Abstract
Bifidobacteria are commonly used as probiotics in dairy foods. Select bifidobacterial species are also early colonizers of the breast-fed infant colon; however, the mechanism for this enrichment is unclear. We previously showed that Bifidobacterium longum subsp. infantis is a prototypical bifidobacterial species that can readily utilize human milk oligosaccharides as the sole carbon source. MS-based glycoprofiling has revealed that numerous B. infantis strains preferentially consume small mass oligosaccharides, abundant in human milks. Genome sequencing revealed that B. infantis possesses a bias toward genes required to use mammalian-derived carbohydrates. Many of these genomic features encode enzymes that are active on milk oligosaccharides including a novel 40-kb region dedicated to oligosaccharide utilization. Biochemical and molecular characterization of the encoded glycosidases and transport proteins has further resolved the mechanism by which B. infantis selectively imports and catabolizes milk oligosaccharides. Expression studies indicate that many of these key functions are only induced during growth on milk oligosaccharides and not expressed during growth on other prebiotics. Analysis of numerous B. infantis isolates has confirmed that these genomic features are common among the B. infantis subspecies and likely constitute a competitive colonization strategy used by these unique bifidobacteria. By detailed characterization of the molecular mechanisms responsible, these studies provide a conceptual framework for bifidobacterial persistence and host interaction in the infant gastrointestinal tract mediated in part through consumption of human milk oligosaccharides.
Introduction
Human milk plays a key role in supporting the survival and development of offspring and is a good example of a nutrient that has been shaped by evolution (1). Strong selective pressures have influenced the composition of this fluid in which factors such as the nutritional and protective needs of the newborn are balanced against the energy cost of milk production by the mother (2).
The WHO defines breastfeeding as the “normal way of providing young infants with the nutrients they need for healthy growth and development” (3, 4). Human milk is considered a gold standard for nutrition (5) and is characterized by high amounts of essential nutrients (6), such as lactose, fatty acids, and proteins, as well as macronutrients, such as vitamins and minerals. Human milk is compositionally different from mother to mother, from time to time during lactation, and also during different breastfeeding periods (7, 8).
Human milk also contains a wide variety of nonessential nutrients that are not consumed by the infant but display complex and potent bioactive functions (9–11). For example, human milk oligosaccharides (HMO)7 are an array of complex carbohydrates that do not provide direct nutritional value for the infant. Among other putative roles, these molecules act as prebiotics, serving as growth substrates for specific colonic bacteria in breast-fed infants, mainly belonging to the Bifidobacterium genus. In this review, we address recent advances in our understanding of the bifidogenic effect of HMO and how specific intestinal bifidobacteria have developed strategies for gaining access to the energy content of HMO.
Human milk oligosaccharides
Oligosaccharides are present in human milk at 5–15 g/L, being the third largest component after lactose and lipids (12). HMO are free soluble carbohydrates that contain 3–15 monosaccharides, linked through a variety of glycosidic bonds. Their concentration is severalfold compared with other mammalian milks (13). HMO composition and abundance progressively change during lactation, with significant variations among individual mothers (14). The question of why these indigestible molecules are present at high concentrations in milk has challenged researchers for decades. HMO are synthesized solely in the mammary gland during lactation, and the complex pathways that result in all the structures observed have not yet been completely elucidated. HMO are minimally affected by transit through the stomach and small intestine, reaching a high concentration in the infant colon (14–16).
Breastfeeding has been associated with a lower incidence of diarrhea and gastrointestinal infections caused by bacterial and viral pathogens (17–20). Many milk oligosaccharides contain structural elements that are homologous to glycoconjugates present in the intestinal mucosa used by pathogens for adherence and invasion. For these reasons, HMO are proposed to act as soluble receptor analogs and inhibit the adhesion of pathogens (19, 21).
Neutral HMO are composed of galactose (Gal), N-acetylglucosamine (GlcNAc), fucose, and a lactose core, whereas acidic HMO can contain these monosaccharides in addition to one or more molecules of N-acetylneuraminic acid or sialic acid (22). A variety of glycosyltransferases are active in HMO synthesis, creating 13 potential glycosidic bonds, which results in more than a thousand possible combinations (12, 23). However, only a few hundred have been identified. The Gal unit of the lactose core is linked to subunits of lacto-N-biose I (LNB) (Galβ1–3GlcNAc, type 1 chain) and/or N-acetyllactosamine (type 2 chain, Galβ1–4GlcNAc) originating specific HMO isomer classes. Fucose and sialic acid residues are generally found attached to these backbones in terminal positions. A high-throughput strategy based on high-accuracy MS combined with nanoliquid chromatography (24) was recently used to annotate the molecular and structural complexity of HMO.
Establishment of the infant microbiota: impact of breast milk on bifidobacteria
Immediately after birth, the newborn is exposed to a nonsterile environment, and microorganisms colonize mucosal surfaces including the gastrointestinal tract, initiating what will be a long-term microbe-host relationship. The initial microbial colonization patterns present in the distal colon have an impact on several aspects of the infant health, and, moreover, they are thought to have a long-term impact on human health (25, 26). However, the mechanisms by which some microorganisms are responsible for these health effects are not well understood.
The process of colonization is dependent on the mode of birth. Vaginal delivery allows direct contact of the neonate with several rich bacterial niches including the vaginal and the fecal maternal microbiota (27–30). Others have shown that some infant gut bacterial clades are also present in human milk, suggesting that this could represent another reservoir for microorganisms colonizing the infant gut (31–34). Conversely, the main sources of bacteria for cesarean-born infants are found in the hospital environment (35) and the skin microbiome (29), and a delay in colonization by prominent members of the intestinal microbiota such as Bifidobacterium, Bacteroides, and Escherichia coli have been observed (36, 37). Moreover bifidobacterial counts have been shown to be lower in cesarean-born infants (38, 39).
The type of infant feeding, either breast or formula milk, is a main factor contributing to the development of the infant gut microbiota. Infant formulas are usually based on cow’s milk, supplemented with vegetal fatty acids, minerals, and vitamins. Despite recent advances and new ingredients (40), it is difficult to exactly replicate the wide array of nonessential nutrients present in breast milk, such as oligosaccharides, antibodies, and bioactive proteins.
Breast milk guides the development of the infant gut microbiota by preventing pathogen colonization and also by selecting bacteria that can use HMO as a growth substrate, such as Bifidobacterium species. This genus is the most abundant in the infant gut microbiome in both breast-fed and formula-fed infants (33, 38, 41, 42). However, the bifidobacterial species recovered from infants subject to both diets differ. In general, a small number of Bifidobacterium species are routinely recovered from the feces of breast-fed infants including Bifidobacterium longum subsp. longum, B. longum subsp. infantis, Bifidobacterium bifidum, and Bifidobacterium breve (43–45). The bifidobacterial species found in feces from bottle-fed infants is more diverse and include the aforementioned species and also Bifidobacterium adolescentis and Bifidobacterium pseudocatenulatum, which are also commonly found in adults (46).
Molecular adaptations of bifidobacteria to utilize HMO
The inherent complexity of HMO renders them inaccessible to digestive enzymes; therefore, they can reach high concentrations in the distal colon. To grow on HMO as a substrate, a microorganism requires specific transporters and/or enzymatic machinery to process these molecules. Recent advances in our understanding of how bifidobacteria utilize HMO correlates also with a general notion of bifidobacteria providing health benefits to the host, given the probiotic or antipathogenic activities shown for certain strains.
From the early studies of Moro in 1905 (47) and Gyorgy in 1954 (48), a relationship between human milk and the selective growth of Bifidobacterium was determined. Growth factors in human milk, or bifidus factor, were thought to be GlcNAc containing oligosaccharides and glycoproteins that stimulated the growth of bifidobacteria specifically in breast-fed infants. Bifidobacterium species are gram positive and strictly anaerobic bacteria, with a fermentative metabolism that produces acetate and lactate as end products (49). Bifidobacteria are often dominant microorganisms in the breast-fed infant gut microbiota and also are significant members of the adult gut microbiome.
Ward et al. (50), first showed that bifidobacteria can consume HMO as the sole carbon source. The extent of growth observed by B. infantis was higher compared with other species in the genus. Later work demonstrated that B. infantis preferentially consumed short-chain HMO (degree of polymerization <7), but longer chains were used when the total concentration of HMO was reduced (51).
The genome sequence of B. infantis provided an explanation for this phenotype (52). Similar to other bifidobacterial genomes, B. infantis possessed a number of genes involved in consumption of complex carbohydrates (53, 54). However, gene clusters dedicated to the metabolism of plant polysaccharides in the closely related B. longum were replaced in B. infantis with gene functions related to HMO consumption. Of particular interest was a 43-kb gene cluster (termed HMO cluster I), which so far has been only found in B. infantis strains. It contained several genes predicted to be involved in the import and metabolism of HMO, such as glycosyl hydrolases and oligosaccharide transport proteins, all within a single locus (52). The HMO cluster I, as well as other potentially HMO-associated clusters, were shown to be conserved among different strains of B. infantis (55, 56). Importantly, a B. infantis isolate in which the HMO cluster I was partially deleted could only weakly utilize HMO, suggesting a correlation of the presence of the HMO cluster I with vigorous growth on HMO (57). These observations also confirmed the genetic and functional divergence from B. longum strains, which lacked several of these HMO-related clusters.
Several observations suggest that B. infantis imports and degrades HMO intracellularly. For example, glycolytic enzymes potentially active on HMOs encoded within this microorganism lack signal peptide sequences, indicating a cytoplasmic localization. In addition, the B. infantis genome contains several family 1 solute-binding proteins (SBPs; pfam01547) with predicted affinity for oligosaccharides, suggesting a link to HMO transport. Recently Garrido et al. (58) determined that 10 of 20 SBPs encoded by B. infantis exhibit a binding preference for prominent mammalian glycans. The affinities of these SBPs covered great part of the spectrum of HMO linkages, including type 1 and 2 HMO (Table 1), also matching the substrates that B. infantis is able to consume in vitro (51). Genes encoding SBPs that bind type 1 and 2 chains were specifically expressed during growth on HMO (Table 1), but not on fructooligosaccharides and galactooligosaccharides (58). The consumption of specific HMO such as lacto-N-tetraose (LNT), lacto-N-neotetraose, and certain fucosylated HMO by B. infantis was recently observed, and it was suggested that ABC importers are associated with their import (59).
Table 1.
Genes in Bifidobacterium infantis upregulated during bacterial growth on HMO, associated with its consumption1
| HMO transport | |
| Blon_2344-Blon_2347 | Import of type 2 HMO, such as lacto-N-neotetraose or LacNAc containing oligosaccharides; also bind glycans found in colonic mucins |
| Blon_2350-Blon_2351 | Import of galacto-N-biose |
| Blon_2177 | Import of lacto-N-tetraose and other type 1 HMO; constitutive expression |
| Blon_0883 | Import of lacto-N-biose, galacto-N-biose,and certain fucosylated blood sugar oligosaccharides. |
| Glycosyl hydrolases | |
| Blon_2348 | Exo α-sialidase, active on α2–3/6 linkages |
| Blon_2355 | β-hexosaminidase; active on GlcNAcβ1–3Gal linkages |
| Blon_0732-Blon_0459 | β-hexosaminidase; active on GlcNAcβ1–3/6Gal linkages |
| Blon_2016 | β-galactosidase specific for type 1 HMO (Galβ1–3GlcNAc); constitutive expression |
| Blon_2334 | β-galactosidase specific for type 2 HMO (Galβ1–4GlcNAc); constitutive expression |
| Blon_2335 | α-fucosidase, with preference for Fucα1–2 linkages found in HMO but also active on Fucα1–3/4 |
| Blon_2336 | α-fucosidase, specific for Fucα1–3/4 linkages found in HMO |
Gal, galactose; GlcNAc, HMO, human milk oligosaccharides; -acetylglucosamine; LacNAc.
The enzymatic deconstruction of HMO within B. infantis appears to occur sequentially via an array of glycosyl hydrolases (60), and some of them have been associated to HMO consumption given their gene expression patterns (Table 1). The ability to consume sialylated HMO such as sialyl-LNT is likely mediated by Blon_2348, one of two α-sialidase genes in B. infantis. Only Blon_2348 is upregulated during growth on HMO, and the encoded enzyme more effectively cleaves both α2–3 and α2–6 linkages found in acidic HMO compared with Blon_0646 (61). Fucosylated HMO are highly abundant in breast milk, and glycoprofiling of HMO consumption revealed that B. infantis readily utilizes lacto-N-fucopentaoses and lacto-N-difucohexaoses, however, only after LNT is consumed first. Two α-fucosidases encoded in the HMO cluster I, Blon_2335 and Blon_2336, are expressed during growth on HMO, and both release fucose from 2′- and 3′-fucosyllactose, Lewis a, Lewis x, and fucosylated HMO such as lacto-N-fucopentaose I and III (62). Other fucosidases in the B. infantis genome, albeit showing high kinetic rates and activity on certain HMO species, did not show induction during bacterial growth on HMO.
In addition, two β-galactosidases, Blon_2016 and Blon_2334, were recently shown to be active on type 1 and 2 HMO linkages, respectively (63). The genes encoding these enzymes showed similar gene expression levels between glucose and HMO (63). Finally, two β-hexosaminidases in B. infantis are constitutively expressed during growth on HMO and lactose. Blon_2355 seems to be specific for linear GlcNAcβ1–3Gal linkages, whereas Blon_0732 and Blon_0459 can additionally release GlcNAc from branched HMO characterized by GlcNAcβ1-6Gal (64). Together, these observations indicate that glycosyl hydrolases in B. infantis are expressed during grown on HMO and can cleave all the different linkages found in these molecules. A summary of the molecular determinants within B. infantis associated to HMO import and deconstruction is presented in Table 1.
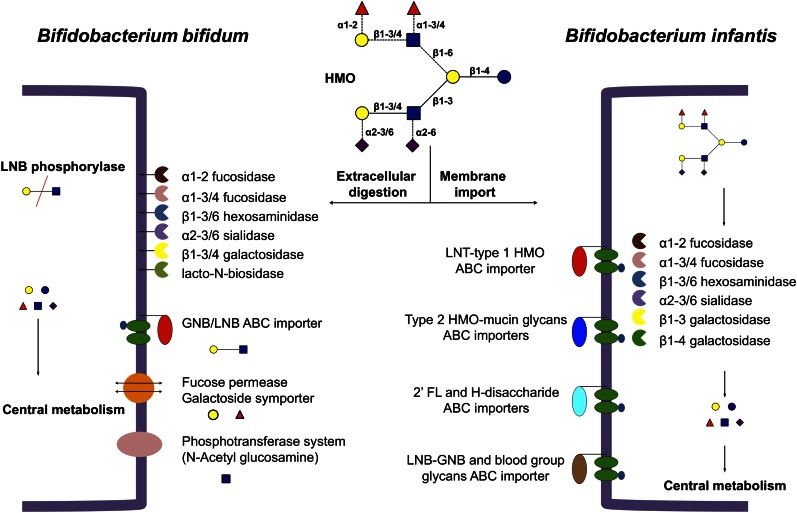
The utilization of HMO by B. bifidum has also been described in detail (59). This infant-borne bacterium possesses a wide array of extracellular glycosyl hydrolases that can cleave linkages found in HMO and mucin oligosaccharides (54, 65, 66). A single F1SBP is thought to participate in the import of LNB and galacto-N-biose, as well as other importers that can import monosaccharides (67). The induction of several of these enzymes has been also determined during growth in vitro on HMO or pig mucin (54). HMO consumption by B. bifidum is different from that of B. infantis in that it deploys many extracellular glycosyl hydrolases to digest HMO into components, some of which are consumed, whereas others, such as fucose, are left behind (50). It remains to be determined whether these two different HMO consumption strategies undertaken by these two species enable colonization of different niches within the infant colon. A comparison of HMO consumption between these two species is presented in Figure 1.
Figure 1.
Possible strategies for human milk oligosaccharides (HMO) consumption in Bifidobacterium bifidum and Bifidobacterium infantis. Yellow circles indicate galactose; red triangles, fucose; blue squares, GlcNAc; purple diamonds, blue circles, glucose. Dashed lines in the HMO figure represent potential linkages. GNB, galacto-N-biose; LNB, lacto-N-biose.
B. longum, as recently described, is routinely found in both infant and adult microbiota (53, 55, 68, 69). However, unlike B. bifidum and B. infantis, the number of enzymes and transporters involved in the metabolism of HMO in B. longum appears to be limited. A membrane-associated endo-N-acetylgalactosaminidase has been described in certain B. longum strains, suggesting possible mucin oligosaccharide release (70, 71). B. longum, as well as several infant-associated bifidobacteria, possesses a gene cluster dedicated to the metabolism of LNB and galacto-N-biose, linking type 1 HMO and mucin oligosaccharide consumption (55, 67).
If the enrichment of bifidobacteria in the infant colon is the result of coevolution of specific bifidobacteria and milk components, one might predict that the host-microbe interface is similarly influenced by milk components. Numerous researchers have demonstrated a beneficial impact of bifidobacterial probiotics on the host in both animal models (72) and human studies (73–75). Recently Fukuda et al. (76) demonstrated that production of acetate, a main end product of bifidobacterial metabolism, is a protective factor modulating intestinal permeability in a mouse model. In that work, production of acetate by certain bifidobacterial strains was linked to specific sugar transporters, suggesting that select sugar consumption is a driving factor for protective colonization of the host.
If milk glycans evolved as a selective substrate for specific bifidobacterial strains commonly found in infants, it is tempting to speculate that the resultant acetate production by those infant-borne bacteria is 1 mechanism by which milk-driven enrichment of a bifidobacteria protects the infant. However, other HMO-induced protective interfaces are at play as well. Recently, Chichlowski et al. (77) demonstrated that growth on HMO increases intestinal cell binding and enhances protective modulation of tight junction proteins and cytokines. In aggregate, these results advance a concept of a unique relationship between milk glycans, enrichment of specific bifidobacteria, and protection of the infant host.
Although a full mechanistic understanding of how specific human milk components promote infant growth, development, and protection remains elusive, the application of new approaches in analytical chemistry, glycobiology, and genomics have advanced our understanding tremendously. It is now clear that specific structural elements of milk oligosaccharides are crucial for their ability to selectively enrich beneficial bifidobacteria while inhibiting or acting as poor growth substrates for undesirable and pathogenic bacteria. Moreover, the genetic and enzymatic determinants that enable specific bifidobacteria to deconstruct and grow on these unique substrates are increasingly being identified and characterized, providing an emerging mechanistic picture of this enrichment. The consequences of this enrichment for the infant, however, are still relatively unclear. It can be predicted that systems biology tools such as metabolomics and next-generation sequencing of intestinal metagenomes or transcriptomes will help to identify, at a more global level, how breast milk is selective for beneficial microbes, how the target microbes respond to this stimulus, and how they interface with the host. Moreover, it is likely that these approaches will help in the design of more specific nutritional formulations, primed to drive enrichment in the infant gut of specific bacterial strains with a proven and understood health benefit.
Acknowledgments
All authors read and approved the final manuscript.
Footnotes
Published in a supplement to Advances in Nutrition. Presented at the conference “The First International Conference on Glycobiology of Human Milk Oligosaccharides” held in Copenhagen, Denmark, May 16–17, 2011. The conference was supported by Glycom A/S, Lyngby, Denmark. The supplement coordinators for this supplement were Clemens Kunz, University of Giessen, Germany, and Sharon M. Donovan, University of Illinois, Urbana, IL. Supplement Coordinator disclosures: Sharon Donovan has received human milk oligosaccharides from Glycom through their HMO biology donations program. Clemens Kunz has received human milk oligosaccharides from Glycom through their HMO biology donation program. The supplement is the responsibility of the Guest Editor to whom the Editor of Advances in Nutrition has delegated supervision of both technical conformity to the published regulations of Advances in Nutrition and general oversight of the scientific merit of each article. The Guest Editor for this supplement was Mark A. McGuire, University of Idaho. Guest Editor disclosure: Mark A. McGuire consults with Abbott Nutrition, a division of Abbott Laboratories, and provides professional opinions on matters related to human lactation and infant nutrition. Further, he collaborates with Dr. Lars Bode, University of California, San Diego, in research related to human milk oligosaccharides. The opinions expressed in this publication are those of the authors and are not attributable to the sponsors or the publisher, Editor, or Editorial Board of Advances in Nutrition.
Supported by the University of California Discovery Grant Program, the California Dairy Research Foundation and National Institutes of Health awards R01HD059127, R01HD065122, R01HD061923, and R21AT006180.
Author disclosures: D. Garrido, D. Barile, and D.A. Mills, no conflicts of interest.
Abbreviations used: Gal, galactose; HMO, human milk oligosaccharides; GlcNAc, N-acetylglucosamine; LNB, lacto-N-biose; LNT, lacto-N-tetraose; SBP, solute-binding protein.
Literature Cited
- 1.German JB, Freeman SL, Lebrilla CB, Mills DA. Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:205–18, discussion 18–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuthbertson WF. Evolution of infant nutrition. Br J Nutr. 1999;81:359–71 [PubMed] [Google Scholar]
- 3.Kramer MS, Kakuma R. Optimal duration of exclusive breastfeeding. Cochrane Database Syst Rev. 2002; (1):CD003517. [DOI] [PubMed] [Google Scholar]
- 4.Fewtrell MS, Morgan JB, Duggan C, Gunnlaugsson G, Hibberd PL, Lucas A, Kleinman RE. Optimal duration of exclusive breastfeeding: what is the evidence to support current recommendations? Am J Clin Nutr. 2007;85:635S–8S [DOI] [PubMed] [Google Scholar]
- 5.Walker A. Breast milk as the gold standard for protective nutrients. J Pediatr. 2010; 156(2, Suppl)S3–7 [DOI] [PubMed] [Google Scholar]
- 6.Kent JC. How breastfeeding works. J Midwifery Womens Health. 2007;52:564–70 [DOI] [PubMed] [Google Scholar]
- 7.Neville MC, Allen JC, Archer PC, Casey CE, Seacat J, Keller RP, Lutes V, Rasbach J, Neifert M. Studies in human lactation: milk volume and nutrient composition during weaning and lactogenesis. Am J Clin Nutr. 1991;54:81–92 [DOI] [PubMed] [Google Scholar]
- 8.Allen JC, Keller RP, Archer P, Neville MC. Studies in human lactation: milk composition and daily secretion rates of macronutrients in the first year of lactation. Am J Clin Nutr. 1991;54:69–80 [DOI] [PubMed] [Google Scholar]
- 9.Walker WA. Mead Johnson Symposium: functional proteins in human milk: role in infant health and development. J Pediatr. 2010; 156(2, Suppl)S1–2 [DOI] [PubMed] [Google Scholar]
- 10.Vidal K, Labeta MO, Schiffrin EJ, Donnet-Hughes A. Soluble CD14 in human breast milk and its role in innate immune responses. Acta Odontol Scand. 2001;59:330–4 [DOI] [PubMed] [Google Scholar]
- 11.Lönnerdal B. Nutritional roles of lactoferrin. Curr Opin Clin Nutr Metab Care. 2009;12:293–7 [DOI] [PubMed] [Google Scholar]
- 12.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722 [DOI] [PubMed] [Google Scholar]
- 13.Tao N, Wu S, Kim J, An HJ, Hinde K, Power ML, Gagneux P, German JB, Lebrilla CB. Evolutionary glycomics: characterization of milk oligosaccharides in primates. J Proteome Res. 2011;10:1548–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaturvedi P, Warren CD, Altaye M, Morrow AL, Ruiz-Palacios G, Pickering LK, Newburg DS. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology. 2001;11:365–72 [DOI] [PubMed] [Google Scholar]
- 15.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71:1589–96 [DOI] [PubMed] [Google Scholar]
- 16.Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. Human milk oligosaccharides are minimally digested in vitro. J Nutr. 2000;130:3014–20 [DOI] [PubMed] [Google Scholar]
- 17.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–20 [DOI] [PubMed] [Google Scholar]
- 18.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58 [DOI] [PubMed] [Google Scholar]
- 19.Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, Farkas T, Chaturvedi P, Pickering LK, Newburg DS. Human milk oligosaccharide blood group epitopes and innate immune protection against campylobacter and calicivirus diarrhea in breastfed infants. Adv Exp Med Biol. 2004;554:443–6 [DOI] [PubMed] [Google Scholar]
- 20.Coppa GV, Zampini L, Galeazzi T, Facinelli B, Ferrante L, Capretti R, Orazio G. Human milk oligosaccharides inhibit the adhesion to Caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr Res. 2006;59:377–82 [DOI] [PubMed] [Google Scholar]
- 21.Imberty A, Varrot A. Microbial recognition of human cell surface glycoconjugates. Curr Opin Struct Biol. 2008;18:567–76 [DOI] [PubMed] [Google Scholar]
- 22.Gopal PK, Gill HS. Oligosaccharides and glycoconjugates in bovine milk and colostrum. Br J Nutr. 2000;84: Suppl 1:S69–74 [DOI] [PubMed] [Google Scholar]
- 23.Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–80 [DOI] [PubMed] [Google Scholar]
- 24.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Huërou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23:23–36 [DOI] [PubMed] [Google Scholar]
- 26.Conroy ME, Shi HN, Walker WA. The long-term health effects of neonatal microbial flora. Curr Opin Allergy Clin Immunol. 2009;9:197–201 [DOI] [PubMed] [Google Scholar]
- 27.Tannock GW, Fuller R, Pedersen K. Lactobacillus succession in the piglet digestive tract demonstrated by plasmid profiling. Appl Environ Microbiol. 1990;56:1310–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumiya Y, Kato N, Watanabe K, Kato H. Molecular epidemiological study of vertical transmission of vaginal Lactobacillus species from mothers to newborn infants in Japanese, by arbitrarily primed polymerase chain reaction. J Infect Chemother. 2002;8:43–9 [DOI] [PubMed] [Google Scholar]
- 29.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino H, Kushiro A, Ishikawa E, Muylaert D, Kubota H, Sakai T, Oishi K, Martin R, Ben Amor K, Oozeer R, et al. Transmission of Intestinal Bifidobacterium longum subsp. longum Strains from Mother to Infant, Determined by Multilocus Sequencing Typing and Amplified Fragment Length Polymorphism. Appl Environ Microbiol. 2011;77:6788–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martín R, Langa S, Reviriego C, Jiminez E, Marin ML, Xaus J, Fernandez L, Rodriguez JM. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143:754–8 [DOI] [PubMed] [Google Scholar]
- 32.Solís G, de Los Reyes-Gavilan CG, Fernandez N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16:307–10 [DOI] [PubMed] [Google Scholar]
- 33.Grönlund MM, Gueimonde M, Laitinen K, Kociubinski G, Gronroos T, Salminen S, Isolauri E. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy. 2007;37:1764–72 [DOI] [PubMed] [Google Scholar]
- 34.Collado MC, Delgado S, Maldonado A, Rodriguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol. 2009;48:523–8 [DOI] [PubMed] [Google Scholar]
- 35.Martirosian G, Kuipers S, Verbrugh H, van Belkum A, Meisel-Mikolajczyk F. PCR ribotyping and arbitrarily primed PCR for typing strains of Clostridium difficile from a Polish maternity hospital. J Clin Microbiol. 1995;33:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adlerberth I, Lindberg E, Aberg N, Hesselmar B, Saalman R, Strannegard IL, Wold AE. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59:96–101 [DOI] [PubMed] [Google Scholar]
- 37.Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A. Fecal microflora of Greek healthy neonates. Anaerobe. 2008;14:94–101 [DOI] [PubMed] [Google Scholar]
- 38.Boesten R, Schuren F, Ben Amor K, Haarman M, Knol J, de Vos WM. Bifidobacterium population analysis in the infant gut by direct mapping of genomic hybridization patterns: potential for monitoring temporal development and effects of dietary regimens. Microb Biotechnol. 2011;4:417–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Cai W, Feng Y. Development of intestinal bifidobacteria and lactobacilli in breast-fed neonates. Clin Nutr. 2007;26:559–66 [DOI] [PubMed] [Google Scholar]
- 40.Koletzko B. Innovations in infant milk feeding: from the past to the future. Nestle Nutr Workshop Ser Pediatr Program. 2010;66:1–17 [DOI] [PubMed] [Google Scholar]
- 41.Magne F, Hachelaf W, Suau A, Boudraa G, Mangin I, Touhami M, Bouziane-Nedjadi K, Pochart P. A longitudinal study of infant faecal microbiota during weaning. FEMS Microbiol Ecol. 2006;58:563–71 [DOI] [PubMed] [Google Scholar]
- 42.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–7 [DOI] [PubMed] [Google Scholar]
- 43.Sakata S, Tonooka T, Ishizeki S, Takada M, Sakamoto M, Fukuyama M, Benno Y. Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol Lett. 2005;243:417–23 [DOI] [PubMed] [Google Scholar]
- 44.Rinne MM, Gueimonde M, Kalliomaki M, Hoppu U, Salminen SJ, Isolauri E. Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota. FEMS Immunol Med Microbiol. 2005;43:59–65 [DOI] [PubMed] [Google Scholar]
- 45.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology. 2010;156:3329–41 [DOI] [PubMed] [Google Scholar]
- 46.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, Cagnasso P, Bizzarri B, de'Angelis GL, Shanahan F, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moro E. Morphologische und biologische Untersuchung ueber die Darmbakterien des Siiuglings. Jahrb Kinderh. 1905;61:687–734 [Google Scholar]
- 48.Gyorgy P, Norris RF, Rose CS. Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch Biochem Biophys. 1954;48:193–201 [DOI] [PubMed] [Google Scholar]
- 49.Lee JH, O'Sullivan DJ. Genomic insights into bifidobacteria. Microbiol Mol Biol Rev. 2010;74:378–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51:1398–405 [DOI] [PubMed] [Google Scholar]
- 51.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–9 [DOI] [PubMed] [Google Scholar]
- 52.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A. 2002;99:14422–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107:19514–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76:7373–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108: Suppl 1:4653–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Locascio RG, Ninonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol. 2009;2:333–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS ONE. 2011;6:e17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of the consumption of human milk oligosaccharides by infant-gut associated bifidobacteria. J Biol Chem. 2011. Aug 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286:11909–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–8 [DOI] [PubMed] [Google Scholar]
- 64.Garrido D, Ruiz-Moyano S, Mills DA. Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H, Yamamoto K. Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology. 2010;20:1402–9 [DOI] [PubMed] [Google Scholar]
- 66.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71:3158–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parche S, Amon J, Jankovic I, Rezzonico E, Beleut M, Barutcu H, Schendel I, Eddy MP, Burkovski A, Arigoni F, et al. Sugar transport systems of Bifidobacterium longum NCC2705. J Mol Microbiol Biotechnol. 2007;12:9–19 [DOI] [PubMed] [Google Scholar]
- 69.Hinz SW, Pastink MI, van den Broek LA, Vincken JP, Voragen AG. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl Environ Microbiol. 2005;71:5501–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol. 2008;74:1936–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ashida H, Maki R, Ozawa H, Tani Y, Kiyohara M, Fujita M, Imamura A, Ishida H, Kiso M, Yamamoto K. Characterization of two different endo-alpha-N-acetylgalactosaminidases from probiotic and pathogenic enterobacteria, Bifidobacterium longum and Clostridium perfringens. Glycobiology. 2008;18:727–34 [DOI] [PubMed] [Google Scholar]
- 72.McCarthy J, O'Mahony L, O'Callaghan L, Sheil B, Vaughan EE, Fitzsimons N, Fitzgibbon J, O'Sullivan GC, Kiely B, Collins JK, et al. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Isolauri E, Arvola T, Sutas Y, Moilanen E, Salminen S. Probiotics in the management of atopic eczema. Clin Exp Allergy. 2000;30:1604–10 [DOI] [PubMed] [Google Scholar]
- 74.Saavedra JM, Bauman NA, Oung I, Perman JA, Yolken RH. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet. 1994;344:1046–9 [DOI] [PubMed] [Google Scholar]
- 75.Saavedra JM. Use of probiotics in pediatrics: rationale, mechanisms of action, and practical aspects. Nutr Clin Pract. 2007;22:351–65 [DOI] [PubMed] [Google Scholar]
- 76.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7 [DOI] [PubMed] [Google Scholar]
- 77.Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastroenterol Nutr. 2012; In press [DOI] [PMC free article] [PubMed] [Google Scholar]