Abstract
Analysis of genetic differences (gene presence/absence and nucleotide polymorphisms) among strains of a bacterial species is crucial to understanding molecular mechanisms of bacterial pathogenesis and selecting targets for novel antibacterial therapeutics. However, lack of genome-wide association studies on large and epidemiologically well-defined strain collections from the same species makes it difficult to identify the genes under positive selection and define adaptive polymorphisms in those genes. To address this need and to overcome existing limitations, we propose to create a “microbial variome"—a species-specific resource database of genomic variations based on molecular evolutionary analysis. Here, we present prototype variome databases of Escherichia coli and Salmonella enterica subspecies enterica (http://depts.washington.edu/sokurel/variome, last accessed March 26, 2013). The prototypes currently include the point mutations data of core protein-coding genes from completely sequenced genomes of 22 E. coli and 17 S. enterica strains. These publicly available databases allow for single- and multiple-field sorting, filtering, and searching of the gene variability data and the potential adaptive significance. Such resource databases would immensely help experimental research, clinical diagnostics, epidemiology, and environmental control of human pathogens.
Keywords: microbial variome, adaptive evolution, nucleotide polymorphisms, database
In the microbial world, organisms belonging to same species exhibit a diverse set of phenotypic traits, both quantitative and qualitative, resulting in extensive heterogeneity in appearance, metabolism, ability to occupy specific habitats, cause disease, or to resist antimicrobial agents (Orr and Smith 1998; Anisimova and Liberles 2007). Such diversity, in many cases, is defined not only by the presence/absence of certain genes or large gene clusters (Brookfield 2005; Pallen and Wren 2007; Keeling and Palmer 2008) but also by mutational variations in the shared genes, often as single-nucleotide polymorphisms (SNPs) (Feil 2004; Knight et al. 2006; Henriques-Normark et al. 2008). The gene pool of a species is represented by the combination of all genes and their variants that occur in individuals belonging to the given species. The pool is continuously enriched by mutation, sifting and shifting of representative genes, either by random genetic drift or by various selective pressures (Morjan and Rieseberg 2004; Sokurenko et al. 2006; Perron et al. 2008; Rocha 2008). The field of population genetics allows assessment of structure, organization, and dynamics of these gene pools by comparative sequence analysis of several genetic loci across different populations (Kassen and Rainey 2004; Mes 2008). Previously, however, population genetics has been traditionally restricted by the use of limited numbers of loci, due to sequencing and labor expenses (Butlin 2010). This, in turn, has led to major limitations in our ability to detect genome-wide, positively selected variations and to associate them with specific phenotypic diversity that defines fitness in specific environments (Conrad and Hurles 2007). Moreover, lack of experimental knowledge of the genetic basis for most phenotypic traits is another key problem, even for the best studied organisms.
From this perspective, the benefits of whole-genome sequencing and genome-wide search for functional sequence variations are now well recognized. With the flood of new genomic data arising from affordable, rapid, high-quality sequencing technologies, a new field of population genomics has now emerged. It is focused on population-level genetic analysis of variations of individual organisms across the gene pool for a given species (DeLong 2004; Whitaker and Banfield 2006; Brinkman and Parkhill 2008; Stinchcombe and Hoekstra 2008) with the ultimate goal of linking gene function to specific physiological traits (Fournier et al. 2007). We have recently reported a novel analytical approach, Zonal Phylogeny, that detects phylogenetically unlinked mutations at the same amino acid positions, termed hotspot mutations (Sokurenko et al. 2004; Chattopadhyay et al. 2007). These mutations represent a strong indicator of adaptive evolution via molecular convergence of protein variants. Our recent work employing this analytical tool (Chattopadhyay et al. 2009, 2012), along with other studies (Lefebure and Stanhope 2007; Petersen et al. 2007; Soyer et al. 2009), represented initial attempts to provide the research community with a set of positively selected loci that presumably are important in defining the pathogenic potential of organisms, even though the functions of those loci are not yet fully understood.
Here, we present a unique approach to incorporate all results of the molecular evolutionary, selection-based analysis for a microbial species under the umbrella of a free publicly available database. We propose to designate such species-specific resources of genome-wide variations, and their potential adaptive significance, as a “microbial variome,” by analogy with the human variome database (http://www.humanvariomeproject.org/, last accessed March 26, 2013). In this article, we announce variome databases for Escherichia coli and Salmonella enterica subspecies enterica, based on the core protein-coding genes from 22 and 17 genomes (supplementary tables S1 and S2, Supplementary material online), respectively. Both for E. coli and Salmonella, we primarily selected the strains that were clonally distinct, that is, having different alleles of housekeeping genes used for multilocus sequence typing analysis. The only exceptions were two pairs of clonally identical strains—extraintestinal pathogenic strains S88 and UTI89 for E. coli, and serovar Paratyphi A strains ATCC 9150 and AKU_12601 for Salmonella. As a next step, we will expand the list of strains for both species, along with incorporating all protein-coding genes—the core (included in the present version of the database) and the noncore ones.
Database Description
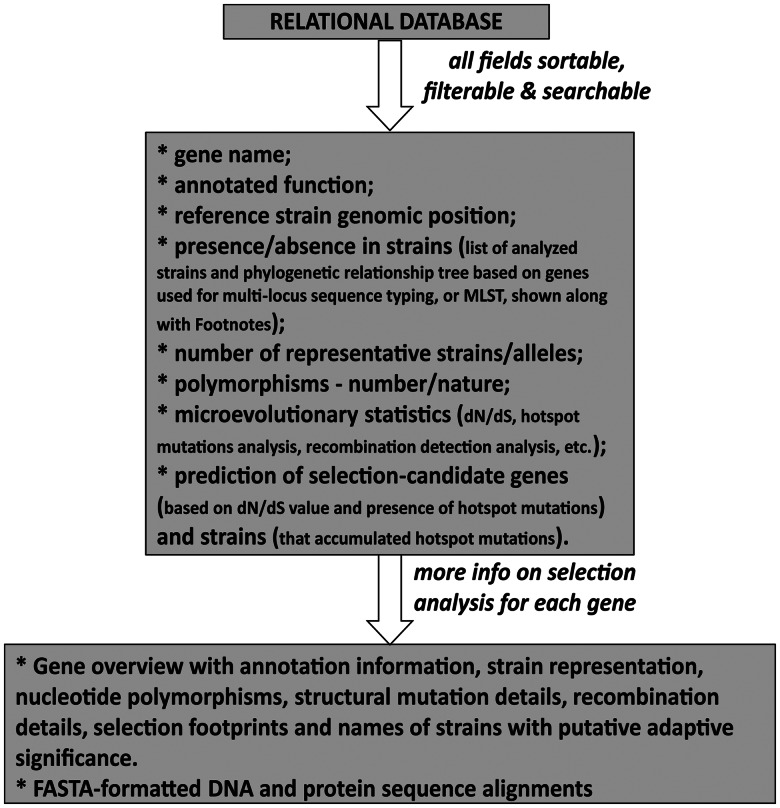
Microbial variome databases are built with relational database software, with a platform-agnostic, browser-based public front end, to curate species-specific project information and analyzed data. The benefits of a relational database are many and well documented (Codd 1970). The basic idea is to present the genomic data in a way that makes it easy to start from a bird's-eye view of the genomic diversity of an entire species or of specific groups/clones and to drill down, in intuitive ways, to individual genomes, genes, and polymorphisms (fig. 1).
Fig. 1.
Schematic architecture of microbial variome database.
The prototype core variome databases of E. coli and Salmonella are focused on innovative, clear visual presentations of data depicting the core genes’ polymorphism diversity. We present the information for both species within a single, unified webpage structure. This makes the relevant data of interest (such as synonymous and nonsynonymous variability, recombination signals, DNA and protein sequence alignments, list of amino acid mutations, and footprints of positive selection based on hotspot mutations and dN/dS statistics) easily accessible at every stage. Although the present version does not offer user any option to enter sequences for comparing with the ones in the database, the user can easily track the gene(s) of interest via sort/filter/search facility and can get access to both DNA and protein sequence data for the corresponding gene(s) as explained below.
For each species database, the website has a pair of overview layouts:
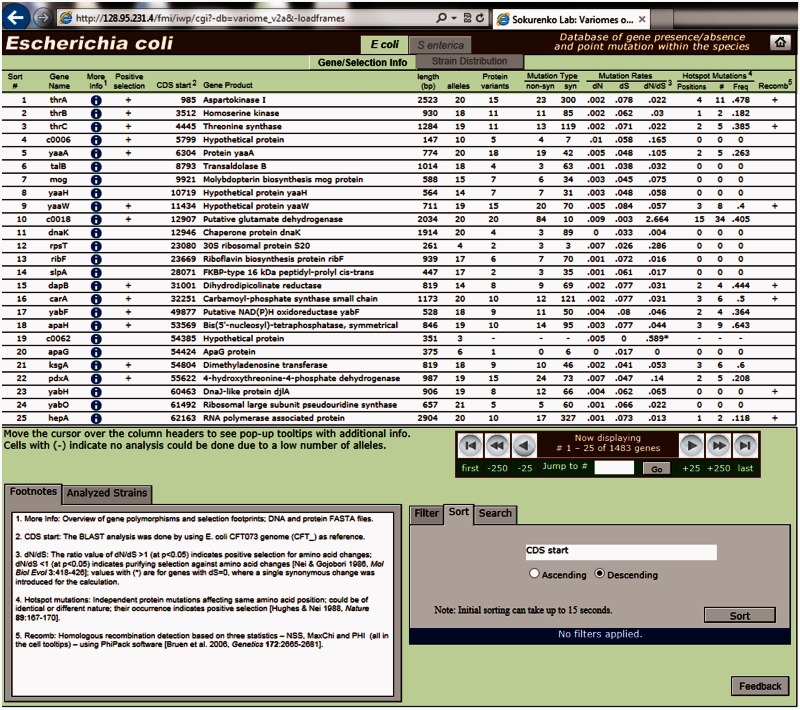
“Gene/Selection Info,” which includes reference-genome annotation of the gene, number of representative strains and alleles, number and nature (synonymous/nonsynonymous) of polymorphisms, analysis results of dN/dS statistics, hotspot mutations, and recombination detection, along with prediction of positive selection effects based on dN/dS values and presence/absence of hotspot mutations (fig. 2).
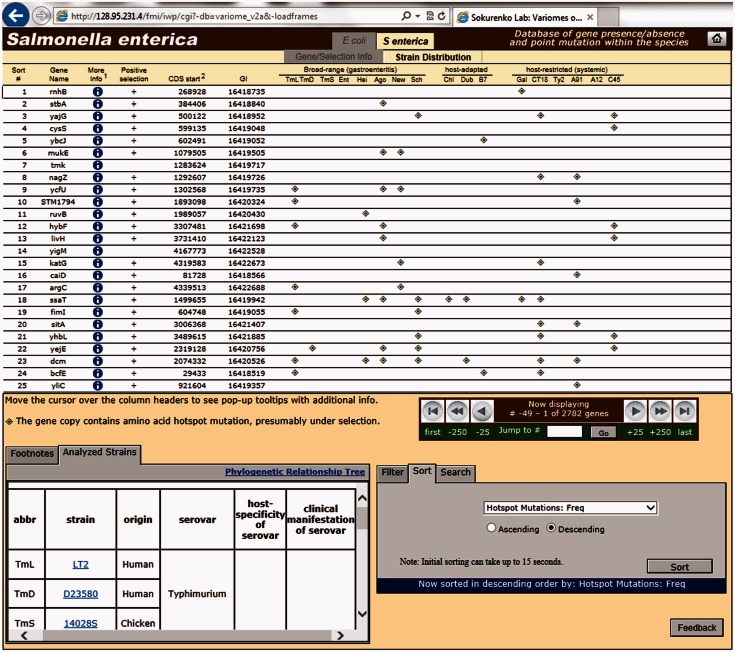
“Strain Distribution,” which indicates strains that accumulate hotspot mutations in positively selected genes (fig. 3).
Fig. 2.
Snapshot of Escherichia coli variome “Gene/Selection Info” layout, showing default sorted list of genes in ascending order of the genomic location in reference genome.
Fig. 3.
Snapshot of Salmonella variome “Strain Distribution” layout, showing sorted list of genes in descending order of the frequency of hotspot mutations.
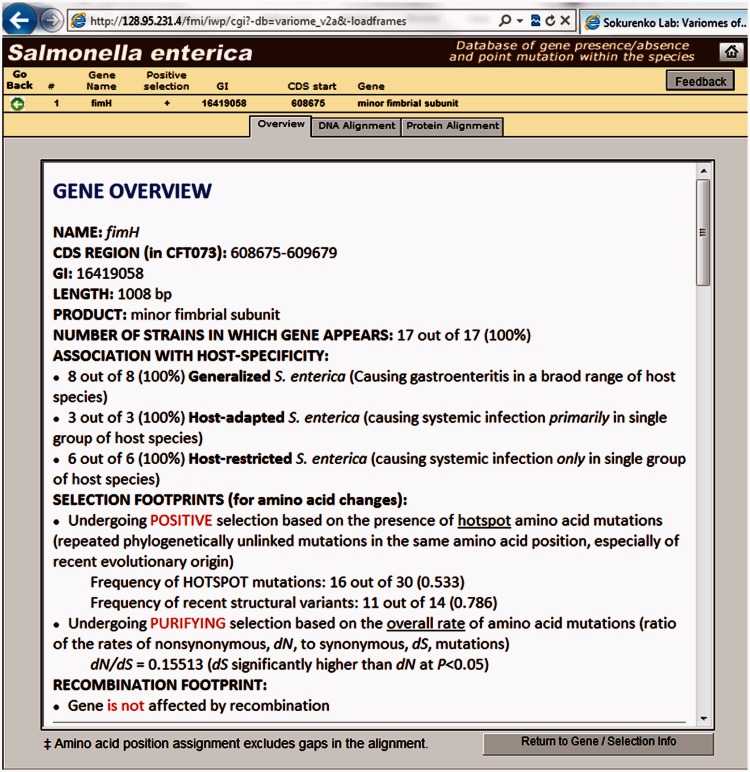
Each layout offers optional single- and multiple-field sorting, filtering, and keyword-based searching. These layouts enable the user to rapidly search or scan the entire set, or a filtered subset of genes, for data of particular interest. The “More Info" button links to an analysis overview for each gene, along with the corresponding FASTA-formatted DNA/protein sequence alignment data. As an example, figure 4 represents a snapshot of a portion of “Gene Overview" for Salmonella gene fimH, encoding the type 1 fimbrial adhesin protein. The results suggest that the gene evolves under positive selection via accumulation of hotspot mutations, in the absence of any intragenic recombination (as detected by three recombination detection statistics—pairwise homoplasy index, maximum χ2, and neighbor similarity score—incorporated in PhiPack software package [Bruen et al. 2006]). This prediction is validated by our recent work on the fimH gene with a larger set of Salmonella strains, demonstrating adaptive convergent evolution of the adhesive protein (Kisiela et al. 2012). It is worth mentioning here that we assess for only intragenic recombination (i.e., exchange of small internal regions in genes) to detect the presence of hotspot mutations of non-recombinant origin. In contrast, if the polymorphisms result from intergenic recombination (i.e., exchange of larger segments of the chromosome and spanning multiple genes), the parent and recombinant alleles of a single gene would be phylogenetically linked and thus not appear as hotspot change in our analysis. Therefore, the identification of positively selected genes via hotspot mutations will not be affected by the presence of intergenic recombination. However, in such cases, the list of SNPs might be mischaracterized, along with the calculations of dN and dS (rate of nonsynonymous and synonymous mutations). Also, we applied high threshold of 95% for both nucleotide sequence identity and length coverage to find an ortholog of each reference gene across the strains. We believe that this selection of high cutoff value minimizes the incorporation of (highly diverse) intergenic recombinants as orthologs.
Fig. 4.
Snapshot of a portion of “Gene Overview” showing the analysis results of Salmonella fimH gene.
As proximate goals, the presently available core variome databases enable the E. coli and Salmonella research communities to easily obtain 1) basic population genetics data on core protein-coding genes across the genomes (based on the reference genome annotation) and 2) information on naturally occurring mutations that are potentially (patho)adaptive for the microorganisms of interest.
Availability
The core E. coli and Salmonella variome databases are hosted by the Sokurenko’s laboratory via the Department of Microbiology, University of Washington, and can be accessed via web browser at http://depts.washington.edu/sokurel/variome, last accessed March 26, 2013. The databases are developed with FileMaker Pro Advanced. They are hosted by FileMaker Server Advanced, which enables access by both FileMaker Pro clients and via any modern web browsers.
The home page of microbial variome databases features a “Downloads" section where Excel files containing the core variome data for E. coli and S. enterica are available for downloading. Also, each page of the variome databases includes a “Feedback" button. By clicking the button from any page, users are welcome to enter their questions, comments, concerns, and recommendations, along with their email addresses if a response is desired, or anonymously if they so choose. As part of maintenance of the databases, we will regularly respond to the feedbacks from the users.
Future Directions
This work presents the first attempt to create microbial variome databases. Besides maintaining the existing database, we plan to periodically update the databases to make them more user-friendly and more informative, based on both user feedback and availability of newly sequenced genomes for these species. With each inclusion of additional data, we need to perform all phylogenetic analyses anew via our newly developed software package TimeZone (Chattopadhyay et al. 2013), followed by the incorporation of new results in the database. As a next step, we also conceptualize the development of a cross-species microbial variome database of closely related species (e.g., E. coli and S. enterica subspecies enterica, or E. coli and Klebsiella pneumoniae) commonly inhabiting similar host compartments as human microbiota. This potentially could enlighten us about the variety of selection pressures in response to host–pathogen, pathogen–pathogen, and pathogen–commensal interactions.
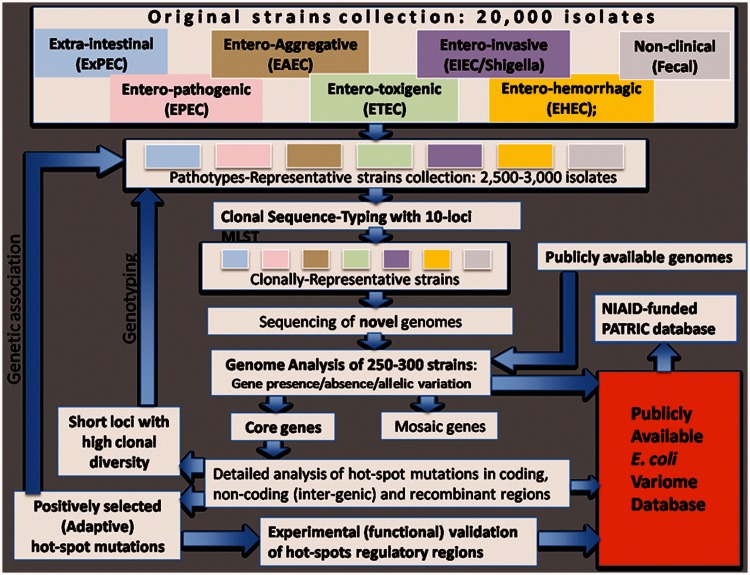
We believe that the establishment of microbial variome databases signals a paradigm shift in microbial genomics, both quantitatively and qualitatively. These tools can be used to retrieve and analyze detailed information of all genetic variations in a given sequence, their population dynamics, and, most importantly, the action of various types of selection pressures. Along with information on strains’ source of isolation, clonal identity, and pathotype, as well as the position of potentially adaptive changes along the affected proteins, this database will offer broad applicability of population genomics tools to experimental research, clinical diagnostics, epidemiology, and environmental control of pathogens. Overall, this knowledge will aid the detection of genotype/phenotype associations and deepen our understanding of bacterial evolution. Our planned layout of microbial variome database (fig. 5, using E. coli as an example) will on the one hand provide information on potential targets for vaccines, antibiotics, and other therapeutic development, while on the other hand, will enable a global surveillance system that can identify newly emerging or re-emerging pathogenic clones and the genetic mechanisms behind such emergence events.
Fig. 5.
Layout plan for Escherichia coli variome database.
Acknowledgments
This work was supported by the National Institute of Health (NIH) grants R01 GM084318 and RC4 AI092828.
Supplementary Material
Supplementary tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
References
- Anisimova M, Liberles DA. The quest for natural selection in the age of comparative genomics. Heredity (Edinb) 2007;99:567–579. doi: 10.1038/sj.hdy.6801052. [DOI] [PubMed] [Google Scholar]
- Brinkman FS, Parkhill J. Population genomics: modeling the new and a renaissance of the old. Curr Opin Microbiol. 2008;11:439–441. doi: 10.1016/j.mib.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Brookfield JF. The ecology of the genome—mobile DNA elements and their hosts. Nat Rev Genet. 2005;6:128–136. doi: 10.1038/nrg1524. [DOI] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK. Population genomics and speciation. Genetica. 2010;138:409–418. doi: 10.1007/s10709-008-9321-3. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Dykhuizen DE, Sokurenko EV. ZPS: visualization of recent adaptive evolution of proteins. BMC Bioinformatics. 2007;8:187. doi: 10.1186/1471-2105-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Paul S, Dykhuizen DE, Sokurenko EV. Tracking recent adaptive evolution in microbial species using TimeZone. Nat Protoc. 2013;8:652–665. doi: 10.1038/nprot.2013.031. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Paul S, Kisiela DI, Linardopoulou EV, Sokurenko EV. Convergent molecular evolution of genomic cores in Salmonella enterica and Escherichia coli. J Bacteriol. 2012;194:5002–5011. doi: 10.1128/JB.00552-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Weissman SJ, Minin VN, Russo TA, Dykhuizen DE, Sokurenko EV. High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection. Proc Natl Acad Sci U S A. 2009;106:12412–12417. doi: 10.1073/pnas.0906217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd EF. A relational model of data for large shared data banks. Commun ACM. 1970;13:377–387. [PubMed] [Google Scholar]
- Conrad DF, Hurles ME. The population genetics of structural variation. Nat Genet. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. Microbial population genomics and ecology: the road ahead. Environ Microbiol. 2004;6:875–878. doi: 10.1111/j.1462-2920.2004.00668.x. [DOI] [PubMed] [Google Scholar]
- Feil EJ. Small change: keeping pace with microevolution. Nat Rev Microbiol. 2004;2:483–495. doi: 10.1038/nrmicro904. [DOI] [PubMed] [Google Scholar]
- Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infect Dis. 2007;7:711–723. doi: 10.1016/S1473-3099(07)70260-8. [DOI] [PubMed] [Google Scholar]
- Henriques-Normark B, Blomberg C, Dagerhamn J, Battig P, Normark S. The rise and fall of bacterial clones: Streptococcus pneumoniae. Nat Rev Microbiol. 2008;6:827–837. doi: 10.1038/nrmicro2011. [DOI] [PubMed] [Google Scholar]
- Kassen R, Rainey PB. The ecology and genetics of microbial diversity. Annu Rev Microbiol. 2004;58:207–231. doi: 10.1146/annurev.micro.58.030603.123654. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Kisiela DI, Chattopadhyay S, Libby SJ, et al. (14 co-authors) Evolution of Salmonella enterica virulence via point mutations in the fimbrial adhesin. PLoS Pathog. 2012;8:e1002733. doi: 10.1371/journal.ppat.1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CG, Zitzmann N, Prabhakar S, Antrobus R, Dwek R, Hebestreit H, Rainey PB. Unraveling adaptive evolution: how a single point mutation affects the protein coregulation network. Nat Genet. 2006;38:1015–1022. doi: 10.1038/ng1867. [DOI] [PubMed] [Google Scholar]
- Lefebure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mes TH. Microbial diversity—insights from population genetics. Environ Microbiol. 2008;10:251–264. doi: 10.1111/j.1462-2920.2007.01449.x. [DOI] [PubMed] [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Mol Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MR, Smith TB. Ecology and speciation. Trends Ecol Evol. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449:835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- Perron GG, Gonzalez A, Buckling A. The rate of environmental change drives adaptation to an antibiotic sink. J Evol Biol. 2008;21:1724–1731. doi: 10.1111/j.1420-9101.2008.01596.x. [DOI] [PubMed] [Google Scholar]
- Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17:1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. Evolutionary patterns in prokaryotic genomes. Curr Opin Microbiol. 2008;11:454–460. doi: 10.1016/j.mib.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, Chattopadhyay S, Johnson JR, Dykhuizen DE. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol. 2004;21:1373–1383. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Gomulkiewicz R, Dykhuizen DE. Source-sink dynamics of virulence evolution. Nat Rev Microbiol. 2006;4:548–555. doi: 10.1038/nrmicro1446. [DOI] [PubMed] [Google Scholar]
- Soyer Y, Orsi RH, Rodriguez-Rivera LD, Sun Q, Wiedmann M. Genome wide evolutionary analyses reveal serotype specific patterns of positive selection in selected Salmonella serotypes. BMC Evol Biol. 2009;9:264. doi: 10.1186/1471-2148-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity (Edinb) 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Banfield JF. Population genomics in natural microbial communities. Trends Ecol Evol. 2006;21:508–516. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.