Abstract
Escherichia coli K12 is a commensal bacteria and one of the best-studied model organisms. Salmonella enterica serovar Typhimurium, on the other hand, is a facultative intracellular pathogen. These two prokaryotic species can be considered related phylogenetically, and they share a large amount of their genetic material, which is commonly termed the “core genome.” Despite their shared core genome, both species display very different lifestyles, and it is unclear to what extent the core genome, apart from the species-specific genes, plays a role in this lifestyle divergence. In this study, we focus on the differences in expression domains for the orthologous genes in E. coli and S. Typhimurium. The iterative comparison of coexpression methodology was used on large expression compendia of both species to uncover the conservation and divergence of gene expression. We found that gene expression conservation occurs mostly independently from amino acid similarity. According to our estimates, at least more than one quarter of the orthologous genes has a different expression domain in E. coli than in S. Typhimurium. Genes involved with key cellular processes are most likely to have conserved their expression domains, whereas genes showing diverged expression are associated with metabolic processes that, although present in both species, are regulated differently. The expression domains of the shared “core” genome of E. coli and S. Typhimurium, consisting of highly conserved orthologs, have been tuned to help accommodate the differences in lifestyle and the pathogenic potential of Salmonella.
Keywords: gene expression, expression divergence, expression conservation, Salmonella, pathogenesis, Escherichia coli
Introduction
Escherichia coli K12 is a commensal bacteria and one of the best-studied model organisms. Salmonella enterica serovar Typhimurium, on the other hand, is a facultative intracellular pathogen. Current estimates put the divergence of the E. coli and S. Typhimurium strains approximately 100–160 Ma, which is about the same time frame as the divergence of the placental mammals (Kumar and Hedges 1998). These two prokaryotic species can be considered related phylogenetically (Ochman and Wilson 1987; Doolittle et al. 1996). They share a large amount of their genetic material, which has often been termed the “core genome" and typically varies between 2,500 and 3,100 orthologous genes (between 50% and 70% of the whole genome, depending on the used methodology) for the class of enterobacteria (McClelland et al. 2001; Dobrindt et al. 2003; Le Gall et al. 2005). Despite their shared core genome, both species display very different lifestyles, and it is unclear to what extent the core genome, apart from the species-specific genes, plays a role in this lifestyle divergence. Although these genes in the core genome are highly similar at the sequence level, it is uncertain whether they have retained the same function in both species (Callister et al. 2008) as they reside in a different genomic background and are thus are not functionally independent of the species-specific genes. Furthermore, gene expression is known to be optimized throughout evolution toward the changes in an organism's lifestyle and the niche that it occupies (Dekel and Alon 2005; Mandel and Silhavy 2005; López-Maury et al. 2008; Cooper et al. 2009). This often occurs by changing the regulatory programs for specific genes with the loss and gain of regulatory elements that impact the organisms regulatory network (Winfield and Groisman 2004; Isalan et al. 2008). Divergence of expression domains are the most direct manifestations of such network rewiring, as it is difficult to identify the impact of certain changes relying on the sequence alone. Even minor changes in the regulatory network, such as a single point mutation in a transcription factor (TF), can result in radical changes of the phenotype (Mandel et al. 2009; Hindré et al. 2012). In this study, we want to explore if there are any indications that orthologous gene pairs have different expression domains in E. coli and S. Typhimurium and whether these unique expression domains might be related to the differences in lifestyle and biological niche.
In this article, we analyzed the conservation and divergence of expression domains of the E. coli and S. Typhimurium core genome to better understand the impact of expression divergence on organism-specific tuning toward a specific environment. We used the cross-platform expression compendia of E. coli and S. Typhimurium available on COLOMBOS (Engelen et al. 2011) and relied on the “iterative comparison of coexpression" methodology (Dutilh et al. 2006; Tirosh and Barkai 2007) to compare the expression data for the orthologs of both species. We demonstrate that within the core genome, several genes have diverged expression wise, whereas others seem to have been strongly conserved. To further assess the functional and transcriptional characteristics of the core genes, we identify sets of functional expression classes. These classes show different levels of expression conservation (EC) and can be related to the differences in lifestyle between both bacteria.
Results
Orthologous gene mapping of E. coli and S. Typhimurium resulted in 2,886 unambiguous gene pairs that we shall term the core genome for the purposes of this article (Li et al. 2003). The degree to which the expression of each orthologous gene pair was conserved was estimated using the “iterative comparison of co-expression (ICC)" methodology. This method evaluates the EC of a single orthologous gene pair from microarray compendia from different species, despite the compendia consisting of different conditions. The output for each gene pair is a value termed the EC score, which is calculated by estimating the retention of the similarity in expression domains to all other genes in the core genome. Analyses that compare these EC scores with protein and gene promoter similarities indicate a difference between sequence similarity and EC (see supplementary material S1, Supplementary Material online). This has already been observed in past studies of eukaryotic species (Wagner 2000; Dutilh et al. 2006), and it supports our primary hypothesis that the core genome, despite being similar in amino acid content, could have altered its function by divergence of expression regulation. In the following sections, we will further explore which genes are most diverged or conserved expression wise.
Bimodal Distribution Reflects EC and Divergence
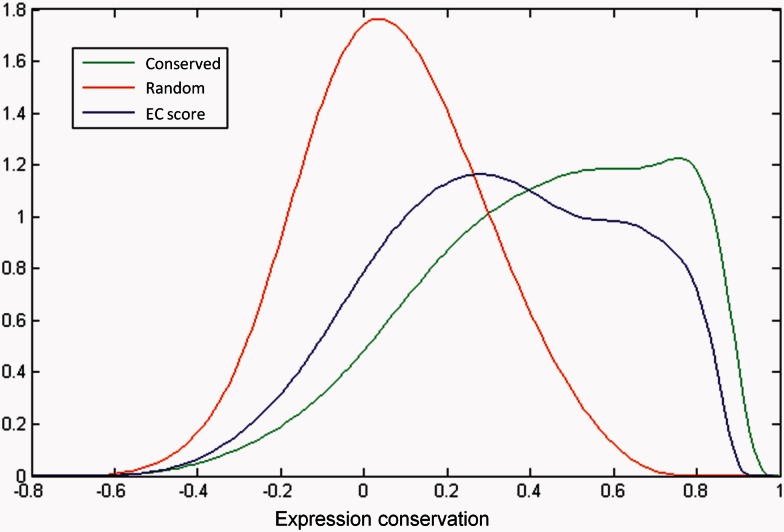
The distribution of the EC scores of the orthologous gene pairs is given in figure 1. The distribution consists of two peaks (or “modes"), one at 0.3 and a smaller one at 0.6. There are also several gene pairs whose EC score is negative, indicating orthologs whose expression domains tend to be reversed in these two compendia. The full listing of the orthologous genes and their scores can be found in supplementary data set S1, Supplementary Material online. To facilitate comparison between the EC scores, we quantified the level of variability that can arise when the conservation and divergence is known. To this end, we constructed background distributions both for the case of expression divergence and for the case of perfect EC. The divergence background distribution is shown in figure 1. As can be seen, the EC scores of gene pairs with permutated expression values vary between −0.6 and 0.7. Additionally, we also created a background distribution for the case of conserved gene expression domains by splitting the largest expression compendium, namely E. coli, into two equal halves (multiple times), with each half containing a different set of microarray experiments. The two E. coli compendia are then compared against each other (fig. 1). In effect, we are simulating perfect conservation by comparing a species to itself but accounting for the presence of different experimental setups and conditions in both expression compendia. Surprisingly, the correlation score of a perfectly conserved gene pair can vary between −0.5 and 1. The fact that the score can be so low, even when expression should be perfectly conserved, can be attributed to the condition dependency of the EC score. Indeed, when cross-comparing the split compendium where the experimental conditions are similar in either half, the resulting EC scores are much higher (see supplementary material S2, Supplementary Material online). Although the EC score was specifically developed to compare expression compendia that do not necessarily survey the same biological conditions, a different set of conditions does affect the observed correlations between genes of the same species and hence also the EC scores.
Fig. 1.
Distribution of the EC score between the orthologous genes of Escherichia coli and Salmonella enterica serovar Typhimurium depicted by its kernel smoothed density estimate (blue line). The distribution of the EC scores for gene pairs with randomized expression values, which represent the estimated score given no conservation of expression, is shown as a red line. The distribution of the EC scores resulting from comparison of the E. coli compendium to itself with data from different experiments is shown as a green line and represents the estimated score given perfect conservation of expression.
Taken together, both background distributions provide an explanation for the bimodal nature of the EC scores' distribution: The peaks represent overall expression divergence and conservation levels. The large overlap between the two background distributions also shows that for the majority of gene pairs, it will be difficult to reliably estimate the degree of conservation based on the EC scores beyond the coarse distinction of “divergent" or “conserved" for gene pairs with the most extreme EC scores. Using these background distributions, we can estimate the number of genes we expect to have diverged expression. We expect the found EC distribution to be a mix between genes that have conserved their expression domains and those that have diverged. As each background represents either conserved or diverged expression domains, the most likely combination of the two background distributions into the found EC distribution between the two compendia can be used as a measure for the fraction of diverged genes. This analysis (supplementary material S3, Supplementary Material online) estimated that approximately 25% of the genes have divergent expression domains between these two species.
The Basal Cellular Machinery Has the Most Conserved Expression Domains
There is significant overlap between the conserved and the divergent background distribution, thus only for gene pairs with extreme EC scores can we judge if the expression domains have remained conserved or not. To evaluate the relationship between EC and biological function, we selected a restrictive set of genes with strongly conserved expression and a set with low EC scores.
The estimated background distribution for nonconserved genes never achieves a score higher than 0.7. It can therefore be reasonable to assume that gene pairs with a higher EC score are very likely to have conserved expression domains, so we define a restrictive set of genes with conserved expression at a cutoff of 0.7. This results in a set of 335 genes, which are enriched in the gene ontology (GO) for a number of not only key cellular processes (table 1), such as protein translation, ribosome biogenesis, and gene transcription, but also metabolic processes, such as the biosynthesis of nucleotides and fatty acids. Genes involved in key cellular processes seem to also have strongly conserved expression between these two compendia.
Table 1.
GO Enrichment of Conserved and Divergent Genes.
| GO | P | |
|---|---|---|
| Genes with divergent expression | ||
| Phospholipid biosynthetic process | 4.87 × 10−6 | |
| Lipid A biosynthetic process | 1.81 × 10−5 | |
| Biosynthetic process | 3.15 × 10−5 | |
| Catabolic process | 1.44 × 10−5 | |
| Metabolic process | 6.55 × 10−6 | |
| Genes with conserved expression | ||
| Translation | 1.42 × 10−52 | |
| Regulation of translation | 6.79 × 10−7 | |
| Translational termination | <1 × 10−60 | |
| Translational elongation | <1 × 10−60 | |
| Protein metabolic process | 4.99 × 10−33 | |
| Gene expression | 8.67 × 10−19 | |
| Transcription termination | <1 × 10−60 | |
| tRNA metabolic process | 4.65 × 10−5 | |
| tRNA aminoacylation for protein translation | 1.37 × 10−6 | |
| ncRNA metabolic process | 9.04 × 10−7 | |
| tRNA aminoacylation | 2.68 × 10−6 | |
| Ribosome biogenesis | 3.80 × 10−5 | |
| Macromolecule metabolic process | 1.85 × 10−16 | |
| Macromolecular complex subunit organization | 1.01 × 10−7 | |
| Primary metabolic process | 1.23 × 10−19 | |
| Metabolic process | 2.82 × 10−23 | |
| Nucleotide biosynthetic process | 1.25 × 10−6 | |
| Nucleoside biosynthetic process | 1.80 × 10−6 | |
| Purine ribonucleotide biosynthetic process | 3.65 × 10−5 | |
| Purine ribonucleoside biosynthetic process | 4.13 × 10−5 | |
| Ribonucleotide biosynthetic process | 8.55 × 10−7 | |
| Ribonucleoprotein complex biogenesis | 3.8 × 10−5 | |
| Amino acid derivative metabolic process | 1.87 × 10−5 | |
| Fatty acid biosynthetic process | 1.24 × 10−7 | |
As an additional comparison, we collected a list of essential genes for E. coli and S. Typhimurium: genes that when knocked-out do not allow growth under normal laboratory conditions. This is the case for 300 E. coli genes, of which 272 are in the core genome. In the restrictive set of genes with conserved expression (score > 0.7), there are 104 essential genes, a significant enrichment with a P value of 5.58 × 10−12. For S. Typhimurium, 253 genes have been designated essential, and of these, 162 are in the core genome. In this case, 40 of the conserved genes are labeled as essential (enrichment P value of 4.42 × 10−7). Based on the orthologous gene pair mapping between these two species, the overlap of the essential core genes in E. coli and those of S. Typhimurium is a list of 72 genes (see supplementary table S1, Supplementary Material online). From this list, 32 genes can be found in our conserved expression gene set (enrichment P value of 2.25 × 10−12).
In a similar manner, we could attempt to define a set of diverged genes. In this case, it is, however, less evident as the conserved distribution was able to achieve scores almost as low as the found EC values for the E. coli–S. Typhimurium comparison. Given the two background distributions and the 25% divergent genes that we estimated before, we take an arbitrary cutoff of −0.1, for which we can expect a false-positive rate of 0.42, that is, 42% of the gene pairs with an EC score lower than −0.1 can be expected to be conserved. This is a set of 173 orthologous pairs that are enriched for biosynthesis of phospholipids and lipid A (table 1). The latter is of specific interest as lipid A has been postulated to cause toxicity in Salmonella infections (Khan et al. 1998). Furthermore, genes involved in the synthesis of lipid A have been associated with antibiotic resistance. For example; the E. coli genes arnABCD and eptA are all assigned a score less than −0.1 and are involved in lipid A biosynthesis and polymyxin resistance. It has already been noticed that there is a difference in polymyxin resistance between E. coli and S. Typhimurium and that this is indeed due to divergent transcription regulation of exactly these genes (Marchal et al. 2004; Winfield and Groisman 2004; Monsieurs et al. 2005). The diverged set was also significantly depleted (P value: 0.0018) in essential E. coli genes, containing only six essential E coli genes. This set also contains only three essential S. Typhimurium genes (depletion P value: 0.0094). Interestingly, despite being essential in both organisms, both the ftsZ and thrS orthologous gene pairs have a very low EC score (−0.326 and −0.310, respectively).
Functional Expression Classes
The cutoffs used earlier to define a set of diverged and conserved genes were very strict and did not provide any information on the majority of the core genes (the ones that reside in the region where both background distributions overlap). For a more comprehensive analysis of the entire core genome, we first created “functional expression classes" for each organism and then evaluated how these correspond with the calculated EC scores. Functional expression classes were defined based on a hierarchical clustering of the correlation matrices that were constructed for each species. In this way, genes are not grouped together based on the similarity of expression profiles under specific conditions (as one would in a normal clustering or biclustering approach) but based on a similar expression correlation toward the other genes in the compendium. This has the advantage of finding broad common expression relationships present in the entire compendium. As we will show, these functional expression classes are biologically relevant and can be directly related to the divergence of the expression domains. Functional expression classes could also be constructed for the full genome, instead of only the core genome, of each species and these results are discussed in supplementary material S4, Supplementary Material online.
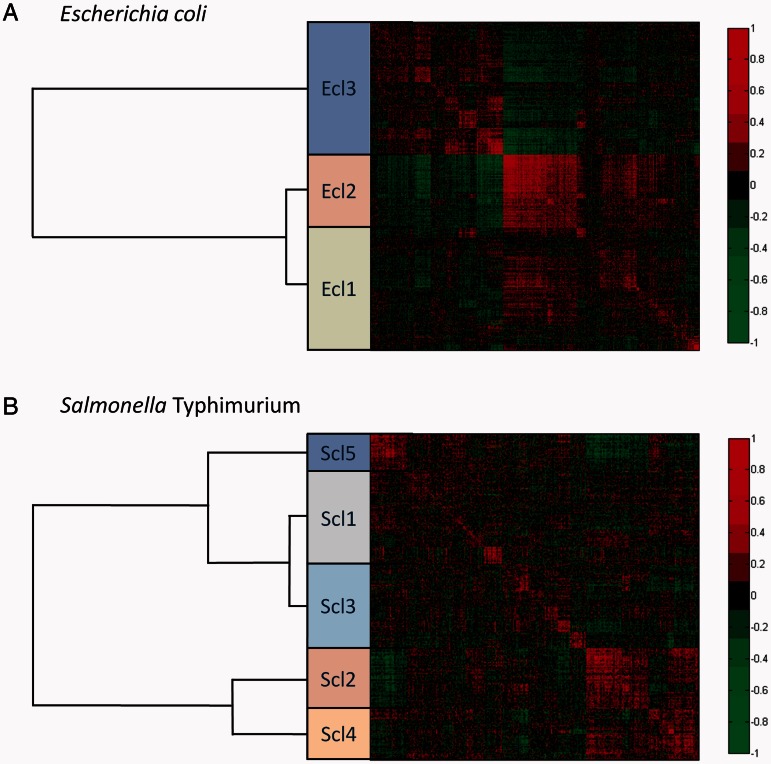
In E. coli, three major functional expression classes appear. As can be seen in figure 2a, the correlation profiles of Ecl1 (1,094 genes) and Ecl2 (734 genes) are more similar than that of Ecl3 (1,058 genes) based on the clustering. See supplementary data set S2, Supplementary Material online, for the full listing of genes. The most striking observation is that the Ecl2 genes are internally highly similar (indicating that the genes of Ecl2 are consistently up- and downregulated under similar conditions) and are also generally anticorrelated to the Ecl3 genes.
Fig. 2.
Expression correlation matrices of the genes in the core genome of Escherichia coli (A) and Salmonella enterica serovar Typhimurium (B). Each value presented in the heatmap is the Pearson correlation coefficient between the expression profile of the gene in the row and the expression profile of the gene in the column from the compendium of the given species. The rows and columns are sorted according to a hierarchical clustering, and functional expression classes were created at a cutoff of 110 distance units. The classes are represented by colored boxes with their hierarchical relationship given in the tree to the left. Each class is labeled E. coli class (Ecl) or S. Typhimurium class (Scl) appended by a number.
To estimate the functional roles of the genes that are present in each of the three classes, we relied on the annotation of these genes from various heterogeneous data sources. As can be seen from the summary in table 2, Ecl1 seems to contain several genes related to anabolism, energy metabolism, and cell motility. Also present in Ecl1 are several genes related to very specific stresses, such as cation and osmotic stress. The Ecl2 genes mostly encode for proteins responsible for cellular structure and key cellular processes, such as protein translation, gene transcription, and cell division. Thus, most essential genes are present in this expression class. Also, Ecl2 contains various biosynthesis genes of nucleotides and carbohydrates. The Ecl3 genes are mostly involved in nutrient catabolism and stress responses. This is further supported by the enrichment for targets of various global TFs known to be active in these processes and for the targets of σ38, the general stress response sigma factor (SF). This difference in function might explain the general trend of anticorrelation between the Ecl2 genes and the Ecl3 genes, as this seems to represent the switch between growth in a beneficial environment (Ecl2) versus survival in a more hostile environment (Ecl3).
Table 2.
Functional Evaluation of the E. coli Expression Classes.
| E. coli expression class | Ecl1 | Ecl2 | Ecl3 |
|---|---|---|---|
| No. genes | 1,094 genes | 734 genes | 1,058 genes |
| GO enrich.a | Chemotaxis | Cell division | Multiorganism processes |
| Energy metabolism | Cell wall assembly | Cell adhesion | |
| Amino acid metabolism | Carbohydrate biosynthesis | Carbohydrate catabolism | |
| Nucleotide metabolism | Nucleotide biosynthesis | Transport proteins | |
| Cation/osmotic stress | Transcription | Acidity stress | |
| Translation | Starvation stress | ||
| Toxin stress | |||
| Oxidative stress | |||
| Funct. div.b | Anabolism | Anabolism | Catabolism |
| 2.43E-12 enrich. p-val | 2.2E-3 enrich. p-val | 8.51E-6 enrich. p-val | |
| Central metabolism | |||
| 8.51E-6 enrich. p-val | |||
| Ess. genesc | 60 essential genes | 204 essential genes | 8 essential genes |
| 3.3E-9 depl. p-val | 2.4E-12 enrich. p-val | 9.8E-45 depl. p-val | |
| TF targetsd | FlhDC | LexA | CRP |
| (TrpR) | SoxS | IHF | |
| (TyrR) | DnaA | FhlA | |
| (Lrp) | PurR | NarP | |
| GadE | CysB | ||
| SF targetsd | σ28 | σ70 | σ38 |
| (σ70) | (σ24) | ||
| SF presente | σ28 | σ54 | σ24 |
| σ32 | σ38 | ||
| σ70 |
aSummary of GO enrichment results, full listing available in supplementary table S2, Supplementary Material online.
bEnriched functional divisions using the annotation provided by Seshasayee et al. (2009).
cEssential E. coli genes.
dTarget genes for given TF or SF enriched in cluster, TFs in parenthesis were not significant according to multiple testing criterion, full listing available in supplementary table S3, Supplementary Material online.
eGene encoding for SF present in cluster.
For S. Typhimurium, the division into clusters is not as obvious, and we end up with five major functional expression classes (fig. 2b). Here, the correlation profiles of Scl1 (833 genes) and Scl3 (741 genes) seem to be the most similar (MS) while displaying only weak correlation or anticorrelation to most other functional expression classes. The genes of Scl5 (321 genes) also display a similar pattern to that observed for the two classes Scl1 and Scl3 but seem to have a more pronounced correlation with their own cluster of genes and have a strong anticorrelation with Scl2 (545 genes) and Scl4 (445 genes). Finally, Scl2 and Scl4 also cluster together with similar correlation patterns. The S. Typhimurium functional expression classes can be found in supplementary data set S3, Supplementary Material online.
The functional roles of these gene clusters based on various gene annotation sources are summarized in table 3. The amount of information available for S. Typhimurium is limited, with several important biological processes being heavily underrepresented in the employed annotation, such as anaerobic respiration (six genes) and biofilm formation (one gene). Also as most pathogenic genes in S. Typhimurium have no orthologs in E. coli, these genes could not be included in this analysis. However, in the comparison with the full genome functional expression classes, it was clear that the genes that were found to be part of the same class as these pathogenic genes are here present in the Scl3 class (see supplementary material S4, Supplementary Material online). We relied on additional gene characterizations collected from the literature to bridge the gap in annotation information (Lawley et al. 2006; Evans et al. 2011) and included target gene predictions for 48 TFs in S. Typhimurium. In summary, we found that Scl1 seemed to be enriched for sulfur compound and vitamin metabolism genes. The genes in the Scl2 class code for most of the key cellular processes and cellular components, and represent the largest fraction of essential genes. Many Scl3 genes can be associated to pathogenesis as we find a number of enriched infection-related biological processes, such as cell adhesion. Scl3 is possibly also related to anaerobic respiration based on the ArcA target enrichment. Scl4 is mostly annotated with aerobic respiration, nitrogen compound biosynthesis, and cell motility. Lastly, the Scl5 class seems to contain a number of stress response genes as is supported by the enrichment of genes associated with the stress response ontology and targets of Fis, a global stress response TF.
Table 3.
Functional Evaluation of the S. Typhimurium Expression Classes.
| S. Typhimurium expression class | Scl1 | Scl2 | Scl3 | Scl4 | Scl5 |
|---|---|---|---|---|---|
| No. genes | 833 genes | 545 genes | 741 genes | 445 genes | 321 genes |
| GO enrich.a | Amino acid biosynthesis | Cell cycle | Transport proteins | Aerobic respiration | Response to stress |
| Sulfur metabolism | Cellular component biosynthesis | Cell adhesion | Nitrogen compound biosynthesis | ||
| Vitamin biosynthesis | Lipid biosynthesis | Cell motility | |||
| Transcription | |||||
| Translation | |||||
| Infection genesb | 11 inf. genes | 5 inf. genes | 28 inf. genes | 5 inf. genes | 7 inf. genes |
| Essential genesc | 30 ess. genes | 64 ess. genes | 28 ess. genes | 28 ess. genes | 12 ess. genes |
| TF pred. targetsd | (FadR) | (ArgP) | IclR | IscR | Fis |
| (TyrR) | (Fur) | NanR | FlhDC | (PhoP) | |
| (GntR) | (DnaA) | (FNR) | (GalR) | (FruR) | |
| (CRP) | (MelR) | (GlpR) | |||
| (ArcA) | |||||
| (H-NS) | |||||
| (SoxS) | |||||
| ArcA pot. targetse | 36 targets | 27 targets | 87 targets | 46 targets | 25 targets |
| SF presentf | σ24 | σ28 | σ32 | ||
| σ54 | σ38 | ||||
| σ70 |
aSummary of GO enrichment results, full listing available in supplementary table S4, Supplementary Material online.
bGenes required for long term infection as identified by Lawley et al. (2006).
cEssential S. Typhimurium genes.
dTarget genes for given TF enriched in cluster, TFs in parenthesis were not significant according to multiple testing criterion, full listing available in supplementary table S5, Supplementary Material online.
eGenes directly or indirectly regulated by ArcA as identified by Evans et al. (2011).
fGene coding for SF present in cluster.
Expression Divergence Reflects Differences in Lifestyle
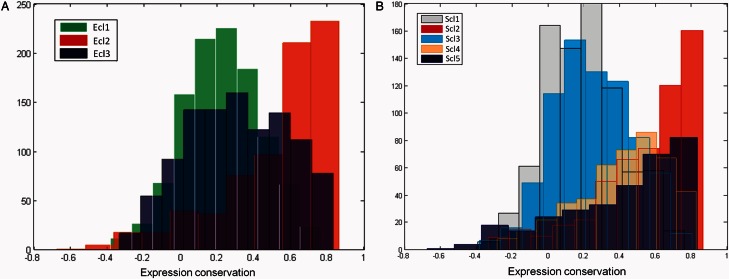
Now that we have defined functional expression classes, they can be related back to the EC between E. coli and S. Typhimurium. The EC scores of the genes in each expression class are shown in figure 3. Interestingly, almost every functional expression class can be characterized as either being more conserved or more diverged than average: The distribution of the EC scores are not bimodal but, with only one exception, follow the same pattern as either the conserved or divergent background distributions. The conserved classes have a distribution with a strong peak at higher EC scores with a tail to the left, whereas the diverged classes follow a normal distribution centered around a low EC score. Even when accounting for the variation of the EC score, these classes seem to be conserved or diverged. Regarding the E. coli functional expression classes in figure 3A, the genes from Ecl1 seem to be the most diverged in their expression domains with only very few members being assigned a high conservation score. This seems to indicate that at least some of the anabolism and central metabolism-related genes have a different expression pattern in S. Typhimurium. On the other hand, the EC scores for Ecl2 are on average the highest. This is in line with our previous observations that most essential E. coli genes are present in this class and that the expression patterns of these genes are more conserved than average. The distribution of the Ecl3 scores seems slightly bimodal and is thus the only exception to the general observation. This might indicate that although part of this class has conserved its expression domains between E. coli and S. Typhimurium, there is another part that has diverged. From figure 3B, we can conclude that the functional expression classes of S. Typhimurium also greatly differ in their expression correlation values. Most of the genes in both Scl1 and Scl3 have a below average conservation score. This is interesting given the relationship that we identified Scl3 as the expression class most related to pathogenesis, with the correlation profiles of Scl1 being very similar to those of Scl3 according to the dendrogram of figure 3B. Again the class most enriched for essential genes has the strongest EC, namely Scl2. Given the overlap between the essential genes of E. coli and S. Typhimurium, it is likely that many genes from this class have retained their function across these two species and are thus mapped to a similar functional expression class. Many of the Scl5 genes also have a high conservation score but the distribution has a heavy tail, possibly indicating that there is a small set of genes within this class whose expression might have diverged. The genes of Scl4 also tend to be conserved in their expression but less outspoken than Scl2 or Scl5.
Fig. 3.
Histogram of the EC score distribution for the orthologous genes split by the functional expression classes found in the expression compendia of the core genomes. (A) The distribution of EC scores for each of the three Escherichia coli classes (Ecl1: green, Ecl2: red, and Ecl3: blue). (B) The distribution of EC scores for each of the five Salmonella enterica serovar Typhimurium classes (Scl1: gray, Scl2: red, Scl3: cyan, Scl4: orange, and Scl5: blue).
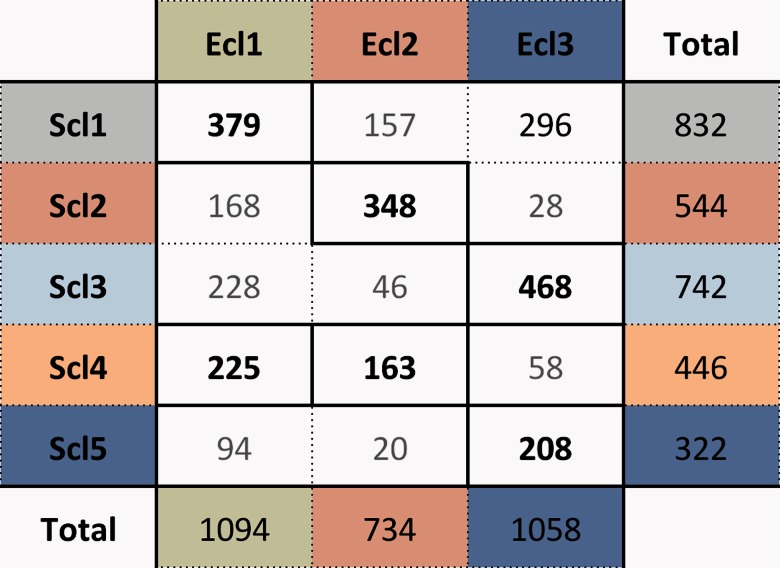
Figure 4 visualizes the overlap in gene content of the different functional expression classes between both organisms. Each functional expression class can be found to be enriched for the genes of at least one class of the other organism, thus indicating that these clusters are in some way preserved across evolution. The S. Typhimurium genes from Scl2 and Scl4 correspond to the E. coli genes in Ecl1 and Ecl2. Indeed, both were assigned similar functional roles in the previous analysis. Both Scl2 and Ecl2 contain genes involved in key cellular processes, such as the synthesis of the cellular components, and both are enriched in essential genes. Also Ecl1 and Scl4 share many similarities: Both include genes for cell motility and are thus regulated by FlhDC in each case, and both also include genes for aerobic respiration. They also share very similar correlation profiles; a high overall correlation to the essential classes (Ecl2 and Scl2, respectively) and a strong inner correlation, which is in line with the high EC scores of Scl2 and Scl4. There is also a strong overlap between the genes of Ecl3 and Scl5. In this case, both Scl5 and Ecl3 were reported to be involved in general stress response. These two classes also display a very similar set of correlation profiles, with a high inner correlation and anticorrelated to the essential class, meaning that the set of genes in this overlap of these two classes have likely been conserved in their functionality, which is further supported by the high EC scores of Scl5. The equivalent class for the pathogen-associated Scl3 seems to be Ecl3 as almost half of the Ecl3 genes map to those of Scl3. Here, both classes seem to be enriched for transporter proteins and targets for various global regulators, such as CRP and IHF. There is a clear difference in the correlation profiles of these two classes though: where all genes of Ecl3 had high inner correlation and were clearly anticorrelated with the essential gene class, this is much less outspoken for Scl3. Furthermore, the correlation between Scl3 and Scl5, where a large segment of the other Ecl3 genes mapped to, is very low. This indicates that the genes of Scl3 have a different expression profile than those of the stress-response cluster unlike the equivalent genes in E. coli and explains the poor conservation score of the Scl3 genes and the bimodal distribution of the Ecl3 scores. The other divergent class, namely Scl1, is not only enriched for mapping to Ecl1 but also contains many genes mapping to Ecl3 (neither depleted nor enriched). The main similarity between Scl1 and Ecl1 is that they were enriched for both amino acid metabolism and vitamin biosynthesis. Although Ecl1 was strongly correlated to the essential gene class Ecl2, Scl1 is not and its expression profiles are more related with Scl3. As Scl1 represents the largest mapping to Ecl1, it accounts for the low EC score of both these classes.
Fig. 4.
Overlap between the functional expression classes of Escherichia coli (columns) and Salmonella enterica serovar Typhimurium (rows). Reported is the number of orthologous gene pairs in each combination of classes. Numbers printed in bold are overlaps between classes that are significantly enriched (P value < 0.01) and those that are faded out are significantly depleted for each other (P value < 0.01).
The functional expression classes that were overall diverged in their expression warrant further study. Given that these represent a subset of the core compendium, selected independently from the EC scores, their fraction of conserved genes may differ from the original estimation. Indeed, a re-evaluation of this fraction reveals that for the combination of Scl1 and Scl3, the percentage of conserved genes can be estimated at 42%. This signifies that for these classes, the false-positive rate is only 21% at our earlier cutoff of −0.1. A closer look at the genes in both the Scl1 and Scl3 classes that were assigned an EC score of −0.1 reveals a number of interesting genes. The full listing of these genes can be found in supplementary tables S6 and S7, Supplementary Material online, for Scl1 and Scl3, respectively. The most diverged genes of these classes include many involved with cell adhesion, such as the E. coli sfmA, sfmC, sfmF, sfmH, flhC, flhD, and fimI. The divergent expression of these genes can be linked to the diversity in lifestyles as cell adhesion is an essential component of many host–cell interactions (Kline et al. 2009). The sfm operon is known to be poorly expressed in the E. coli K12 strain but is known to promote the adhesion to eukaryotic epithelial cells when they are constitutively expressed (Korea et al. 2010). The importance of this operon is further demonstrated by the fact that the S. Typhimurium homolog of sfmF has been shown to direct host–cell-specific recognition (Guo et al. 2009). Further, the divergent Scl3 genes include the TF cysB. Unlike E. coli, S. Typhimurium is able to use thiosulphate as an alternative receptor. This provides a growth advantage for Salmonella in an inflamed gut (Winter et al. 2010) and cysB, as a regulator of sulfur metabolism, could play a role in this process. Additionally, the anti-SF rseB gene is also part of the diverged Scl3 set. This anti-SF regulates SF E, which has been known to regulate several virulence genes (Cano et al. 2001; Osborne and Coombes 2009). There are also several pseudouridine synthases present, which are known to modulate macrophage caspase-1 activation in Francisella tularensis (Ulland et al. 2013). Other genes of interest include pagP, a PhoPQ-regulated palmitoyl transferase for lipid A; sdiA, a regulator of quorum sensing and virulence (Ahmer et al. 1998; Volf et al. 2002); permeases of oligopeptides, such as oppF and oppB (Goodell and Higgins 1987; Orchard and Goodrich-Blair 2004); and several genes involved in drug resistance, such as emrE and nudF. Both the Scl1 and Scl3 sets also include many genes with divergent expression, which have not yet been characterized and that might be interesting targets for further study.
Co-Ortholog EC
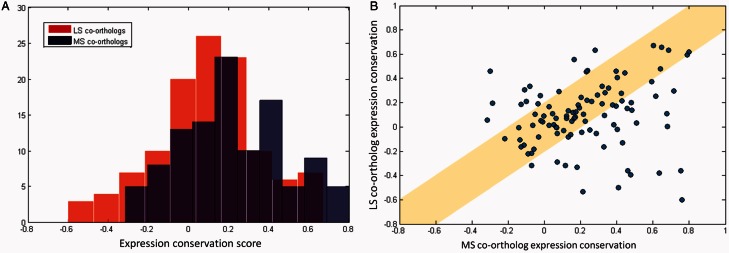
The previous analyses were limited to the gene pairs for which there is only one clear ortholog in both species, and thus, the “core genome" as used here was very strict. The ortholog mapping identified a number of instances where one species had a single copy of a gene, whereas the other had two or more. These genes were not included in the core genome list to avoid ambiguity in the gene mapping. Nevertheless, it would be interesting to investigate to what extent the EC of these genes corresponds to our previous findings. Multiple gene copies in a single organism can be the result of gene duplication and/or gene loss events after these two species diverged or a recent horizontal gene transfer (Kunin and Ouzounis 2003; Price et al. 2008; Touchon et al. 2009). We shall simply consider all instances as co-orthologs without further distinction, as the goal of this article is to study expression adaptation to the environment and not the evolutionary origin of the genes. In each set of co-orthologs, there is always one copy whose protein sequence is MS to that of the unique copy in the other species. This copy will be referred to as the MS co-ortholog and at face value the most likely to have retained its biological function, in which case it could be expected to show a higher EC than the least similar (LS) co-ortholog. Thus, for each of these instances, the EC score is calculated as before. The EC score can be calculated for every co-ortholog against the unique copy of the other species. The results indicate that on average, the gene identified as the MS co-ortholog has a higher EC score than the LS co-orthologs (with a P value of 5.2E-3 in a KS-test), as shown in figure 5A. The more similar protein copy tends to retain more of its expression characteristics, supporting the notion that it is more likely conserve its function. Another way to visualize this is by plotting the EC score for the MS co-orthologs and the LS co-ortholog from the same duplication pair against each other (fig. 5B, more detailed results can be found in supplementary table S8, Supplementary Material online). A large number of co-ortholog pairs have similar EC values for both co-orthologs. There is a small segment of pairs where the LS co-ortholog has a higher conservation score than the MS co-ortholog (fig. 5B above the diagonal). However, this seems to be more the exception than the rule and is most often case for co-orthologs where both copies have poor EC. The most extreme example is the S. Typhimurium ydcR gene, which has two co-orthologs in the E. coli, namely yjiR and ydcR, with an EC score of 0.46 and −0.29, respectively. Unfortunately, nothing is known about these genes in either organism except that they are predicted to be GntR-type TFs. Protein sequence alignment shows that the E. coli ydcR copy is almost identical to the S. Typhimurium gene (data not shown). Although it is not opportune to draw conclusions on their functional divergence based on this analysis alone, these are certainly prime candidates for further investigation. A much larger set of gene pairs follows the expected trend where the MS co-ortholog has a better EC than the LS co-ortholog (fig. 5B under the diagonal). The most extreme case here is for the S. Typhimurium pitA gene where the E. coli pitA and pitB co-orthologs have an EC score of 0.76 and −0.60, respectively. Indeed, expression regulation of the MS co-ortholog pitA is very different from that of the LS co-ortholog pitB: Although both pitA and pitB encode for a phosphate transporter, pitA is constitutively expressed, and pitB is only expressed under conditions of phosphate stress (Harris et al. 2001).
Fig. 5.
EC score of the co-orthologous gene pairs of Escherichia coli and Salmonella enterica serovar Typhimurium, which were not included in our core genome. EC scores were calculated by integrating each co-ortholog gene pair in turn into the expression correlation matrices and recalculating the EC. The resulting score assigned to the co-ortholog gene pair is considered as its EC score. (A) Histogram of the distribution of the EC scores of the MS co-orthologs (blue), which have the highest protein similarity of the gene pairs between the two species in the same co-ortholog cluster, and the LS co-orthologs (red), which are the remainder of the co-ortholog gene pairs. (B) Direct comparison of the MS co-ortholog EC (x axis) and the LS co-ortholog (y axis) EC of the same co-ortholog cluster. The yellow band along the diagonal indicates the segment of the plot where both co-orthologs seem to have diverged their expression at the same rate (within an error margin of 0.2).
Discussion
In this article, we attempted to address the question if the orthologous genes between E. coli and S. Typhimurium display the same expression domains. We compared the orthologous genes between these species using the ICC methodology on two publicly available microarray expression compendia. Using background distributions, we estimated that a quarter of the genes in the core genome have divergent expression domains. However, from the same analysis, it was clear that the EC score is very susceptible to changing conditions as we were able to show by comparing the E. coli compendia to itself. It is likely that this finding is not unique to this article but will also be present in prior studies that use similar methodologies. The effect of expression variation between different experimental conditions has been studied in the past and found to be not a significant factor in the comparison of expression domains in similar experimental setups as those performed in this article (Dutilh et al. 2006). However, the size of the used compendia and their consistent condition annotation allowed us to accurately estimate the effect of comparing expression data from different experiments under different conditions. As previously discussed, the potential EC score can be very low even in the case of perfect conservation. This signifies that there is a large error intrinsic to this approach and that EC scores alone are insufficient to conclude if a gene pair has diverged. In this article, we circumvented this weakness by not treating the EC score as an absolute truth but rather as an indication that a gene or a set of genes have likely retained their expression domains or not. In this manner, we saw strong EC for most genes that were essential for growth in both organisms. A result that was not unexpected as it has been noted in prior studies on other organisms (Stuart et al. 2003; Tirosh and Barkai 2008; Zarrineh et al. 2011).
Further, we were able to show that the expression divergence was largely independent from the protein similarity between gene pairs, as was also found in studies on other data sets (Le Gall et al. 2005; Khaitovich et al. 2005; Tirosh and Barkai 2008). Similar results were found when dealing with co-orthologs, where the divergent gene also tended to have less protein similarity, but again this relationship is far from absolute as we found several exceptions. This too had been observed in other organisms (Wagner 2000; Tirosh and Barkai 2007). Although it is possible that the bias intrinsic to the EC score due to the variation into the compendia had an effect on these results, they still support the theory that evolution of EC and protein similarity can happen independently.
In a more comprehensive analysis, we identified functional expression classes in each species and related those back to the EC scores for the core genome. These expression classes were functionally consistent and had a common EC. For E. coli, we found three clusters that seemed to represent a trade-off between genes responsible for growth and those gene enabling survival under less than ideal conditions. The classes in E. coli were also clearly enriched for regulatory programs such as sigma and TFs that are known to play an important role in such conditions. In fact, the correlation profiles of these genes might group together exactly because they share a regulatory program. Indeed, large-scale expression clustering approaches have been known to have a strong relationship to the underlying regulatory network (Treviño et al. 2012). The correlation profiles of the orthologous genes in S. Typhimurium also feature three classes that correspond to the three classes of E. coli, with which they not only share a similar functional annotation but were also estimated to have strong EC. The genes in these classes were primarily involved in growth, biosynthesis of cellular components, aerobic respiration, cell motility, nucleotide biosynthesis, and stress response. Salmonella enterica serovar Typhimurium also has two additional classes, Scl1 and Scl3, that show diverged expression domains. Our analysis indicates that the genes in these classes, although representing various “backbone” metabolic pathways, are used in different ways compared with E. coli and may help accommodate the virulent life style of S. Typhimurium. The involved metabolic pathways cover a variety of biological processes, such as cell adhesion, anaerobic respiration, amino acid metabolism, and sulfur metabolism. Although seemingly disparate categories, it is interesting to note that most enriched amino acid processes were those related to sulfur containing amino acids and that certain sulfur compounds are known to be involved in Salmonella virulence. In an inflamed gut, Salmonella can rely on thiosulphate as an alternative respiratory electron acceptor to support its growth and gain a competitive advantage, whereas E. coli cannot (Hensel et al. 1999; Price-carter et al. 2001; Winter et al. 2010). Additionally, we could directly relate the expression behavior of Scl3 to those of known species-specific (i.e., not part of the core genome) virulence genes of S. Typhimurium (see supplementary material S3, Supplementary Material online). It seems that being a pathogen is a major cause of the expression variation of S. Typhimurium when compared with the commensal E. coli. These findings match previous reports where it was found that the transcriptomes of different prokaryotes group together based on lifestyle (Le Gall et al. 2005). Furthermore, the pathogenesis of S. Typhimurium has been linked to several sigma and TFs (Groisman and Mouslim 2006), which supports our finding of a “pathogenesis" functional expression class in S. Typhimurium. The genes in this class would be interesting targets for further research into pathogenic traits of S. Typhimurium, as they are also present in the nonpathogenic E. coli but seem to be functionally different as attested to by their diverging expression.
Materials and Methods
Data Sets
For the purposes of this article, we worked with the genomes of E. coli K12 (NC_000913.2) and S. Typhimurium LT2 (NC_003197.1), as these are the strains that have been best characterized for these two species.
The orthologous genes were identified using the OrthoMCL v1.4 algorithm with the default settings on the protein sequences of both strains (Li et al. 2003). In this manner, we found 2,944 genes with only a single homolog in either species (one-to-one mapping). We also found 75 genes with a single copy in E. coli and multiple copies in S. Typhimurium and 49 genes in the reverse direction (one-to-many mapping). We disregarded the many-to-many results (48 gene clusters).
The microarray data sets were retrieved from the cross-platform expression compendia COLOMBOS v1.9 (Engelen et al. 2011). This data consisted of 1,570 measured conditions for E. coli and 925 conditions for S. Typhimurium. The 58 genes for which expression values were measured in less than half of the conditions present in either compendia were excluded from further analysis. The resulting list of 2,886 genes is what we refer to as the core genome, that is, a set of genes that have been conserved in both species without any additional duplications. This list of gene pairs was used throughout the entire article unless specifically mentioned otherwise.
EC Calculation
The EC score is calculated using the ICC methodology (Dutilh et al. 2006; Tirosh and Barkai 2007). In brief, we construct a correlation matrix of 2,886 × 2,886 for each organism by calculating the Pearson correlation coefficient between the expression profiles of each pair of core genes with a single organism. Thus, every element of the matrix is the correlation value of the gene on the row versus the gene on the column across every measured condition. This correlation matrix is symmetrical across the diagonal, and the diagonal consists of perfect correlation values. Each row can thus be seen as the correlation profile of a given gene, which lists the correlation of this gene against all the other genes. These correlation matrices are now ordered based on the orthology information, so that the equivalent rows of the two matrices correspond to the correlation profiles of a pair of orthologs. As these matrices have similar dimensionality, we can compare the equivalent rows by calculating their Pearson correlation. To correct for the influence of orthologous genes whose expression has diverged on the final score, the correlation is recalculated giving larger weights to genes of which expression has been conserved between the two species. The weight assigned to each orthologous gene pair is equal to the correlation calculated in the last round and is used in the calculation through a standard weighted Pearson correlation methodology. This process is iterated at least 10 times until an optimum is reached (subsequent iterations do not change the correlation values significantly). For every orthologous gene pair (i.e., every row in both matrices), we acquire a score that measures the conservation of its expression in the two compendia. This score is based on the correlations of the expression values with other orthologous pairs whose expression has been conserved. In effect, every orthologous gene pair is assigned a single value between −1 and 1. In this case, a value of 1 signifies perfect conservation of expression with respect to the correlation with all other genes. Although 0 signifies no conservation in the correlation values between the given gene and the other genes of the compendium. A value of −1 signifies a reverse of expression, that is, the genes that were correlated with the expression of the orthologous gene in a single compendium are anticorrelated with the ortholog in the other compendium and vice versa.
Background EC Distribution
In prior studies, the background distribution in case of no conservation was calculated by swapping the correlation profiles for two genes in a single correlation matrix while keeping the remainder the same and recalculating the conservation score. However, this can create unrealistic situations in the correlation matrix as one might end up with a gene that is poorly correlated with itself within a single compendium. We therefore permute the expression values of a single gene in one of the compendia, recreate the entire correlation matrix, and recalculate the EC score. This process is iterated for every gene pair, and the score for the permutated gene is kept.
The background distribution simulated in the case of conserved expression domains was accomplished by splitting a compendium into two equal halves and applying the ICC methodology on each half as if they were separate species. The background distribution shown in this article is derived from 10 random divisions of the E. coli compendium. Similar results were achieved with the S. Typhimurium compendium (results not shown). More information about the calculation of the background distribution can be found in supplementary material S2, Supplementary Material online.
Gene Ontology
The GO information for both species was downloaded from the UniProt-GO Annotation database (Dimmer et al. 2011). Enrichment calculations were achieved by applying a one-sided hypergeometric distribution to each biological process ontology present in the relevant gene set, where the algorithm will propagate up the GO tree as long as no enrichment is found. The P value is then adjusted for multiple testing using the Bonferroni correction for all tested ontologies. Any ontology with less than five assigned genes is not shown in the results, so that listings remain brief and only report general ontology categories. Please note that for the gene pair sets (lists of genes from both species), we used the E. coli ontology mapping as it is much more comprehensive.
Essential Genes
The list of essential genes was extracted from the Database of Essential Genes (Zhang et al. 2004). For E. coli, we used the data from the Keio collection (Baba et al. 2006) (300 essential genes), and for S. Typhimurium, the data collected by Knuth et al. (2004) (243 essential genes). A list of the orthologous gene pairs essential in both species can be found in supplementary table S1, Supplementary Material online.
Protein Similarity
The protein similarity as used in this article was calculated by Protein–Protein Blast 2.2.23 as applied on the protein sequences of the orthologous gene pairs (Altschul et al. 1990). The score reported as positive amino acid hits in the alignment (sequence identity) was then extracted from the results and divided by the alignment length. Using other measures (Blast E value, bit score, and coding sequence nucleotide alignment score) gave similar results (data not shown).
Intergenic Nucleotide Sequence Alignment
The intergenic regions were acquired for both species by extracting the nucleotide sequence upstream from the translation start site of every gene toward the edge of the previous gene. To only compare promoter regions, this was only done for genes that are assigned as being first in their operon in the E. coli RegulonDB database v7.2 (Gama-Castro et al. 2011). The sequences of each orthologous gene pair were then aligned with the Needleman–Wunsch algorithm. The settings used were 1 for a match, −1 for a mismatch, and −2 for the gap penalty. The end gap penalty was set at 0, so that sequences at different lengths are not penalized. The eventual alignment score is then divided by the length of the shortest nucleotide sequence.
Functional Expression Classes
To identify the main expression classes present in each species, the expression correlation matrices are constructed as described earlier for the ICC methodology. The gene classes are then constructed by calculating the Euclidean distance between each row of the correlation matrix, and the tree is built based on the inner squared distance of the clusters (Minimum variance algorithm of Matlab 2008a). The constructed tree is cut at 110 distance units for both organisms and the resulting gene sets are then grouped into classes.
Regulatory Network and Enrichment
The regulatory interactions for the SFs and TFs for E.coli were both downloaded from RegulonDB v7.2 (Gama-Castro et al. 2011). This database contains targets for 176 TFs and 7 SFs. As there is no equivalent public collection for S. Typhimurium, the regulatory interactions for this species were predicted using the CRoSSeD motif detection algorithm (Meysman et al. 2010). The known binding sites from RegulonDB were used to train models for all E. coli TFs with more than four binding sites, which also had a clear ortholog in S. Typhimurium with a conserved DNA-binding domain (protein similarity ≥ 90%). This resulted in 48 binding site models, of which 42 are for local TFs and 6 are for global TFs. The regulatory targets were then predicted by screening the S. Typhimurium intergenic region with these 48 binding site models and assigning the highest score found in a region to the adjacent genes while accounting for directionality. For our purposes, it was sufficient to assign the regulatory interactions to the top 30 potential target genes of the local TFs and the top 100 genes for the global TFs. For S. Typhimurium, this procedure resulted in 3,000 regulatory interactions, which can be found in supplementary data set S4, Supplementary Material online.
The calculation of TF enrichment on any set of genes is performed by calculating the hypergeometric statistic based on the known interactions for E. coli or the predicted interactions for S. Typhimurium. With Bonferroni multiple testing corrections, the P value cutoff for enrichment is 2.7 × 10−4 in E. coli (0.05/176 TFs + 7 SFs) and 1 × 10−3 in S. Typhimurium (0.05/48 TFs).
Additional Annotation Sources
A high-level functional division of E. coli genes was used from Seshasayee et al. (2009), where several genes were characterized as involved in catabolism (186 genes), anabolism (339 genes), or central metabolism (109 genes) based on a metabolic network constructed from the literature.
The findings of Lawley et al. (2006) were used to identify a set of genes related to pathogenesis. In this study, a mutant library of S. Typhimurium genes was screened on its effect on long-term systemic infection in mice. A total of 120 genes were identified in this manner, of which 57 could be found in the core genome.
The direct and indirect regulatory targets of ArcA, an anaerobic and virulence regulator, for S. Typhimurium were taken from Evans et al. (2011). This study identified 392 genes whose expression changed in an arcA knock-out mutant of S. Typhimurium. Of these genes, only 221 were present in our core genome.
Co-Ortholog EC
To find the MS orthologs in the one-to-many cases we had ignored for the core genome, we used the one-to-one ortholog mapping from the results of Moreno-Hagelsieb and Latimer (2008), who classified the orthologs of E. coli and S. Typhimurium, among others, based on reciprocal best blast hits combined with a soft filtering by a Smith–Waterman alignment. This mapping consists of 3,125 ortholog pairs and is used to determine the MS co-ortholog in a set of co-orthologs (the one-to-many mappings). Other mappings based on other measures were also evaluated, and although some specific cases did change, the general conclusions remained the same. The EC score was then again calculated using the ICC methodology, with the exception that each core genome compendia is expanded by a single row of expression measurements for a single gene. This gene is the unique ortholog copy for one species and one of the co-orthologs in the other species. This entire procedure is iterated to get a score for every co-ortholog pair.
Supplementary Material
Supplementary material S1–S4, tables S1–S8, figures S2 and S3, and data sets S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Hans Steenackers and Sandra Vanpuyvelde for their valuable comments. This work was supported by the KU Leuven Research Council (GOA/011/2008, PF/10/010:NATAR, CREA/08/023); the Agency for Innovation by Science and Technology (SB-83297, SBO-BioFrame); Ghent University (Multidisciplinary Research Partnership “M2N”); and Research Foundation-Flanders (FWO-G.0329.09).
References
- Ahmer BM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F. Salmonella Typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister S, McCue L, Turse J, Monroe M. Comparative bacterial proteomics: analysis of the core genome concept. PLoS One. 2008;3:e1542. doi: 10.1371/journal.pone.0001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano D, Martínez-Moya M, Graciela Pucciarelli M, Groisman E, Casadesus J, Garcia-Del Portillo F. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect Immun. 2001;69:6463–6474. doi: 10.1128/IAI.69.10.6463-6474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MB, Loose M, Brookfield JFY. The evolutionary influence of binding site organisation on gene regulatory networks. Biosystems. 2009;96:185–193. doi: 10.1016/j.biosystems.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- Dimmer EC, Huntley RP, Alam-Faruque Y, et al. (55 co-authors) The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2011;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Agerer F, Michaelis K, Janka A, Buchrieser C, Samuelson M, Svanborg C, Gottschalk G, Karch H, Hacker J. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol. 2003;185:1831–1840. doi: 10.1128/JB.185.6.1831-1840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF, Feng DF, Tsang S, Cho G, Little E. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- Dutilh BE, Huynen MA, Snel B. A global definition of expression context is conserved between orthologs, but does not correlate with sequence conservation. BMC Genomics. 2006;7:10. doi: 10.1186/1471-2164-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen K, Fu Q, Meysman P, Sánchez-Rodríguez A, De Smet R, Lemmens K, Fierro AC, Marchal K. COLOMBOS: access port for cross-platform bacterial expression compendia. PLoS One. 2011;6:e20938. doi: 10.1371/journal.pone.0020938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, Hassan HM. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol. 2011;11:58. doi: 10.1186/1471-2180-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, Salgado H, Peralta-Gil M, et al. (28 co-authors) RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell EW, Higgins CF. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J Bacteriol. 1987;169:3861–3865. doi: 10.1128/jb.169.8.3861-3865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Mouslim C. Sensing by bacterial regulatory systems in host and non-host environments. Nat Rev Microbiol. 2006;4:705–709. doi: 10.1038/nrmicro1478. [DOI] [PubMed] [Google Scholar]
- Guo A, Cao S, Tu L, Chen P, Zhang C, Jia A, Yang W, Liu Z, Chen H, Schifferli DM. FimH alleles direct preferential binding of Salmonella to distinct mammalian cells or to avian cells. Microbiology. 2009;155:1623–1633. doi: 10.1099/mic.0.026286-0. [DOI] [PubMed] [Google Scholar]
- Harris R, Webb D, Howitt S, Cox G. Characterization of PitA and PitB from Escherichia coli. J Bacteriol. 2001;183:5008–5014. doi: 10.1128/JB.183.17.5008-5014.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. The genetic basis of tetrathionate respiration in Salmonella Typhimurium. Mol Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- Hindré T, Knibbe C, Beslon G, Schneider D. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol. 2012;10:352–365. doi: 10.1038/nrmicro2750. [DOI] [PubMed] [Google Scholar]
- Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, Garriga-Canut M, Serrano L. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khan SA, Everest P, Servos S, Foxwell N, Zähringer U, Brade H, Rietschel ET, Dougan G, Charles IG, Maskell DJ. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998;29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Knuth K, Niesalla H, Hueck CJ, Fuchs TM. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol Microbiol. 2004;51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- Korea C-G, Badouraly R, Prevost M-C, Ghigo J-M, Beloin C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ Microbiol. 2010;12:1957–1977. doi: 10.1111/j.1462-2920.2010.02202.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Kunin V, Ouzounis CA. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall T, Darlu P, Escobar-páramo P, Picard B, Denamur E. Selection-driven transcriptome polymorphism in Escherichia coli/Shigella species. Genome Res. 2005;15:260–268. doi: 10.1101/gr.2405905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- Mandel MJ, Silhavy TJ. Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability. J Bacteriol. 2005;187:434–442. doi: 10.1128/JB.187.2.434-442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal K, De Keersmaecker S, Monsieurs P, van Boxel N, Lemmens K, Thijs G, Vanderleyden J, De Moor B. In silico identification and experimental validation of PmrAB targets in Salmonella typhimurium by regulatory motif detection. Genome Biol. 2004;5:R9. doi: 10.1186/gb-2004-5-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, et al. (26 co-authors) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Meysman P, Dang TH, Laukens K, De Smet R, Wu Y, Marchal K, Engelen K. Use of structural DNA properties for the prediction of transcription-factor binding sites in Escherichia coli. Nucleic Acids Res. 2010;39:e6. doi: 10.1093/nar/gkq1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang FC, De Moor B, Vanderleyden J, Marchal K. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J Mol Evol. 2005;60:462–474. doi: 10.1007/s00239-004-0212-7. [DOI] [PubMed] [Google Scholar]
- Moreno-hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- Ochman H, Wilson AC. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Orchard SS, Goodrich-Blair H. Identification and functional characterization of a Xenorhabdus nematophila oligopeptide permease. Appl Environ Microbiol. 2004;70:5621–5627. doi: 10.1128/AEM.70.9.5621-5627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne SE, Coombes BK. RpoE fine tunes expression of a subset of SsrB-regulated virulence factors in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2009;9:45. doi: 10.1186/1471-2180-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome Biol. 2008;9:R4. doi: 10.1186/gb-2008-9-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica Serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshasayee ASN, Fraser GM, Babu MM, Luscombe NM. Principles of transcriptional regulation and evolution of the metabolic system in E. coli. Genome Res. 2009;19:79–91. doi: 10.1101/gr.079715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8:R50. doi: 10.1186/gb-2007-8-4-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Evolution of gene sequence and gene expression are not correlated in yeast. Trends Genet. 2008;24:109–113. doi: 10.1016/j.tig.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, et al. (41 co-authors) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treviño S, Sun Y, Cooper TF, Bassler KE. Robust detection of hierarchical communities from Escherichia coli gene expression data. PLoS Comput Biol. 2012;8:e1002391. doi: 10.1371/journal.pcbi.1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland TK, Janowski AM, Buchan BW, Faron M, Cassel SL, Jones BD, Sutterwala FS. Francisella tularensis LVS folate metabolism and pseudouridine synthase gene mutants modulate macrophage caspase-1 activation. Infect Immun. 2013;81:201–208. doi: 10.1128/IAI.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volf J, Sevcik M, Havlickova H, Sisak F, Damborsky J, Rychlik I. Role of SdiA in Salmonella enterica serovar Typhimurium physiology and virulence. Arch Microbiol. 2002;178:94–101. doi: 10.1007/s00203-002-0424-4. [DOI] [PubMed] [Google Scholar]
- Wagner A. Decoupled evolution of coding region and mRNA expression patterns after gene duplication: implications for the neutralist-selectionist debate. Proc Natl Acad Sci U S A. 2000;97:6579–6584. doi: 10.1073/pnas.110147097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MD, Groisman EA. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc Natl Acad Sci U S A. 2004;101:17162–17167. doi: 10.1073/pnas.0406038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, et al. (12 co-authors) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrineh P, Fierro AC, Sánchez-Rodríguez A, De Moor B, Engelen K, Marchal K. COMODO: an adaptive coclustering strategy to identify conserved coexpression modules between organisms. Nucleic Acids Res. 2011;39:e41. doi: 10.1093/nar/gkq1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Ou H-Y, Zhang C-T. DEG: a database of essential genes. Nucleic Acids Res. 2004;32:D271–D272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.