Abstract
Intestinal colonization of bifidobacteria is important for the health of infants. Human milk oligosaccharides (HMO) have been identified as growth factors for bifidobacteria. Recently, a bifidobacterial enzymatic system to metabolize HMO was identified. 1,3-β-Galactosyl-N-acetylhexosamine phosphorylase (GLNBP, EC 2.4.1.211), which catalyzes the reversible phosphorolysis of galacto-N-biose (GNB) (Galβ1→3GalNAc)] and lacto-N-biose I (LNB) (Galβ1→3GlcNAc), is a key enzyme to explain the metabolism of HMO. Infant-type bifidobacteria possess the intracellular pathway to specifically metabolize GNB and LNB (GNB/LNB pathway). Bifidobacterium bifidum possesses extracellular enzymes to liberate LNB from HMO. However, Bifidobacterium longum subsp. infantis imports intact HMO to be hydrolyzed by intracellular enzymes. Bifidobacterial enzymes related to the metabolism of HMO are useful tools for preparing compounds related to HMO. For instance, LNB and GNB were produced from sucrose and GlcNAc/GalNAc in 1 pot using 4 bifidobacterial enzymes, including GLNBP. LNB is expected to be a selective bifidus factor for infant-type strains.
Introduction
Bifidobacterium is the genus of gram-positive anaerobic bacteria. Some of its species are typical inhabitants in the human gut. Their intestinal growth is considered to be beneficial for health, and they are often used as probiotics.
Until the early 20th century, formula-fed infants often had pathogenic bacterial infections that caused diarrhea and other infectious diseases (1). Bifidobacteria were first identified in 1899 by Tissier (2) from the feces of a breast-fed infant. Shortly after this discovery, it was found that bifidobacteria were rarely isolated from feces of formula-fed infants; however, they were the predominant intestinal bacteria in breast-fed infants. Intestinal colonization of bifidobacteria is considered to prevent the intestinal growth of pathogenic bacteria and maintain health in breast-fed infants (1).
Investigations have been conducted since then to identify growth-stimulating factors for bifidobacteria (bifidus factor) present in human milk. In 1950s, a GlcNAc-containing sugar was assumed to be the bifidus factor, but subsequent studies revealed that the originators of this idea were misled because of the use of a particular GlcNAc-auxotrophic strain of Bifidobacterium bifidum var. pennsylvanicus (3, 4). Human milk oligosaccharides (HMO)4 have been considered as the most promising candidates for the real bifidus factor in human milk since then (1).
The predominant carbohydrate in human milk is lactose, which is found at a concentration of 60–70 g/L. Human milk also contains HMO possessing degrees of polymerization that are ≥3 at a concentration of 22–24 g/L in colostrum and 12–13 g/L in mature milk (5, 6). Lactose is digested by lactase in the small intestine and serves as a nutrient for infants, but HMO are not digestible by intestinal enzymes and reach the large intestine intact, where they are potentially used by bifidobacteria.
Later research revealed that various nondigestible oligosaccharides acted as bifidus factors, and in the early 1980s, prebiotic oligosaccharides were developed in Japan (7). These oligosaccharides reach the large intestine intact and are used there by bifidobacteria to enhance their intestinal growth. Modern milk formulas are supplemented with prebiotic oligosaccharides such as fructooligosaccharides, galactooligosaccharides, and lactulose to allow the growth of intestinal bifidobacteria, and infant health with regard to infectious diseases improves. However, the intestinal microbiota of bottle-fed infants is different from that of breast-fed infants, with a larger population of Enterobacteriaceae (8). Thus, an understanding of the bifidus factor in human milk is still necessary.
The complex composition of HMO, which consists of >130 different oligosaccharides (9, 10), has made it difficult to understand how HMO acts as a bifidus factor (9, 10). Each component of HMO can be assigned 1 of 12 core structures with or without the modifications of fucosylation and/or sialylation (9, 10). All the core structures possess a lactose unit at the reducing end. Core structures are classified into types I and II based on the disaccharide unit at their nonreducing ends, i.e., containing lacto-N-biose I (LNB) (Galβ1→3GlcNAc) and N-acetyllactosamine (Galβ1→4GlcNAc) structures, respectively. Type I oligosaccharides are predominant in HMO, and type II oligosaccharides are minor components. 2′-Fucosyllactose (Fucα1→2Galβ1→4Glc), lacto-N-tetraose (LNT) (Galβ1→3GlcNAcβ1→3Galβ1→4Glc), and lacto-N-fucopentaose I (Fucα1→2Galβ1→3GlcNAcβ1→3Galβ1→4Glc) are the most abundant components in HMO (11), and the latter 2 are type I oligosaccharides. Such type I predominance in milk oligosaccharides is a specific feature of milk in humans and not in other mammals including anthropoids (12). The Galβ1→3GlcNAc bond in LNB is resistant to most β-galactosidases (13–15). Thus, it is important to discover how bifidobacteria digest this bond to adequately understand the prebiotic effect of HMO.
Two species, B. bifidum and Bifidobacterium longum subsp. infantis can grow in a medium with HMO as the sole carbon source (16–18). In this review, bifidobacterial enzymes related to the cleavage of the core structures of HMO as well as the release of fucosyl and sialyl residues are discussed, especially of the 2 species.
Current status of knowledge
The intracellular galacto-N-biose/LNB pathway
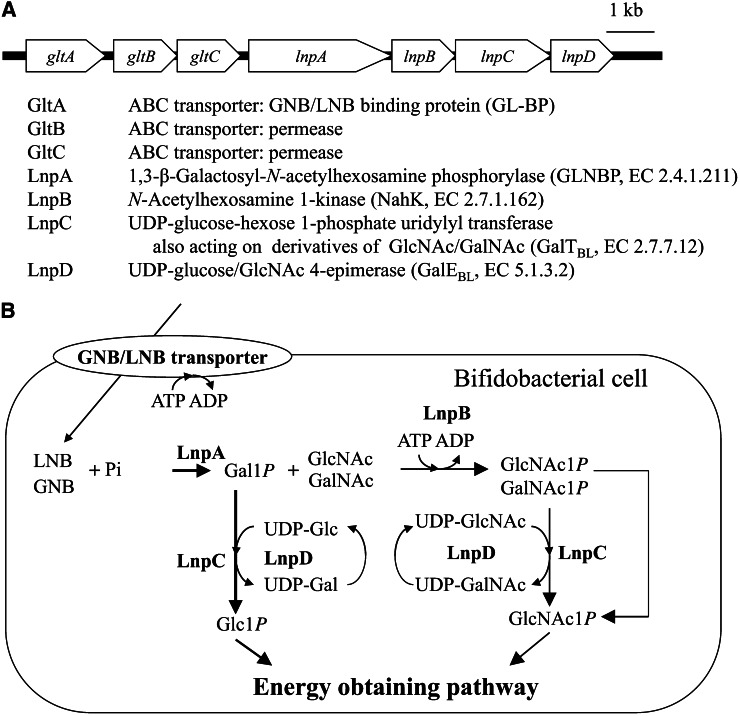
Since the discovery of 1,3-β-galactosyl-N-acetylhexosamine phosphorylase (19) in the cell-free extract of B. bifidum, the intracellular sugar-metabolic pathway specific to galacto-N-biose (GNB) (Galβ1→3GalNAc) and LNB, the GNB/LNB pathway (20, 21), has been identified from several species of bifidobacteria. The genes encoding the components of the pathway are clustered as shown in Figure 1. As explained in the following, the pathway fully describes the metabolism of GNB and LNB.
Figure 1.
The galacto-N-biose (GNB)/lacto-N-biose I (LNB) pathway found in bifidobacteria. A, The gene cluster encoding the pathway. B, Schematic representation of the pathway. GalNAc1P, α-GalNAc 1-phosphate; Gal1P, α-galactose 1-phosphate; GlcNAc1P, α-GlcNAc 1-phosphate; Glc1P, α-glucose 1-phosphate.
GNB/LNB-specific transporter.
The upstream genes gltA–C encode components of an ATP-binding cassette sugar transporter. A prediction was made that gltA would encode a solute-binding protein that determines the specificity of the transporter. Research showed that the GltA protein [named GNB/LNB binding protein (GL-BP)] of B. longum subsp. longum JCM1217 strongly bound GNB (Kd = 10 nM) and LNB (Kd = 87 nM), but did not bind with LacNAc, indicating that gltA–C encodes the specific GNB/LNB transporter that allows intake of GNB and LNB generated outside the cell (22, 23). GL-BP also showed the capacity to bind to LNT with a higher Kd value (11 μM), suggesting that LNT may also be transported in the absence of GNB and LNB. The residues involved in binding GNB and LNB were identified through a structural analysis of GL-BP (22). Genes encoding for proteins sharing >30% identity with GL-BP have not been found in genomic sequences of intestinal bacteria other than bifidobacteria.
GNB/LNB phosphorylase.
1,3-β-Galactosyl-N-acetylhexosamine phosphorylase phosphorolyzes GNB and LNB into α-galactose 1-phosphate (Gal1P) and the corresponding HexNAc (GalNAc and GlcNAc, respectively). It is categorized into 3 groups based on the activity on GNB and LNB: GNB phosphorylase (specific to GNB) (24–26), LNB phosphorylase (LNBP, specific to LNB) (27), and GNB/LNB phosphorylase (GLNBP, acting on both GNB and LNB equally) (19, 26, 28–31). The bifidobacterial enzymes are members of GLNBP, and lnpA (Fig. 1) encodes GLNBP.
In the CAZy database (32), GLNBP is classified into the glycoside hydrolase family (GH) 112 (33) as well as GNB phosphorylase, LNBP, and 1,4-β-D-galactosyl-L-rhamnose phosphorylase (29, 34), based on structural similarity with β-galactosidase GH42 (35). Mutational analysis of the GLNBP of B. longum subsp. longum revealed the residues that determined the substrate specificities of GH112 enzymes (36).
The presence of a phosphorylase often suggests that the substrate sugar plays an important role for the energy source especially for anaerobic bacteria (37). Because the phosphorolytic reaction by phosphorylase directly produces a phosphorylated sugar without consuming ATP, the energy obtained from the substrate sugar is considerably higher than that from other sugars, especially under anaerobic conditions in which only 3 molecules of ATP are available via the glycolytic pathway from glucose 6-phosphate.
N-Acetylhexosamine 1-kinase.
The LnpB protein was shown to produce α-GlcNAc 1-phosphate (GlcNAc1P) from GlcNAc and ATP, indicating that LnpB is a novel anomeric kinase (20). This protein had similar activity on GalNAc, yielding α-GalNAc 1-phosphate (GalNAc1P), and weak activities on several monosaccharides. The enzyme was named N-acetylhexosamine 1-kinase, with a new EC number, EC 2.7.1.162. LnpB does not have a signal peptide sequence and is considered to be an intracellular enzyme.
Most kinases acting on hexoses phosphorylate their substrates at the sixth position. Three enzymes—galactokinase (EC 2.7.1.6), N-acetylgalactosamine kinase (EC 2.7.1.157), and fucokinase (EC 2.7.1.52)—are known to phosphorylate the anomeric hydroxyl group. An anomeric kinase on a gluco-type sugar had not been reported before N-acetylhexosamine 1-kinase, which has been used for preparing various analogs of GlcNAc1P and GalNAc1P (38, 39).
UDP-glucose-hexose 1-phosphate uridylyl transferase (GalT) and UDP-glucose 4-epimerase (GalE).
LnpC and LnpD were predicted to be a GalT (EC 2.7.7.12) and GalE (EC 5.1.3.2), respectively, based on their amino acid sequences. GalT and GalE are the enzymes involved in the Leloir pathway for galactose metabolism (40). They act in concert to transfer Gal1P to α-glucose 1-phosphate (Glc1P), enabling them to enter the glycolytic pathway. Because Gal1P is generated from GNB and LNB through phosphorolysis by GLNBP, the presence of GalT and GalE suggested that the galactose part of GNB and LNB is transformed into Glc1P, which is then sent to the energy-obtaining pathway within the enzymes encoded in the gene cluster.
GalT and GalE activities were proven experimentally with LnpC and LnpD, and both proteins possessed additional activities (20). LnpC transfers the UMP unit of UDP-Glc to both GlcNAc1P and GalNAc1P. LnpC is a novel type of GalT, having wide substrate specificity, as no other GalT has been found to recognize GlcNAc1P or GalNAc1P (20). LnpC was named GalTBL. LnpD demonstrated UDP-GlcNAc 4-epimerase activity as well as UDP-glucose 4-epimerase and was named GalEBL (20). These activities explain the transformation of GalNAc1P into GlcNAc1P by way of the Leloir pathway. Because GalNAc must be transformed to GlcNAc before entering an energy-obtaining pathway, the enzymes encoded by lnpB-D explain the metabolism of the HexNAc part of GNB and LNB.
Distribution of the GNB/LNB pathway.
The distribution of the GNB/LNB pathway was examined with various species of bifidobacteria by detecting the presence of GLNBP (41). All strains of B. longum subsp. longum, B. longum subsp. infantis, Bifidobacterium. breve, and B. bifidum possess GLNBP. It should be noted that the 4 species are often isolated from infant feces. However, GLNBP has not been found in any strains of Bifidobacterium adolescentis and Bifidobacterium catenulatum, the major bifidobacterial species in the adult intestine.
It is valuable to find the origins of GNB and LNB in the human intestine to understand the role of the GNB/LNB pathway in the intestinal growth of bifidobacteria. GNB exists as a structural component of O-linked glycoproteins in mucous membranes. An enzyme that liberates GNB (endo-α-N-acetylgalactosaminidase) was found in B. longum subsp. longum (42, 43). In case of LNB, HMO are the serious candidate for the source of LNB because it would be in agreement with the fact that the infant type of bifidobacteria possess the GNB/LNB pathway. The presence of the LNB structure in HMO explains the predominant growth of bifidobacteria if they possess an extracellular enzymatic system to liberate LNB from HMO (LNB hypothesis) (28).
Extracellular enzymes hydrolyzing HMO isolated from B. bifidum
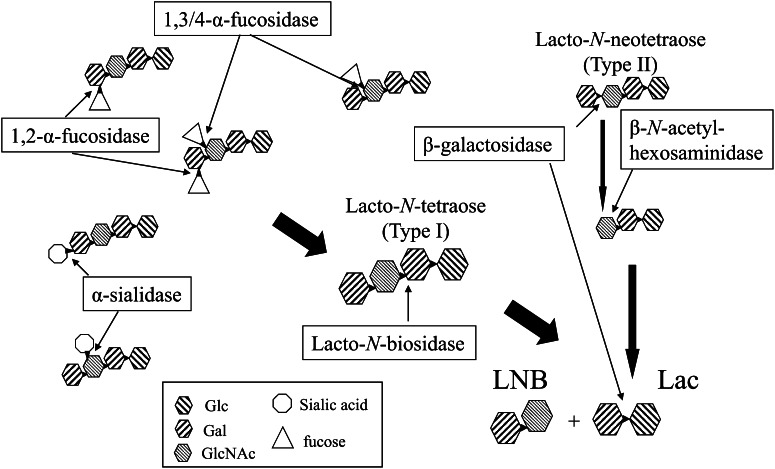
An extracellular enzymatic system to liberate LNB from HMO was elucidated from B. bifidum JCM1254. Seven enzymes are involved in the extracellular hydrolyses. The possible pathway is illustrated in Figure 2. Two α-fucosidases are involved in the cleavage of HMO. One is 1,2-α-fucosidase belonging to GH95 (AfcA) (44, 45), which hydrolyzes the fucosyl unit bound to the second position of the galactosyl residue at the nonreducing end of HMO. The specificity was also confirmed with the selective syntheses of 1,2-α-fucoside catalyzed by a glycosynthase mutant of AfcA (46). The other is 1,3/4-α-fucosidase belonging to GH29 (AfcB) (47), which hydrolyzes the fucosyl unit neighboring the β-linked galactose. The enzyme hydrolyzes 1,4-α-fucoside in type I sugar and 1,3-α-fucoside in type II sugar. Two 2,3/6-α-sialidases belonging to GH33 (SiaBB1 and SiaBB2) (48, 49) were also isolated. Because major modifications in the core structures of HMO are α-1,2/3/4-fucosylation and α-2,3/6-sialylation, the presence of these enzymes is beneficial for bifidobacteria to remove the branched sugar from the core structure of HMO.
Figure 2.
Pathway of the extracellular enzymatic hydrolysis of HMO by Bifidobacterium bifidum. Lac, lactose; LNB, lacto-N-biose I.
Extracellular lacto-N-biosidase belonging to GH20 (LnbB) (50) was also isolated from B. bifidum. The LnbB exhibited 38% amino acid identity to the lacto-N-biosidase protein from Streptomyces sp. (51, 52), which was the only lacto-N-biosidase isolated before LnbB. The presence of LnbB indicates that B. bifidum produces LNB from HMO by the extracellular enzymes. Furthermore, a β-galactosidase belonging to GH2 (Bbg3) and a β-N-acetylglucosaminidase (Bbh1) belonging to GH20 were shown to digest type II sugars (53). It should be noted that Bbg3 hydrolyzes neither LNB nor LNT (53).
All the enzymes listed possess a membrane-anchoring motif at the C-terminal, suggesting that they are cell-bound enzymes. Genes encoding these enzymes are also found in the genomic sequences of 2 B. bifidum strains (54, 55). They may be advantageous for the use of HMO by the B. bifidum that hydrolyzes HMO on the surface of the cells.
Metabolism of HMO in B. longum subsp. infantis
The genomic sequence of B. longum subsp. infantis, a consumer of HMO, revealed that the strain possesses a 43-kb gene cluster specific for HMO degradation that is not found in other bifidobacterial species (56, 57). In the cluster, homologous genes of 1,2-α-fucosidase, 1,3/4-α-fucosidase, and 2,3/6-α-sialidase are present in addition to the genes for β-galactosidase and β-N-acetylhexosaminidase, but they are considered to be intracellular proteins because of the lack of signal peptides. The cluster includes genes encoding several sugar transporters that are considered to import HMO intact (58). Although the strain also possesses the GNB/LNB pathway, neither a gene predicted to encode lacto-N-biosidase (56) nor lacto-N-biosidase activity (50) has been identified. The β-galactosidase in the gene cluster (Bga2A) hydrolyzed type II sugars, but did not hydrolyze type I sugars. Yoshida et al. (59) and Garrido et al. (60) independently identified an intracellular β-galactosidase belonging to GH42 (Bga42A) that hydrolyzed type I sugars. Bga42A hydrolyzes LNT 40 times more effectively than LNB (59). It is concluded that B. longum subsp. infantis incorporates HMO intact and then degrades them exowisely from the nonreducing ends without using the GNB/LNB pathway.
Practical preparation of LNB using GLNBP
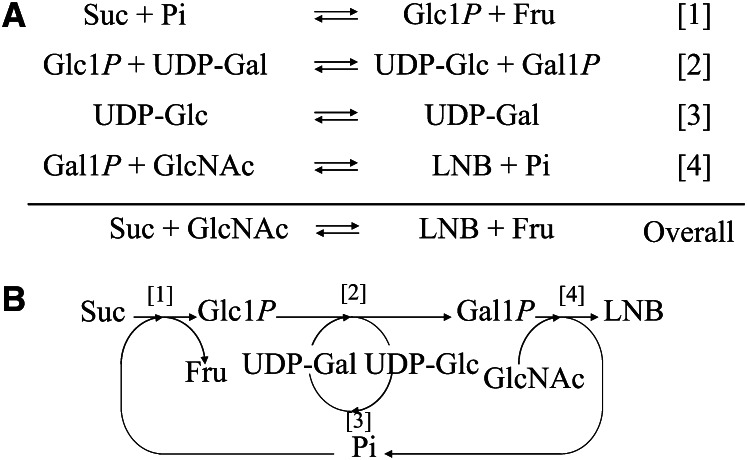
The presence of the GNB/LNB pathway in infant-type bifidobacteria suggests that LNB acts as a promoting factor for the growth of bifidobacteria. A practical preparation method was reported (61) that uses a 1-pot enzymatic reaction to produce LNB and is summarized in Figure 3. In this method, sucrose and GlcNAc are converted into LNB and fructose by the concerted reactions of 4 enzymes—sucrose phosphorylase (EC 2.4.1.7), GalT, GalE, and GLNBP—in the presence of catalytic amounts of UDP-glucose and inorganic phosphate. Sucrose is phosphorolyzed to Glc1P and fructose by sucrose phosphorylase, and then Glc1P is converted to UDP-Glc concomitantly with the conversion of UDP-Gal into Gal1P by GalT. Then UDP-Glc is converted to UDP-Gal by GalE, and the resulting UDP-Gal is consumed in the reaction of GalT. Finally, LNB is synthesized from Gal1P and GlcNAc by GalHexNAcP.
Figure 3.
One-pot enzymatic production of lacto-N-biose I (LNB). A, Principle of the reaction. Four enzymatic reactions are carried out in 1 pot, and the compounds appearing on both sides are recycled in the overall reaction. Thus, the overall reaction is to produce LNB and fructose from sucrose and GlcNAc in the presence of 4 enzymes and catalytic amounts of UDP-Glc and Pi. B, Schematic description of the reaction. Catalyzed by sucrose phosphorylase [1], DP-glucose-hexose 1-phosphate uridylyl transferase (GalT) [2], UDP-glucose 4-epimerase (GalE) [3], and Galacto-N-biose/LNB phosphorylase (GLNBP) [4]. Gal1P, α-galactose 1-phosphate; Glc1P, α-glucose 1-phosphate.
Starting from a 10-L reaction mixture consisting of 660 mmol/Lsucrose and 600 mmol/L GlcNAc, the concentration of LNB produced reached 500 mmol/L (190 g/L). The yield was 83% based on the amount of GlcNAc used. LNB was isolated by crystallization after fermentation of the reaction mixture by yeast to remove the resultant fructose and unreacted sucrose to obtain 1.8 kg of crystalline LNB (95% purity). Recrystallization of LNB yielded 1.4 kg of crystalline LNB (99.6% purity). This method was carried out with unit processes that were ready for scaling up. The process for the isolation of LNB does not require any chromatography step, which is often difficult to scale up. This method can be easily used in the production of GNB by substituting GlcNAc for GalNAc (62, 63).
LNB as a growth-promoting factor
Because large-scale production of LNB is feasible, experiments that require large amounts of LNB can easily be conducted. Kiyohara et al. (64) examined the in vitro growth-promoting activity of LNB in various intestinal bacteria. LNB showed such activity in several bifidobacteria, including B. bifidum, B. breve, B. longum subsp. longum, B. longum subsp. infantis, and Bifidobacterium scardovii, but not in other strains of bifidobacteria or in other intestinal bacteria. Xiao et al. (41) examined the effect of LNB on 203 strains of bifidobacteria and found that LNB promoted the growth of bifidobacterial strains possessing the GNB/LNB pathway except for strains of Bifidobacterium pseudocatenulatum. All the infant-type strains of bifidobacteria showed growths on LNB, suggesting that LNB is a possible candidate to be used as a prebiotic for certain strains of bifidobacteria.
Conclusions
Discovery of the GNB/LNB pathway opens up the possibility to systematically investigate HMO metabolism in bifidobacteria. The GNB/LNB pathway was found to be distributed in infant-type bifidobacterial species such as B. longum subsp. longum (65–68), B. longum subsp. infantis (56, 69), B. bifidum (54, 55), and B. breve (70).
The extracellular enzymatic system of B. bifidum that digests HMO to generate LNB has been fully identified. However, the enzymatic system of B. longum subsp. infantis suggests that the strain uptake of intact HMO followed by intracellular exowise hydrolyses does not use the GNB/LNB pathway in the metabolism of HMO. All the bifidobacterial enzymes related to the metabolism of HMO have finally been identified, as listed in Table 1.
TABLE 1.
Location of the enzymes related to the metabolism in HMO by Bifidobacterium bifidum JCM1254 and Bifidobacterium longum subsp. infantis ATCC156971
| Enzyme | Family | B. bifidum | B. longum subsp. infantis |
| 1,2-α-Fucosidase | GH95 | Cell bound | Cytosol |
| 1,3/4-α-Fucosidase | GH29 | Cell bound | Cytosol |
| Sialidase | GH33 | Cell bound | Cytosol |
| Lacto-N-biosidase | GH20 | Cell bound | Does not exist |
| β-Galactosidase | GH2 | Cell bound | Cytosol |
| β-N-acetylhexosaminidase | GH20 | Cell bound | Cytosol |
| GNB/LNB transporter | Transmembrane | Transmembrane | |
| GLNBP | GH112 | Cytosol | Cytosol |
| GNB/LNB pathway | Cytosol | Cytosol | |
| Transporters in HMO cluster | Does not exist | Transmembrane | |
| LNT β-galactosidase | GH42 | Cytosol | Cytosol |
GLNBP, galacto- -biose/lacto-N-biose I phosphorylase; GNB/LNB, galacto-N-biose/lacto-N-biose I; HMO, human milk oligosaccharides; LNT, lacto-N-tetraose.
It had often been reported that B. longum subsp. infantis ferments HMO in vitro but B. bifidum does not (17, 18), even though the species possesses the enzymes to use HMO. A recent study has revealed that the result is probably because of a deficiency of the GNB/LNB transporter in the type strain of B. bifidum, and that other strains of B. bifidum were able to ferment HMO (16). The consumption pattern of HMO by B. longum subsp. infantis and B. bifidum reflect their pathways. B. longum subsp. infantis consumed HMO without the accumulation of any oligosaccharides as the intermediate (16). B. bifidum temporarily accumulates disaccharides such as lactose and LNB in the medium during the fermentation (16).
It should be noted that B. breve is the species most abundantly isolated from infant feces (8, 71). However, it has been reported that B. breve is incapable of fermenting HMO in vitro, and fucosidase, sialidase, and lacto-N-biosidase have not been isolated from B. breve. Some mutualism should be assumed to understand the growth of B. breve in the gut of breast-fed infants (72). B. bifidum may be a candidate for mutualism because it accumulates LNB to be consumed by other bacteria (16).
LNB is expected to function as a specific prebiotic for infant-type bifidobacteria that possess the GNB/LNB pathway. The production of LNB at an industrial scale may allow its use as a functional food ingredient and, perhaps, as a supplement for infant formula milk.
Acknowledgments
The sole author had responsibility for all parts of the manuscript.
Footnotes
Published in a supplement to Advances in Nutrition. Presented at the conference “The First International Conference on Glycobiology of Human Milk Oligosaccharides” held in Copenhagen, Denmark, May 16–17, 2011. The conference was supported by Glycom A/S, Lyngby, Denmark. The supplement coordinators for this supplement were Clemens Kunz, University of Giessen, Germany, and Sharon M. Donovan, University of Illinois, Urbana, IL. Supplement Coordinator disclosures: Sharon Donovan has received human milk oligosaccharides from Glycom through their HMO biology donations program. Clemens Kunz has received human milk oligosaccharides from Glycom through their HMO biology donation program. The supplement is the responsibility of the Guest Editor to whom the Editor of Advances in Nutrition has delegated supervision of both technical conformity to the published regulations of Advances in Nutrition and general oversight of the scientific merit of each article. The Guest Editor for this supplement was Mark A. McGuire, University of Idaho. Guest Editor disclosure: Mark A. McGuire consults with Abbott Nutrition, a division of Abbott Laboratories, and provides professional opinions on matters related to human lactation and infant nutrition. Further, he collaborates with Dr. Lars Bode, University of California, San Diego, in research related to human milk oligosaccharides. The opinions expressed in this publication are those of the authors and are not attributable to the sponsors or the publisher, Editor, or Editorial Board of Advances in Nutrition.
Supported by a grant from the Promotion of Basic Research Activities for Innovative Biosciences of Japan.
Author disclosures: M. Kitaoka, no conflicts of interest.
Abbreviations used: Gal1P, α-galactose 1-phosphate; GalE, UDP-glucose 4-epimerase; GalNAc1P, α-GalNAc 1-phosphate; GalT, UDP-glucose-hexose 1-phosphate uridylyl transferase; GH, glycoside hydrolase family; GL-BP, galacto-N-biose/lacto-N-biose binding protein; GlcNAc1P, α-GlcNAc 1-phosphate; GLNBP, galacto-N-biose/lacto-N-biose I phosphorylase; GNB, galacto-N-biose; HMO, human milk oligosaccharides; LNB, lacto-N-biose I; LNT, lacto-N-tetraose.
Literature Cited
- 1.Bezkorovainy A. Ecology of bifidobacteria. In: Bezkorovainy A, Miller-Catchpole R, editors. Biochemistry and physiology of bifidobacteria. Cleveland: CRC Press; 1989. p. 29–72.
- 2.Tissier H. Recherches sur la flore intestinale normale et pathologique du nourisson. Thesis. Paris: University of Paris; 1900.
- 3.Gyorgy P, Rose CS, Springer GF. Enzymatic inactivation of bifidus factor and blood group substances. J Lab Clin Med. 1954;43:543–52 [PubMed] [Google Scholar]
- 4.Veerkamp JH. Uptake and metabolism of determinatives of 2-deoxy-2-amino-D-glucose in Bifidobacterium bifidum var. pennsylvanicus. Arch Biochem Biophys. 1969;129:248–56 [DOI] [PubMed] [Google Scholar]
- 5.Newburg DS, Neubauer SH. Carbohydrate in milks: analysis, quantities, and significance. : Jensen RG, editor Handbook of Milk Composition. San Diego: Academic Press; 1995. p. 273–349 [Google Scholar]
- 6.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. In: McCormick DB, Bier DM, Cousins RJ, editors. Annual Review of Nutrition. Palo Alto: Annual Reviews; 2000. p. 699–722. [DOI] [PubMed]
- 7.Nakakuki T. Present status and future prospects of functional oligosaccharide development in Japan. Trends Glycosci Glycotechnol. 2005;52:267–71 [Google Scholar]
- 8.Benno Y, Sawada K, Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28:975–86 [DOI] [PubMed] [Google Scholar]
- 9.Kobata A. Structures and application of oligosaccharides in human milk. Proc Jpn Acad, Ser B, Phys Biol Sci. 2010;86:731–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urashima T, Fukuda K, Kitaoka M, Ohnishi M, Terabayashi T, Kobata A. Milk oligosaccharides. In: Gordon NS, editor. Oligosaccharides: Sources, Properties and Applications. New York: Nova Biochemical Books; 2011. p. 183–203.
- 11.Asakuma S, Urashima T, Akahori M, Obayashi H, Nakamura T, Kimura K, Watanabe Y, Arai I, Sanai Y. Variation of major neutral oligosaccharides levels in human colostrum. Eur J Clin Nutr. 2008;62:488–94 [DOI] [PubMed] [Google Scholar]
- 12.Urashima T, Odaka G, Asakuma S, Uemura Y, Goto K, Senda A, Saito T, Fukuda K, Messer M, Oftedal OT. Chemical characterization of oligosaccharides in chimpanzee, bonobo, gorilla, orangutan, and siamang milk or colostrum. Glycobiology. 2009;19:499–508 [DOI] [PubMed] [Google Scholar]
- 13.Goulas T, Goulas A, Tzortzis G, Gibson GR. Expression of four beta-galactosidases from Bifidobacterium bifidum NCIMB41171 and their contribution on the hydrolysis and synthesis of galactooligosaccharides. Appl Microbiol Biotechnol. 2009;84:899–907 [DOI] [PubMed] [Google Scholar]
- 14.Goulas T, Goulas A, Tzortzis G, Gibson GR. Comparative analysis of four β-galactosidases from Bifidobacterium bifidum NCIMB41171: purification and biochemical characterisation. Appl Microbiol Biotechnol. 2009;82:1079–88 [DOI] [PubMed] [Google Scholar]
- 15.Goulas TK, Goulas AK, Tzortzis G, Gibson GR. Molecular cloning and comparative analysis of four β-galactosidase genes from Bifidobacterium bifidum NCIMB41171. Appl Microbiol Biotechnol. 2007;76:1365–72 [DOI] [PubMed] [Google Scholar]
- 16.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286:34583–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51:1398–405 [DOI] [PubMed] [Google Scholar]
- 19.Derensy-Dron D, Krzewinski F, Brassart C, Bouquelet S. β-1,3-Galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol Appl Biochem. 1999;29:3–10 [PubMed] [Google Scholar]
- 20.Nishimoto M, Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl Environ Microbiol. 2007;73:6444–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fushinobu S. Unique sugar metabolic pathways of bifidobacteria. Biosci Biotechnol Biochem. 2010;74:2374–84 [DOI] [PubMed] [Google Scholar]
- 22.Suzuki R, Wada J, Katayama T, Fushinobu S, Wakagi T, Shoun H, Sugimoto H, Tanaka A, Kumagai H, Ashida H, et al. Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J Biol Chem. 2008;283:13165–73 [DOI] [PubMed] [Google Scholar]
- 23.Wada J, Suzuki R, Fushinobu S, Kitaoka M, Wakagi T, Shoun H, Ashida H, Kumagai H, Katayama T, Yamamoto K. Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:751–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakajima M, Nihira T, Nishimoto M, Kitaoka M. Identification of galacto-N-biose phosphorylase from Clostridium perfringens ATCC13124. Appl Microbiol Biotechnol. 2008;78:465–71 [DOI] [PubMed] [Google Scholar]
- 25.Nakajima M, Nishimoto M, Kitaoka M. Characterization of β-1,3-galactosyl-N-acetylhexosamine phosphorylase from Propionibacterium acnes. Appl Microbiol Biotechnol. 2009;83:109–15 [DOI] [PubMed] [Google Scholar]
- 26.Chao C, Wim S, Tom D. Characterization of β-galactoside phosphorylases with diverging acceptor specificities. Enzyme Microb Technol. 2011;49:59–65 [DOI] [PubMed] [Google Scholar]
- 27.Nakajima M, Kitaoka M. Identification of lacto-N-Biose I phosphorylase from Vibrio vulnificus CMCP6. Appl Environ Microbiol. 2008;74:6333–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71:3158–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakajima M, Nishimoto M, Kitaoka M. Characterization of three β-galactoside phosphorylases from Clostridium phytofermentans: discovery of D-galactosyl-beta1→4-L-rhamnose phosphorylase. J Biol Chem. 2009;284:19220–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimoto M, Kitaoka M. Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211). Biosci Biotechnol Biochem. 2007;71:1587–91 [DOI] [PubMed] [Google Scholar]
- 31.Yu H, Thon V, Lau K, Cai L, Chen Y, Mu S, Li Y, Wang PG, Chen X. Highly efficient chemoenzymatic synthesis of β1–3-linked galactosides. Chem Commun (Camb). 2010;46:7507–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hidaka M, Nishimoto M, Kitaoka M, Wakagi T, Shoun H, Fushinobu S. The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: a large deformation of a TIM barrel scaffold. J Biol Chem. 2009;284:7273–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakajima M, Nishimoto M, Kitaoka M. Characterization of D-galactosyl-β1→4-L-rhamnose phosphorylase from Opitutus terrae. Enzyme Microb Technol. 2010;46:315–9 [Google Scholar]
- 35.Hidaka M, Fushinobu S, Ohtsu N, Motoshima H, Matsuzawa H, Shoun H, Wakagi T. Trimeric crystal structure of the glycoside hydrolase family 42 β-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J Mol Biol. 2002;322:79–91 [DOI] [PubMed] [Google Scholar]
- 36.Nishimoto M, Hidaka M, Nakajima M, Fushinobu S, Kitaoka M. Identification of amino acid residues determining substrate preference of 1,3-β-galactosyl-N-acetylhexosamine phosphorylase. J Mol Catal, B Enzym. 2012;74:97–102 [Google Scholar]
- 37.Kitaoka M, Hayashi K. Carbohydrate processing phosphorolytic enzymes. Trends Glycosci Glycotechnol. 2002;14:35–50 [Google Scholar]
- 38.Cai L, Guan W, Kitaoka M, Shen J, Xia C, Chen W, Wang PG. A chemoenzymatic route to N-acetylglucosamine-1-phosphate analogues: substrate specificity investigations of N-acetylhexosamine 1-kinase. Chem Commun (Camb). 2009;2944–6 [DOI] [PubMed] [Google Scholar]
- 39.Cai L, Guan W, Wang W, Zhao W, Kitaoka M, Shen J, O'Neil C, Wang PG. Substrate specificity of N-acetylhexosamine kinase towards N-acetylgalactosamine derivatives. Bioorg Med Chem Lett. 2009;19:5433–5 [DOI] [PubMed] [Google Scholar]
- 40.Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–8 [DOI] [PubMed] [Google Scholar]
- 41.Xiao JZ, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K, Kitaoka M. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. 2010;76:54–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita K, Oura F, Nagamine N, Katayama T, Hiratake J, Sakata K, Kumagai H, Yamamoto K. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem. 2005;280:37415–22 [DOI] [PubMed] [Google Scholar]
- 43.Suzuki R, Katayama T, Kitaoka M, Kumagai H, Wakagi T, Shoun H, Ashida H, Yamamoto K, Fushinobu S. Crystallographic and mutational analyses of substrate recognition of endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J Biochem. 2009;146:389–98 [DOI] [PubMed] [Google Scholar]
- 44.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol. 2004;186:4885–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagae M, Tsuchiya A, Katayama T, Yamamoto K, Wakatsuki S, Kato R. Structural basis of the catalytic reaction mechanism of novel 1,2-α-L-fucosidase from Bifidobacterium bifidum. J Biol Chem. 2007;282:18497–509 [DOI] [PubMed] [Google Scholar]
- 46.Wada J, Honda Y, Nagae M, Kato R, Wakatsuki S, Katayama T, Taniguchi H, Kumagai H, Kitaoka M, Yamamoto K. 1,2-α-L-Fucosynthase: a glycosynthase derived from an inverting α-glycosidase with an unusual reaction mechanism. FEBS Lett. 2008;582:3739–43 [DOI] [PubMed] [Google Scholar]
- 47.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–7 [DOI] [PubMed] [Google Scholar]
- 48.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2011;21:437–47 [DOI] [PubMed] [Google Scholar]
- 49.Katayama T, Fujita K, Yamamoto K. Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J Biosci Bioeng. 2005;99:457–65 [DOI] [PubMed] [Google Scholar]
- 50.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sano M, Hayakawa K, Kato I. Purification and characterization of an enzyme releasing lacto-N-biose from oligosaccharides with type 1 chain. J Biol Chem. 1993;268:18560–6 [PubMed] [Google Scholar]
- 52.Sano M, Hayakawa K, Kato I. An enzyme releasing lacto-N-biose from oligosaccharides. Proc Natl Acad Sci U S A. 1992;89:8512–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H, Yamamoto K. Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology. 2010;20:1402–9 [DOI] [PubMed] [Google Scholar]
- 54.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107:19514–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhurina D, Zomer A, Gleinser M, Brancaccio VF, Auchter M, Waidmann MS, Westermann C, van Sinderen D, Riedel CU. Complete genome sequence of Bifidobacterium bifidum S17. J Bacteriol. 2011;193:301–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108:4653–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS ONE. 2011;6:e17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–8 [DOI] [PubMed] [Google Scholar]
- 60.Garrido D, Sela D, Wu S, Jimenez-Espinoza R, Eom H-J, German J, Block D, Lebrilla C, Mills D. Characterization of glycosyl hydrolases in Bifidobacterium longum subsp. infantis active on human milk oligosaccharides. Book of Abstracts of 1st International Conference on the Glycobiology of Human Milk Oligosaccharides. 2011. p.17.
- 61.Nishimoto M, Kitaoka M. Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci Biotechnol Biochem. 2007;71:2101–4 [DOI] [PubMed] [Google Scholar]
- 62.Nishimoto M, Kitaoka M. One-pot enzymatic production of β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-D-galactose (galacto-N-biose) from sucrose and 2-acetamido-2-deoxy-D-galactose (N-acetylgalactosamine). Carbohydr Res. 2009;344:2573–6 [DOI] [PubMed] [Google Scholar]
- 63.Inoue K, Nishimoto M, Kitaoka M. One-pot enzymatic production of 2-acetamido-2-deoxy-D-galactose (GalNAc) from 2-acetamido-2-deoxy-D-glucose (GlcNAc). Carbohydr Res. 2011;346:2432–6 [DOI] [PubMed] [Google Scholar]
- 64.Kiyohara M, Tachizawa A, Nishimoto M, Kitaoka M, Ashida H, Yamamoto K. Prebiotic effect of lacto-N-biose I on bifidobacterial growth. Biosci Biotechnol Biochem. 2009;73:1175–9 [DOI] [PubMed] [Google Scholar]
- 65.Hao Y, Huang D, Guo H, Xiao M, An H, Zhao L, Zuo F, Zhang B, Hu S, Song S, et al. Complete genome sequence of Bifidobacterium longum subsp. longum BBMN68, a new strain from a healthy chinese centenarian. J Bacteriol. 2011;193:787–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A. 2002;99:14422–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei YX, Zhang ZY, Liu C, Zhu YZ, Zhu YQ, Zheng H, Zhao GP, Wang S, Guo XK. Complete genome sequence of Bifidobacterium longum JDM301. J Bacteriol. 2010;192:4076–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad Conservation of Milk Utilization Genes in Bifidobacterium longum subsp infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76:7373–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A. 2011;108:11217–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]