Abstract
Transposable elements (TEs) are mobile genetic sequences that can jump around the genome from one location to another, behaving as genomic parasites. TEs have been particularly effective in colonizing mammalian genomes, and such heavy TE load is expected to have conditioned genome evolution. Indeed, studies conducted both at the gene and genome levels have uncovered TE insertions that seem to have been co-opted—or exapted—by providing transcription factor binding sites (TFBSs) that serve as promoters and enhancers, leading to the hypothesis that TE exaptation is a major factor in the evolution of gene regulation. Here, we critically review the evidence for exaptation of TE-derived sequences as TFBSs, promoters, enhancers, and silencers/insulators both at the gene and genome levels. We classify the functional impact attributed to TE insertions into four categories of increasing complexity and argue that so far very few studies have conclusively demonstrated exaptation of TEs as transcriptional regulatory regions. We also contend that many genome-wide studies dealing with TE exaptation in recent lineages of mammals are still inconclusive and that the hypothesis of rapid transcriptional regulatory rewiring mediated by TE mobilization must be taken with caution. Finally, we suggest experimental approaches that may help attributing higher-order functions to candidate exapted TEs.
Keywords: exaptation, enhancer, mobile element, gene expression
Introduction
An important observation of the genomic era is that a great part of vertebrate genomes is comprised of transposable elements (TEs). In mammals, TEs are classified as 1) autonomous long interspersed nucleotide elements (LINEs), 2) short interspersed nucleotide elements (SINEs), which depend on LINEs for their propagation, 3) endogenous retrovirus (ERV)-like elements with long terminal repeat (LTRs) sequences, and 4) DNA transposons. The first three of these TE classes are the most abundant in mammals and reproduce via a RNA intermediate, being thus called retrotransposons (see Böhne et al. 2008 for review), whereas DNA transposons jump around the genome by a cut-and-paste mechanism. In mammals, 30–50% of the genome comprised TEs, chiefly retrotransposons, where most family members have a narrow taxonomic distribution. Alu and B1 elements, for instance, are SINEs only found in primates and rodents, respectively. Although TEs are in essence parasitic DNA elements with no intrinsic function for the host (Doolittle and Sapienza 1980; Orgel and Crick 1980), early studies indicated that particular instances of TE insertions contributed with new genes, exons, or regulatory regions (Brosius 2003). In 1992, Brosius and Gould (1992) suggested the term “exaptation” for the phenomenon of “junk” DNA sequences such as TEs acquiring a novel function in the genome. Because regulatory innovation is expected to drive evolution (Carroll 2008), and considering the huge TE content of genomes, exaptation of TEs into new promoters, enhancers, chromatin barriers, and other regulatory elements is likely to have contributed to the evolution of regulatory networks (Medstrand et al. 2005).
Since the times of Brosius and Gould, much evidence has accumulated suggesting exaptation of TEs as transcriptional regulatory regions; indeed, this idea has reached genomic proportions with several whole-genome studies showing that TFs can bind to thousands of TE instances in the genome. This has fueled the hypothesis that TE insertions are responsible for an extremely rapid transcriptional rewiring in mammals and other vertebrates (Feschotte 2008; Bourque 2009), reminiscent of Barbara McClintock's original idea that transposons acted as “controlling elements” of several loci in the genome (McClintock 1956). Based on the correlation between past bursts of TE activity and the emergence of certain vertebrate lineages, it has even been hypothesized that TEs have crucially contributed to the origin and radiation of mammals (Okada et al. 2010) and specific mammalian groups such as anthropoid primates, rodents, and microbats (Oliver and Greene 2009, 2011; Britten 2010).
Where do we stand regarding transcriptional regulation by TE insertions? Here, we review some recent developments in the exaptation of TEs into new promoters, enhancers, and insulators from both gene-centered and genome-wide studies. We conclude that, although it can be safely ascertained that TE exaptation has been an important evolutionary drive in transcriptional networks in the past, genomic searches for transcription factor binding sites (TFBSs) have been of limited value in uncovering TE exaptations in recent lineages, including humans, and more work is necessary before the global contribution of TE exaptation to transcriptional network evolution can be fully appreciated.
Promoters Derived from TEs
LINEs and TEs carrying LTRs have intrinsic ability to recruit RNA polymerase II and thus have great potential to be exapted as promoters. That TE insertions in the near vicinity of genes can be detrimental to their function is evidenced by the lower-than-expected frequency of LINEs and LTR TEs within or close to transcriptional units (Medstrand et al. 2005). In spite of this, whole-genome analyses have shown that up to 25% of genes have TEs into their promoter and/or untranslated regions (Jordan et al. 2003; van de Lagemaat et al. 2003). In addition, transcriptome analyses indicate that 18% and 31% of cap-selected RNA transcripts initiate within repetitive elements in mouse and human cell lines, respectively, showing that transcription start sites (TSS) embedded within TEs are not an uncommon phenomenon (Faulkner et al. 2009).
Even though the results of genome-wide expression surveys are interesting, the functional impact of TE-derived TSSs is still unknown. Faulkner et al. (2009) observed that these RNA variants are rare, with only 2.8% and 5.2% of the most abundant RNAs deriving from retrotransposon TSSs in mice and humans, respectively. The same authors observed that more than half of TE-derived transcripts stay in the nucleus, which may indicate a function as noncoding regulatory RNAs or that most of them are simply nonfunctional (Faulkner et al. 2009). Indeed, detailed, gene-centered studies of protein-encoding genes have shown that TEs usually act as alternative promoters that originate minor mRNA variants of uncertain functional significance (reviewed in Cohen et al. 2009).
For certain genes, however, most or all transcriptions initiate within a TE, a hallmark for authentic exaptation. For instance, mouse Naip genes, which function in apoptosis, are expressed exclusively from an ORR1E LTR-derived promoter with ubiquitous activity (Romanish et al. 2007). The human homolog of mouse Naip harbors an unrelated LTR element (MER21C) that drives expression to testis but in this case as a minor (12%) mRNA species (Romanish et al. 2009). In the teleost fish medaka, the master regulator gene dmrt1bγ has changed its position in the hierarchy of sex determination genes after the insertion of a novel DNA transposon in its promoter (Herpin et al. 2010). In some mammalian cases, the insertion of a TE drives expression to a novel tissue, usually the placenta, where LTR retroviruses are known to be most active (Cohen et al. 2009). For instance, the insulin-like 4 (INSL4) gene only exists in anthropoid primates and its only known expression site is the placenta, where all of its transcripts are derived from an LTR promoter, a strong indication of exaptation of this TE in humans and our close relatives (Bièche et al. 2003). Recently, it has been shown that different TEs have been independently recruited by the prolactin genes of anthropoid primates (LTR MER39), rodents (LTR MER77), and African elephants (LINE L1-2_LA) as promoters with specific activity in the placenta, where prolactin is known to have an effect in sustaining pregnancy, at least in rodents (Emera et al. 2012).
Enhancers Derived from TEs
Enhancers are arrays of TFBS that alter transcriptional rates of genes. These regions, usually 150–500 bp in length, may be located far away from the transcriptional start site making their discovery and confirmation more difficult (Visel et al. 2007). Many enhancers are under purifying selection and can be identified by phylogenetic footprinting strategies that search for conserved noncoding regions in related genomes (Visel et al. 2007).
In contrast to enhancers, most TE-derived sequences in vertebrate genomes are expected to have no function and thus to evolve at a neutral rate. Indeed, analysis of ancestral TEs shared by humans and mice using a neutral insertion–deletion model indicate that over 95% of ancient TE insertions in the human genome are evolving at a neutral rate (Lunter et al. 2006; Meader et al. 2010). However, genome-wide analyses have uncovered a large number of TE-derived sequences that are under purifying selection and are located in noncoding, conserved regions of vertebrate genomes (e.g., see table 2). A recent comparison of 29 mammalian genomes found a staggering more than 280,000 conserved, TE-derived noncoding elements with potential regulatory function (Lindblad-Toh et al. 2011; Lowe and Haussler 2012). The high level of conservation of these elements strongly suggests functionality, and many could be enhancers.
Table 2.
Genome-Wide Evidence for Noncoding, Phylogenetically Conserved TE Exaptations.
| TE | Number of TE Instances Found and Functionality Criteria | Evolutionary Conservation | References |
|---|---|---|---|
| Mainly MIR and L2 | Two conserved instances per gene in intergenic regions | Human and mouse | Silva et al. (2003) |
| LF-SINE | >200 noncoding conserved instances in human genome | Tetrapods (degree of conservation not specified) | Bejerano et al. (2006) |
| Many | >10,000 nonexonic conserved instances | Primates, rodents, and dog | Lowe et al. (2007) |
| Many | >33,000 noncoding conserved instances | Human, rodents, and dog | Mikkelsen et al. (2007) |
| Many | >2,500 conserved instances in putative regulatory regions (CRMs) | Marsupial and placental mammals | Gentles et al. (2007) |
| CORE-SINE | Four intergenic and three intronic conserved instances | Marsupial and placental mammals | Santangelo et al. (2007) |
| AmnSINE1 | 124 conserved instances in human genome | Marsupial and placental mammals | Sasaki et al. (2008) |
| MER20 (DNA) | >6,900 conserved Instances near endometrium-specific expressed genes | Placental mammals | Lynch et al. (2011) |
| Many | >280,000 conserved noncoding instances in placental mammals | 29 placental mammals | Lindblad-Toh et al. (2011) |
| Many | >280,000 conserved in placentals; 25% overlap DNase I HS (opened chromatin) | 29 placental mammals | Lowe and Haussler (2012) |
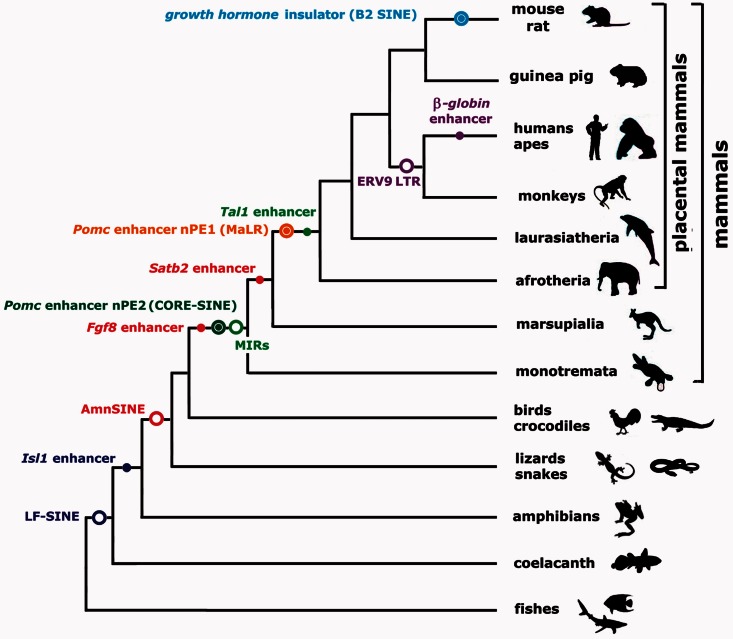
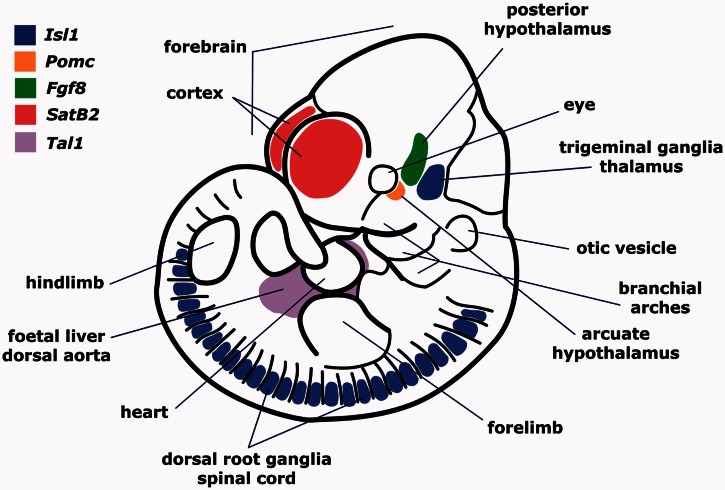
Few putative cases of TE exaptation into transcriptional enhancers have been experimentally tested in transgenic mouse models (table 1 and figs. 1 and 2). Bejerano et al. (2006) used transgenesis to show that a conserved noncoding sequence derived from an LF-SINE was in fact a developmental enhancer of the Isl1 gene in mice. LF-SINEs are an ancient repeat family, and phylogenetic conservation analysis suggests that this TE-derived sequence works as an Isl1 enhancer in all tetrapods (fig. 1; Bejerano et al. 2006). Sasaki et al. (2008) identified two different AmnSINE1-derived sequences that function as distal transcriptional enhancers in mouse embryos, controlling Fgf8 and SATB2 (Sasaki et al. 2008, Tashiro et al. 2011, Nakanishi et al. 2012). Although AmnSINEs are also an ancient TE family, phylogenetic conservation indicates that exaptation apparently happened in a mammalian ancestor (fig. 1). Looking for the evolutionary origin a conserved neuronal enhancer of the Pomc gene named nPE2 (de Souza et al. 2005), we found that it was derived from a member of the CORE-SINE family (Santangelo et al. 2007). Recently, we also found that a second neuronal enhancer of POMC—named nPE1—is derived from a mammalian apparent LTR retrotransposon (Franchini et al. 2011). Interestingly, the TEs that gave rise to the Pomc enhancers were exapted at different time points: nPE2 originated from an earlier exaptation of in the lineage leading to mammals sometime before the Prototheria/Theria split (∼170 Ma), whereas nPE1 is a placental novelty that originated after the Metatheria/Eutheria split (∼150 Ma) and before the wide mammalian radiation that occurred 90 Ma (fig. 1). Thus, two different neuronal enhancers driving expression to an identical set of hypothalamic neurons are the result of two independent exaptation events, a first example of convergent evolution of transcriptional enhancers (Franchini et al. 2011, 2012).
Table 1.
Evidence for Conserved and Nonconserveda TEs Exapted as Enhancers and Insulators.
| TE | Function | Genes Controlled | Technique(s) | Evolutionarily Conserved? | References |
|---|---|---|---|---|---|
| LINE1 | Enhancer | Human apoa | YAC transgenic mice, reporter assays in cell culture | No (humans, primates?) | Yang et al. (1998) |
| Alu | Insulator, barrier element | Human K18 | Transgenic mice | No (humans, primates?) | Willoughby et al. (2000) |
| LF-SINE | Enhancer | Mammalian Isl1 | Conservation, mouse transgenesis | Mammals | Bejerano et al. (2006) |
| CORE-SINE | Enhancer (nPE2) | Mammalian Pomc | Conservation, mouse transgenesis | All mammals | Santangelo et al. (2007) |
| B2 SINE | Insulator, barrier element | Mouse growth hormone | Mouse BAC transgenesis, reporter assays in cell culture | No (only mouse/rodents) | Lunyak et al. (2007) |
| MIR SINE | Enhancer “boost" | Mammalian Tal1 | Conservation, reporter assays stable cell culture, embryoid bodies | Placental mammals | Smith et al. (2008) |
| AmnSINE | Enhancer | Mammalian SATB2 | Conservation, mouse transgenesis | Mammals | Sasaki et al. (2008) and Tashiro et al. (2011) |
| AmnSINE | Enhancer | mammalian Fgf8 | conservation, mouse transgenesis | Mammals | Sasaki et al. (2008) and Nakanishi et al. (2012) |
| ERV9 (LTR) | Enhancer | β-globin locus | BAC transgenic mice, ChIP, 3C | Great apes and humans only | Pi et al. (2010) |
| MaLR (LTR) | Enhancer | Mammalian Pomc | Conservation, mouse transgenesis | Placental mammals | Franchini et al. (2011) |
| Hopscotch (DNA) | Enhancer | Maize/teosinte tb1 | QTL positional cloning | No (polymorphic in teosinte) | Studer et al. (2011) |
Note.—MaLR, mammalian apparent LTR; QTL, quantitative trait locus.
aConservation in restricted lineages such as primates and rodents was counted as “non-conserved."
Fig. 1.
Scheme of vertebrate phylogenetic tree showing well-characterized transposable elements (TE)-exaptation events into enhancers and insulators. Different TE exaptation events are indicated with different colors. Circles indicate the branch where the corresponding TE subfamily originated and/or diversified before exaptation, whereas filled circles indicate the branch where a particular TE instance was exaptated as a regulatory element. References for TE origins are LF-SINE, Bejerano et al. (2006); AmnSINE, Sasaki et al. (2008); MIR/CORE-SINE, Gilbert and Labuda (1999, 2000); mammalian apparent LTR (MaLR), Smit (1993); ERV9, Costas and Naveira (2000); and B2 SINE, Churakov et al. (2010). Other references can be found in table 1. Branches not drawn to scale.
Fig. 2.
Scheme of a mouse embryo at approximately 11 days of gestation showing the expression patterns driven by well-characterized, TE-derived enhancers. Expression territories of each gene and some anatomical features are indicated. See text and table 1 for references and details.
An exapted SINE element with a somewhat different activity has been described by Smith et al. (2008), who found that an MIR element (a member of the CORE-SINE family) located near the hematopoietic gene Tal1 acts as a booster of gene expression. This MIR was inserted around the time of the Metatheria/Eutheria branchpoint (fig. 1) and is located in a noncoding conserved element located at +18 kb of the Tal1 gene. Although it does not behave as a classical enhancer in transgenic mouse experiments, sophisticated cell culture experiments (including stable transfection assays and targeted-transgene insertion into mouse embryonic stem (ES) cells followed by differentiation into hematopoietic precursors) indicate that the +18-kb element acts as a modulator of another Tal1 enhancer located nearby at +19 kb (Smith et al. 2008).
Although the examples mentioned earlier are from ancient TE insertions, there are a few particularly convincing examples of recent TE insertions that were exapted into enhancers. Near the locus control region of the higher primate β-globin locus, there is an LTR of an ERV9 element that can act as an enhancer in cell culture assays and transgenic zebrafish (Ling et al. 2002; Pi et al. 2004). This ERV9 LTR is found in orangutans, humans, and close relatives but not other mammals (Ling et al. 2002). Recently, Pi et al. (2010) created bacterial artificial chromosomes (BACs) carrying the human β-globin locus with a floxed (flanked with loxP sites) ERV9 LTR allele and inserted the BACs into transgenic mice. Deletion of the ERV9 LTR by cre recombinase caused a 50–80% drop in β-globin transcription in adult erythroid mouse cells, whereas, interestingly, transcription of fetal γ-globin gene was spuriously increased in adult erythroid cells (Pi et al. 2010). In addition, chromatin immunoprecipitation (ChIP) experiments with erythroid cells from transgenic mice showed that the ERV9 LTR is responsible for recruiting transcription factors (TFs) and RNA polymerase II to the β-globin promoter, and chromosome conformation capture (3C) assays with transgenic spleen erythroid cells showed that the ERV9 LTR loops with the β-globin promoter and that this looping depends, to a great extent, on the presence of the LTR (Pi et al. 2010). Thus, conversions of TE sequences into transcriptional enhancers have happened at various levels of the vertebrate phylogenetic tree (fig. 1).
Another notable example of an even more recent, functional TE exaptation into an enhancer comes from maize, where a quantitative trait locus controlling stem branching was mapped to a Hopscotch transposon (Studer et al. 2011). The TE insertion increases 2-fold the expression of the gene tb1 (teosinte branched 1, located over 60 kb away) and is a major factor in the branching pattern of domesticated maize. The TE got inserted into this location approximately 20,000 years ago, and this modified locus was selected by farmers when teosinte—the wild ancestor of maize—was domesticated 10,000 years ago (Studer et al. 2011).
TE-Derived Insulators
Insulators are DNA sequences that can act either as a barrier element preventing the spread of heterochromatin (i.e., condensed chromatin) from one locus to another or as an enhancer blocker preventing enhancers from spuriously influencing other loci (Gaszner and Felsenfeld 2006). In mammals, a well-known insulator-binding protein is the zinc-finger CCCTC-binding factor (CTCF), which participates in the organization of chromatin loops and in the creation of transcriptional domains (Phillips and Corces 2009). Bourque et al. (2008) found that approximately 33% of CTCF-binding elements present in mouse ES cells are derived from the B2 subfamily of SINEs, which is restricted to a few rodent groups. As expected, these CTCF-binding regions were not conserved in the human genome, suggesting that the chromatin architecture in ES cells may be different from that of other mammals (Bourque et al. 2008). Recently, Román et al. (2011) showed that another mouse-specific SINE subfamily, the B1, has insulator activity in cell culture assays and in transgenic zebrafish also via CTCF binding.
That TEs can become insulators of a nearby transcriptional unit (table 1) is best evidenced by the work of Lunyak et al. (2007) with the murine growth hormone (GH) locus. During the organogenesis of the pituitary, a boundary is established between open and closed chromatin domains 5 kb upstream of the GH locus. Interestingly, this boundary coincides with a B2 SINE, a retroposon restricted to mice, rats, and close relatives (fig. 1; Serdobova and Kramerov 1998; Churakov et al. 2010). Detailed studies showed that the B2 element is transcribed in both orientations by RNA polymerases II and III and that it can act as an enhancer blocker in cell culture. Notably, Lunyak et al. showed using BAC transgenesis that GH expression depends on the presence of the B2 element and that in its absence the GH locus remains transcriptionally silent, presumably because heterochromatin spreads in from neighboring loci. The results of Lunyak et al. are reminiscent of an earlier work by Willoughby et al. (2000) that found an Alu-rich region upstream of the human keratin-18 (K18) gene that has typical insulator properties such as chromatin barrier activity (although not enhancer blocking activity) in transgene mouse analysis.
TEs as a Source of New TFBS
Promoters and enhancers work primarily by the binding of TF proteins, which ultimately interact with the basal transcriptional machinery. For over 20 years, TFBSs have been found within TE sequences (Brosius 2003). In these earlier studies, evidence for functionality of TE insertions and embedded TFBSs was usually assessed by reporter gene expression in transient transfection assays in cell culture and, more recently, by ChIP, which can reveal in vivo binding of TFs to TEs (for recent examples see table 3 and for older examples see Brosius 2003). It is important to note, however, that TF binding by itself does not necessarily mean that a TE should work as a regulatory region.
Table 3.
Recent Experimental Evidence for Gene Regulation by TFBSs Present in TEs.
| TE | TF Bound | (Putative) Genes Controlled | Technique(s) | Cell Type(s) | References |
|---|---|---|---|---|---|
| AluSx (primate SINE) | Vitamin D receptor (VDR) | CAMP (cathelicidin microbial peptide) | Reporter assay in cell culture | NB4 (human myeloid line) | Gombart et al. (2009) |
| AluS (primate SINE) | Retinoic acid receptor (RAR) | RAI1, GPRC5A, SMYD5, RARRES1, RARRES3 | ChIP | SCC25 (human squamous cell carcinoma) | Laperriere et al. (2007) |
| LTR10, MER61 (class I ERV LTR) | p53 | DHX37, neogenin, PTPRM, TMEM12, others | ChIP, reporter assay in cell culture | HCT116 (human colorectal carcinoma) | Wang et al. (2007) |
| B1 (murine SINE) | Dioxin receptor (AhR) and Slug | Lpp, Tbc1d1, Dad1, others | ChIP (overexpressed TFs), reporter assay in cell culture | Hepa 1 (mouse hepatoma) | Román et al. (2008) |
| B1 (murine SINE) | Dioxin receptor (AhR), Slug, CTCF | Dad1, Lpp, Cabin1, others | Insulator assays in cell culture and zebrafish; ChIP | Hepa1 cells, zebrafish embryos | Román et al. (2011) |
| AluS (primate SINE), MIRb (SINE), L2A (LINE), others | Estrogen receptor α (ESR1) | Many, undetermined | Genome-wide ChIP, reporter assay in cell culture | MCF7 (human breast cancer) | Mason et al. (2010) |
| Alu (primate SINE) | LXR, PPARα, RXR | Myeloperoxidase (MPO) | Reporter assay in cell culture, transgenic mice assays | CV-1 (monkey kidney fibroblasts), transgenic mouse macrophages | Reynolds et al. (2006) |
| AluSx (primate SINE), MLT2b2 (ERV LTR) | NF-κB | Interferon λ1 | Reporter assay in cell culture | Modified HEK-293 (human embryonic kidney) | Thomson et al. (2009) |
| Mostly MIR (ESR1); ERV1 (p53); ERVK (Oct4-Sox2) and murine B2 SINE (CTCF) | ESR1, p53, c-Myc, RELA, POU5F1 (Oct4), Sox2, CTCF | Many, undetermined | Genome-wide ChIP-PET and ChIP-seq | MCF7, HCT116, Burkitt's human lymphoma, human leukemia T cells, mouse ES cells | Bourque et al. (2008) |
| Alu (primate SINE), L2 (LINE) | REST | CACNA1A (calcium channel subunit), others | ChIP | HeLa (human cervical cancer) | Johnson et al. (2006) |
| Mainly ERV1 (LTR), others | Nanog, Oct4 | Many, undetermined | Genome-wide ChIP-PET | mouse and human ES cells | Kunarso et al. (2010) |
| Mainly B2, B4 (SINE), ERVK | Nanog, Oct4, CTCF, others | Csd4, Mtf2, others | Correlation between variations in mRNA levels and genome-wide ChIP-seq | Mouse zygotes, morulae, blastocysts | Xie et al. (2010) |
| MER5B (DNA transposon) | Foxa1, p53, Sma2/4 | α-fetoprotein (afp) | Reporter assays in cell culture, ChIP | Mouse ES cells | Taube et al. (2010) |
| EnSpmN6_DR (DNA transposon) | p53 | trim8, others | Reporter assays in human (HeLa) cell culture | Zebrafish | Micale et al. (2012) |
In the last years, ChIP coupled to microarray hybridization (ChIP-chip), high-throughput sequencing (ChIP-seq) or paired-end diTag (ChIP-PET) have been employed to determine the genome regions bound by TFs in an unbiased way (Farnham 2009). To estimate the prevalence of TF binding to TEs, Bourque et al. (2008) reassessed seven genome-wide ChIP studies of six different TFs and found that these factors frequently bind to sites embedded within TEs. Thus, for instance, approximately 20% of estrogen receptor α (ESR1) binding regions overlap SINEs of the MIR family, 24% of Oct4-Sox2 binding regions overlap endogenous retrovirus K (ERVK) repeats, and approximately 40% of p53 binding occur within ERV1 elements (Bourque et al. 2008). Similarly, Kunarso et al. (2010) found that, in human ES cells, 21% and 14.6% of binding sites for Oct4 and Nanog, respectively, reside in TEs, and, interestingly, only approximately 5% of the binding regions for each factor are conserved in mouse ES cells, indicating a possible divergence in the transcriptional network between mammalian ES cells. Xie et al. (2010) found that the nearby presence of murine TEs carrying TFBSs correlates with increased mRNA expression in mouse in relation to human and bovine embryos. Interestingly, this correlation was significant only with TEs harboring two or more TFBSs, suggesting that the TEs with potential to influence gene expression are preferentially those that carry a full cis-regulatory module (Xie et al. 2010). These and other similar studies (Wang et al. 2007; Román et al. 2008, see table 1) led to the idea that TE insertion can rapidly change the regulatory landscape of mammalian genomes during evolution (Bourque 2009).
TEs and the Birth of New Regulatory Regions
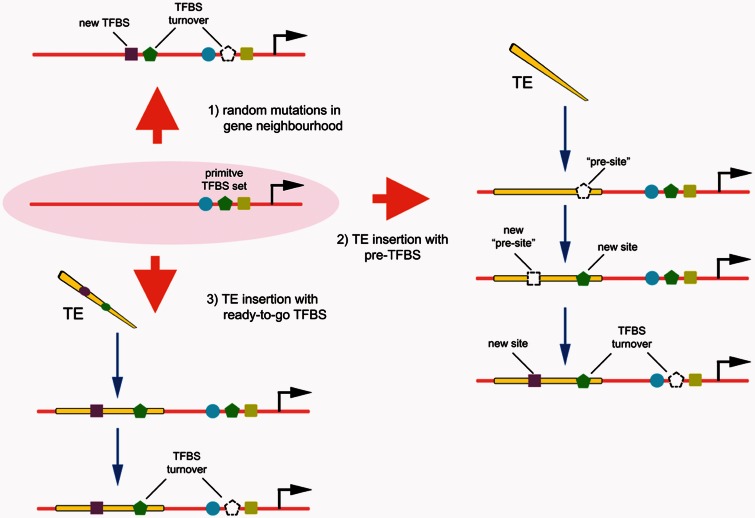
As evolutionary time goes by, new TF-binding motifs are expected to appear in promoters or enhancers by random mutation. These new motifs can be eventually lost from the population, become fixed by genetic drift or, in case they have a large positive effect on fitness, might be kept by natural selection (Lynch 2007). New motifs may also replace pre-existent ones (turnover) by stabilizing selection, without significantly changing the expression of the gene (Ludwig 2002). In this context, TEs might contribute to the birth of new transcriptional regulatory elements using two alternative routes (fig. 3):
-
-
A new TFBS appears as soon as a TE that inherently carries one or more functional motif(s) is inserted near a gene. In this case, the change in transcription levels is immediate, implying that some TEs may act as “jumping regulatory regions” and might rewire the network of genes controlled by a TF in a short evolutionary time (Bourque et al. 2008).
-
-
New TFBSs appear by random mutation on a TE previously inserted in the vicinity of a transcriptional unit. In this case, TEs act as raw material for the appearance of novel active TFBSs, and, given the abundance of TEs, it is a mechanism that deserves serious consideration. It is also possible that some TE classes have “pre-sites” that only require few mutations to originate functional TFBSs.
Fig. 3.
Scheme of the evolution of new TFBS in the vicinity of genes. Primitively, a hypothetical gene (pink oval) is controlled by a set of three TFBS (blue circle, green pentagon, and yellow square). 1) New sites may appear in the vicinity by random mutation, possibly leading to turnover of previously present TFBS (green pentagon) or a new TFBS appearing (violet square). 2) Alternatively, the insertion of a TE nearby initially has no influence on transcription, but random mutation leads to TFBS turnover and/or new TFBS. Some sites might be just a few mutations from acquiring functionality (presites). 3) In some instances, a TE carrying functional TFBS may insert near a promoter, leading to an immediate change in transcription. TE-derived TFBS may eventually cause turnover of primitive TFBS.
It must be noted that regulatory regions such as enhancers carry complex arrays of TFBSs, making it unlikely that TEs might work as a full enhancer when inserted near a gene, at least in most cases. Thus, in both scenarios above, the first functional site(s) to appear may work as a “seed” for the appearance of additional functional sites nearby, eventually originating a regulatory block with several TFBSs that works as a bona fide enhancer or promoter (a process called “epistatic capture” by Emera and Wagner [2012]). For regulatory elements that appeared long ago and have changed significantly from their ancestral TEs, it is difficult to reconstruct what was the mechanism of exaptation, but recent exaptation events may be more informative. In a recent work, Emera and Wagner (2012) describe the exaptation of MER20 and MER68 elements as a prolactin (prl) alternative promoter with activity in endometrial cells of the ape uterus. A functional comparison of TE-derived prl promoter from apes with that of nonape primates—which possess the same MER insertions in prl but lacking promoter activity—revealed a ETS1 site that was present in the ancestral MER element, as well as new TFBSs that appeared in the ape lineage (Emera and Wagner 2012). Thus, the creation of an alternative promoter for prl in apes involved both a “ready-to-go” site (ETS1) and new sites that appeared subsequently (Emera and Wagner 2012).
TE Insertion and Changes in Regulatory Function
Although the examples reviewed above suggest that TEs have contributed to the creation of functional regulatory regions, each example differs in the techniques used and in the depth of experimental evidence collected to assign function. Thus, we believe that it is necessary to compare the relative strength of the different experimental designs to properly evaluate the putative regulatory functions of TE-derived sequences. The ENCODE Project Consortium (2007) uses the idea of 1) biochemical function as the detection of a certain biochemical behavior of a particular sequence, for example, ChIP analyses showing that a TF can bind to a particular TE-derived sequence. A second concept, also used in the ENCODE project, is that of 2) regulatory role, defined as the transcriptional consequence of TE insertions on genes, for instance, showing that a particular TFBS present on a given TE can increase transcription of a nearby gene. Usually, these studies are done by transient transfection assays in cell culture with artificial reporter gene constructs harboring wild-type or mutated forms of TEs. A third level is 3) physiological function, understood as the higher-order physiological and/or morphological changes induced by TE-derived sequences and that may create variation or innovation. One example is the change in branching pattern in maize and teosinte due to the insertion of a TE-derived enhancer, which increased the morphological variation of teosinte and was later selected by early farmers (Studer et al. 2011). The ultimate functional demonstration is provided by the 4) fitness effect of a TE insertion, which needs an evaluation of the impact of the TE-derived transcriptional regulatory change on the viability and reproductive success (i.e., the fitness) of the organism.
These four functional levels are obviously related, but one should not assume that a TE-mediated effect at a lower level would translate on an impact at a higher level, as the fitness effect of the TE-mediated perturbation may be near neutral. This is a significant concern because it is expected from population genetics that mutations in the genomes of species with small effective population sizes (such as vertebrates and higher plants) will only be selected against if the fitness disadvantage of the mutation is very high (Lynch and Conery 2003; Lynch 2007). Genome-wide and many gene-centered studies that suggest TE-derived exaptation events (i.e., most studies listed in table 3) are based on ChIP experiments showing that TFs can bind to TE instances, complemented by functional experiments in transient transfection cell culture assays (e.g., Wang et al. 2007; Bourque et al. 2008; Román et al. 2008, 2011; Kunarso et al. 2010; Xie et al. 2010). These experiments correspond to the “lower” levels of functional characterization (biochemical and regulatory), and it is not known if the transcriptional effect observed in these studies might lead to any significant effect at “higher” levels (physiological and fitness). In addition, almost all these genome-wide studies deal with recent, lineage-specific TE insertions (such as primate Alus, primate LTR retrotransposons, or rodent B1 and B2 SINEs; see table 3), precluding the use of phylogenetic conservation as a criterion for functionality, as employed successfully to identify TE-derived enhancers (table 2).
Genome-wide studies certainly indicate a potential for TE expansions in particular lineages to change or “rewire” transcriptional networks, but the physiological significance of this rewiring, if any, is still unknown. In this regard, the work of Naito et al. (2009) analyzing the ongoing colonization of a rice strain by the mPing DNA transposon offers a note of caution. mPing mobilization generates approximately 40 new TE insertions per plant per generation, providing a unique opportunity to study the consequences of a TE burst as it happens in a complex organism. Although few inserts (∼1%) happen within exons, minimizing deleterious effects, many mPing insertions occur close to genes, and quantitative comparisons between the transcriptomes of different strains revealed that most insertions either upregulate or have no effect on the transcription of nearby genes (Naito et al. 2009). Notably, in some cases, the insertions confer inducibility to cold and high salinity. The work by Naito et al. (2009) shows that a complex genome can be quite resistant to massive TE colonization without an overt loss in fitness and, although many insertions influence transcription of nearby genes, natural selection has had no time to act and no “real” function can be assigned to them. This illustrates the difficulty of assuming an important regulatory function for the transcriptional influence of recent TE insertions, as suggested in the genome-wide studies listed in table 3.
Although the highest levels of functionality (3 and 4 above) might be considered the most relevant, they are seldom addressed experimentally due to intrinsic technical limitations of most available models. A good proxy for functionality is, nevertheless, evolutionary conservation. Thus, an important fitness effect of some TE-derived mammalian enhancers (Bejerano et al. 2006; Santangelo et al. 2007; Sasaki et al. 2008; Smith et al. 2008; Franchini et al. 2011) is clearly inferred from their deep conservation among mammals, indicating strong purifying selection for over 100 My. In addition, most of these works use transgenic mice to show that these elements display enhancer activity compatible with the expression of nearby genes. The mere existence of thousands of conserved-noncoding elements derived from ancient TE insertions (Lowe et al. 2007; Lindblad-Toh et al. 2011; Lynch et al. 2011) suggests that these well-studied cases are but the tip of an iceberg. Loss of function studies of TE-derived enhancers in knockout mice have not been performed yet and would provide an ultimate experimental proof of their importance in fitness. It must be borne in mind, however, that expression of genes may be controlled by several enhancers with overlapping transcriptional activity, as in the case of TE-derived Pomc enhancers nPE1 and nPE2 (de Souza et al. 2005; Franchini et al. 2011) and that even enhancer knockout studies may fail to show a clear fitness effect (Ahituv et al. 2007), unless the experimental mutants are raised in challenging environmental conditions (Frankel et al. 2010). As for TE-derived promoters, several of them are also clear examples of exaptation, when transcription starts exclusively within a TE-derived sequence (see Cohen et al. 2009 for a review).
In addition to the identification of conserved sequences, it will be interesting to explore the overlap between TEs and active enhancers identified by chromatin marks (e.g., H3K27ac and H3K4me3) or transcription cofactor binding (e.g., p300), which are a better proxy to functionality than TF-binding alone. Recently, Huda et al. (2011) have done just that using data from two human cell lines analyzed by the ENCODE project and were able to find hundreds of regions with epigenetic markers typical for active enhancers that overlap TE insertions. Interestingly, the largest proportion of TEs with enhancer marks are MIR-SINE and L2-LINE subfamilies (Huda et al. 2011), two ancient mammalian TE groups, indicating that exaptation may have happened before the mammalian radiation. Another work linking ancient TE insertions with epigenetic marks and cofactors was done by Lynch et al. (2011), in this case studying the contribution of the DNA-transposon MER20 to the differentiation of the endometrium during mammalian pregnancy. They found that the neighborhood of genes expressed in progesterone-induced, differentiating human endometrium is enriched in MER20 instances that are conserved in placental mammals. With an array of ChIP assays, selected MER20 instances could be roughly divided into two groups, an “enhancer-repressor type," which binds p300, progesterone receptor, and others, whereas the second group, the “insulator-type," bound CTCF, USF1, and other insulator proteins (Lynch et al. 2011). Although detailed functional studies are still lacking, the evolutionary conservation and the ChIP data indicate that MER20 exaptation had a role in rewiring the endometrial gene expression network when pregnancy evolved in placental mammals, over 100 Ma (Lynch et al. 2011), consistent with the hypothesis that TE mobilization played a role in the early evolution of mammals (Okada et al. 2010).
Detecting Exaptation Events by Sequence Conservation
As discussed earlier, the identification of evolutionary conserved noncoding elements (CNEs) derived from TEs has been successfully used to identify candidates for exaptation events in the mammalian lineage. Although this may seem straightforward, there are some limitations involved. First, it can be difficult to recognize an exapted element if the sequence has evolved too fast since the TE insertion. If a TE-derived sequence retains less than 70% nucleotide identity with the original TE, it is very possible that it will not be recognized by alignment algorithms such as BLASTN (Altschul et al. 1990) or BLAT (Kent 2002). Second, the exapted area of the TE may be too small. In general, searches at the genome level use threshold levels in the length of the overlapping area and retain only elements that cover the majority of the conserved element as a TE (Lowe and Haussler 2012). It has been observed that some regions within TEs are more likely to be exapted than others, either because TE insertion is generally incomplete (e.g., LINEs tend to be truncated at the 5′-region) or because some TE sequences may carry functional binding sites for TFs (Lowe et al. 2007; Lowe and Haussler 2012). Third, the TE that gave origin to a regulatory sequence may have been inactivated long ago, making it very difficult to identify exaptation. Current estimates indicate that repeat families can be recognized if they are younger than 200 My (Brosius 2003) suggesting that some exaptation events cannot be recognized as such. However, several reports have reported ancient exaptation events (Bejerano et al. 2006; Xie et al. 2006; Santangelo et al. 2007). In all these cases, the identification was facilitated because close relatives of the TE that originated the conserved noncoding sequence is active in some other organisms such as the LF-SINE in the coelacanth (Bejerano et al. 2006), the MAR1 in the opossum (Santangelo et al. 2007), and the SINE3 in the zebrafish (Xie et al. 2006). The fact that the element was still active in other organisms provided many instances of the element that allowed the generation of consensus sequences to improve detection based on sequence similarity.
The limitations in the identification of TE exaptation events are exemplified by Lowe and Haussler (2012) who worked with aligned sequences of 29 mammals and compared them with the RepeatMasker annotation in the human genome. Although they show that at least 11% of CNEs in the human genome have most probably originated from TEs, they have only identified 133 exaptation events that predate the speciation of ray-finned fishes from the human lineage from a total set of 284,857 exapted elements (Lowe and Haussler 2012). These 133 exapted elements are identifiable because they have evolved in a very slow rate and are large enough that they can still be aligned to the TE consensus sequences. On the other hand, in this approach, sequences that are not annotated as human repetitive elements are missed. To overcome this difficulty, other researchers (Xie et al. 2006) compared all CNEs not overlapping with known human TEs with the whole set of repeats deposited in the RepBase database (Jurka et al. 2005) that includes TEs from all kinds of organisms. This approach allowed them to discover a family of CNEs derived from a SINE3, an element still active in the zebrafish genome. Thus, the difficulties in identifying ancient exaptation events, together with the problematic identification of lineage or species-specific exaptation events, suggest that the impact of TEs in the evolution of gene regulatory networks in vertebrates is underestimated.
Evaluating the Function of Recent TE Insertions
Although evolutionary conservation is harder to be employed for sequences that are shared between few species, recent TE exaptation might still be identified by comparative genomics provided a good number of sequences of related species is available. For Alu elements, for instance, a possibility is to use “phylogenetic shadowing," which involves the comparison of several primate sequences (Boffelli et al. 2003). Another possibility is to infer selective pressure within human populations using polymorphism variation data from the increasing number of sequenced human genomes, as has been done to evaluate evolutionary pressure in nonconserved parts of the human genome (Ward and Kellis 2012).
One constraint in the study of primate-specific TEs is that the generation of transgenic or knockout animals is unrealistic. An alternative is to study transgenic mouse models, ideally using large BAC transgenes to better simulate the genomic environment of the locus under study. Using BAC constructs, Lunyak et al. (2007) and Pi et al. (2010) provided very convincing evidence for exaptation of two recent TE insertions that act as a barrier element and an enhancer, respectively. In another work, yeast artificial chromosomes (YACs) were engineered to harbor a LINE 1-derived enhancer of the human apolipoprotein a (apoa) gene flanked by cre-loxP sites (Yang et al. 1998; Huby et al. 2003). Deletion of the enhancer in the context of the 270-kb YAC construct in transgenic mice indicated that it was responsible for approximately 30% of the transcriptional activity of the human gene in the mouse liver. The observed effect was relatively small considering previous results obtained in cell culture, showing the importance of confirming transient transfection assays in more physiological contexts (Huby et al. 2003).
Over large evolutionary distances, for example, zebrafish and mice, conserved enhancers may not exhibit the same regulatory activity (see, e.g., Ariza-Cosano et al. 2012), but the examples above indicate that mice are suitable models to analyze TE exaptation events in the primate and human lineage. These few examples show the feasibility of transgenic mice as tools to explore the functional significance of recent TE insertions in mammalian genomes. In addition, traditional transient transgene assays in cell lines, which is a staple in most studies testing the functionality of TFBSs in TEs, might be complemented by more sophisticated cell culture experiments. For example, Smith et al. (2008) employed stable transfection assays, which presumably better simulates the genomic environment of the locus under study, to determine whether a TE was exapted as an hematopoietic enhancer of the Tal1 gene. Taking advantage of the possibility of obtaining blood progenitors from ES cells, they further showed the enhancing function of the TE-derived sequence by testing transgenes inserted by homologous recombination into ES followed by differentiation in culture. Although Smith et al. (2008) studied an ancient mammalian TE exaptation, the techniques they used might also be employed to functionally test recent TE insertions. Finally, ChIP studies with several TFs and transcriptional cofactors might also provide a strong indication of recent exaptation, as was done by Lynch et al. (2011) to show functionality of ancient TE insertions.
Final Remarks
Although the results stemming from genome-wide studies are still difficult to be fully interpreted, the available evidence from gene-centered studies indicates that TEs provide essential raw material for the process of regulatory evolution in mammals and other organisms. Even if, as predicted by population genetics, the majority of the transcriptional influence exerted by recent TE insertions in mammals and other complex organisms is nearly neutral from an evolutionary point of view, they must definitely contribute to the array of regulatory variation that allows the exploration of the mutational “neutral network” that drives molecular and morphological evolution (Wagner 2008). Thus, what today may be a minor effect of a TE insertion, for instance, a new TFBS leading to a small increase in transcription or a minor alternative promoter that drives expression of a gene to a new tissue, may constitute the first step in a major regulatory reshaping and/or in the co-option of that gene to a new crucial function. Although recent TE exaptation is difficult to test experimentally, the remnants of ancient exaptation events show that this area of research bears great promise for illuminating the mechanisms of genome evolution.
While this manuscript was on final revision and proof stage, critiques on the ENCODE project by Niu and Jiang (2013), Graur et al (2013), and Doolittle (2013) have appeared that, together with a previous comment by Eddy (2012), essentially agree with us that functionality should not be lightly attributed to biochemical activities on the genome, including transposable elements, without proper experimental evidence.
Acknowledgments
The authors thank Daniela Orquera for help with figures. This work was supported by NIH grant DK068400 (M.R.), Agencia Nacional de Promoción Científica y Tecnológica, Argentina PICT 2010–2105 (M.R., F.J.S.d.S.) and 2008–1071 (L.F.F.), and UBACYT 01/Y043 (M.R.).
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ariza-Cosano A, Visel A, Pennacchio LA, Fraser HB, Gómez-Skarmeta JL, Irimia M, Bessa J. Differences in enhancer activity in mouse and zebrafish reporter assays are often associated with changes in gene expression. BMC Genomics. 2012;13:713. doi: 10.1186/1471-2164-13-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Bièche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo JL, Olivi M, Fausser JL, Evain-Brion D, Vidaud M. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod. 2003;68:1422–1429. doi: 10.1095/biolreprod.102.010322. [DOI] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff J-N. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008;16:203–215. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19:607–612. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, et al. (11 co-authors) Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci U S A. 2010;107:19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- Brosius J, Gould SJ. On “genomenclature": a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.”. Proc Natl Acad Sci U S A. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Churakov G, Sadasivuni MK, Rosenbloom KR, Huchon D, Brosius J, Schmitz J. Rodent evolution: back to the root. Mol Biol Evol. 2010;27:1315–1326. doi: 10.1093/molbev/msq019. [DOI] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Costas J, Naveira H. Evolutionary history of the human endogenous retrovirus family ERV9. Mol Biol Evol. 2000;17:320–330. doi: 10.1093/oxfordjournals.molbev.a026312. [DOI] [PubMed] [Google Scholar]
- de Souza FS, Santangelo AM, Bumaschny VF, Avale ME, Smart J, Low MJ, Rubinstein M. Identification of neuronal enhancers of the proopiomelanocortin gene by transgenic mouse analysis and phylogenetic footprinting. Mol Cell Biol. 2005;25:3076–3086. doi: 10.1128/MCB.25.8.3076-3086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A. 2013;110:5294–5300. doi: 10.1073/pnas.1221376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. The C-value paradox, junk DNA and ENCODE. Curr Biol. 2012;22:R898–9. doi: 10.1016/j.cub.2012.10.002. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, Wagner GP. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol Biol Evol. 2012;29:239–247. doi: 10.1093/molbev/msr189. [DOI] [PubMed] [Google Scholar]
- Emera D, Wagner GP. Transformation of a transposon into a derived prolactin promoter with function during human pregnancy. Proc Natl Acad Sci U S A. 2012;109:11246–11251. doi: 10.1073/pnas.1118566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, et al. (22 co-authors) The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini LF, de Souza FS, Low MJ, Rubinstein M. Positive selection of co-opted mobile genetic elements in a mammalian gene: if you can't beat them, join them. Mob Genet Elements. 2012;2:106–109. doi: 10.4161/mge.20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini LF, López-Leal R, Nasif S, Beati P, Gelman DM, Low MJ, de Souza FS, Rubinstein M. Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons. Proc Natl Acad Sci U S A. 2011;108:15270–15275. doi: 10.1073/pnas.1104997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Wakefield MJ, Kohany O, Gu W, Batzer MA, Pollock DD, Jurka J. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Labuda D. CORE-SINEs: eukaryotic short interspersed retroposing elements with common sequence motifs. Proc Natl Acad Sci U S A. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Labuda D. Evolutionary inventions and continuity of CORE-SINEs in mammals. J Mol Biol. 2000;298:365–377. doi: 10.1006/jmbi.2000.3695. [DOI] [PubMed] [Google Scholar]
- Gombart AF, Saito T, Koeffler HP. Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics. 2009;10:321. doi: 10.1186/1471-2164-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. On the immortality of television sets: “Function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5:578–90. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Braasch I, Kraeussling M, Schmidt C, Thoma EC, Nakamura S, Tanaka M, Schartl M. Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet. 2010;6:e1000844. doi: 10.1371/journal.pgen.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huby T, Afzal V, Doucet C, Lawn RM, Gong EL, Chapman MJ, Thillet J, Rubin EM. Regulation of the expression of the apolipoprotein(a) gene: evidence for a regulatory role of the 5' distal apolipoprotein(a) transcription control region enhancer in yeast artificial chromosome transgenic mice. Arterioscler Thromb Vasc Biol. 2003;23:1633–1639. doi: 10.1161/01.ATV.0000084637.01883.CA. [DOI] [PubMed] [Google Scholar]
- Huda A, Tyagi E, Mariño-Ramírez L, Bowen NJ, Jjingo D, Jordan IK. Prediction of transposable element derived enhancers using chromatin modification profiles. PLoS One. 2011;6:e27513. doi: 10.1371/journal.pone.0027513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Gamblin RJ, Ooi L, Bruce AW, Donaldson IJ, Westhead DR, Wood IC, Jackson RM, Buckley NJ. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Laperriere D, Wang TT, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, et al. (90 co-authors) A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Pi W, Bollag R, Zeng S, Keskintepe M, Saliman H, Krantz S, Whitney B, Tuan D. The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol. 2002;76:2410–2423. doi: 10.1128/jvi.76.5.2410-2423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D. Thousands of humanmobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Haussler D. 29 mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS One. 2012;7:e43128. doi: 10.1371/journal.pone.0043128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ. Functional evolution of noncoding DNA. Curr Opin Genet Dev. 2002;12:634–639. doi: 10.1016/s0959-437x(02)00355-6. [DOI] [PubMed] [Google Scholar]
- Lunter G, Ponting CP, Hein J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Núñez E, et al. (14 co-authors) Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- Mason CE, Shu FJ, Wang C, Session RM, Kallen RG, Sidell N, Yu T, Liu MH, Cheung E, Kallen CB. Location analysis for the estrogen receptor-alpha reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res. 2010;38:2355–2368. doi: 10.1093/nar/gkp1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- Meader S, Ponting CP, Lunter G. Massive turnover of functional sequence in human and other mammalian genomes. Genome Res. 2010;20:1335–1343. doi: 10.1101/gr.108795.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, Mager DL. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet Genome Res. 2005;110:342–352. doi: 10.1159/000084966. [DOI] [PubMed] [Google Scholar]
- Micale L, Loviglio MN, Manzoni M, et al. (11 co-authors) A fish-specific transposable element shapes the repertoire of p53 target genes in zebrafish. PLoS One. 2012;7:e46642. doi: 10.1371/journal.pone.0046642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, et al. (64 co-authors) Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- Nakanishi A, Kobayashi N, Suzuki-Hirano A, Nishihara H, Sasaki T, Hirakawa M, Sumiyama K, Shimogori T, Okada N. A SINE-derived element constitutes a unique modular enhancer for mammalian diencephalic Fgf8. PLoS One. 2012;7:e43785. doi: 10.1371/journal.pone.0043785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu DK, Jiang L. Can ENCODE tell us how much junk DNA we carry in our genome? Biochem Biophys Res Commun. 2013;430:1340–3. doi: 10.1016/j.bbrc.2012.12.074. [DOI] [PubMed] [Google Scholar]
- Okada N, Sasaki T, Shimogori T, Nishihara H. Emergence of mammals by emergency: exaptation. Genes Cells. 2010;15:801–812. doi: 10.1111/j.1365-2443.2010.01429.x. [DOI] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. Bioessays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. Mobile DNA and the TE-Thrust hypothesis: supporting evidence from the primates. Mob DNA. 2011;2:8. doi: 10.1186/1759-8753-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;288:645–646. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi W, Yang Z, Wang J, et al. (11 co-authors) The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc Natl Acad Sci U S A. 2004;101:805–10. doi: 10.1073/pnas.0307698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi W, Zhu X, Wu M, Wang Y, Fulzele S, Eroglu A, Ling J, Tuan D. Long-range function of an intergenic retrotransposon. Proc Natl Acad Sci U S A. 2010;107:12992–12997. doi: 10.1073/pnas.1004139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds WF, Kumar AP, Piedrafita FJ. The human myeloperoxidase gene is regulated by LXR and PPARalpha ligands. Biochem Biophys Res Commun. 2006;349:846–854. doi: 10.1016/j.bbrc.2006.08.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román AC, Benitez DA, Carvajal-Gonzalez JM, Fernandez-Salguero PM. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc Natl Acad Sci U S A. 2008;105:1632–1637. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román AC, González-Rico FJ, Moltó E, et al. (12 co-authors) Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21:422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS One. 2009;4:e5761. doi: 10.1371/journal.pone.0005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo AM, de Souza FS, Franchini LF, Bumaschny VF, Low MJ, Rubinstein M. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–1826. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Nishihara H, Hirakawa M, et al. (12 co-authors) Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci U S A. 2008;105:4220–4225. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serdobova IM, Kramerov DA. Short retroposons of the B2 superfamily: evolution and application for the study of rodent phylogeny. J Mol Evol. 1998;46:202–214. doi: 10.1007/pl00006295. [DOI] [PubMed] [Google Scholar]
- Silva JC, Shabalina SA, Harris DG, Spouge JL, Kondrashovi AS. Conserved fragments of transposable elements in intergenic regions: evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet Res. 2003;82:1–18. doi: 10.1017/s0016672303006268. [DOI] [PubMed] [Google Scholar]
- Smit AF. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Sanchez MJ, Follows GA, Kinston S, Donaldson IJ, Green AR, Göttgens B. A novel mode of enhancer evolution: the Tal1 stem cell enhancer recruited a MIR element to specifically boost its activity. Genome Res. 2008;18:1422–1432. doi: 10.1101/gr.077008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro K, Teissier A, Kobayashi N, et al. (13 co-authors) A mammalian conserved element derived from SINE displays enhancer properties recapitulating Satb2 expression in early-born callosal projection neurons. PLoS One. 2011;6:e28497. doi: 10.1371/journal.pone.0028497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem. 2010;285:16135–16144. doi: 10.1074/jbc.M109.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SJ, Goh FG, Banks H, Krausgruber T, Kotenko SV, Foxwell BM, Udalova IA. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci U S A. 2009;106:11564–11569. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Visel A, Bristow J, Pennacchio LA. Enhancer identification through comparative genomics. Semin Cell Dev Biol. 2007;18:140–152. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Neutralism and selectionism: a network-based reconciliation. Nat Rev Genet. 2008;9:965–974. doi: 10.1038/nrg2473. [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science. 2012;337:1675–1678. doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby DA, Vilalta A, Oshima RG. An Alu element from the K18 gene confers position-independent expression in transgenic mice. J Biol Chem. 2000;275:759–768. doi: 10.1074/jbc.275.2.759. [DOI] [PubMed] [Google Scholar]
- Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20:804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kamal M, Lander ES. A family of conserved noncoding elements derived from an ancient transposable element. Proc Natl Acad Sci U S A. 2006;103:11659–11664. doi: 10.1073/pnas.0604768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem. 1998;273:891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]