Abstract
Zymoseptoria tritici is an important fungal pathogen on wheat that originated in the Fertile Crescent. Its closely related sister species Z. pseudotritici and Z. ardabiliae infect wild grasses in the same region. This recently emerged host–pathogen system provides a rare opportunity to investigate the evolutionary processes shaping the genome of an emerging pathogen. Here, we investigate genetic signatures in plant cell wall degrading enzymes (PCWDEs) that are likely affected by or driving coevolution in plant-pathogen systems. We hypothesize four main evolutionary scenarios and combine comparative genomics, transcriptomics, and selection analyses to assign the majority of PCWDEs in Z. tritici to one of these scenarios. We found widespread differential transcription among different members of the same gene family, challenging the idea of functional redundancy and suggesting instead that specialized enzymatic activity occurs during different stages of the pathogen life cycle. We also find that natural selection has significantly affected at least 19 of the 48 identified PCWDEs. The majority of genes showed signatures of purifying selection, typical for the scenario of conserved substrate optimization. However, six genes showed diversifying selection that could be attributed to either host adaptation or host evasion. This study provides a powerful framework to better understand the roles played by different members of multigene families and to determine which genes are the most appropriate targets for wet laboratory experimentation, for example, to elucidate enzymatic function during relevant phases of a pathogen’s life cycle.
Keywords: adaptive evolution, host adaptation, natural selection, coevolution
Introduction
Coevolution makes a major contribution to generating and maintaining biodiversity (Thompson 1999). Signatures of coevolution are pre-eminent in host–pathogen systems because of the strong selective pressures that the pathogen and host exert on each other. Host–pathogen coevolution is a ubiquitous phenomenon that likely affects all organisms, but much of the research effort has been concentrated in systems of medical or economic importance. For example, human diseases such as malaria, AIDS, or influenza are caused by coevolving pathogens. Host–pathogen coevolution is equally important in agriculture, where livestock is attacked by coevolving parasites and crops are affected by an armada of rapidly coevolving bacterial, viral, and fungal pathogens. The development of effective human vaccines or sustainably resistant crops hinges on understanding the underlying genetic basis of coevolutionary adaptation.
Although traditional studies mainly sought phenomenological evidence for coevolution and adaptation, a key problem in modern evolutionary biology is to connect the observed phenotypes with a genotype. Recent advances in sequencing technologies and steadily dropping costs are making it possible to obtain whole-genome sequences from many nonmodel organisms, leading to the rapidly emerging fields of ecological and population genomics (e.g., Luikart et al. 2003). One important outcome of this development is the possibility to compare large numbers of candidate genes or to detect new target genes of selection without prior knowledge of the underlying phenotypic variation (Ellegren 2008).
Next-generation sequencing technologies also provide new approaches to study coevolution of pathogenic species allowing the comparison of multiple gene families at the genomic and transcriptomic level. Here, we focus on a group of key enzymes of plant pathogenic fungi that target plant cell walls. In addition to its structural function (Sarkar et al. 2009), the plant cell wall provides the first line of defense against pathogens and offers a significant source of pathogen nutrition (De Lorenzo et al. 1997). More than 120 years ago, De Bary (1886) proposed that pathogen-secreted proteins now called plant cell wall degrading enzymes (PCWDEs) play an important role in pathogenesis by destroying plant cells during infection by fungal pathogens. Plant pathogenic fungi exhibit complex life cycles that can include many different kinds of interactions with their hosts (see review by Horbach et al. 2011 and references therein). For example, hemibiotrophic fungi display a three-stage life cycle. During the first biotrophic stage, the fungus grows asymptomatically within the living plant, whereas the second necrotrophic stage is characterized by death of the plant tissue. During the final saprotrophic stage, the fungus completes its life cycle on the dead plant tissue. Over the last decades, it has become clear that fungal plant pathogens have evolved an arsenal of substrate-specific PCWDEs that cleave the various components of the plant cell wall, with different PCWDEs likely being utilized during different phases of the pathogen life cycle (Cooper 1983; Cooper et al. 1988; Xu et al. 2006). Thus, PCWDEs play a crucial role both in host cell destruction and as providers of pathogen nutrition, and it is likely that selection has had a significant impact on PCWDEs due to these vital functions. For example, adaptation to specific substrates (i.e., components of the plant cell wall) will reduce the diversity at nonsynonymous codon sites compared with synonymous sites due to functional optimization of enzymatic activity. This process will leave a genetic signature of purifying selection on some PCWDEs.
PCWDEs or some of their degradation products may also act as effector molecules that elicit defense responses in plants (Esquerré-Tugayé et al. 2000). In recent years, a growing number of plant proteins have been isolated that inhibit the activity of PCWDEs and were proposed to form part of the plant immune system (Debyser et al. 1999; McLauchlan et al. 1999; Federici et al. 2006). The growing evidence for a complex array of PCWDE inhibitors provides compelling evidence for a plant–microbe coevolutionary arms race between the pathogen-produced enzymes and these inhibitors (Juge 2006). In this case, selection will favor mutations that allow the pathogen to avoid recognition or inactivation of the affected PCWDE, resulting in genetic signatures of diversifying selection.
The wheat pathogen Z. tritici and its close relatives provide a powerful model system to differentiate the evolutionary processes affecting PCWDEs in plant pathogenic fungi. Zymoseptoria tritici is a filamentous fungus with a small complement of PCWDEs compared with other fungi (Ohm et al. 2012), and it is, therefore, less likely that a high redundancy in PCWDE functions exists. Zymoseptoria tritici has two closely related ancestral species, Z. pseudotritici and Z. ardabiliae, that were recently discovered infecting uncultivated grasses in the Fertile Crescent. It was hypothesized that Z. tritici emerged as a pathogen specialized to infect wheat during the domestication of wheat in the Fertile Crescent approximately 10,000 years ago (Stukenbrock et al. 2007). The genomes of several strains in each species were sequenced and compared, allowing the identification of the full complement of PCWDEs in each species and assignment of orthologs across species (Stukenbrock et al. 2011).
Although sequences are available for all PCWDEs in Z. tritici, it is not known when these genes are expressed during the host–pathogen interaction. Hemibiotrophic fungi such as Z. tritici display a distinctive three-stage life cycle, and we hypothesized that different PCWDEs may have evolved to have an optimized enzymatic activity during different phases of the pathogen life cycle. Life-cycle-dependent transcription patterns for members of three multigene families were found for the human malaria parasite Plasmodium falciparum revealed (Le Roche et al. 2003). Thus, we combined the population genomic data sets with transcription data from an RNA-Seq experiment to investigate the evolutionary forces acting on each PCWDE. We found that the evolution of PCWDEs in plant pathogenic fungi has been significantly affected by life cycle specialization and by varying degrees of purifying and diversifying selection. On the basis of these results, we formulate specific hypotheses regarding the evolutionary processes operating on candidate genes to gain insight into their specialized functions during the host–pathogen interaction. This information can then be used to determine which members of each family are the most appropriate targets for detailed wet laboratory experimentation to elucidate enzymatic function during relevant phases of the pathogen life cycle.
Results
Our data set included 48 PCWDEs from the reference genome Z. tritici IPO323 that were distributed among 21 Carbohydrate-Active EnZymes database (CAZy) families according to structurally related and shared functional domains (supplementary table S1, Supplementary Material online). All these genes were identified in all nine resequenced genomes of Z. tritici with an average amino acid (aa) identity of 98%. Although orthologs for all 48 PCWDEs were detected also in Z. pseudotritici (94% average aa identity), 46 genes were found in Z. ardabiliae (92% average aa identity) and 34 were found in the more distant outgroup species Z. passerinii (84% average aa identity).
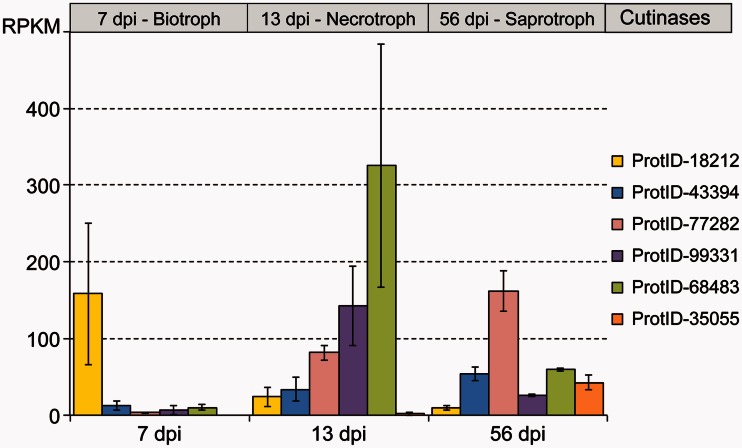
The transcription analyses are summarized in supplementary table S2, Supplementary Material online. By comparing transcription profiles among members of the same CAZy family, we were able to identify 28 genes displaying life-cycle-specific expression. For example, five of the six genes belonging to the CAZy family carbohydrate esterases 5-encoding cutinases displayed life-cycle-specific expression. ProtID 18212 was preferentially expressed during the biotrophic stage but only marginally expressed during the necrotrophic and saprotrophic stages in the life cycle. ProtID 68483 and ProtID 99331 were expressed mainly in the necrotrophic stage, and ProtID 77282 was expressed mainly during the saprotrophic stage (fig. 1). Similar patterns of life-cycle-specific expression were found in other CAZy families (supplementary fig. S1 and table S2, Supplementary Material online), consistent with the hypothesis of life cycle specialization.
Fig. 1.
Standardized transcription values (RPKM) from RNA-Seq experiments in Zymoseptoria tritici indicate differential in planta expression for cutinase genes during the biotrophic, necrotrophic, and saprotrophic life cycle stages. The gene expression bars represent the average from three biological replicates and error bars are one standard deviation from the mean; dpi, days post-inoculation. Detailed values for all cell wall degrading enzymes are given in supplementary table S2, Supplementary Material online.
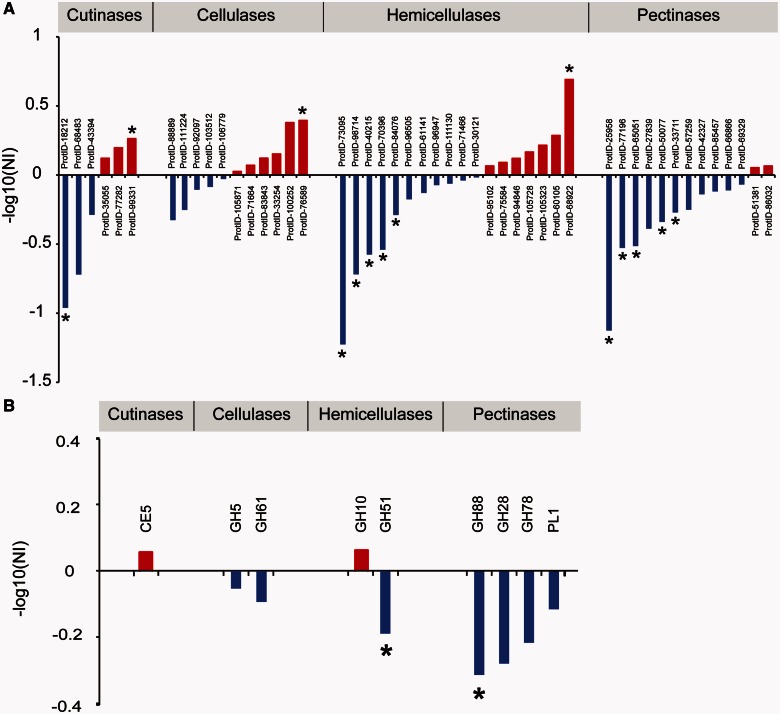
All PCWDEs showed nucleotide variability within and/or between species. Neutrality indices estimated from McDonald–Kreitman tests (MKTs) were significantly different from zero for 14 genes, suggesting that selection played an important role during the evolution of these PCWDEs (fig. 2; supplementary tables S3a and S3b, Supplementary Material online). With the exception of cellulases, significant purifying selection was indicated for 11 genes distributed over all classes. Accordingly, three genes were identified as being under positive selection, with one each attributed to the cutinases, cellulases, and hemicellulases.
Fig. 2.
Plot of McDonald–Kreitman test neutrality indices (NI) estimated from the combined pairwise species comparisons Zymoseptoria tritici–Z. pseudotritici, Z. tritici–Z. ardabiliae, and Z. pseudotritici–Z. ardabiliae. Values above zero (red) indicate diversifying selection, whereas values below zero (blue) indicate purifying selection. Significance values of P ≤ 0.01 are indicated with asterisks. (a) NIs for all 48 cell wall degrading enzymes organized according to CAZy families. (b) Combined NIs for multigene CAZy families. Detailed values are given in supplementary tables S3a and S3b, Supplementary Material online. CE, carbohydrate esterases; GH, glycoside hydrolases; PL, polysaccharide lyases.
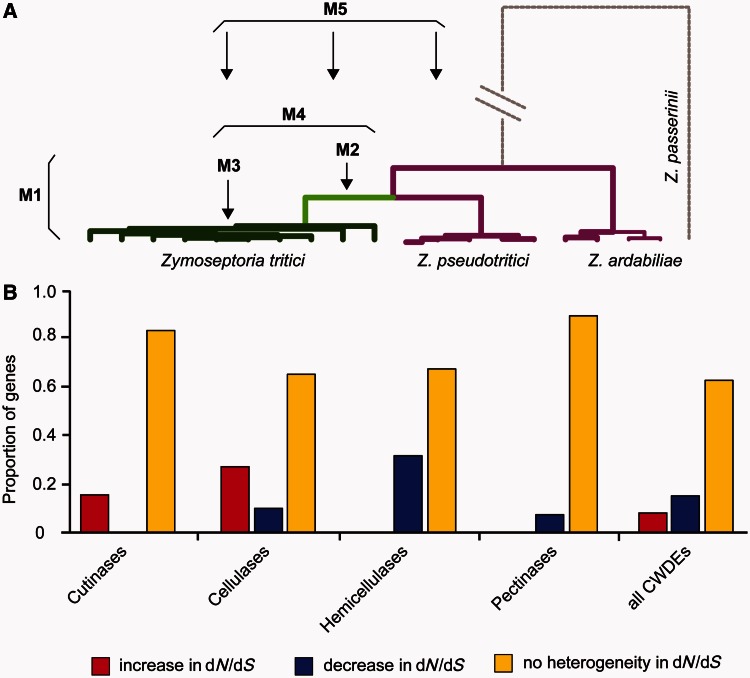
We used phylogenetic analyses to assess whether significant differences in selection pressure, as measured by dN/dS ratios, were operating on the PCWDEs during the evolution of Z. tritici. Models 1 and 2, which assume the same dN/dS ratios for Z. tritici and its ancestors (i.e., no selection heterogeneity), were the best models for 36 genes. However, 12 genes showed significant heterogeneity in dN/dS at P ≤ 0.01, supporting our hypothesis that changing selection pressure has affected the evolution of PCWDEs (fig. 3; supplementary table S4, Supplementary Material online). Of these, eight genes showed significantly lower dN/dS, suggestive of an increase in purifying selection, and four genes showed a significantly higher dN/dS, suggestive of an increase in diversifying selection in Z. tritici, for example, due to the new environment/host species (supplementary table S4, Supplementary Material online). A detailed summary of the main findings for each class of PCWDE is provided as supplementary results, Supplementary Material online.
Fig. 3.
Likelihood ratio tests to detect selection heterogeneity across the phylogeny of Zymoseptoria tritici against its ancestors for all 48 cell wall degrading enzymes. (a) Phylogeny and models used to assess heterogeneity in dN/dS. Models M2–M5 assume heterogeneity on different levels and are compared against the null-model M1 assuming a constant dN/dS value. (b) Gene frequencies of significantly higher dN/dS ratios (red) or significantly lower dN/dS ratios (blue) for Z. tritici at P ≤ 0.01. Nonsignificant selection heterogeneity is indicated in yellow. Detailed values are given in supplementary table S4, Supplementary Material online.
Discussion
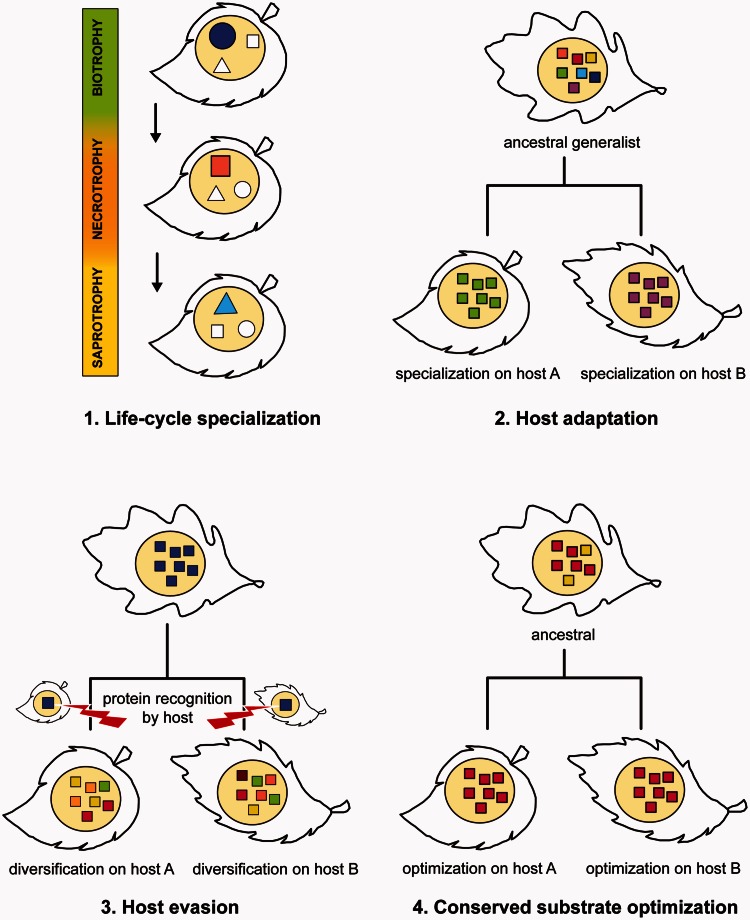
Parasites must adapt to their hosts during all stages of their life cycle including initial infection and host-dependent growth and reproduction. A crucial first step toward understanding the processes underlying host–pathogen coevolution is to identify the relevant genes and determine their roles during different phases of the infection cycle. Our findings revealed that the evolution of PCWDEs in plant pathogenic fungi has been significantly affected by life cycle specialization and by varying degrees of purifying and diversifying selection. In the following sections, we combine our results and propose specific but simplified hypotheses regarding the evolutionary forces acting on each gene and their possible cellular functions as illustrated in figure 5.
Fig. 5.
Schematic visualization of the four main evolutionary scenarios postulated to affect the evolution of PCWDEs in plant pathogenic fungi (yellow circles). Different hosts are represented by different leaf shapes. Different symbols indicate PCWDEs belonging to the same gene family and colors represent different alleles (protein variants) of a specific gene. 1) Life cycle specialization: Members of the same gene family are preferentially expressed at different stages of the pathogen life cycle. 2) Host adaptation: Purifying selection for different protein variants of the same gene on different hosts. 3) Host evasion: Recognition of a specific protein (blue) by the host and subsequent diversifying selection on the same protein to evade recognition. 4) Conserved substrate optimization: Purifying selection for the same optimized protein variant on different hosts.
PCWDEs Involved in Life Cycle Specialization
Many genes within a species can be grouped into multigene families based on similar enzymatic functions. However, the existence of gene families does not necessarily imply redundancy for the associated metabolic functions. We hypothesized that some PCWDEs belonging to the same enzymatic family have been selected for optimized function during different phases of the life cycle of a hemibiotrophic pathogen. Hemibiotrophic pathogens undergo different types of growth and metabolism during their life cycle in the host and therefore propagate through changing conditions in the plant, such as different pH environments that are likely to occur during the biotrophic, necrotrophic, and saprotrophic phases of the life cycle (scenario 1 in fig. 5). Under this evolutionary scenario, that we will call “life cycle specialization,” we did not necessarily expect to find genetic signatures of natural selection; however, we expected to detect differential expression patterns for different members of each class of PCWDE.
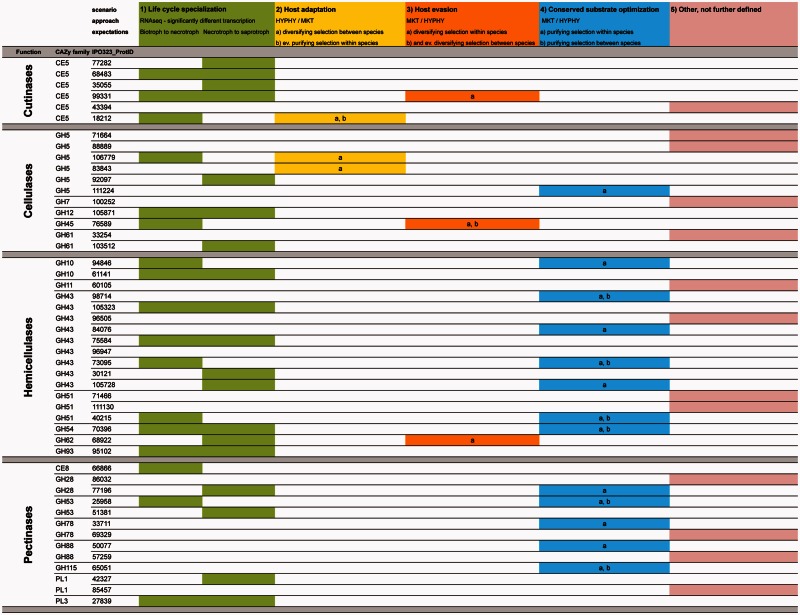
Twenty-eight of the 48 PCWDEs showed expression levels significantly different in distinct stages and, thus, can be classified as life cycle specific (fig. 4). Several examples were found in the cutinase family. Cutinases are not considered to be PCWDEs in the strict sense, but they are important enzymes for plant pathogenic fungi that need to penetrate the cuticle to initiate a successful infection. The role of cutinases in pathogenicity remains contradictory and circumstantial (Sweigard et al. 1992; Rogers et al. 1994; Yao and Koller 1995), and some authors proposed that cutinases are more important during the saprophytic growth of the fungus (Stahl and Schäfer 1992). Zymoseptoria tritici is thought to penetrate the plant exclusively through stomata (e.g., Kema et al. 1996), though several studies indicated that direct penetration through the cuticle also could occur (e.g., Weber 1922; Hilu and Bever 1957; Dancer et al. 1999). Given the consensus of stomatal infection, it was surprising to find six cutinase genes in Z. tritici. Our findings suggest that cutinase activity is important during all stages of the life cycle of Z. tritici, but different cutinases are active during different stages of the life cycle (fig. 1).
Fig. 4.
Categorization of PCWDEs according to the four main evolutionary scenarios postulated to affect their evolution (see also fig. 5). Only significant differences in transcript abundances among the different life stages are highlighted. The fifth column identifies the 13 enzymes that could not be categorized.
A common approach to understand the role of different PCWDEs during the pathogen life cycle (especially during the biotrophic or necrotrophic phases) has been to delete individual genes and compare the resulting phenotype on the host. The outcomes of many of these experiments have been inconclusive, which the investigators often attribute to functional redundancy within these multigene families (e.g., García-Maceira et al. 2000; Idnurm and Howlett 2001; Dobinson et al. 2004). However, because most of these experiments were conducted before fungal genome data were available, the investigators generally did not know how many genes were in each PCWDE family nor when a gene was expressed in planta during the pathogen life cycle. For example, although several studies, including expression analyses in Z. tritici, suggested that xylanases are virulence factors (Douaiher et al. 2007), disruption experiments, including analysis of a triple xylanase mutant, did not find an effect on virulence (e.g., Apel et al. 1993; Scott-Craig et al. 1998; Gόmez-Gόmez et al. 2002). However, our results indicated that the xylanase genes (classified in CAZy families GH10, GH11, and GH43; fig. 4) were differentially transcribed during the life cycle stages. Thus, we hypothesize that effects on virulence in Z. tritici will be found only for hemicellulases that are highly or exclusively expressed during the necrotrophic phase, for example, ProtID 61141 or ProtID 30121 (Supplementary table S2, Supplementary Material online).
PCWDEs Involved in Host Adaptation
The evolution of pathogens and their hosts are often tightly connected. For example, field and laboratory studies showed that infections of Daphinia spp. by the bacterium Pasteuria ramosa were highly dependent on the host species, on the host clones, on the population from which the hosts and parasites were collected, and on the ecological setting in which the infection occurred (Ebert 2005 and references therein). The evolution of pathogen PCWDEs is also likely to be affected by host specialization. Under this evolutionary scenario, which we will call “host adaptation,” we expected that diversifying selection will generate significant differences among orthologous enzymes in pathogens infecting different hosts, but we could expect to find purifying selection to optimize enzymatic function within species (scenario 2 in fig. 5). Zymoseptoria tritici has only recently emerged as a pathogen specialized to infect domesticated wheat (Triticum aestivum and T. durum), whereas the more ancestral species Z. pseudotritici and Z. ardabiliae are found on a diversity of wild grasses, including Lolium, Dactylus, and Agropyron (Stukenbrock et al. 2007). Three enzymes of Z. tritici fit our scenario of host adaptation (fig. 4): one cutinase (ProtID 18212) and two cellulases (ProtIDs 106779 and 83843). Interestingly, the two cellulases have their highest expression during the biotrophic phase of the life cycle. We speculate that these host-adapted cellulases in Z. tritici have evolved to digest a cellulose component unique to wheat and available to support pathogen growth during the biotrophic phase of its life cycle.
PCWDEs That Evolved to Evade Host Recognition
The outcome of an infection is determined by complex interactions between the host and pathogen. On the host side, specialized proteins have evolved to detect the pathogen and activate subsequent defense responses. On the pathogen side, proteins have coevolved to avoid host recognition and to suppress the host’s defense responses. Examples of diversifying selection often not only include host genes involved in pathogen recognition—such as the mammalian MHC and TLR genes (Hughes and Nei 1988; White et al. 2003) but also include pathogen genes encoding proteins that are targets of host recognition (Endo et al. 1996).
Some PCWDEs can be recognized by the plant immune system and will become targets of inhibitory proteins (Federici et al. 2006; Juge 2006) produced by the host plant during the biotrophic and/or necrotrophic stages in the life cycle (scenario 3 in fig. 5). Under this evolutionary scenario, which we will call “host evasion,” our expectation was that diversifying selection will operate on an enzyme within the pathogen species to avoid recognition, similar to what has been observed for pathogen effectors (e.g., Schürch et al. 2004; Liu et al. 2005; Stukenbrock and McDonald 2007).
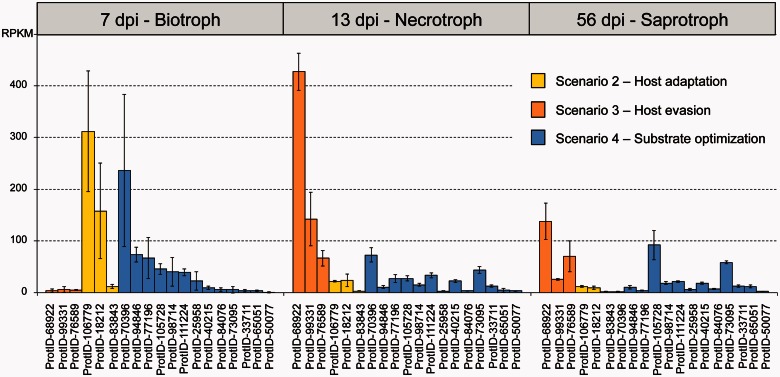
Zymoseptoria enzymes that fit this scenario included a cutinase, a cellulase, and a hemicellulase (fig. 4). This is the first reported case of diversifying selection in a fungal cutinase gene. Most interestingly, all three of these genes also show very low expression during the biotrophic phase, consistent with selection to avoid recognition by the plant immune system, but their transcription increased approximately 10 to 100-fold during the necrotrophic stage (fig. 6). In contrast with the general belief that cellulases are less important during the host–pathogen interaction, we found that the cellulase gene GH54-ProtID 76589 was under significant diversifying selection; dN/dS values were four times higher in Z. tritici compared with its ancestors, and the gene also had very low reads per kilobase per million mapped reads (RPKM) values during the biotrophic phase. We speculate that this pattern could reflect a novel host interaction that emerged during the domestication of the pathogen on wheat.
Fig. 6.
Standardized transcription values (RPKM) for cell wall degrading enzymes in Zymoseptoria tritici that showed significant values of diversifying or purifying selection according to McDonald–Kreitman tests. Genes are color-coded according to the evolutionary scenario to which they were assigned (see also figs. 4 and 5). The gene expression bars represent the average from three biological replicates and error bars are one standard deviation from the mean; dpi, days post-inoculation. Detailed values are given in supplementary table S2, Supplementary Material online.
PCWDEs That Evolved Toward Optimized Function on Conserved Substrates
Most PCWDEs exist as multigene families that likely originated through gene duplication (Wapinski et al. 2007), with several members of each family contributing to digestion of the same plant polymer. If the product of a gene duplication continues to be functionally expressed, it will be subjected to purifying selection because most nonsynonymous substitutions will have a deleterious effect and because the corresponding protein is evolving toward functional optimization (reviewed in Hughes 1994).
Some components of the plant cell wall will be highly conserved between different plant species. We hypothesized that fungal PCWDEs targeting these conserved components have been under purifying selection within and between species for optimized enzymatic function (scenario 4 in fig. 5). Examples of enzymes that exhibited significant purifying selection within and between species include the hemicellulases GH43-ProtID 98714 and GH51-ProtID 40215 and several pectinases (fig. 4). Pectinases are thought to play an important role during the initial biotrophic infection stage of Z. tritici. During this stage, hyphal growth is strictly intercellular without symptoms on the plant, and it is thought that the fungus is relying mainly on pectin as a source of nutrients. Concordant with this early role in infection was the observation that pectinases are among the first PCWDEs produced by many plant pathogenic fungi including Z. tritici (Cooper 1983; Douaiher et al. 2007). In our study, the overall pattern of pectinase evolution was consistent with purifying selection rather than diversifying selection as would be expected under a host evasion scenario. Five of 13 pectinase genes showed significant purifying selection based on MKTs (fig. 2) and two of these also showed a decrease in dN/dS, consistent with our scenario of evolution for optimized function on highly conserved substrates (fig. 4).
We were not able to assign 13 of the 48 genes to one of the four evolutionary scenarios that we postulate have affected the evolution of PCWDEs in pathogens (column 5 in fig. 4). We consider it likely that other evolutionary mechanisms not included among our four scenarios will affect the evolution of PCWDEs. For example, switching to a new host may render a formerly useful gene worthless if the corresponding substrate does not exist in the new host, leading to relaxation of purifying selection and possibly to pseudogenization. Pseudogenization has often been associated with gene duplication (Lynch and Conery 2000). The gene ProtID 33254 is one of two members in the cellulase family GH61 and was under significant diversifying selection (fig. 2; supplementary table S3a, Supplementary Material online). However, the RNA-Seq experiment revealed that this gene was not transcribed in any of the three life cycle stages, in contrast to its sister gene GH61-ProtID 103512 that showed high RPKM values in the saprotrophic stage (supplementary table S2, Supplementary Material online). This suggests that the molecular signal of diversifying selection may be due to relaxed purifying selection operating on a pseudogenized gene. This example illustrates well the advantage of combining population genomics, comparative genomics, and RNA-Seq to infer evolutionary processes affecting enzymes.
Conclusion
Our findings demonstrate life-cycle-dependent expression of key enzymes in a plant pathogenic fungus. We furthermore show that complex evolutionary processes have shaped the sequence composition of these PCWDE in the emerging wheat pathogen Z. tritici. The evolution of PCWDEs in Zymoseptoria has been significantly affected by varying degrees of purifying and diversifying selection. Brown and Tellier (2011) proposed a wealth of theoretical coevolutionary processes and patterns and showed that coevolutionary dynamics at the molecular level might be quite complex. In this regard, our four proposed scenarios are “coarse-grained” and may represent oversimplifications of complex, multistep evolutionary processes. Most coevolutionary processes are also likely to be too interwoven to claim that our scenarios are exclusive and independent from each other. For example, figure 4 shows that the scenario of “life-cycle specialization” can be caused by one of the other proposed scenarios, for example, substrate optimization during a specific stage of the life cycle. However, the four simplified evolutionary scenarios provide a useful first-level classification that can be applied to subsequent, more detailed research on individual proteins. This study provides a powerful framework to better understand the roles played by different members of multigene families and to identify genes that are the most appropriate targets for subsequent wet laboratory experimentation. Our approach allows us to test specific predictions about the enzymatic function and role of specific categories of enzymes that are active during relevant phases of a pathogen’s life cycle.
Materials and Methods
Comparative Genomics
The Zymoseptoria tritici isolate IPO323 genome sequence and annotations are deposited at the Joint Genome Institute (http://genome.jgi-psf.org/Mycgr3/Mycgr3.info.html; Mycosphaerella graminicola v2.0, last accessed March 2013). The CAZy (Cantarel et al. 2009) pipeline was used to identify and annotate predicted PCWDE proteins for this isolate (Goodwin et al. 2011). We retrieved the complete sequences of all PCWDEs of Z. tritici IPO323 predicted to degrade cellulose, hemicellulose, pectin, and cutin. We used a combination of LASTZ (Harris 2007) and Basic Local Alignment Search Tool (Altschul et al. 1990) to identify PCWDE homologs and orthologs in 19 additional whole-genome assemblies of Z. tritici and its closest relatives. These previously reported genome assemblies included nine genomes of Z. tritici (National Center for Biotechnology Information; NCBI BioProject PRJNA178194), five genomes of Z. pseudotritici, four genomes of Z. ardabiliae, and one isolate of Z. passerinii (NCBI BioProject PRJNA63131) used as an outgroup (Stukenbrock et al. 2011; Torriani et al. 2011). Exonic sequences from each PCWDE found in IPO323 were mapped to genomic scaffolds of the resequenced isolates with LASTZ. We used a nucleotide similarity cutoff of 60% and custom Perl scripts to screen low scoring hits. Putative ortholog sets were aligned with MAFFT (Katoh et al. 2005) using the iterative refinement option (–maxiterate 1,000). We visually inspected all putative ortholog sets for 1) unambiguous alignment of the coding and flanking sequences and 2) for obvious signs of pseudogenization (1–2 bp indels). We tested all ortholog sets to determine whether a BLASTN search identified the orthologs as best reciprocal matching pairs between the most distant species pair available for a given ortholog set (e.g., Z. tritici and Z. passerinii).
Phylogeny
The phylogenetic relationship among the different species was reconstructed following the proposed fungal barcoding standard (James et al. 2006). Full-length sequences were extracted from all 20 genomes for the following six genes; 18S and 28S ribosomal genes, the internal transcribed spacer, elongation factor EF1-alpha, RNA polymerase II largest subunit (RPB1), and second largest subunit (RPB2). Unambiguous multiple sequence alignment of the concatenated data set was conducted on the aa level using MAFFT (Katoh et al. 2005). jModelTest (Posada 2008) was used to fit the data to the best general-time-reversible model, and the phylogeny was inferred using MrBayes 3.2 (Ronquist et al. 2012) for 9 × 106 generations sampling every 500 generations.
Transcription Analyses
Recently, it has become possible to determine which enzyme-encoding gene is transcribed during each phase in the pathogen life cycle using next-generation sequencing tools such as RNA-Seq (Morin et al. 2008). The hemibiotrophic pathogen Z. tritici displays a life cycle composed of an initial biotrophic and asymptomatic phase spanning 10–12 days post-inoculation (dpi), followed by a necrotrophic period lasting 1–4 days during which the leaf tissue is killed. Afterward, the fungus survives as a saprotroph on dead wheat leaves. The timing of the transitions between different life stages varies depending on the virulence of the strain. The highly virulent Z. tritici strain ST99CH3D7 was used to determine PCWDE expression profiles during the three life cycle stages following infection on Triticum aestivum. Three-week-old T. aestivum var. Drifter plants were inoculated with 120 ml of a spore suspension (106 spores/ml) containing 50 µl Tween. Total RNA was extracted using TRIzol (Invitrogen) from the second leaf of three different inoculated plants at each time point. The sampled plants were removed from the experiment to prevent resampling of the same plant. Sampling dates were at 7 dpi (biotrophic stage), 13 dpi (necrotrophic stage), and 56 dpi (saprotrophic stage). The quality of the isolated RNA was determined with a Qubit (1.0) Fluorometer (Life Technologies, CA) and a Bioanalyzer 2100 (Agilent, Waldbronn, Germany). All samples had a 260 nm/280 nm ratio between 1.8 and 2.1 and a 28S/18S ratio within 1.5–2.0. The TruSeq RNA Sample Prep Kit v2 (Illumina, Inc., CA) was used in the subsequent steps. Briefly, total RNA samples (1 μg) were poly-A enriched and then reverse transcribed into double-stranded cDNA. TruSeq adapters were ligated to double-stranded cDNA. Fragments with an average fragment size of approximately 260 bp containing TruSeq adapters on both ends were selectively enriched using a polymerase chain reaction. The quality and quantity of the enriched libraries were validated using the Qubit (1.0) Fluorometer and the Caliper GX LabChip GX (Caliper Life Sciences, Inc.). The libraries were normalized to 10 nM in Tris-Cl 10 mM, pH 8.5 with 0.1% Tween 20.
The TruSeq PE Cluster Kit v3-cBot-HS (Illumina, Inc., CA) was used for cluster generation using 2 pM of pooled normalized libraries on the cBOT. Sequencing was performed on the Illumina HiSeq 2000 paired end at 2 × 101 bp using the TruSeq SBS Kit v3-HS (Illumina Inc., CA).
RNA-Seq reads were quality checked using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed March 2013). Reads were aligned to the reference genome and transcriptome with TopHat v1.3.3 (Trapnell et al. 2009). We used the reference genome build Mgraminicolav2, and the transcriptome reference was the filtered gene models provided in Mgraminicolav2.FilteredModels1.gff (http://genomeportal.jgi-psf.org/Mycgr3/Mycgr3.home.html, last accessed March 2013). Before mapping, low-quality ends of the reads were clipped (3 bases from the read start and 10 bases from the read end). TopHat was run with default options. The fragment length parameter was set to 100 bases with a standard deviation of 100 bases. The distribution of reads across genomic features was assessed based on these alignments. CLC Genomics Workbench v5.0.1 (CLC Bio, Aarhus, Denmark) was then used to visualize the results. Data were normalized by calculating the reads per kilobase per million mapped reads (RPKM = total exon reads/mapped reads in millions × exon length in kb; Mortazavi et al. 2008). We tested for significant differences in transcript abundances among the different samples using the Cufflinks assembler v. 2.0.2 (Trapnell et al. 2010). Differential expression among samples was analyzed as a time series, and transcript abundance was normalized using fragment bias correction and upper quartile normalization (Trapnell et al. 2010; Roberts et al. 2011).
Selection Analyses
We used the MKT to investigate selection on the DNA level (McDonald and Kreitman 1991). The MKT approach compares the amount of nucleotide variation within a species (i.e., “polymorphism”) with the amount of variation between species (i.e., “divergence”). We applied the MKTs on all pairwise combinations of the three species Z. tritici, Z. pseudotritici, and Z. ardabiliae. MKTs were conducted on each PCWDE individually. The Mantel–Haenszel method provides a pooled odds ratio across the strata of 2 × 2 contingency tables. When this test indicated homogeneity across individual data sets, combined data sets of CAZy multigene families were also analyzed using the MKT. Significance of deviations from neutral expectations was calculated using Fisher’s exact tests from contingency tables of divergence corrected by Jukes and Cantor (1969). The amount and direction of inferred selection was quantified using the neutrality index (NI) and its equivalent for multilocus data sets, the Mantel–Haenszel estimator (Rand and Kann 1996). For better visualization, NI values were −log10 transformed, resulting in negative values being indicative of purifying (negative) selection and positive values being indicative of diversifying (positive) selection.
We have reasons to expect that evolutionary pressures acting on the PCWDEs have differed significantly between Z. tritici and its two relatives because they originate from distinct host species, domesticated wheat versus wild grasses, and they show evidence of host specialization (Brunner et al. 2009; Stukenbrock et al. 2011). We used phylogenetic analyses based on maximum likelihood as implemented in the HyPhy software package (Kosakovsky Pond et al. 2005) to determine whether significant differences in selection pressure, as measured by dN/dS ratios, were operating on these PCWDEs during the evolution of Z. tritici. First, the best nucleotide substitution model was determined through a hierarchical testing procedure, and the aligned sequences were tested for recombination before selection analyses using the implemented options on the web server of the HyPhy package (http://www.datamonkey.org/, last accessed March 2013). The phylogenetic trees for each gene were reconstructed with MEGA5 (Tamura et al. 2011) using the neighbor-joining method and the maximum composite likelihood model. We used the HyPhy batch file “SelectionLRT” to compare the plausibility of evolutionary models, where dN estimates were either independent or constrained to be equal in different combinations of three specified partitions within the phylogeny. The specified partitions were the Z. tritici clade (i.e., the domesticated, host-specialized pathogen), the ancestral clade (i.e., the unspecialized Z. pseudotritici and Z. ardabiliae progenitors on wild grasses), and their separating branch. The models tested were M1, uniform global dN estimate; M2, constrained dN estimate for Z. tritici and the ancestral clade with independent estimate for the separating branch; M3, constrained dN estimate for Z. tritici plus separating branch and independent estimate for the ancestral clade; M4, constrained dN estimate for the ancestral clade plus separating branch and independent estimate for Z. tritici; and M5, independent dN estimates for Z. tritici, ancestral clade, and the separating branch (Frost et al. 2005). Akaike Information Criterion (AIC) was used to determine the relative fit of each model (Burnham and Anderson 2002).
Supplementary Material
Supplementary results, figure S1, and tables S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The RNA-Seq data set was generated at the Functional Genomics Center Zurich (FGCZ) with the help of Marcello Zala and Dee Carter and analyzed with assistance from the Genetic Diversity Center (GDC) at ETH Zurich. Maria Anisimova provided comments on an earlier draft of the manuscript. This work was supported by ETH Zurich and by Swiss National Science Foundation grant no. 31003A_134755 and a grant from the Danish Research Council to E.H.S.
References
- Altschul S, Gish W, Miller W, Myers E, Lipman D. A basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apel PC, Panaccione DG, Holden FR, Walton JD. Cloning and targeted gene disruption of XYL1, a beta 1,4-xylanase gene from the maize pathogen Cochliobolus carbonum. Mol Plant Microbe Interact. 1993;6:467–473. doi: 10.1094/mpmi-6-467. [DOI] [PubMed] [Google Scholar]
- Brunner PC, Keller N, McDonald BA. Wheat domestication accelerated evolution and triggered positive selection in Mycosphaerella graminicola cell-wall degrading enzymes. PLoS One. 2009;4:e7884. doi: 10.1371/journal.pone.0007884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JKM, Tellier A. Plant-parasite coevolution: bridging the gap between genetics and ecology. Annu Rev Phytopathol. 2011;49:345–367. doi: 10.1146/annurev-phyto-072910-095301. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. 2nd ed. New York: Springer-Verlag; 2002. [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RM. The mechanisms and significance of enzymatic degradation of host cell walls by parasites. In: Callow JA, editor. Biochemical plant pathology. New York: John Wiley; 1983. pp. 101–135. [Google Scholar]
- Cooper RM, Longman D, Campell A, Henry M, Lees PE. Enzymic adaptation of cereal pathogens to the monocotyledonous primary wall. Physiol Mol Plant Pathol. 1988;32:33–47. [Google Scholar]
- Dancer J, Daniels A, Cooley N, Foster S. Septoria tritici and Stagonospora nodorum as model pathogens for fungicide discovery. In: Lucas JA, Bowyer P, Anderson HM, editors. Septoria on cereals: a study of pathosystems. New York: CABI Publishing; 1999. pp. 316–331. [Google Scholar]
- De Bary A. Über einige Sclerotinien und Sclerotienkrankheiten. Bot Zeit. 1886;44:376–480. [Google Scholar]
- De Lorenzo G, Castoria R, Bellincampi D, Cervone F. 1997. Fungal invasion enzymes and their inhibition. In: Carroll GC, Tudzynski P, editors. The mycota. V. Plant relationships, Part B. Berlin (Germany): Springer-Verlag. p. 61–83. [Google Scholar]
- Debyser W, Pneumans WJ, Van Damme EJM, Delcour JA. Triticum aestivum xylanase inhibitor (TAXI), a new class of enzyme inhibitor affecting breadmaking performance. J Cereal Sci. 1999;30:39–43. [Google Scholar]
- Dobinson KF, Grant SJ, Kang S. Cloning and targeted disruption, via Agrobacterium tumefaciens-mediated transformation, of a trypsin protease gene from the vascular wilt fungus Verticillium dahliae. Curr Genet. 2004;45:104–110. doi: 10.1007/s00294-003-0464-6. [DOI] [PubMed] [Google Scholar]
- Douaiher M-N, Nowak E, Durand R, Halama P, Reignault P. Correlative analysis of Mycosphaerella graminicola pathogenicity and cell-wall degrading enzymes produced in vitro: the importance of xylanase and polygalacturonase. Plant Pathol. 2007;56:79–86. [Google Scholar]
- Ebert D. Ecology, epidemiology, and evolution of parasitism in Daphnia. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2005. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books, last accessed March 2013. [Google Scholar]
- Ellegren H. Comparative genomics and the study of evolution by natural selection. Mol Ecol. 2008;17:4586–4596. doi: 10.1111/j.1365-294X.2008.03954.x. [DOI] [PubMed] [Google Scholar]
- Endo T, Ikeo K, Gojobori T. Large-scale search for genes on which positive selection may operate. Mol Biol Evol. 1996;1996:13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- Esquerré-Tugayé M-T, Boudart G, Dumas B. Cell wall degrading enzymes, inhibitor proteins, and oligosaccharides participate in the molecular dialogue between plants and pathogens. Plant Physiol Biochem. 2000;38:15–163. [Google Scholar]
- Federici L, Di Matteo A, Fernandez-Recio J, Tsernoglou D, Cervone F. Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 2006;11:65–70. doi: 10.1016/j.tplants.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Frost SDW, Liu Y, Kosakovsky Pond SL, Chappey C, Wrin T, Petropoulos CJ, Little SJ, Richman DD. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol. 2005;79:6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Maceira FI, Di Pietro A, Roncero MIG. Cloning and disruption of pgx4 encoding an in planta expressed exopolygalacturonase from Fusarium oxysporum. Mol Plant Microbe Interact. 2000;13:359–365. doi: 10.1094/MPMI.2000.13.4.359. [DOI] [PubMed] [Google Scholar]
- Gόmez-Gόmez E, Ruíz-Roldán MC, Di Pietro A, Roncero MI, Hera C. Role in pathogenesis of two endo-beta-1,4-xylanase genes from the vascular wilt fungus Fusarium oxysporum. Fungal Genet Biol. 2002;35:213–222. doi: 10.1006/fgbi.2001.1318. [DOI] [PubMed] [Google Scholar]
- Goodwin SB, Ben M’Barek S, Dhillon B, et al. (57 co-authors) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011;7(6):e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. Improved pairwise alignment of genomic DNA. University Park: Pennsylvania State University; 2007. [Google Scholar]
- Hilu HM, Bever WM. Inoculation, oversummering, and suscept-pathogen relationship of Septoria tritici on Triticum species. Phytopathology. 1957;57:474–480. [Google Scholar]
- Horbach R, Rocio Navarro-Quesada A, Knogge W, Deising HB. When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J Plant Physiol. 2011;168:51–62. doi: 10.1016/j.jplph.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functional novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide selection at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Howlett BJ. Pathogenicity genes of phytopathogenic fungi. Mol Plant Pathol. 2001;4:241–255. doi: 10.1046/j.1464-6722.2001.00070.x. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. (70 co-authors) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Juge N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 2006;11:359–367. doi: 10.1016/j.tplants.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–123. [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kema GHJ, Yu D, Rijkenberg FHJ, Shaw MW, Baayen RP. Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology. 1996;86:777–786. [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Le Roche KG, Zhou Y, Blair PL, et al. (11 co-authors) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Liu Z, Bos JI, Armstrong M, et al. (11 co-authors) Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophtora infestans. Mol Biol Evol. 2005;22:659–672. doi: 10.1093/molbev/msi049. [DOI] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McLauchlan WR, Garcia-Conesa MT, Williamson G, Roza M, Ravestein P, Maat J. A novel class of protein from wheat which inhibits xylanases. Biochem J. 1999;338:441–446. [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh TJ, McDonald H, Varhol R, Jones SJM, Marra MA. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Ohm RA, Feau N, Henrissat B, et al. (28 co-authors) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 2012;8:e1003037. doi: 10.1371/journal.ppat.1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LM, Flaishman MA, Kolattukudy PE. Cutinase gene disruption in Fusarium solani f sp pisi decreases its virulence on pea. Plant Cell. 1994;6:935–945. doi: 10.1105/tpc.6.7.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar P, Bosneaga E, Auer M. Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J Exp Bot. 2009;60:3615–3635. doi: 10.1093/jxb/erp245. [DOI] [PubMed] [Google Scholar]
- Schürch S, Linde CC, Knogge W, Jackson LF, McDonald BA. Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol Plant Microbe Interact. 2004;17:1114–1125. doi: 10.1094/MPMI.2004.17.10.1114. [DOI] [PubMed] [Google Scholar]
- Scott-Craig JS, Cheng YQ, Cervone F, De Lorenzo G, Pitkin JW, Walton JD. Targeted mutants of Cochliobolus carbonum lacking the two major extracellular polygalacturonases. Appl Environ Microbiol. 1998;64:1497–1503. doi: 10.1128/aem.64.4.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DJ, Schäfer W. Cutinase is not required for fungal pathogenicity on pea. Plant Cell. 1992;4:621–629. doi: 10.1105/tpc.4.6.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Banke S, Javan-Nikkha M, McDonald BA. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol Biol Evol. 2007;24:398–411. doi: 10.1093/molbev/msl169. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, Zala M, McDonald BA, Jun W, Schierup MH. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 2011;21:2157–2166. doi: 10.1101/gr.118851.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, McDonald BA. Geographic variation and positive diversifying selection in the host specific toxin SnToxA. Mol Plant Pathol. 2007;8:321–323. doi: 10.1111/j.1364-3703.2007.00396.x. [DOI] [PubMed] [Google Scholar]
- Sweigard A, Chumley FG, Valent B. Disruption of a Magnaporthe grisea cutinase gene. Mol Gen Genet. 1992;232:183–190. [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. Specific hypotheses on the geographic mosaic of coevolution. Am Nat. 1999;153:S1–S14. [Google Scholar]
- Torriani SFF, Brunner PC, McDonald BA. Evolutionary history of the mitochondrial genome in Mycosphaerella populations infecting bread wheat, durum wheat and wild grasses. Mol Phylogenet Evol. 2011;58:192–197. doi: 10.1016/j.ympev.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Weber G. Septoria diseases of wheat. Phytopathology. 1922;12:537–585. [Google Scholar]
- White SN, Taylor KH, Abbey CA, Gill CA, Womack JE. Haplotype variation in bovine Toll-like receptor 4 and computational prediction of a positively selected ligand-binding domain. Proc Natl Acad Sci U S A. 2003;100:10364–10369. doi: 10.1073/pnas.1333957100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J-R, Peng Y-L, Dickman MB, Sharon A. The dawn of fungal pathogen genomics. Annu Rev Phytopathol. 2006;44:337–366. doi: 10.1146/annurev.phyto.44.070505.143412. [DOI] [PubMed] [Google Scholar]
- Yao C, Koller W. Diversity of cutinases from plant pathogenic fungi: different cutinases are expressed during saprophytic and pathogenic stages of Alternaria brassicicola. Mol Plant Microbe Interact. 1995;8:122–130. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.