. 2004;279:50829–39 [DOI] [PubMed] [Google Scholar]
- 63.Su N, Kilberg MS. C/EBP homology protein (chop) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J Biol Chem. 2008;283:35106–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopez AB, Wang C, Huang CC, Yaman I, Li Y, Chakravarty K, Johnson PF, Chiang CM, Snider MD, Wek RC, et al. A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem J. 2007;402:163–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Averous J, Bruhat A, Jousse C, Carraro V, Thiel G, Fafournoux P. Induction of CHOP Expression by Amino Acid Limitation Requires Both ATF4 Expression and ATF2 Phosphorylation. J Biol Chem. 2004;279:5288–97 [DOI] [PubMed] [Google Scholar]
- 66.Bruhat A, Cherasse Y, Maurin AC, Breitwieser W, Parry L, Deval C, Jones N, Jousse C, Fafournoux P. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res. 2007;35:1312–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaveroux C, Jousse C, Cherasse Y, Maurin AC, Parry L, Carraro V, Derijard B, Bruhat A, Fafournoux P. Identification of a novel amino acid response pathway triggering ATF2 phosphorylation in mammals. Mol Cell Biol. 2009;29:6515–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruhat A, Jousse C, Carraro V, Reimold AM, Ferrara M, Fafournoux P. Amino acids control mammalian gene transcription: activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol Cell Biol. 2000;20:7192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu L, Balasubramanian M, Shan J, Dudenhausen EE, Kilberg MS. Auto-activation of c-JUN gene by amino acid deprivation of hepatocellular carcinoma cells reveals a novel c-JUN-mediated signaling pathway. J Biol Chem. 2011;286:36724–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gachon F, Devaux C, Mesnard JM. Activation of HTLV-I transcription in the presence of Tax is independent of the acetylation of CREB-2 (ATF-4). Virology. 2002;299:271–8 [DOI] [PubMed] [Google Scholar]
- 71.Lassot I, Estrabaud E, Emiliani S, Benkirane M, Benarous R, Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J Biol Chem. 2005;280:41537–45 [DOI] [PubMed] [Google Scholar]
- 72.Greene LA, Lee HY, Angelastro JM. The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem. 2009;108:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al Sarraj J, Vinson C, Thiel G. Regulation of asparagine synthetase gene transcription by the basic region leucine zipper transcription factors ATF5 and CHOP. Biol Chem. 2005;386:873–9 [DOI] [PubMed] [Google Scholar]
- 74.Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, Kimura N, Hirose H, Takahashi S, Takahashi Y. Stress-induced translation of ATF5 mRNA is regulated by the 5'-untranslated region. J Biol Chem. 2008;283:2543–53 [DOI] [PubMed] [Google Scholar]
- 75.Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–73 [DOI] [PubMed] [Google Scholar]
- 76.Yamazaki T, Ohmi A, Kurumaya H, Kato K, Abe T, Yamamoto H, Nakanishi N, Okuyama R, Umemura M, Kaise T, et al. Regulation of the human CHOP gene promoter by the stress response transcription factor ATF5 via the AARE1 site in human hepatoma HepG2 cells. Life Sci. 2010;87:294–301 [DOI] [PubMed] [Google Scholar]
- 77.Li G, Li W, Angelastro JM, Greene LA, Liu DX. Identification of a novel DNA binding site and a transcriptional target for activating transcription factor 5 in c6 glioma and mcf-7 breast cancer cells. Mol Cancer Res. 2009;7:933–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rousseau J, Gagne V, Labuda M, Beaubois C, Sinnett D, Laverdiere C, Moghrabi A, Sallan SE, Silverman LB, Neuberg D, et al. ATF5 polymorphisms influence ATF function and response to treatment in children with childhood acute lymphoblastic leukemia. Blood. 2011;118:5883–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr. 2010;15:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med. 2009;87:1053–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan YX, Chen H, Siu F, Kilberg MS. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple ATF3 mRNA species which, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J Biol Chem. 2003;278:38402–12 [DOI] [PubMed] [Google Scholar]
- 82.Pan Y, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3′-untranslated region regulates its amino acid limitation-induced stabilization. J Biol Chem. 2005;280:34609–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yaman I, Fernandez J, Sarkar B, Schneider RJ, Snider MD, Nagy LE, Hatzoglou M. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J Biol Chem. 2002;277:41539–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wolfgang CD, Liang G, Okamoto Y, Allen AE, Hai T. Transcriptional autorepression of the stress-inducible gene ATF3. J Biol Chem. 2000;275:16865–70 [DOI] [PubMed] [Google Scholar]
- 85.Zhou D, Zhang Y, Pan YX, Chen H. Dickkopf homolog 1, a Wnt signaling antagonist, is transcriptionally up-regulated via an ATF4-independent and MAPK/ERK-dependent pathway following amino acid deprivation. Biochim Biophys Acta. 2011;1809:306–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–79 [DOI] [PubMed] [Google Scholar]
- 87.Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–32 [PMC free article] [PubMed] [Google Scholar]
- 88.Kimball SR, Jefferson LS. Molecular mechanisms through which amino acids mediate signaling through the mammalian target of rapamycin. Curr Opin Clin Nutr Metab Care. 2004;7:39–44 [DOI] [PubMed] [Google Scholar]
- 89.Wilson FA, Suryawan A, Gazzaneo MC, Orellana RA, Nguyen HV, Davis TA. Stimulation of muscle protein synthesis by prolonged parenteral infusion of leucine is dependent on amino acid availability in neonatal pigs. J Nutr. 2010;140:264–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y, Bevilacqua E, Chiribau CB, Majumder M, Wang C, Croniger CM, Snider MD, Johnson PF, Hatzoglou M. Differential control of the CCAAT/enhancer-binding protein beta (C/EBPbeta) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J Biol Chem. 2008;283:22443–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiribau CB, Gaccioli F, Huang CC, Yuan CL, Hatzoglou M. Molecular symbiosis of CHOP and C/EBP beta isoform LIP contributes to endoplasmic reticulum stress-induced apoptosis. Mol Cell Biol. 2010;30:3722–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zinszner H, Kuroda M, Wang XZ, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Price BD, Calderwood SK. Gadd45 and Gadd153 messenger RNA levels are increased during hypoxia and after exposure of cells to agents which elevate the levels of the glucose-regulated proteins. Cancer Res. 1992;52:3814–7 [PubMed] [Google Scholar]
- 94.Jousse C, Bruhat A, Harding HP, Ferrara M, Ron D, Fafournoux P. Amino acid limitation regulates CHOP expression through a specific pathway independent of the unfolded protein response. FEBS Lett. 1999;448:211–6 [DOI] [PubMed] [Google Scholar]
- 95.Bruhat A, Jousse C, Wang XZ, Ron D, Ferrara M, Fafournoux P. Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and post-transcriptional levels. J Biol Chem. 1997;272:17588–93 [DOI] [PubMed] [Google Scholar]
- 96.Abcouwer SF, Schwarz C, Meguid RA. Glutamine deprivation induces the expression of GADD45 and GADD153 primarily by mRNA stabilization. J Biol Chem. 1999;274:28645–51 [DOI] [PubMed] [Google Scholar]
- 97.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pohjanpelto P, Holtta E. Deprivation of a single amino acid induces protein synthesis-dependent increases in c-jun, c-myc, and ornithine decarboxylase mRNAs in Chinese hamster ovary cells. Mol Cell Biol. 1990;10:5814–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li J, Liu J, Song J, Wang X, Weiss HL, Townsend CM, Jr, Gao T, Evers BM. mTORC1 inhibition increases neurotensin secretion and gene expression through activation of the MEK/ERK/c-Jun pathway in the human endocrine cell line BON. Am J Physiol Cell Physiol. 2011;301:C213–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gietzen DW, Ross CM, Hao S, Sharp JW. Phosphorylation of eIF2alpha is involved in the signaling of indispensable amino acid deficiency in the anterior piriform cortex of the brain in rats. J Nutr. 2004;134:717–23 [DOI] [PubMed] [Google Scholar]
- 101.Shan J, Lopez MC, Baker HV, Kilberg MS. Expression profiling after activation of the amino acid deprivation response in HepG2 human hepatoma cells. Physiol Genomics. 2010;41:315–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lopez-Bergami P, Lau E, Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer. 2010;10:65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Dam H, Duyndam M, Rottier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb AJ. Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 1993;12:479–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morooka H, Bonventre JV, Pombo CM, Kyriakis JM, Force T. Ischemia and reperfusion enhance ATF-2 and c-Jun binding to cAMP response elements and to an AP-1 binding site from the c-jun promoter. J Biol Chem. 1995;270:30084–92 [DOI] [PubMed] [Google Scholar]
- 105.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin C, Ugai H, Song J, Murata T, Nili F, Sun K, Horikoshi M, Yokoyama KK. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett. 2001;489:34–41 [DOI] [PubMed] [Google Scholar]
- 108.Jin C, Li H, Murata T, Sun K, Horikoshi M, Chiu R, Yokoyama KK. JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Mol Cell Biol. 2002;22:4815–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chérasse Y, Chaveroux C, Jousse C, Maurin AC, Carraro V, Parry L, Fafournoux P, Bruhat A. Role of the repressor JDP2 in the amino acid-regulated transcription of CHOP. FEBS Lett. 2008;582:1537–41 [DOI] [PubMed] [Google Scholar]
- 110.Weidenfeld-Baranboim K, Hasin T, Darlyuk I, Heinrich R, Elhanani O, Pan J, Yokoyama KK, Aronheim A. The ubiquitously expressed bZIP inhibitor, JDP2, suppresses the transcription of its homologue immediate early gene counterpart, ATF3. Nucleic Acids Res. 2009;37:2194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weidenfeld-Baranboim K, Bitton-Worms K, Aronheim A. TRE-dependent transcription activation by JDP2-CHOP10 association. Nucleic Acids Res. 2008;36:3608–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Matsukawa T, Inoue Y, Oishi Y, Kato H, Noguchi T. Up-regulation of upstream stimulatory factors by protein malnutrition and its possible role in regulation of the IGF-binding protein-1 gene. Endocrinology. 2001;142:4643–51 [DOI] [PubMed] [Google Scholar]
- 113.Kokkinakis DM, Liu X, Chada S, Ahmed MM, Shareef MM, Singha UK, Yang S, Luo J. Modulation of gene expression in human central nervous system tumors under methionine deprivation-induced stress. Cancer Res. 2004;64:7513–25 [DOI] [PubMed] [Google Scholar]
- 114.Jiang HY, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, Wek RC. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23:5651–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiang HY, Wek RC. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J. 2005;385:371–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–6 [PubMed] [Google Scholar]
- 117.Imae M, Inoue Y, Fu Z, Kato H, Noguchi T. Gene expression of the three members of hepatocyte nuclear factor-3 is differentially regulated by nutritional and hormonal factors. J Endocrinol. 2000;167:R1–5 [DOI] [PubMed] [Google Scholar]
- 118.Su N, Thiaville MM, Awad K, Gjymishka A, Brant JO, Yang TP, Kilberg MS. Protein or amino acid deprivation differentially regulates the hepatic forkhead box protein A (FOXA) genes through an activating transcription factor-4-independent pathway. Hepatology. 2009;50:282–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Deval C, Chaveroux C, Maurin AC, Cherasse Y, Parry L, Carraro V, Milenkovic D, Ferrara M, Bruhat A, Jousse C, et al. Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 2009;276:707–18 [DOI] [PubMed] [Google Scholar]
- 120.Taylor PM. Amino acid transporters: éminences grises of nutrient signalling mechanisms? Biochem Soc Trans. 2009;37:237–41 [DOI] [PubMed] [Google Scholar]
- 121.Conigrave AD, Hampson DR. Broad-spectrum L-amino acid sensing by class 3 G-protein-coupled receptors. Trends Endocrinol Metab. 2006;17:398–407 [DOI] [PubMed] [Google Scholar]