Abstract
Modifying the sense strand of nuclease-resistant siRNA with 3’-cholesterol (Chol-*siRNA) increases mRNA suppression after i.v. administration but with relatively low efficacy. We previously found evidence in vitro that suggests complexation of Chol-siRNA with PLL-PEG(5K), a block copolymer of poly-L-lysine and 5 kDa polyethylene glycol, may increase the efficacy of Chol-siRNA in vivo in a PLL block length-dependent manner. In this study, the extent that polyplexes of PLL10-PEG(5K), PLL30-PEG(5K), and PLL50-PEG(5K) protect complexed Chol-siRNA in high concentrations of murine serum and affect the activity of Chol-*siRNA in murine 4T1 breast tumor epithelial cells in vitro and in primary orthotopic tumors of 4T1 was compared. PLL-PEG(5K) required 3’-Chol to protect full-length siRNA from nuclease degradation in 90% (v/v) murine serum and protection was increased by increasing PLL block length and nuclease resistance of Chol-siRNA. Polyplexes of Chol-*siLuc suppressed stably expressed luciferase in 4T1-Luc cells to different levels in vitro where PLL30>PLL50>PLL10. In contrast, only polyplexes of Chol-*siLuc and PLL30-PEG(5K) or PLL50-PEG(5K) suppressed high levels of luciferase in primary orthotopic tumors of 4T1-Luc after i.v. administration, whereas polyplexes of Chol-*siLuc and PLL10-PEG(5K), inactive Chol-*siCtrl polyplexes of PLL-PEG(5K), or Chol-*siLuc alone had no detectable activity. As a whole, these results indicate that polyplexes of PLL-PEG(5K) increase the efficacy of nuclease-resistant Chol-siRNA in primary breast tumors after i.v. administration in a PLL block length-dependent manner. Thus, complexation of Chol-siRNA with PLL-PEG(5K) may be a promising approach to increase the efficacy of Chol-siRNA in a wide range of primary tumors, metastases, and other tissues but likely requires a PLL block length that balances polymer-related adverse effects, Chol-siRNA bioavailability, and subsequent activity in the target cell.
Keywords: siRNA delivery, drug delivery, RNAi, RNA interference, tumor delivery, systemic delivery, gene therapy, siRNA polyplexes, polymer siRNA complexes
1. Introduction
Small interfering RNA (siRNA) is a naturally occurring dsRNA molecule (21–23 nucleotides with 2 nucleotide overhangs on 3’ end of the sense and antisense strands) that inhibits protein expression through the sequence-specific degradation of target mRNA [1, 2]. As such, siRNA has tremendous potential in the treatment of medical conditions such as cancer where the suppression of a single or multiple proteins can produce a therapeutic effect [3].
Despite major advances in the administration of siRNA to primary tumors through local delivery [3], systemic delivery of siRNA is still required to treat diffuse or inaccessible primary tumors as well as distal metastases. The efficacy of siRNA after i.v. administration, however, is limited by its relatively short plasma half-life (0.03 h) [4], minimal cellular uptake, and inability to escape the endosomes / lysosomes into its site of action, the cytosol [5–7].
Modifying the sense strand of nuclease-resistant siRNA with 3’-cholesterol (Chol-siRNA) increases that activity of siRNA in the liver and jejunum after i.v. administration but requires a relatively high dose of Chol-siRNA to achieve a therapeutic effect (50 mg/kg) [8]. We previously found that forming a polymer complex (polyplex) between Chol-siRNA and PLL-PEG(5K), a block copolymer of poly-L-lysine and 5 kDa poly(ethylene glycol), protects a dsDNA model of Chol-siRNA against DNase activity as well as increases the efficacy of mRNA suppression by Chol-siRNA in primary murine MVEC in vitro [9]. Furthermore, increasing the PLL block length of PLL-PEG(5K) from 10 to 50 increases protection of complexed model siRNA against nuclease activity but decreases siRNA activity in conditionally immortalized murine mammary MVEC [9]. Thus, we hypothesized that Chol-siRNA polyplexes of PLL-PEG(5K) can increase the efficacy of Chol-siRNA after i.v. administration in a PLL block length-dependent manner. To test this hypothesis, the extent that polyplexes of PLL10-PEG(5K), PLL30-PEG(5K), and PLL50-PEG(5K) protect complexed Chol-siRNA in high concentrations of murine serum and affect the activity of Chol-siRNA against stably expressed luciferase in murine breast tumor epithelial cells (4T1-Luc) in vitro and in primary orthotopic tumors of 4T1-Luc after i.v. administration was compared in this study.
2. Materials and methods
2.1 Polymer
PLL-PEG(5K): Block copolymers of methoxy-poly(ethylene glycol)-b-poly(L-lysine hydrochloride) with 5 kDa polyethylene glycol (PEG) and PLL blocks of 10 (PLL10-PEG(5K); MW: 6,600 Da ), 30 (PLL30-PEG(5K); MW: 9,900 Da ), or 50 (PLL50-PEG(5K); MW: 13,200 Da) poly-L-lysine groups and were purchased from Alamanda Polymers (Huntsville, AL). The polydispersity index of each polymer was between 1 and 1.1.
2.2. siRNA
All siRNA (Thermo Fisher Scientific Biosciences, Waltham, MA) were resuspended in provided siRNA buffer according to the manufacturer’s instructions and stored in aliquots at −80°C. siRNA and Chol-siRNA: siRNA were 19 bp with 3’-UU overhangs on the sense and antisense strands. siCtrl (Murine non-targeting siRNA, D-001810-01: 5’- UGG UUU ACA UGU CGA CUA A - 3’); siLuc (Custom anti-luciferase siRNA generated against CpG-free Luc::Sh (InvivoGen) with the Dharmacon siDESIGN center), 5’- AGA AGG AGA UUG UGG ACU A - 3’); Chol-siCtrl (siCtrl modified with 3’-cholesterol on the sense strand through a 6 carbon hydroxyproline linker and purified by standard desalting); Chol-siLuc (siLuc modified with 3’-cholesterol as described for Chol-siCtrl). Nuclease-resistant Chol-siRNA: Nuclease-resistant siRNA (designated with an asterisk) [8, 10] were 19 bp with a blunt 3’-end on the sense strand and a UU overhang on the 3’-end of the antisense strand. The sense strand was modified with 3’-cholesterol as described for Chol-siCtrl and purified by HPLC for in vivo administration. Chol-*siCtrl: sense 5’- UGG UUU ACA UGU CGA CUA A^chol - 3’, antisense 5’- U UAG UCG ACA UGU AAA CCa^(u^U) - 3’; Chol-*siLuc: sense 5’- AGA AGG AGA UUG UGG ACU A^chol - 3’; antisense 5’- U AGU CCA CAA UCU CCU UCu^(u^U) where “^” indicates phosphorothioate linkages and lower case letters indicate 2’-O-methyl modification of the ribose sugar.
2.3 Minimum N/P ratio for complexation of siRNA and Chol-siRNA with PLL-PEG(5K)
N/P molar ratios were calculated using moles PLL-PEG(5K) primary amines [PLL10-PEG(5K): 1.5 mmol 1’ amine / g polymer; PLL30-PEG(5K): 3 mmol 1’ amine / g polymer; PLL50-PEG(5K): 3.8 mmol 1’ amine / g polymer] to moles siRNA phosphates (42 mol phosphate / mol siRNA and Chol-siRNA; 40 mol phosphate / mol nuclease-resistant Chol-siRNA). Polyplexes were prepared by adding siRNA or Chol-siRNA (1.56 µM, 10 µL) in HEPES Buffer (0.1 M HEPES [pH 7.4]) to HEPES Buffer (10 µL, N/P = 0) or HEPES Buffer (10 µL) containing a concentration of PLL-PEG(5K) to provide the indicated N/P ratio, vortexing, and incubating at RT for 30 min [9]. Solutions were then were mixed with 6X DNA loading buffer (120 mg Ficoll Type 400 /mL and 0.003% xylene cyanol in dH20, 4 µL), loaded (10 µL) on a 1% TBE agarose gel (UltraPure™ Agarose-1000, Invitrogen, Grand Island, NY) containing SYBR Green II (Invitrogen) and run at 120V for 15 min. Gels were imaged under UV transillumination using a Molecular Imager® ChemiDoc™ XRS (BioRad, Hercules, CA). The first N/P ratio where polyplexes were completely retained in the well was defined as the minimum N/P ratio required for complexation. Similarities between the concentrations of siRNA and Chol-siRNA in the 1.5 µM stock solutions were confirmed by comparing band intensities of siRNA and Chol-siRNA on the same gel (N/P 0) using Quantity One® software (BioRad). All N/P ratios are representative of two independent experiments.
2.4 Hydrodynamic diameter of Chol-siRNA polyplexes
The hydrodynamic diameters of Chol-siCtrl polyplexes in 0.1 M HEPES [pH 7.4] at 1 mg polymer / mL and indicated N/P ratio were measured by Dynamic Light Scattering (DLS) using a ZetaSizer Nano ZS (Malvern Instruments, Malvern, UK) equipped with He-Ne laser (λ = 633 nm) as the incident beam. Average polyplex diameters of PLL-PEG(5K) (n=3 measurements ±SD) with Choi-model siRNA [9] and Chol-siCtrl were compared by unpaired t-test (P <0.05).
2.5 Degradation of siRNA and Chol-siRNA in serum
Murine serum (Sigma, 4.5 µL) or HEPES Buffer (0.1 M HEPES [pH 7.4], 4.5 µL) was added to siRNA or Chol-siRNA (10 µM in HEPES Buffer, 0.5 µL) and incubated at 37°C for 15 min. To quench serum nuclease activity, RNaseOUT™ (Invitrogen, 0.6 µL) was added to serum-treated samples at 4.6 U/µL serum and HEPES buffer (0.6 µL) was added to HEPES Buffer controls and incubated on ice for 15 min. To resolve Chol-siRNA from serum proteins, water soluble cholesterol (Sigma Aldrich, St. Louis, MO) was additionally added to serum-treated Chol-siRNA (1mM water soluble cholesterol in HEPES Buffer, 5 µL) at a 1:1 molar ratio water soluble cholesterol:Chol-siRNA, whereas HEPES Buffer (5 µL) was added to serum-treated siRNA and HEPES Buffer control samples then incubated on ice for 15 min. Sample volumes were adjusted to 25 µL with HEPES Buffer and 3 µL was run on a 10% TBE/polyacrylamide gel at 120V for 30 min. Gels were post-stained with SYBR Gold (Invitrogen) and imaged under UV trans-illumination using a Molecular Imager® ChemiDoc XRS™ (BioRad, Hercules, CA).
2.6 Protection of siRNA and Chol-siRNA from serum nuclease activity
Polyplexes were prepared by adding siRNA or Chol-siRNA (20 µM, 10 µL) in HEPES Buffer (0.1 M HEPES [pH 7.4]) to HEPES Buffer (10 µL) containing a concentration of PLL-PEG(5K) to provide the indicated N/P ratio and incubating at RT for 30 min. Murine serum (Sigma; 4.5 µL) or HEPES Buffer (4.5 µL) was added to polyplexes (0.5 µL) and incubated at 37°C for the indicated time. At each time point, samples were immediately submerged in liquid N2 and stored at −20°C. On the day of analysis, samples were thawed on ice and treated with RNaseOUT™ (0.6 µL) and water soluble cholesterol (5 µL) as described in Section 2.5. To displace remaining siRNA or Chol-siRNA from polyplexes, heparin (5 mg sodium heparin/mL, 5 µL) was added. Total sample volumes were adjusted to 25 µL with HEPES Buffer and 3 µL was analyzed by gel electrophoresis as described in Section 2.5. The single band of remaining siRNA or Chol-siRNA from serum-treated polyplexes was normalized to the single band of siRNA or Chol-siRNA from buffer-treated polyplexes by densitometry (Quantity One, BioRad). Percent protected siRNA or Chol-siRNA was expressed as [(average density of band from serum treated polyplexes/average density of band from corresponding buffer treated polyplexes)*100] ± SD (n=2). Protection by polyplexes at any given time-point was compared by One-way ANOVA followed by Tukey’s post-test.
2.7 Protection of complexed siRNA and Chol-siRNA from displacement in serum
PLL-PEG(5K) was complexed with siRNA or Chol-siRNA as described in Section 2.3 at the indicated minimum N/P ratio required for complete complexation. RNase activity of murine serum was inactivated by pre-incubating with RNaseOUT™ (4.6 U/µL serum) for 15 min. Duplicates for each PLL-PEG(5K) construct were prepared by adding polyplexes (10µM siRNA in HEPES buffer, 0.5 µL) to RNase-inactivated murine serum (4.5 µL) and incubating at 37° C for the indicated time. After incubation, samples were placed on ice and treated again with RNaseOUT™ (0.6 µL) and water soluble cholesterol (Chol-siRNA samples only, 5 µL) as described under Section 2.5. Heparin was then added to one of the duplicates to displace total siRNA or Chol-siRNA for normalizing the amount of released siRNA and incubated on ice for 30 min. The sample volume was adjusted to 25 µL using HEPES buffer and analyzed by gel electrophoresis as described in Section 2.5. Bands from samples without heparin were normalized to bands from the heparin treated duplicate of the same sample by densitometry (Quantity One® software).
2.8 Cell culture
A murine breast tumor epithelial cell line stably expressing firefly luciferase (4T1-Luc) (CpG-free Luc::Sh, in pCpGvitro-blasti, InvivoGen) was cultured in 10% Complete DMEM (DMEM (Invitrogen, Carlsbad, CA), FBS 10% [Atlanta Biologicals (Atlanta, GA), endotoxin <0.3 EU scale, heat-inactivated by incubation at 56°C for 30 minutes and cooling in an ice bath], 1 mM L-glutamine, 2 mM Glutamax™, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 1X vitamins, 5.0 µg/mL amphotericin B [Fungizone™], 50 µg/mL gentamycin, 100 U/mL penicillin G, 100 µg/mL streptomycin sulfate (Invitrogen, Carlsbad, CA)) and blasticidin hydrochloride (15 µg /mL, Fisher Scientific, Pittsburgh, PA) as a selection agent. To determine luciferase expression levels, sodium D-luciferin (Gold Biotechnology, St. Louis, MO) was added to serial dilutions of cells in 24-well plates at 150 µg D-luciferin/mL (500 µL) and incubated for 5 min. Luminescence from cells was measured using a Xenogen IVIS® 200 Series (Caliper Life Sciences, Hopkinton, MA) and total flux (photons/sec) was quantitated using the Living Image® software (Caliper Life Sciences). The average flux per cell for 4T1-Luc was greater than recommended for in vivo imaging (3620 vs. 500 photons/sec) [11].
2.9 Relative suppression of luciferase activity and cytotoxicity in 4T1-Luc in vitro
To confirm the activity of siLuc and Chol-siLuc, 4T1-Luc were electroporated (Nucleofector, Lonza AG, Bazel, Switzerland) with the indicated siRNA using the Cell Line Nucleofector kit V (VACA-1001, Lonza) on setting T-024 (300nM siRNA or Chol-siRNA, 5 × 106 cells/mL) according to manufacturer’s instructions and plated in 6-well plates. Luciferase activity was measured at 24 h and 48 h as described in Section 2.8 and an average radiance (photons/sec/cm2/sr) was quantitated using the Living Image® software. Luciferase activity at each time-point was expressed as [(avg. radiance from 4T1-Luc electroporated with siRNA /average radiance from 4T1-Luc electroporated without siRNA)*100] ± propagated SD (n=3).
For transfections, 4T1-Luc were seeded in 24-well plates (20,000 cells/well) in antibiotic free 10% FBS Complete DMEM medium and incubated at 37°C 14–16 h before transfection. On the day of transfection, polymers were sterilized under vacuum for 2 h in a desiccator containing a glass dish of 95% alcohol and resuspended in HEPES Buffer (0.1 M HEPES [pH 7.4]) with vortexing for 2 min. Stock solutions of siRNA, Chol-siRNA, and Chol-*siRNA (20 µM in HEPES Buffer) were diluted to 2 µM in HEPES Buffer and added to polymer solutions in equal volumes at the indicated N/P ratios as described in Section 2.3, then diluted in Complete DMEM lacking FBS and antibiotics to a final siRNA concentration of 200 nM. Diluted polyplexes were added to 4T1-Luc (250 µL) for 4 h then an equal volume of 20% FBS Complete DMEM (250 µL) was added and cells were further incubated for 20 h. For untreated cells, Complete DMEM lacking FBS and antibiotics (250 µL) was added to 4T1-Luc for 4 then an equal volume of 20% FBS Complete DMEM (250 µL) was added and cells were further incubated for 20 h. Luciferase expression was measured at 24 h by bioluminescent imaging as described in Section 2.8. Percent relative luciferase activity was expressed as [(avg. radiance from siRNA treated cells / avg. radiance from untreated cells)*100] ± propagated SD (n=3). Differences in relative luciferase activity between cells transfected with different siRNA polyplexes were compared by one way ANOVA and Tukey post-test.
Percent viability and total live cell count relative to untreated 4T1-Luc was determined at 24 h by trypan blue exclusion (Cellometer Auto T4; Nexcelom Biosciences, Lawrence, MA). Percent live cells were calculated as [(avg. total live cells treated with siRNA polyplexes / avg. total live cells without treatment) × 100] ± propagated SD. Differences in the average total percentage of live cells were compared by one-way ANOVA followed by Dunnett’s posttest vs. untreated 4T1-Luc.
2.10 Relative suppression of luciferase activity in primary mammary tumors of 4T1-Luc
All procedures were approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee. Female Balb/c mice (6–8 weeks old, NCI) were maintained under pathogen free conditions. 4T1-Luc cells (1 × 106 cells in 0.1mL sterile PBS) were injected SQ into abdominal mammary fat pad number 4 (http://tvmouse.ucdavis.edu/bcancercd/22/mouse_figure.html). Tumor volume (by calipers using the formula, a*b2/2, where ‘b’ is the shorter diameter) and body weight were measured every other day before treatment, then daily until 48 h after the last treatment. When tumors had grown for 6–7 days or reached 50–100 mm3, whichever came first (treatment Day 0), 10 mM HEPES buffer (filter-sterilized 10mM HEPES/150mM NaCl [pH 7.4], 100 µL) or the indicated Chol-siRNA formulation (2.5. mg Chol-siRNA/kg in 100 µL 10 mM HEPES buffer) was injected into the tail-vein on days 0, 1 and 2 (n=5 animals). On the day of each injection, PLL-PEG(5K) constructs were sterilized under vacuum for 2 h in a desiccator containing a glass dish of 95% alcohol and resuspended in 10 mM HEPES buffer by vortexing for 2 min. Stock solutions of siRNA (100µM) were diluted in 10 mM HEPES buffer and siRNA polyplexes were prepared at the minimum N/P ratio required for complexation as described in Section 2.3.
For bioluminescent imaging, a filter-sterilized (0.2 µm) solution of D-luciferin in PBS (30 mg/mL) was injected i.p. (100 µL) 15 min before measuring luminescence. Mice were anesthetized with an initial dose of 5% isoflurane and a maintenance dose of 1.5% isoflurane. During the course of treatment (Days 0, 1, 2), imaging was performed before i.v. injections. Bioluminescent signal from the primary 4T1-Luc tumors was quantitated using the Living Image software according to manufacturer’s instructions. Average radiance values within each cohort were normalized to the same cohort on the first day of treatment (Day 0) and percent relative luciferase activity was expressed as [(mean average radiance of cohort on a given day/mean average radiance of same cohort on Day 0)*100] ± propagated SEM. Differences in luciferase activity within each cohort were compared by Friedman non-parametric repeated measures ANOVA with Dunn’s multiple comparison test.
3. Results
3.1 Effect of PLL block length and modifying siRNA with 3’-cholesterol on the complexation of siRNA with PLL-PEG(5K)
Complexes of siRNA and polymers (siRNA polyplexes) are conventionally formed by mixing solutions of siRNA and polymer at different molar ratios of positively charged groups (amines-N) on the polymer to negatively charged groups (phosphates-P) on the siRNA (N/P ratio). N/P ratios that produce neutral / electropositive polyplexes frequently have higher stability and activity than N/P ratios that produce electronegative polyplexes [12]. Thus, any change in the minimum N/P ratio required to form neutral / electropositive siRNA polyplexes may affect the subsequent activity of complexed siRNA.
We previously found that modifying the sense strand of a model siRNA (dsDNA analog of siRNA with 3’-AT overhangs) with 3’-cholesterol and increasing the PLL block length of PLL-PEG(5K) from 10 to 50 does not affect the minimum N/P ratio required by PLL-PEG(5K) to form neutral / electropositive polyplexes (N/P = 2 for polyplexes of PLL10-PEG(5K) or PLL50-PEG(5K) and model siRNA or Chol-model siRNA) [9]. It remained unclear, however, whether the same was true for complexation of actual siRNA over the same range of PLL block lengths.
To determine whether PLL block length and modifying siRNA with 3′-cholesterol affects complexation of siRNA with PLL-PEG(5K), the minimum N/P ratios required to neutralize siRNA or Chol-siRNA by PLL-PEG(5K) with PLL block lengths of 10, 30, or 50 were compared by agarose gel electrophoresis (Table 1). Band intensities of siRNA and Chol-siRNA in the absence of PLL-PEG(5K) were statistically similar, indicating that the concentrations of siRNA and Chol-siRNA in the stock solutions were not significantly different (data not shown).
Table 1. Effect of PLL block length and modifying siRNA with 3’-cholesterol on the minimum N/P ratio required to form neutral / electropositive polyplexes of siRNA with PLL-PEG(5K).
siRNA (siCtrl: 5’- UGG UUU ACA UGU CGA CUA A - 3’ with 3’-UU overhangs) or Chol-siRNA (siCtrl modified with 3’-cholesterol on the sense strand) was mixed with PLL-PEG(5K) in 0.1 M HEPES [pH 7.4] over a range of N/P ratios, incubated at room temperature for 30 min, then run on a 1X TBE agarose/SYBR Green II gel. The first N/P ratio where polyplexes were completely retained in the well was defined as the minimum N/P ratio required for complexation. All N/P ratios are representative of two independent experiments.
| siRNA | Chol-siRNA | |||
|---|---|---|---|---|
| Polymer | N/P ratio |
Loading (wt%) |
N/P Ratio |
Loading (wt%) |
| PLL10-PEG(5K) | 7 | (6.2) | 4 | (10.4) |
| PLL30-PEG(5K) | 6 | (13.3) | 3 | (23.5) |
| pLL50-PEG(5K) | 5 | (18.8) | 2 | (36.6) |
Increasing PLL block length from 10 to 50 decreased the minimum N/P ratios required by PLL-PEG(5K) to neutralize siRNA or Chol-siRNA (Table 1). Modifying siRNA with 3’-cholesterol, however, decreased the minimum N/P ratio required by PLL-PEG(5K) at the same PLL block length to neutralize siRNA (Table 1). Thus, in contrast to our original dsDNA model of siRNA [9], increasing PLL block length and modifying the sense strand of siRNA with 3’-cholesterol collectively decrease the minimum N/P ratio required to form neutral / electropositive polyplexes between siRNA and PLL-PEG(5K) and, consequently, increase siRNA loading over the current range of PLL block lengths.
3.2 Effect of PLL block length and Chol-siRNA on polyplex hydrodynamic diameter
We previously found that the hydrodynamic diameter of polyplexes of PLL50-PEG(5K) formed with Chol-model siRNA (dsDNA analog siRNA with 3’-AT overhangs) was ∼12 nm greater than Chol-model siRNA polyplexes of PLL10-PEG(5K) (66 ± 4 (SD) vs. 54 ± 0.4 nm, P = 0.0067) [9]. This suggests that increasing PLL block length increases the hydrodynamic diameter of Chol-siRNA polyplexes of PLL-PEG(5K). It remained unclear, however, whether the same was true for polyplexes formed with actual Chol-siRNA.
To determine whether PLL block length affects the size of Chol-siRNA polyplexes of PLL-PEG(5K), the average hydrodynamic diameters of Chol-siRNA polyplexes at the minimum N/P ratios that form neutral polyplexes (Table 1) were compared by DLS (Table 2). Increasing PLL block length from 10 to 30 or 50 decreased the hydrodynamic diameters of Chol-siRNA polyplexes of PLL-PEG(5K) by 3 nm (38 ± 2 (SD) vs. 35 ± 1 nm, P = 0.04; 38 ± 2 vs. 35.0 ± 0.4 nm, P = 0.03) (Table 2). Forming polyplexes of PLL10-PEG(5K) and PLL50-PEG(5K) with Chol-siRNA instead of Chol-model siRNA also decreased the hydrodynamic diameter by 16 and 31 nm, respectively (54.0 ± 0.4 (SD) vs. 38 ± 2 nm, P <0.0001; 66 ± 4 vs. 35.0 ± 0.4 nm, P = 0.0002) (Table 2). Thus, in contrast to Chol-model siRNA polyplexes of PLL-PEG(5K), increasing PLL block length to between 10 and 30 slightly decreases the hydrodynamic diameter of Chol-siRNA polyplexes and the hydrodynamic diameters of Chol-siRNA polyplexes are less than our original Chol-model siRNA polyplexes [9] with the current N/P ratios and range of PLL block lengths.
Table 2. Comparison of hydrodynamic diameters of Chol-model siRNA and chol-siRNA polyplexes of PLL-PEG(5K).
Chol-model siRNA (19 bp dsDNA analog of siRNA with 3’-AT overhangs [9]) or Chol-siRNA (siCtrl modified with 3’-cholesterol on the sense strand) was mixed with PLL-PEG(5K) in HEPES buffer [pH 7.4] at the indicated N/P ratio required for complexation, incubated at room temperature for 30 min, and measured by DLS. Values are an average ±SD (n=3 measurements).
| Polyplex | Hydrodynamic Diameter (nm ±SD) | |
|---|---|---|
| Chol-model siRNA [9] | Chol-siRNA | |
| PLL10-PEG(5K) | 54.0 ± 0.4 (N/P 2) | 38 ± 2a (N/P 4) |
| PLL30-PEG(5K) | ND | 35 ± 1 (N/P 3) |
| PLL50-PEG)(5K) | 66 ± 4 (N/P 2) | 35.0 ± 0.4b,c (N/P 2) |
P <0.0001 or
P <0.001 vs. Chol-model siRNA polyplexes with the same PLL block length and
P <0.05 vs. Chol-siRNA polyplexes of PLL10-PEG(5K) by two-sided unpaired t-test. Chol-model siRNA diameters taken from [9].
3.3 Effect of PLL block length, modifying siRNA with 3’-cholesterol, and increasing nuclease resistance of Chol-siRNA on the protection of complexed siRNA from nuclease degradation in high concentrations of serum
Given that nuclease activity greatly decreases the plasma half-life of siRNA [13], it is important that PLL-PEG(5K) increases the duration that complexed siRNA is protected from nuclease degradation in the bloodstream to allow time for the siRNA to accumulate in the target cells. We previously found that increasing the PLL block length of PLL-PEG(5K) from 10 to 50 and modifying the sense strand of a dsDNA model of siRNA with 3’-cholesterol collectively increase the protection of complexed model siRNA from DNase 1 activity in a buffered solution [9]. It remained unclear, however, whether the same was true for the protection of actual siRNA in more physiologically relevant, high concentrations of serum.
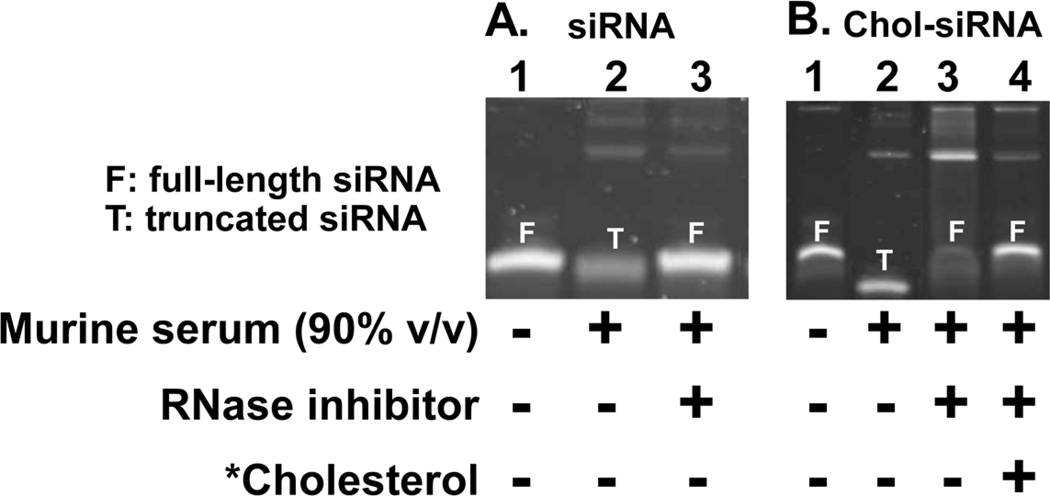
To first establish the pattern of degradation in high concentrations of serum, siRNA and Chol-siRNA were incubated in 90% (v/v) murine serum at 37°C an d compared by polyacrylamide gel electrophoresis (Fig.1). Pre-treatment of murine serum with a broad spectrum inhibitor of RNase A, B, & C [14] protected siRNA and Chol-siRNA from nuclease activity for up to two hours (data not shown) and water solubilized cholesterol (*cholesterol) effectively separated Chol-siRNA from serum proteins (Fig.1B, lane 3 vs. lane 4).
Figure 1. Pattern of free siRNA and Chol-siRNA degradation in 90% (v/v) murine serum.
(A) siRNA (siCtrl: 5’- UGG UUU ACA UGU CGA CUA A - 3’ with 3’-UU overhangs) was incubated at 37°C for 15 min in HEPES buffer (lane 1), 90% v/v m urine serum (lane 2), or 90% v/v murine serum pretreated with a broad spectrum RNase inhibitor (lane 3) then separated and imaged on a 10% polyacrylamide gel post stained with SYBR Gold. (B) Chol-siRNA (siCtrl modified with 3’-cholesterol on the sense strand) was incubated at 37°C for 15 m in in HEPES buffer (lane 1), 90% v/v murine serum (lane 2), or 90% v/v murine serum pretreated with a broad spectrum RNase inhibitor (lanes 3 & 4). HEPES buffer (lanes 1–3) or water solubilized cholesterol (*Cholesterol) at a 1:1 molar ratio cholesterol:Chol-siRNA (lane 4) was added and Chol-siRNA was separated and imaged on a 10% polyacrylamide gel post stained with SYBR Gold.
Both siRNA and Chol-siRNA were undetectable within an hour of incubation in murine serum (data not shown). At an earlier time point (∼15 minutes), however, a single, faster migrating band of siRNA (Fig.1A, lane 2) or Chol-siRNA (Fig.1B, lane 2) was observed that only migrated the same distance as full-length siRNA or Chol-siRNA in buffer after pretreating the serum with a broad spectrum RNase inhibitor (Fig.1A, lane 1 vs. 3 and Fig.1B, lane 1 vs. 4). Thus, the faster migrating band of siRNA or Chol-siRNA is due to truncation by serum RNase activity and not a direct effect of serum on the electrophoresis of siRNA or Chol-siRNA.
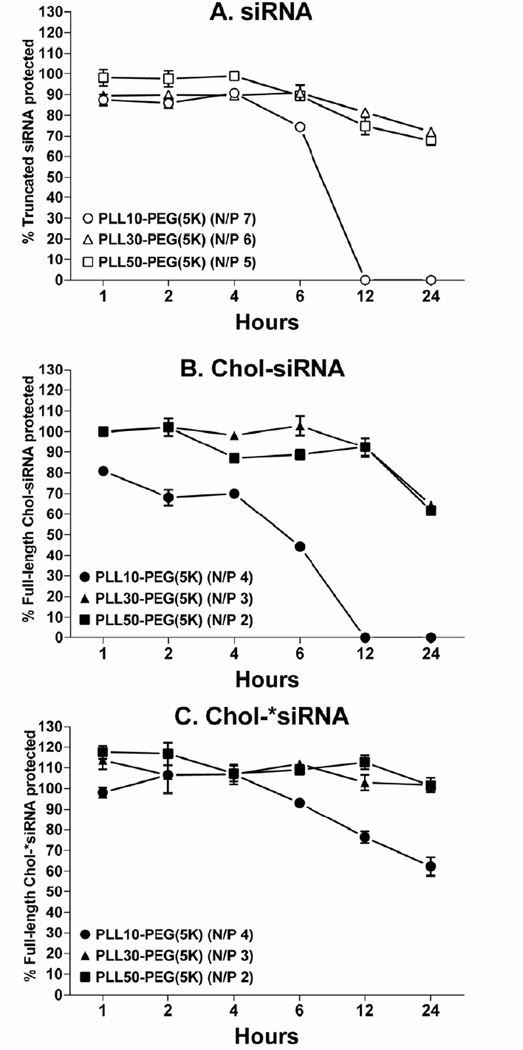
To next determine whether PLL block length and modifying siRNA with 3’-cholesterol affects the ability of PLL-PEG(5K) to protect siRNA from nuclease degradation in high concentrations of serum, PLL-PEG(5K) was complexed with siRNA or Chol-siRNA at the minimum N/P ratios that form neutral polyplexes (Table 1) and protection against nuclease degradation in 90% (v/v) murine serum at 37°C over 24 h was compared by polyacrylamide gel electrophoresis (Fig.2).
Figure 2. Effect of PLL block length, modifying siRNA with 3’-cholesterol, and complexation with nuclease-resistant Chol-siRNA on protection of complexed siRNA from degradation in high serum concentrations by PLL-PEG(5K).
(A) siRNA (siCtrl: 5’- UGG UUU ACA UGU CGA CUA A - 3’ with 3’-UU overhangs), (B) Chol-siRNA (siCtrl modified with 3’-cholesterol on the sense strand), or (C) nuclease-resistant Chol-*siRNA (Chol-siCtrl without 3’-UU overhangs on the sense stand) was incubated with PLL-PEG(5K) in HEPES buffer at room temperature for 30 min at the indicated minimum N/P ratio required for complexation. Polyplexes were then incubated in the presence or absence of 90%v/v murine serum at 37°C for the indicated time, disassembled by heparin, and resolved with or without water solubilized cholesterol (*Cholesterol) as described in Fig.1. The single band for (A) truncated siRNA (open symbols), (B) full-length Chol-siRNA (closed symbols), or (C) full-length Chol-*siRNA (closed symbols) from serum-treated polyplexes was quantified by densitometry and normalized to the respective full-length band from untreated polyplexes at the same N/P ratio. Percent protection ± SD (n=2) is an average of two independent experiments. siRNA and Chol-siRNA were completely degraded within 1 h and Chol-*siRNA was degraded within 1.5 h under the same conditions.
PLL-PEG(5K) protected truncated siRNA but not full-length siRNA from nuclease degradation in high concentrations of murine serum to an extent and duration that was maximized to similar levels by PLL30-PEG(5K) and PLL50-PEG(5K) (Fig.2A). Thus, PLL-PEG(5K) is unable to sufficiently protect full-length siRNA from nuclease degradation in high concentrations of serum with the current N/P ratios and range of PLL block lengths.
In contrast to siRNA, PLL-PEG(5K) protected full-length Chol-siRNA from nuclease degradation in murine serum to an extent and duration that was also maximized to similar levels by PLL30-PEG(5K) and PLL50-PEG(5K) (Fig.2B). Thus, the ability of PLL-PEG(5K) to protect full-length siRNA from nuclease degradation in high concentrations of serum requires modifying siRNA with 3’-cholesterol and is improved by increasing PLL block length.
To next determine whether increasing the nuclease resistance of siRNA can improve the ability of PLL-PEG(5K) to protect Chol-siRNA in high concentrations of serum, polyplexes of PLL-PEG(5K) were alternatively formed with nuclease-resistant Chol-siRNA (Chol-*siRNA) [8, 10] and again compared by polyacrylamide gel electrophoresis (Fig.2C). Chol-*siRNA was undetectable within 1.5 h under the same conditions (data not shown). PLL-PEG(5K) protected Chol-*siRNA to a greater extent and duration (PLL10-PEG(5K)) or duration (PLL30-PEG(5K) & PLL50-PEG(5K)) than Chol-siRNA in high concentrations of murine serum (Fig.2C vs. B). Thus, as a whole, these results indicate that modifying the sense strand of siRNA with 3’-cholesterol, increasing PLL block length, and increasing the nuclease resistance of Chol-siRNA collectively maximize the ability of PLL-PEG(5K) to protect full-length siRNA from nuclease activity in high serum concentrations with the current N/P ratios and range of PLL block lengths.
3.4 Effect of PLL block length and modifying siRNA with 3’-cholesterol on the protection of complexed siRNA from displacement in high concentrations of serum
Polyanions [15] and serum [15–17] disassemble siRNA polyplexes in vitro. Thus, siRNA may be prematurely released from siRNA polyplexes upon i.v. administration through interactions with serum proteins and/or polyanions attached to cell surface proteoglycans. We previously found that increasing PLL block length from 10 to 50 and modifying a dsDNA model of siRNA with 3’-cholesterol does not increase the resistance of PLL-PEG(5K) polyplexes to displacement of siRNA by the polyanion heparin [9]. Whether the same was true with actual siRNA under more physiologically relevant, high concentrations of serum remained unclear.
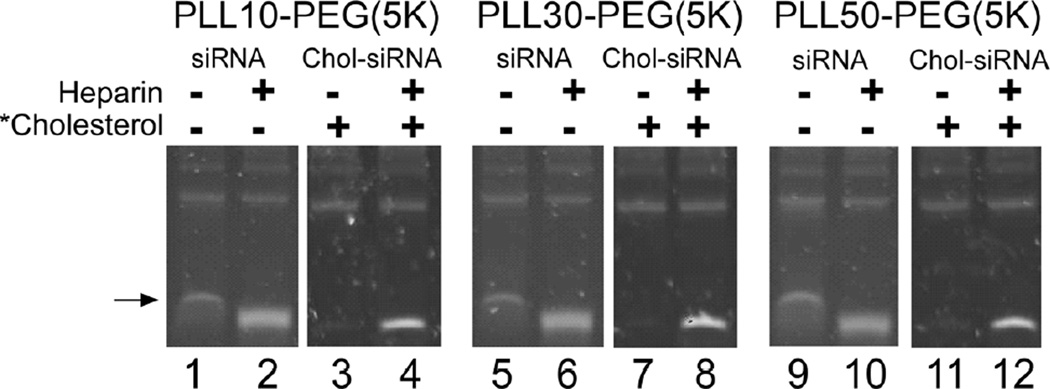
To determine whether PLL block length and modifying siRNA with 3’-cholesterol affect the ability of PLL-PEG(5K) to protect siRNA from displacement in high concentrations of serum, PLL-PEG(5K) was complexed with siRNA or Chol-siRNA at the minimum N/P ratios that form neutral polyplexes (Table 1) and protection against displacement in 90% (v/v) murine serum at 37°C over 2 h was compared by polyacrylamide gel electrophoresis (Fig.3). The possible effect of nuclease activity on siRNA displacement was removed by pretreating the serum with a broad spectrum RNase inhibitor at a concentration that protected siRNA and Chol-siRNA from degradation under the same conditions for up to two hours (data not shown).
Figure 3. Effect of PLL block length and modifying siRNA with 3’-cholesterol on the displacement of siRNA from polyplexes of PLL-PEG(5K) in high serum concentrations.
siRNA (siCtrl: 5’- UGG UUU ACA UGU CGA CUA A - 3’ with 3’-UU overhangs) or Chol-siRNA (siCtrl modified with 3’-cholesterol on the sense strand) was incubated with PLL-PEG(5K) in HEPES buffer at room temperature for 30 min at the minimum N/P ratio required for complexation. Polyplexes were then incubated in the presence or absence of 90%v/v murine serum at 37°C for the indicated time and resolved with or without heparin or water soluble cholesterol (*Cholesterol) as described in Fig.1.
A single band of siRNA (Fig.3 black arrow, lanes 1, 5, and 9) that unexpectedly migrated slower than heparin-released siRNA (Fig.3, lanes 2, 6, and 10) was observed 0, 1, and 2 h after incubating siRNA polyplexes of PLL-PEG(5K) in 90% (v/v) murine serum regardless of PLL block length. The same patterns of siRNA migration in the absence or presence of heparin were observed after incubating siRNA polyplexes of PLL-PEG(5K) in buffer containing bovine serum albumin at a concentration comparable to albumin in murine serum (4.5g / L), whereas neither band was detected after incubating comparable concentrations of PLL-PEG(5K) alone in 90% (v/v) murine serum (data not shown). Thus, the slower migrating band of siRNA occurs through interactions between siRNA polyplexes and serum proteins such as albumin [18] and these interactions may, consequently, interfere with the ability of PLL-PEG(5K) to protect full-length siRNA from truncation in high concentrations of serum (Fig.2A).
In contrast to siRNA polyplexes, bands for released or slower migrating Chol-siRNA were undetected after incubating Chol-siRNA polyplexes up to 2 h in 90% (v/v) murine serum regardless of PLL block length (Fig.3, lanes 3, 7, and 11). Thus, modifying siRNA with 3’-cholesterol is required and sufficient for PLL-PEG(5K) to protect complexed siRNA from interactions with serum proteins in high concentrations of serum with the current N/P ratios and range of PLL block lengths.
3.5 Effect of PLL block length and modifying siRNA with 3’-cholesterol on siRNA activity and cytotoxicity in murine mammary tumor epithelial cells
We previously found that Chol-siRNA polyplexes of PLL10-PEG(5K) suppressed higher levels of native mRNA in conditionally immortalized mammary MVEC than Chol-siRNA polyplexes of PLL50-PEG(5K) (88% vs. 26%) [9]. This suggests that decreasing PLL block length increases mRNA suppression by Chol-siRNA polyplexes of PLL-PEG(5K) in murine mammary MVEC. It remained unclear, however, whether a similar trend exists in other cells important in the therapy of cancer such as the constituent tumor cells.
To determine whether PLL block length and modifying siRNA with 3’-cholesterol affects the activity of complexed siRNA in tumor cells in vitro, the suppression of luciferase in murine tumor epithelial cells that stably express firefly luciferase (4T1-Luc) by siLuc or Chol-siLuc polyplexes of PLL-PEG(5K) at the minimum N/P ratios that form neutral polyplexes (Table 1) was compared by luminescence imaging 24 h after transfection (Fig.4). Luciferase activity is directly proportional to luciferase proteins levels and is, consequently, proportional to luciferase mRNA levels due to the short intracellular half-life of luciferase protein (∼2h). Also, by being constitutively expressed in tumor cells, luciferase expression mimics the expression of oncogenes and drug resistance genes in primary tumors more closely than genes expressed transiently by plasmids [19].
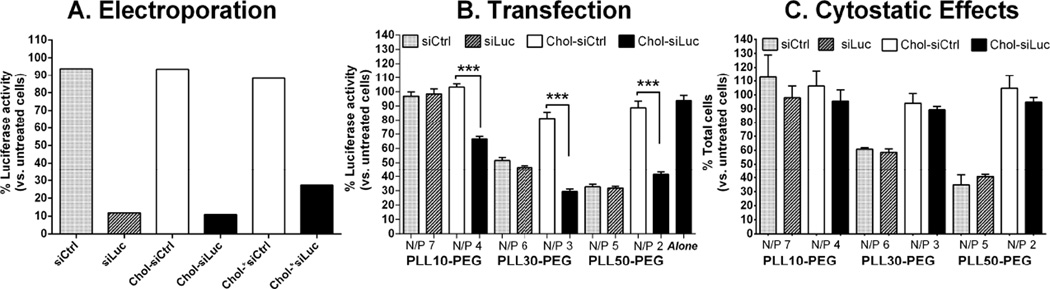
Figure 4. Effect of PLL block length and modifying siRNA with 3’-cholesterol on the suppression of luciferase activity in 4T1-Luc by polyplexes of PLL-PEG(5K).
(A) Electroporation: A murine mammary tumor epithelial cell line stably expressing firefly luciferase (4T1-Luc) was electroporated with 300 nM control siRNA (siCtrl), anti-luciferase siRNA (siLuc), Chol-siCtrl (3’-sense), Chol-siLuc or Chol-*siLuc (3’-sense) and luciferase activity was measured by bioluminescent imaging after 24 h. Radiance from 4T1-Luc electroporated with the indicated siRNA was normalized to radiance from 4T1-Luc electroporated without siRNA on the same plate and expressed as percent luciferase activity. (B) Transfection: Serum- and antibiotic-free complete DMEM containing 200 nM of the indicated siRNA complexed with PLL-PEG(5K) at the indicated N/P ratio or 200 nM Chol-siLuc alone (Alone) was added to 4T1-Luc for 4 h then an equal volume of complete DMEM containing 20% FBS was added and luciferase activity was measured 20 h later by bioluminescent imaging. Average radiance from transfected 4T1-Luc was normalized to average radiance from untreated cells on the same plate and expressed as percent luciferase activity ± SD (n=2, ***P < 0.001 by one way ANOVA and Tukey post-test). Results are representative of two independent experiments. (C) Cytostatic Effects: The viability of 4T1-Luc in all treatment groups relative to untreated 4T1-Luc was between 98%–100% by trypan blue exclusion (data not shown). The average number of live cells from treated 4T1-Luc was normalized to the average number of live cells from untreated 4T1-Luc on the same plate and expressed as percent live cells ± propagated SD (n=2).
Electroporation of 4T1-Luc with siLuc, Chol-siLuc, or nuclease-resistant Chol-siLuc (Chol-*siLuc) greatly decreased luciferase activity relative to electroporated 4T1-Luc (up to 90%), whereas inactive siCtrl, Chol-siCtrl, or Chol-*siCtrl had little effect (up to 12%) (Fig.4A). Thus, siLuc is active against luciferase and unaffected by modifications that increase nuclease resistance. Furthermore, in contrast to conditionally immortalized murine mammary MVEC [9], modifying siRNA with 3’-cholesterol does not significantly decrease the activity of siRNA administered to 4T1 cells by electroporation.
Transfection of 4T1-Luc was not cytotoxic across all treatment groups under the current conditions as determined by trypan blue exclusion relative to untreated 4T1-Luc (data not shown). siCtrl or siLuc polyplexes of PLL10-PEG(5K) had little effect on luciferase activity (Fig.4B) or growth of 4T1-Luc (Fig.4C). Although siLuc polyplexes of PLL30-PEG(5K) and PLL50-PEG(5K) decreased luciferase activity in 4T1-Luc (Fig.4B), their respective inactive siCtrl polyplexes also decreased luciferase activity to similar levels (Fig.4B). Furthermore, the extent that all siLuc and siCtrl polyplexes decreased luciferase activity directly correlated with the extent that each inhibited the growth of 4T1-Luc (Fig.4C vs. B). Thus, given that cellular luciferase activity is proportional to the number of cells that express luciferase [11], the inhibition of luciferase activity by siRNA polyplexes of PLL-PEG(5K) is due primarily to the inhibition of 4T1-Luc growth and not mRNA suppression by complexed siLuc.
In contrast to siCtrl polyplexes (Fig.4B, grey bars), none of the inactive Chol-siCtrl polyplexes of PLL-PEG(5K) decreased luciferase activity (Fig.4B, white bars) or growth of 4T1-Luc (Fig.4C, white bars) regardless of PLL block length. Furthermore, unlike siLuc polyplexes of PLL30-PEG(5K) and PLL50-PEG(5K), all Chol-siLuc polyplexes decreased luciferase activity (Fig.4B, black bars) without affecting 4T1-Luc growth (Fig.4C, black bars), where Chol-siLuc polyplexes of PLL30-PEG(5K) decreased luciferase activity 37% more than PLL10-PEG(5K) (71 ± 2 (SD) vs. 34 ± 2%, P <0.001) and 12% more than PLL50-PEG(5K) (71 ± 2 (SD) vs. 59 ± 2%, P <0.05) (Fig.4B). Chol-siLuc alone also had little effect on luciferase activity (Fig.4B, Alone). Thus, modifying siRNA with 3’-cholesterol increases the efficacy of mRNA suppression by complexed Chol-siRNA in 4T1 cells in vitro in a PLL block length-dependent manner with the current N/P ratios and range of PLL block lengths. Furthermore, in contrast to murine MVEC [9], Chol-siRNA polyplexes with longer PLL blocks have higher levels of activity in 4T1 murine breast tumor epithelial cells than Chol-siRNA polyplexes with shorter PLL block lengths.
3.6 Effect of PLL block length on the suppression of stably expressed luciferase in primary breast tumors of 4T1-Luc by polyplexes of PLL-PEG(5K)
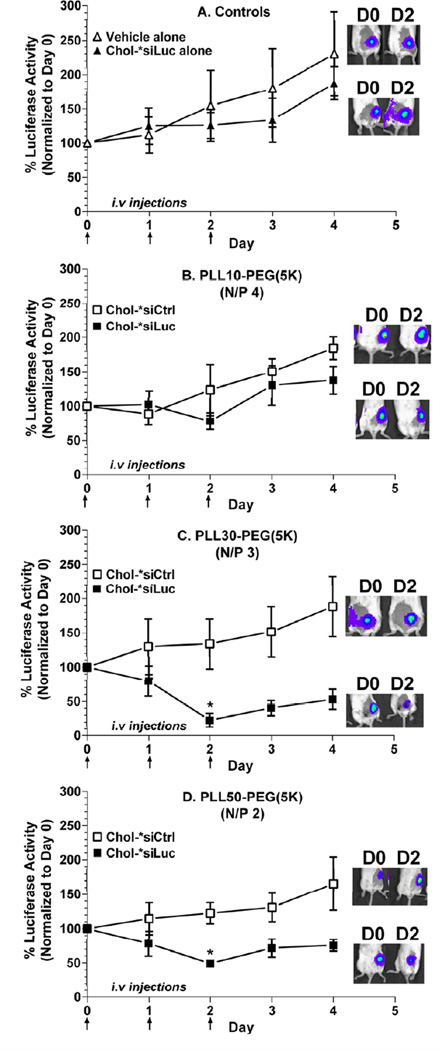
To determine whether complexation with PLL-PEG(5K) increases the efficacy of nuclease-resistant Chol-siRNA (Chol-*siRNA) in primary breast tumors and is affected by PLL block length, relative luciferase activity from primary, orthotopic (mammary) tumors of 4T1-Luc was compared by bioluminescent imaging (Fig.5) after i.v. administration of Chol-*siLuc polyplexes of PLL-PEG(5K) (Table 1). Chol-*siRNA was used for these studies because, unlike Chol-siRNA, PLL-PEG(5K) protected Chol-*siRNA from nuclease degradation in 90% (v/v) murine serum to a similar extent regardless of PLL block length (Fig. 2C) and Chol-*siRNA polyplexes (data not shown) had similar activity against luciferase as Chol-siRNA polyplexes in 4T1-Luc in vitro (Fig. 4B).
Figure 5. Effect of PLL block length on the suppression of luciferase expression in primary 4T1-Luc tumor by polyplexes of nuclease-resistant Chol-siRNA and PLL-PEG(5K).
Primary breast tumors were established by injecting 4T1-Luc cells (1 ×106) SQ into the mammary fat pad of female BALB/c, allowing tumors to grow between 60 and 100 mm3, then determining a baseline luciferase signal. (A) HEPES/saline (open triangles), nuclease resistant Chol-*siLuc alone (closed triangles) or (B) PLL10-PEG, (C) PLL30-PEG or (D) PLL50-PEG complexed with Chol-*siCtrl (open squares) or Chol-*siLuc (closed squares) at the indicated N/P ratio was then intravenously injected at 2.5 mg Chol-siRNA/kg on days 0, 1, and 2. Average radiance from 4T1 tumors within the same cohort was normalized to the average radiance on the first day of treatment (Day 0) and expressed as % luminescence ± propagated SEM (n=3–5 animals). Representative images of luciferase activity in primary 4T1-Luc tumors on the first (D0) and third (D2) day of treatment are shown. *P <0.05 vs. average percent luciferase activity on Day 0 within the same treatment group by Friedman non-parametric repeated measures ANOVA and Dunn’s multiple comparison test.
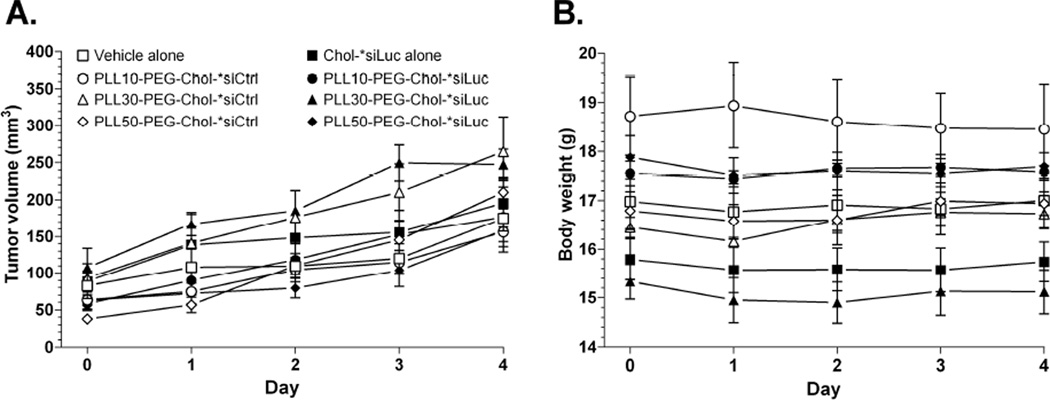
The luciferase activity of primary 4T1-Luc tumors relative to the first day of treatment (Day 0) increased over time after treatment with vehicle alone, Chol-*siLuc alone (Fig.5A), or inactive Chol-*siCtrl polyplexes of PLL-PEG(5K) regardless of PLL block length (Fig.5, open squares). Chol-*siLuc polyplexes of PLL10-PEG(5K) also had no statistical effect on luciferase activity (Day 0 vs. Day 2, P =0.2) (Fig.5B, closed squares). In contrast, Chol-*siLuc polyplexes of PLL30-PEG(5K) and PLL50-PEG(5K) (Fig.5C & D, closed squares) maximally decreased luciferase activity in primary 4T1-Luc tumors on the third day of treatment (Day 0 vs. Day 2, 77 ± 10% (SEM), P = 0.001 and 51 ± 5%, P = 0.03) but were not statistically different (77 ± 10% (SEM) vs. 51 ± 5%, P = 0.1). Furthermore, none of the treatments affected the growth of 4T1-Luc tumors (Fig.6A) or body weight of the animals (Fig.6B) over the course of the study, indicating that differences in luciferase activity are not due to changes in tumor growth or acute toxicity. Thus, complexation with PLL-PEG(5K) increases the efficacy of mRNA suppression by nuclease-resistant Chol-siRNA in primary breast tumors after i.v. administration in a PLL block length-dependent manner with the current N/P ratios and range of PLL block lengths.
Figure 6. Effect of treatments on tumor volume of 4T1-Luc and body weight.
Mice were treated as described in Fig.5 and (A) tumor volume and (B) body weight were measured beginning the first day of treatment (Day 0). Values are expressed as the mean ± SEM (n=3–5 animals).
4. Discussion
This study provides evidence that complexation of nuclease-resistant Chol-siRNA (Chol-*siRNA) with PLL-PEG(5K) increases the efficacy of Chol-*siRNA in primary breast tumors in a PLL block length-dependent manner with the current N/P ratios and range of PLL block lengths. We found that i.v. administration of Chol-*siLuc polyplexes of PLL30-PEG(5K) or PLL50-PEG(5K) decreased high levels of luciferase activity in primary orthotopic (mammary) tumors that stably express luciferase (4T1-Luc) (Fig.5C & D, closed squares), whereas Chol-*siLuc alone (Fig.5A, closed triangles), Chol-*siLuc polyplexes of PLL10-PEG(5K) (Fig.5B, closed squares), or inactive Chol-*siCtrl polyplexes of PLL-PEG(5K) (Fig.5B– D, open squares) had no effect under the current dosage regimen. Furthermore, we found that none of the treatments affected the growth of 4T1-Luc tumors (Fig.6A) or relative body weight (Fig.6B), further corroborating that the inhibition of luciferase activity by Chol-*siRNA polyplexes is due to suppression of luciferase mRNA by complexed Chol-*siRNA and not due to the direct or indirect inhibition of primary 4T1-Luc tumor growth that could alternatively decrease luciferase activity [11].
4.1 Role of PLL block length in the efficacy of polyplexes of nuclease-resistant Chol-siRNA and PLL-PEG(5K) in primary breast tumors
There are at least two possible reasons that may collectively explain why PLL block length affects the efficacy of Chol-*siRNA polyplexes of PLL-PEG(5K) in primary breast tumors after i.v. administration. The first is that longer PLL block lengths increase protection of complexed Chol-*siRNA from RNase degradation and disassembly in the vascular compartment and, consequently, increase the bioavailability of Chol-*siRNA polyplexes of PLL-PEG(5K). The increase in bioavailability then increases subsequent passive targeting of Chol-*siRNA polyplexes to primary breast tumors by the enhanced permeability and retention (EPR) effect [20, 21]. This is supported, in part, by our finding that polyplexes of PLL30-PEG(5K) and PLL50-PEG(5K) protected Chol-*siRNA from degradation in 90% (v/v) murine serum to a greater extent and duration than polyplexes of PLL10-PEG(5K) (Fig.2C). Although we did not detect disassembly of Chol-siRNA polyplexes in 90% (v/v) murine serum under static conditions after 2 hours over the current range of PLL block lengths (Fig.3), it remains possible that longer PLL block lengths also increase bioavailability by decreasing or preventing disassembly by physiological conditions such as shear stress or interactions with cell surfaces within the vascular compartment over a longer period of time.
The second possible reason why PLL block length affects the efficacy of Chol-*siRNA polyplexes of PLL-PEG(5K) in primary breast tumors is that it affects the pharmacological activity of complexed Chol-*siRNA in the tumor cells. This is supported by our finding in vitro that Chol-siLuc polyplexes of PLL30-PEG(5K) suppressed higher levels of luciferase activity in 4T1-Luc (71 ± 2%) than Chol-siLuc polyplexes of PLL50-PEG(5K) (59 ± 2%) or PLL10-PEG(5K) (34 ± 2%) (Fig.4B). A surprisingly similar pattern of luciferase suppression by the same Chol-*siRNA polyplexes was observed in primary tumors of 4T1-Luc after i.v. administration, although there was no statistical difference between suppression by Chol-*siRNA polyplexes of PLL30-PEG(5K) and PLL50-PEG(5K) under the current study power (77 ± 10 (SEM) vs. 51 ± 5% on Day 2, P = 0.1) (Fig.5C & D). Thus, PLL block length likely affects the activity of Chol-*siRNA polyplexes of PLL-PEG(5K) in primary breast tumors through effects on bioavailability and subsequent activity in the target cell.
4.2 Role of PLL block length in the pharmacological activity of Chol-siRNA polyplexes of PLL-PEG(5K)
As previously discussed, the current study shows that Chol-siRNA polyplexes of PLL-PEG(5K) increase the efficacy of mRNA suppression by Chol-siRNA in 4T1 cells in vitro in a PLL block length-dependent manner with the current N/P ratios and range of PLL block lengths. We found that Chol-siLuc polyplexes of PLL-PEG(5K) decreased luciferase activity (Fig.4B, black bars) without affecting 4T1-Luc growth (Fig.4C, black bars), where Chol-siLuc polyplexes of PLL30-PEG(5K) decreased luciferase activity 37% more than PLL10-PEG(5K) (71 ± 2 (SD) vs. 34 ± 2%, P <0.001) and 12% more than PLL50-PEG(5K) (71 ± 2 (SD) vs. 59 ± 2%, P <0.05) (Fig.4B). In contrast, we found that Chol-siLuc alone had little effect on luciferase activity (Fig.4B, Alone).
PLL block length may affect the pharmacological activity of Chol-siRNA polyplexes of PLL-PEG(5K) by affecting the rate of polyplex internalization upon interaction with the target cell and the subsequent that Chol-siRNA is released from PLL-PEG(5K) polyplexes within the endosomes as Chol-siRNA may need to interact with the endosomal membrane and/or, possibly, intracellular cholesterol transporters to escape the endosomes and into its site of action, the cytosol [9]. Thus, it is likely that Chol-siRNA polyplexes of PLL30-PEG(5K) have the best balance of endocytosis rates and intracellular Chol-siRNA release rates in 4T1.
Interestingly, in contrast to the current study where Chol-siRNA polyplexes of PLL10-PEG(5K) had lower activity than Chol-siRNA polyplexes of PLL50-PEG(5K) in 4T1 (67% vs.41% suppression) (Fig.4B), we previously found that Chol-siRNA polyplexes of PLL10-PEG(5K) had significantly higher activity than Chol-siRNA polyplexes of PLL50-PEG(5K) in murine mammary MVEC (88% vs. 12% suppression) [9]. This further suggests that the effect of PLL block length on the pharmacological activity of Chol-siRNA polyplexes of PLL-PEG(5K) is cell-type dependent.
4.3 Role of PLL block length and modifying siRNA with 3’-cholesterol in siRNA loading
The current study shows that increasing PLL block length and modifying the sense strand of siRNA with 3’-cholesterol collectively decrease the minimum N/P ratio required to form neutral polyplexes between siRNA and PLL-PEG(5K) and, consequently, increase siRNA loading over the current range of PLL block lengths (Table 1). Increasing PLL block length may decrease the minimum N/P ratio required to neutralize siRNA by increasing the charge density of PLL-PEG(5K) unimers. This likely increases the affinity of PLL-PEG(5K) for siRNA or Chol-siRNA through an increase in the free energy of binding [22] and, consequently, decreases the amount of polymer required to neutralize the same amount of siRNA or Chol-siRNA. A similar effect was reported for siRNA polyplexes of PEI [23]. The presence of 3’-cholesterol on the sense strand of siRNA may additionally decrease the minimum N/P ratio by forming high molecular weight micellar structures that increase the charge density of siRNA and further increase the affinity of PLL-PEG(5K) for siRNA. This is partially supported by agarose gel data where bands of Chol-siRNA migrated a much shorter distance than unmodified siRNA despite the low MW of cholesterol (∼386 Da) relative to siRNA (∼13 kDa) (data not shown).
4.4 Differences in interactions between PLL-PEG(5K) and siRNA or a dsDNA model of siRNA
Although the current study shows that increasing PLL block length and modifying the sense strand of siRNA with 3’-cholesterol decreases the minimum N/P ratio required to form neutralize polyplexes of PLL-PEG(5K) (Table 1), these changes do not affect the minimum N/P ratio required by PLL-PEG(5K) to form neutral polyplexes with our previous dsDNA model of siRNA [9]. This suggests that PLL-PEG(5K) interacts differently with siRNA (dsRNA) than comparable structures of dsDNA. Another possible difference between the previous and current studies, however, is that our previous dsDNA model of siRNA has 3’-AT overhangs capable of complementary base pairing, whereas the siRNA used in the current studies does not (3’-UU). Complementary base pairing with siRNA possessing longer complementary (“sticky”) overhangs decreases the minimum N/P required by 25kDa PEI to form neutral polyplexes [24]. Furthermore, we found that the minimum N/P ratio required to form neutral polyplexes between the same dsDNA model of siRNA without complementary overhangs (3’-TT) and PLL10-PEG(5K), PLL30-PEG(5K), or PLL50-PEG(5K) (data not shown) were the same as siRNA without complementary overhangs (3’-UU) (Table 1). Thus, the differences in interactions between PLL-PEG(5K) and our previous dsDNA model of siRNA [9] or actual siRNA (dsRNA) are most likely due to complementary overhangs on the dsDNA model of siRNA. Complementary base pairing of our previous dsDNA model of siRNA may also explain why polyplexes of Chol-model siRNA are larger than polyplexes of Chol-siRNA (Table 2).
4.5 Role of PLL block length and modifying siRNA with 3’-cholesterol in the stability of polyplexes of PLL-PEG(5K) in serum
The current study shows that modifying the sense strand of siRNA with 3’-cholesterol, increasing PLL block length, and increasing the nuclease resistance of Chol-siRNA collectively maximize the ability of PLL-PEG(5K) to protect full-length siRNA from nuclease activity and displacement in high serum concentrations with the current N/P ratios and range of PLL block lengths. We found that PLL-PEG(5K) protected only truncated siRNA from degradation in 90% (v/v) murine serum (Fig.2A) but protected full-length Chol-siRNA to an extent and duration that was maximized to similar levels by PLL30-PEG(5K) and PLL50-PEG(5K) vs. PLL10-PEG(5K) (Fig.2B). Furthermore, modifying siRNA with 3’-cholesterol was sufficient for PLL-PEG(5K) to protect complexed Chol-siRNA from interactions with serum proteins (Fig.3).
Modifying siRNA with 3’-cholesterol may increase the ability of PLL-PEG(5K) to protect full-length siRNA from nuclease degradation and displacement in high serum concentrations by increasing the affinity of PLL-PEG(5K) for siRNA as discussed in Section 4.2 as well as through the formation of polyplexes with better defined core-shell morphologies that more effectively decrease the accessibility of complexed siRNA to proteins such as serum nucleases [9]. Increasing PLL block length then further increases the protection of complexed Chol-siRNA by PLL-PEG(5K) by increasing the affinity of PLL-PEG(5K) for Chol-siRNA through an increase in the free energy of siRNA binding and subsequent affinity between PLL-PEG(5K) and siRNA [22]. This is supported, in part, by our finding that siRNA polyplexes of PLL-PEG(5K) interact with serum proteins, whereas Chol-siRNA polyplexes of PLL-PEG(5K) do not (Fig.3).
5. Conclusions
In summary, our results indicate that complexation of nuclease-resistant Chol-siRNA with PLL-PEG(5K) significantly increases the activity of Chol-siRNA in primary breast tumors after i.v. administration in a PLL block length-dependent manner. Thus, polyplexes of Chol-siRNA and PLL-PEG(5K) may be a promising approach to increase the efficacy of Chol-siRNA in wide range of primary tumors, metastases, and other tissues but require a PLL block length that balances polymer-related adverse effects, Chol-siRNA bioavailability, and subsequent activity in the target cell.
Acknowledgements
We are thankful for the support of NIH COBRE grant RR021937 (Nebraska Center for Nanomedicine) (JAV), Susan G. Komen for the Cure Grant KG090860 (RKS), and the University of Nebraska Presidential Graduate Fellowship (VVA). The IVIS instrument was purchased through the Nebraska Tobacco Settlement Biomedical Research Development Fund (NTSBRDF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Bora RS, Gupta D, Mukkur TK, Saini KS. RNA interference therapeutics for cancer: challenges and opportunities (review) Mol Med Report. 2012;6:9–15. doi: 10.3892/mmr.2012.871. [DOI] [PubMed] [Google Scholar]
- 4.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 5.Aigner A. Cellular delivery in vivo of siRNA-based therapeutics. Curr Pharm Des. 2008;14:3603–3619. doi: 10.2174/138161208786898815. [DOI] [PubMed] [Google Scholar]
- 6.Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev. 2009;61:710–720. doi: 10.1016/j.addr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 9.Ambardekar VV, Han HY, Varney ML, Vinogradov SV, Singh RK, Vetro JA. The modification of siRNA with 3′ cholesterol to increase nuclease protection and suppression of native mRNA by select siRNA polyplexes. Biomaterials. 2011;32:1404–1411. doi: 10.1016/j.biomaterials.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan H, Lanting L, Xu ZG, Li SL, Swiderski P, Putta S, et al. Effects of cholesterol-tagged small interfering RNAs targeting 12/15-lipoxygenase on parameters of diabetic nephropathy in a mouse model of type 1 diabetes. Am J Physiol Renal Physiol. 2008;295:F605–F617. doi: 10.1152/ajprenal.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim E, Modi KD, Kim J. In vivo bioluminescent imaging of mammary tumors using IVIS spectrum. J Vis Exp. 2009 doi: 10.3791/1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwoh DY, Coffin CC, Lollo CP, Jovenal J, Banaszczyk MG, Mullen P, et al. Stabilization of poly-L-lysine/DNA polyplexes for in vivo gene delivery to the liver. Biochim Biophys Acta. 1999;1444:171–190. doi: 10.1016/s0167-4781(98)00274-7. [DOI] [PubMed] [Google Scholar]
- 13.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 14.Haupenthal J, Baehr C, Kiermayer S, Zeuzem S, Piiper A. Inhibition of RNAse A family enzymes prevents degradation and loss of silencing activity of siRNAs in serum. Biochem Pharmacol. 2006;71:702–710. doi: 10.1016/j.bcp.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Buyens K, Meyer M, Wagner E, Demeester J, De Smedt SC, Sanders NN. Monitoring the disassembly of siRNA polyplexes in serum is crucial for predicting their biological efficacy. J Control Release. 2010;141:38–41. doi: 10.1016/j.jconrel.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 16.Buyens K, Lucas B, Raemdonck K, Braeckmans K, Vercammen J, Hendrix J, et al. A fast and sensitive method for measuring the integrity of siRNA-carrier complexes in full human serum. J Control Release. 2008;126:67–76. doi: 10.1016/j.jconrel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, et al. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly(ethylene glycol) under in vivo conditions: effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. J Control Release. 2009;138:148–159. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Oupicky D, Konak C, Dash PR, Seymour LW, Ulbrich K. Effect of albumin and polyanion on the structure of DNA complexes with polycation containing hydrophilic nonionic block. Bioconjug Chem. 1999;10:764–772. doi: 10.1021/bc990007+. [DOI] [PubMed] [Google Scholar]
- 19.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 21.Maeda H. Vascular permeability in cancer and infection as related to macromolecular drug delivery, with emphasis on the EPR effect for tumor-selective drug targeting. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:53–71. doi: 10.2183/pjab.88.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouyang D, Zhang H, Herten DP, Parekh HS, Smith SC. Structure, dynamics, and energetics of siRNA-cationic vector complexation: a molecular dynamics study. J Phys Chem B. 2010;114:9220–9230. doi: 10.1021/jp911906e. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Samsonova O, Sproat B, Merkel O, Kissel T. Biophysical characterization of hyper-branched polyethylenimine-graft-polycaprolactone-block-mono-methoxyl-poly(ethylene glycol) copolymers (hy-PEI-PCL-mPEG) for siRNA delivery. J Control Release. 2011;153:262–268. doi: 10.1016/j.jconrel.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci U S A. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]