Abstract
Whole-transcriptome expression profiling provides novel phenotypes for analysis of complex traits. Gene expression measurements reflect quantitative variation in transcript-specific messenger RNA levels and represent phenotypes lying close to the action of genes. Understanding the genetic basis of gene expression will provide insight into the processes that connect genotype to clinically significant traits representing a central tenet of system biology. Synchronous in vivo expression profiles of lymphocytes, muscle, and subcutaneous fat were obtained from healthy Mexican men. Most genes were expressed at detectable levels in multiple tissues, and RNA levels were correlated between tissue types. A subset of transcripts with high reliability of expression across tissues (estimated by intraclass correlation coefficients) was enriched for cis-regulated genes, suggesting that proximal sequence variants may influence expression similarly in different cellular environments. This integrative global gene expression profiling approach is proving extremely useful for identifying genes and pathways that contribute to complex clinical traits. Clearly, the coincidence of clinical trait quantitative trait loci and expression quantitative trait loci can help in the prioritization of positional candidate genes. Such data will be crucial for the formal integration of positional and transcriptomic information characterized as genetical genomics.
Introduction
Quantitative variation in gene expression: the expression phenotype
Normal variation in a complex phenotype, such as susceptibility to obesity and cardiovascular disease (CVD11), is expected to be the result of variation in many genes. An important part of this variation is thought to be in the variability in expression, especially in those genes that are highly conserved (1). Many complex phenotypes, including CVD, are influenced by individual variation in expression of multiple genes. In the past few years, positional genetics (linkage, association) and transcriptomic approaches have been brought together to study the genetic control of gene expression itself (2, 3). This interest derives from the widely held view that phenotypic diversity is at least as likely to come from variation in the levels and/or timing of production of gene products as from functional changes within the genes themselves (2, 4).
Researchers have recently acquired genomewide expression profiles from nontransformed lymphocytes from 1240 individuals in the San Antonio Family Heart Study (SAFHS) (5). Lymphocyte gene expression was analyzed in the Mexican Americans participating in the SAFHS, and heritable variation in expression of >16,000 autosomal genes was found. At least 750 of these showed significant evidence of cis regulation; that is, the strongest linkage signal in a genome scan of each expression phenotype mapped to the physical location of the expressed gene. This strongly suggests that, for these genes, sequence variants in the structural loci are the primary source of individual heritable variation in expression (6). Identification of such variants (e.g., promoter variants) should be especially straightforward, leading to detailed molecular genetic understanding of the sources of heritable variation in expression of each of these genes (7, 8).
Clinical epidemiology: the scope of CVD and metabolic risk in Mexico
In the Third National Health and Nutrition Examination Survey in the United States, the unadjusted and age-adjusted prevalences of the metabolic syndrome, a complex of disorders related to type 2 diabetes mellitus (TSDM), obesity, and the risk of CVD (as defined by the Third Report of The National Cholesterol Education Program Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults) (NCEP-III), were 21.8% and 23.7%, respectively (9). Mexican Americans had the highest age-adjusted prevalence of the metabolic syndrome (31.9%) from a representative sample of U.S. adults (10). By comparison, in a population-based survey in the Republic of Mexico, the prevalence of the metabolic syndrome was 26.6% using the NCEP-III criteria (11).
Today, Mexican Americans make up the largest Hispanic group in the United States (66.9% of the total Hispanic American population), followed by Central and South Americans (14.3%), Puerto Ricans (8.6%), Cubans (3.7%), and other Hispanics (6.5%) (12). As a group, Mexican Americans represent a mixture of several ethnic backgrounds, mainly Spanish-European and Native American (Amerindians).
In Mexico, obesity, T2DM, and CVD now account for more deaths per year than infectious diseases, a change that has been called the epidemiologic transition (13). In 1993, 7.2% of Mexican nationals ages 20 to 69 y were known to have T2DM; by 2000 this prevalence had increased to 10.9%. The prevalence of obesity (BMI ≥30) increased from 21.4–24.4% over the same period (14).
Mexicans share with Mexican Americans a prevalence of CVD risk factors, suggesting shared genetic factors. As the source population, Mexicans are likely to retain more of the allelic diversity resulting from the Spanish conquest of Mexico and subsequent confluence of European and Native American origins. To an even greater degree than Mexican Americans, Mexican nationals have maintained the tradition of large, extended families whose members remain in the same geographic area, providing larger samples of related individuals for genetic analysis. Because much of our interest is in the normal variation of phenotypes lending susceptibility to metabolic disease and CVD risk, it will be interesting to compare the expression of these phenotypes in rapidly developing Mexico with the more economically developed United States. As noted, obesity, T2DM, and CVD are growing public health problems in Mexico. Given historic rates of immigration to the United States, an increase in these risk factors in Mexico will likely affect the U.S. population as well. In summary, the Mexican population is an appropriate and as yet little-studied population for research on the genetic epidemiology of metabolic disease in Hispanics.
Genetic studies of Mexican-American families
The Department of Genetics at Texas Biomedical Research Institute, San Antonio, Texas, has several ongoing genetic epidemiological studies in Mexican-American populations including the SAFHS, the San Antonio Family Diabetes/Gallbladder Study, and the Viva la Familia Study, which is focused on the genetics of obesity and diabetes in Hispanic children (5, 15, 16). These studies have already yielded valuable clues about the genetic basis of metabolic disease via positional information from linkage and genomewide association. The Genética de las Enfermedades Metabólicas en México (Genetics of Metabolic Diseases in Mexico) (GEMM) Family Study will extend these studies to a much larger cohort of people born and living in Mexico (17, 18). This will allow the scientists to compare the effect of a shared genetic background on both sides of the border and will also offer the potential for finding novel genetic variants by examining a larger sample of individuals from different regions in Mexico.
The SAFHS
Although to date there has been little genetic epidemiological study of CVD risk factors in the Republic of Mexico, much research has been done among Hispanics in the United States (Mexican Americans, in particular), motivated by the elevated prevalence of metabolic disease in this population subgroup. The search for genes involved in the expression of obesity has been one of the focal points of the SAFHS, a large, family-based study to examine the genetics of risk of atherosclerosis in Mexican Americans (5). The goal is to dissect genetic signals from the complex interactions of multiple genetic and environmental factors underlying the variation in phenotypes in general populations. The variance components–based statistical methods used in the SAFHS have been developed concurrently with, and often in direct response to, such analysis. In brief, pedigree information and positional marker genotypes (short tandem repeats, single nucleotide polymorphisms, or both) are used to estimate the degree of genetic identity between individuals, and this is compared with their phenotypic similarity. The overall genetic similarity is used to estimate the heritability, or the proportion of total phenotype variance attributable to shared genes. Information from positional markers is used to further decompose the heritability into effects of genetic similarity at specific genomic locations [quantitative trait loci (QTL)] and the main effects of specific markers that may be closely linked to functional genetic variants (19, 20).
To date, genome-scanning efforts have found 2 QTLs located on chromosomes 2 and 8 in this population that have pronounced effects on the expression of a variety of obesity-related phenotypes (e.g., leptin levels, fat mass, and BMI) (21). Arya et al. (22) are currently working to further refine these signals and to identify the genes and allelic variants involved. The same researchers found evidence suggesting that the factor structures for the risk of metabolic syndrome are influenced by multiple distinct genes across the genome. They conducted a principal components analysis using data on 14 phenotypes related to the risk of the development of metabolic syndrome to explore the genetic predispositions of this complex syndrome. The subjects were 566 nondiabetic Mexican Americans in 41 extended families from the SAFHS. Each factor exhibited evidence of either significant or suggestive linkage involving 4 factor-specific chromosomal regions on chromosomes 1, 3, 4, and 6. Significant evidence of linkage of the lipid factor was found on chromosome 4, where the cholecystokinin A receptor (CCKAR) and ADP-ribosyl cyclase 1 (CD38) genes are located. In summary, genetic epidemiological studies have begun to reveal the genetic causes of CVD risk in Mexican Americans, and variance components–based positional gene discovery is an effective tool for understanding the genetic basis of complex phenotypes (23).
The GEMM Family Study
The GEMM study is a newly established, multicenter collaborative study of the genetic epidemiology of metabolic syndrome (17). The overall goal of this project is to identify the genes that are involved in the development of these major public health threats in an effort to better diagnosis and treat those who are afflicted or at risk (18).
Members of the Department of Genetics at Texas Biomedical Research Institute provide oversight and coordination of the GEMM. The 2 participating centers in Mexico have been selected based on their affiliation with a medical university and/or teaching hospital: the Autonomous University of Nuevo León and the Autonomous University of Yucatán. Both centers are currently recruiting individuals in extended families. Volunteers are brought into a dedicated diagnostic facility at each center to provide a medical history, be measured on a variety of anthropometric and other clinical traits relevant to metabolic diseases, provide a blood sample for biochemical analysis and DNA for genotyping, provide biopsy specimens of subcutaneous fat and muscle as a basis for genomewide expression profiling, and provide postprandial measurements of the same traits after a mixed-meal challenge. The entire study is projected to take 5 y. The biochemical analysis and all the genotyping and statistical analyses are carried out at the Texas Biomedical Research Institute (17).
Current status of knowledge
Preliminary data collection in the GEMM
In early 2005, 8 medical institutions in Mexico (Ciudad Obregón, Sonora; Durango, Durango; Monterrey, Nuevo León; San Luis Potosí, S.L.P.; Celaya, Guanajuato; Mexico City; Cuernavaca, Morelos; and Mérida, Yucatán), agreed to recruit a preliminary sample of volunteers as proof of principle for collaborative collection of phenotypes and family data. One to 5e families were recruited at each of the 8 centers. Families were recruited based on their size rather than on any disease status. The clinical examination included basic anthropometric measures [height, BMI (kg/m2) plus a fasting blood draw, waist circumference, and systolic and diastolic blood pressures]. Total serum concentrations of glucose, cholesterol, and triglycerides were measured (17, 18). All the phenotypes examined were significantly heritable.
The centers recruited and examined 381 individuals (200 women and 181 men) in 21 extended families with at least 3 generations represented in each family; 34 women (17.0%) and 29 men (16.0%) in this preliminary cohort of the GEMM had fasting glucose levels ≥126 mg/dL, the cutoff value for T2DM (American Diabetes Association fasting criterion) (24). This measure provided suggestive information about the prevalence of T2DM in this cohort. By comparison, in the SAFHS at initial recruitment, 14.7% of women and 14.9% of men had T2DM according to the American Diabetes Association 1997 criteria (24).
Table 1 presents the distribution of 4 of 5 indicators of the metabolic syndrome (NCEP-III definition) in study participants 20 to 59 y of age (12). It is worth noting the extent of disease risk given that the families were not recruited with respect to any medical condition.
Table 1.
Metabolic syndrome risk factors in women and men aged 20–59 y: Genética de las Enfermedades Metabólicas en México (Genetics of Metabolic Diseases in Mexico) preliminary sample12
| NCEP-III criterion | Women | Men |
| Waist >88 cm (women), >102 cm (men) | 65/162 (40.1) | 41/143 (28.7) |
| Systolic BP ≥130 or diastolic BP ≥85 mm Hg | 52/162 (32.1) | 69/144 (47.9) |
| Triglycerides ≥150 mg/dL | 42/148 (28.4) | 69/133 (52.9) |
| Fasting glucose ≥110 mg/dL | 29/161 (18.0) | 29/143 (20.3) |
Data given as number affected/total number (%).
BP, blood pressure; NCEP-III, Third Report of The National Cholesterol Education Program Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults.
Gene expression phenotype in Mexican Americans
In the SAFHS, genomewide quantitative transcriptional profiles were assayed with lymphocytes from 1431 sample subjects; complete data are available for 1240 subjects (6). These profiles were obtained using an Illumina Bead Station 500 GX platform. High-quality RNA was obtained from frozen lymphocyte samples collected at the first examination period ∼10 y earlier. Of 47,289 transcripts probed by the Sentrix Human-6 Expression BeadChip (Illumina), 20,413 had signal significantly above background. Of these, 20,228 exhibited experiment-wide significant heritabilities.
Using variance component–based quantitative trait linkage analysis, 915 cis-regulated QTLs were identified in the SAFHS data, i.e., loci at which there is experiment-wide significant evidence of variation near the transcript’s genomic location that influences expression level. The average LOD score for these significant cis-acting QTLs is 5 with some LOD scores >40 (6).
Similarly, there is evidence of 1688 trans-acting QTLs that influence expression for T2DM and obesity (3, 4). This data set is extremely useful when prioritizing positional candidate genes because phenotypes showing linkage can be examined for correlations with transcripts within the observed region of linkage. Additionally, the presence of cis-acting QTLs for transcripts within a previously identified linkage region that exhibit correlation with the focal (linked) phenotype can be viewed as independently supported strong candidate genes.
Example: positional-transcriptional candidate gene for HDL levels
To test the utility of the gene expression phenotypes for gene discovery, levels of HDL cholesterol in 1240 SAFHS participants were correlated with levels of the cis-regulated transcripts (6). VNN1 stood out as a gene that showed very strong evidence of both cis regulation and correlation with HDL; bivariate analysis of VNN1 expression and HDL showed genetic correlation of 0.28. VNN1 encodes pantetheinase, an enzyme required for synthesis of the antioxidant cyteamine. Resequencing of the VNN1 promoter identified 4 single nucleotide polymorphisms strongly associated with VNN1 expression and HDL levels (6). Further molecular dissection of this locus is ongoing.
Comparative profiles of lymphocytes, muscle, and adipose tissue in the GEMM: correlation and reliability of synchronous in vivo gene expression in 3 human tissues
Our current recruitment has assayed genomewide gene expression in lymphocytes and biopsy specimens of skeletal muscle and subcutaneous fat in 4 healthy adult men in Monterrey, Mexico, 2 lean (BMI <25) and 2 obese (BMI >30 kg/m2). We obtained RNA samples from lymphocytes and biopsy specimens of quadriceps muscle and subcutaneous fat, 3 tissues relevant to inflammation, obesity, insulin resistance, and T2DM, from these healthy men (25). A similar intervention was done with 4 women in Mérida, Yucatan (unpublished data). The women were 33 to 37 y of age. Inclusion criteria were normotensive, not known to have diabetes or other chronic disease, not taking medication for hypertension or hyperlipidemia, and without symptoms of infection or acute inflammation. They gave informed consent. Recruitment, examination, and biopsies were carried out with approval of the Ethics Committee of the Autonomous University of Nuevo León, Monterrey, Mexico. Analysis of blood and tissue samples was performed at Texas Biomedical Research Institute with approval of the Institutional Review Board of the University of Texas Health Science Center at San Antonio (26). The subjects were examined while in a fasting state. Measurements were made of Blood pressure, height, weight, body composition by bioimpedance, hemoglobin A1c, and fasting plasma glucose measurement were performed. Medical staff collected 3 mL of whole blood and biopsy specimens of subcutaneous abdominal fat and quadriceps muscle under local anesthesia. All subjects underwent follow-up examinations at 2 and 5 d after the procedure, and all recovered as expected (26).
Statistical analyses were performed in R (27). Expression signals were standardized as Z-scores as described (6, 26) to minimize variation due to differences in sample RNA yield. For calculation of intraclass correlation (ICC), the 4 individual measures of each transcript in each tissue type were Z-scored again to minimize the effect of overall differences in mean expression between tissues while maintaining weighted ranks of individual expression levels. ICC, a common measure of reliability of repeated measures (28), is a comparison of within-individual variation with total variation and has a theoretical distribution (−1/(k − 1), 1) where k is the number of repeated measures (29). A negative value of ICC indicates much greater within- than between-individual variance or very poor agreement between repeated measures (in contrast to the Pearson’s correlation, for which −1 = r << 0 implies inverse association).
We used the Illumina Human-6 v2 BeadChip for the Monterrey pilot. All samples provided ample good-quality RNA for amplification and microarray analysis, which required 1.5 μg of amplified RNA per sample (Table 2). Total RNA was queried by microarray for expression of >48,000 transcripts representing validated genes as well as predicted genes and pseudogenes. Expression signals were standardized within each tissue type to remove stochastic variability in sample RNA yield (26). For each tissue type, transcripts with signal above controls at a nominal P < 0.05 in every sample were classed as “detectable” (in the full study, we expect a given transcript might fail to be detectable in some samples). By this criterion, of 48,687 targets queried, detection was 30.9% in lymphocytes, 28.4% in muscle, and 30.8% in adipose tissue. Detection rates per sample were higher (range, 32.8–39.9; Table 2) (25). These results in a small sample compare acceptably to an overall detection rate, by somewhat different criteria, of 43.1% in 1240 lymphocyte samples in the SAFHS (6).
Table 2.
| Clinical measurements |
Total RNA concentration |
% of Detection of transcripts |
||||||||||
| ID | Age, y | Weight, kg | BMI | BP s/d | HbA1c | % Fat | Lymph | Musc | Adip | Lymph | Musc | Adip |
| A | 34 | 63.7 | 20.3 | 100/70 | 4.8 | 12.20 | 3.96 | 75.27 | 43.82 | 35.2 | 34.6 | 39.9 |
| B | 33 | 65.5 | 24.4 | 110/86 | 5.2 | 21.70 | 12.91 | 60.56 | 26.94 | 36.0 | 32.9 | 38.7 |
| C | 33 | 105.2 | 35.6 | 136/90 | 5.0 | 38.30 | 14.66 | 158.40 | 52.60 | 35.7 | 34.5 | 35.6 |
| D | 37 | 132.2 | 41.3 | 126/90 | 5.4 | 36.30 | 9.89 | 179.00 | 32.64 | 35.8 | 32.8 | 39.2 |
Percentage of body fat mass (Fat, %) was measured by bioimpedance. RNA concentration: μg/mL whole blood (lymphocytes) or ng/mg tissue (muscle and adipose); % detection of transcripts at < 0.05 (see text).
BP s/d, blood pressure systolic/diastolic; HbA1c, hemoglobin A1c; ID, identifier; Lymph, lymphocyte; Musc, muscle; Adip, subcutaneous adipose tissue.
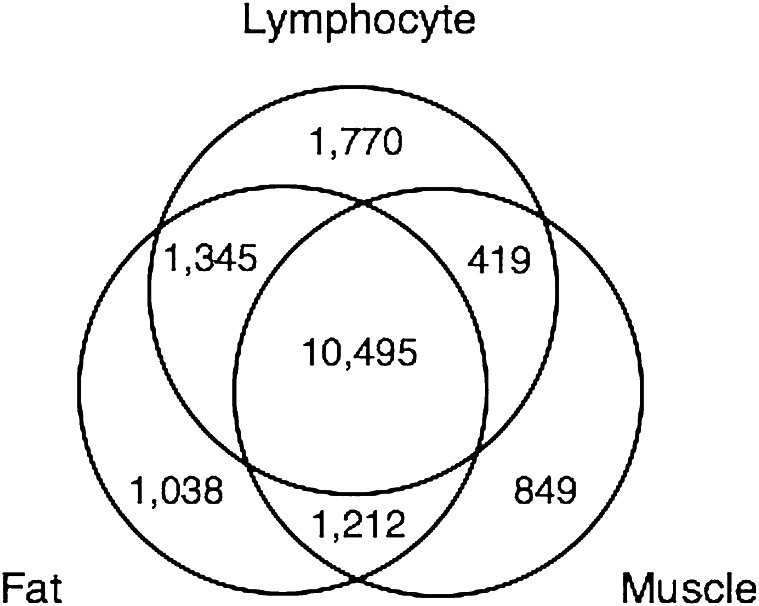
A total of 17,128 gene transcripts were detectable above background in every subject in at least 1 tissue type (25). Of these, 78.6% were detectably expressed in ≥2 tissues, 61.3% in all 3 (Fig. 1). For all detectable transcripts in each pairwise comparison of tissues, Pearson’s correlations of mean transcript expression levels were lymphocyte-fat, r = 0.75 ± 0.006, 11,840 (point estimate ± SE, n transcripts); lymphocyte-muscle, r = 0.61 ± 0.008, 10,914; muscle-fat, r = 0.69 ± 0.007, 11,707. Per transcript, ∼4–6% of genes showed significantly correlated expression (|r| > 0.95 for 4 observations per tissue, 2 df): lymphocyte-fat, 545 (4.60%); lymphocyte-muscle, 634 (5.81%); muscle-fat, 461 (3.94%). Given our sample size, we expect that these are minimum estimates of per-transcript correlation. Mean RNA levels were correlated between tissue types (range, r = 0.61–0.75) (25). For individual genes, we estimated the reliability of expression across tissues as the ICC and found that patterns of individual variation in expression were maintained across tissues for many genes. Transcript reliability was correlated with evidence of cis regulation, suggesting that the effects of proximal sequence variants may be especially consistent in different cellular environments (25, 26).
Figure 1.
The numbers of transcripts detectable above background; most genes were detectable in ≥2 tissue types.
Despite this substantial correlation across tissues, there were some striking examples of tissue specificity in expression. For example, the transcript of the leptin gene LEP was not detectable in lymphocytes, expressed at low levels in muscle, but expressed at high levels in fat; the latter expression was almost perfectly correlated with subject adiposity, increasing ∼1 SD per 10 kg/m2 increase in BMI (26).
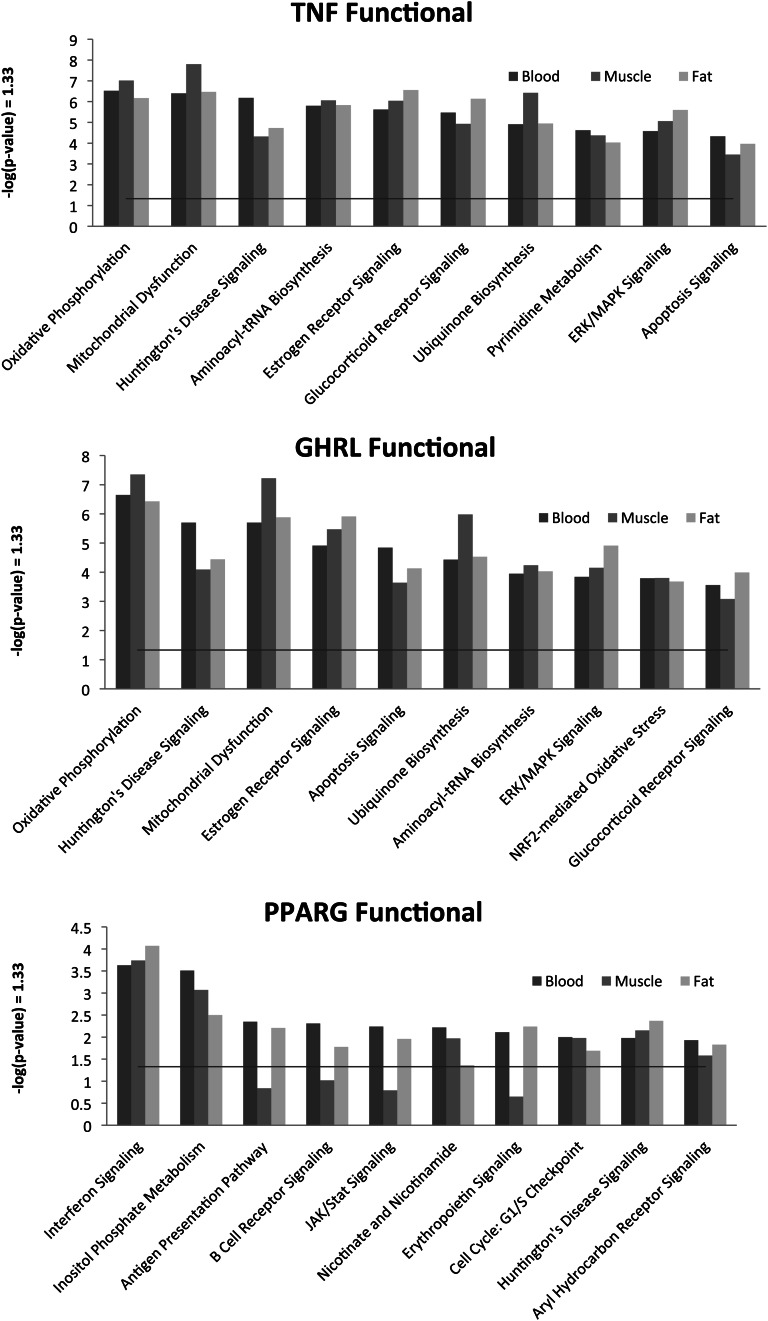
We also examined within- and between-tissue correlation of lymphocyte expression of TNF (TNFA), GHRL (ghrelin), and PPARG, 3 cytokines of interest for their effects on inflammation, and computed Pearson’s correlations with expression in all 3 tissues. Even in this small sample, we found a substantial number of correlations with absolute values ≥80% (Table 3). We then used Ingenuity Pathways analysis to classify the correlated sets by gene function. Figure 2 shows the top 10 functional classes for each focal transcript. There is considerable consistency in functional assignment across tissues. Interestingly, the rank order of functional assignments is quite similar for TNF and ghrelin; lymphocyte expression of these is fully negatively correlated (r = −1), consistent with the function of the gene products (TNF is generally proinflammatory, and ghrelin is anti-inflammatory).
Table 3.
Percentage of detectable transcripts correlated with focal genes (|r| ≥80%) by tissue type1
| Focal transcript | Lymphocyte | Muscle | Adipose |
| TNF | 65.8 | 49.4 | 10.5 |
| GHRL | 68.9 | 77.9 | 9.2 |
| PPARG | 82.4 | 13.8 | 20.2 |
GHRL, ghrelin; PPARG, peroxisome proliferator–activated receptor γ.
Figure 2.
Ingenuity Pathway Analysis functional class assignments for correlated transcripts. Ingenuity Pathway Analysis A is a powerful curated database and analysis system for understanding how proteins work together to effect cellular changes. We use this system to classify the correlated sets by gene function. The top 10 functional classes for each focal transcript are shown. There is considerable consistency in functional assignment across tissues. GHRL, ghrelin; TNF, tumor necrosis factor; PPARG, peroxisome proliferator-activated receptor gamma
The genetic analysis of gene expression naturally leads to the classification of QTLs into cis-acting and trans-acting classes based on the relative genomic locations of the transcript and its QTL (30, 31). For comparison of ICC with the pattern of gene regulation, we used the SAFHS data on cis and trans regulation (6). This comparison was necessarily limited to the subset of transcripts that were detectable in SAFHS and in all 3 tissues in GEMM and that showed nominal evidence (P < 0.05) of heritability in SAFHS. The cis effect size was defined as the proportion of total variance in transcript expression due to allele sharing at the start position of the respective structural gene, with positions established by public genomic data. The trans effect size was defined as the highest positional effect size on any chromosome other than the one bearing the structural gene.
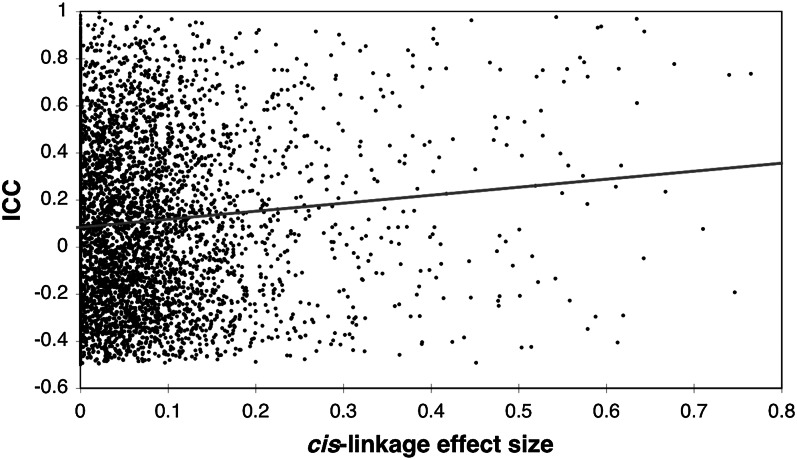
In SAFHS, linkage analysis identified 750 transcripts with nominal genomewide evidence of cis regulation (i.e., the strongest evidence of linkage localized to their structural genes) and 1075 trans-regulated (strongest linkage elsewhere in genome) (6). Results from animal models suggest that effects of cis variants may be especially consistent across tissues, so we hypothesized that cis regulation would enhance reliability (32). In 5443 transcripts that were detectable in both SAFHS and GEMM, transcript reliability in GEMM was positively correlated with the effect size of cis variants in SAFHS (Fig. 3). The pool of highly reliable transcripts (ICC = 0.6, 3 tissues) was enriched for cis-regulated genes relative to the rest (Fisher’s exact test: OR = 2.41, P = 0.0000006) (28). We found no similar enrichment for trans-regulated genes (OR = 1.24, P = 0.19). The latter test is an important control for the possibility that the joint identification of transcripts is reliable and cis-regulated was simply an artifact of signal strength because the level of significance (LOD score = 3, P = 0.0001) was the same for both cis and trans linkage.
Figure 3.
Selected functional comparisons of the top 10% (gray bars) and bottom 10% (black bars) of transcripts ranked by intraclass correlation coefficient. Bar height indicates relative enrichment for genes of a given functional class.
Conclusions
The completion of the Human Genome Project has given researchers the potential to reveal the genetic contribution to major public health problems such as heart disease, diabetes, and obesity. However, finding the individual genetic variants that contribute to the development of disease will be a very difficult task due to the variability in disease genes and their expression in humans (33).
Cardiovascular and metabolic diseases develop as a consequence of a complex cascade of events. An individual inherits a set of alleles (genotype) from his or her parents and, in combination with environmental factors (lifestyle), these determine physiological states such as lipoprotein levels, adiposity, and immune functions. These, in turn, can result in abnormalities such as vessel wall dysfunction, hypertension, and insulin resistance. Over many years, these factors influence the development of chronic diseases such as diabetes, kidney disease, atherosclerosis, and myocardial infarction. Most of these interactions are likely to be influenced by genetic factors.
Most studies of metabolic and cardiovascular disorders to date have used a classic ‘‘one gene at a time’’ approach. This approach has been very successful in clarifying Mendelian traits such as familial hypercholesterolemia. However, complex traits such as the common forms of atherosclerosis present special problems. In particular, it is likely that most complex diseases result from the interactions of multiple genes and, therefore, cannot be realistically modeled by single-gene perturbations. One promising solution involves the combination of natural genetic variation and expression array analysis, sometimes referred to as global gene expression profiling or genetical genomics (34). The measurement of transcript levels in populations allows the identification of coregulated genes and relationships between transcript levels and clinical traits. In addition, this integrative approach relates DNA variation to transcript abundance, allowing identification of primary (cis-acting) and secondary (trans-acting) effects controlling transcript abundance (35).
The GEMM Family Study will provide an opportunity to compare synchronous gene expression in multiple tissues relevant to the metabolic syndrome (17, 25). Although we expect some degree of tissue specificity in expression, our preliminary results suggest that individual variation in gene regulation is consistent across tissues for many genes. Although tissue-specific differences are expected for some genes, many others should exhibit correlated patterns of expression across tissues. This correlation could take 2 forms: 1) mean levels of expression could be correlated across tissues (i.e., a gene that is highly expressed relative to others in one tissue may be similarly ranked in another) or, perhaps of more direct interest to us as genetic epidemiologists, 2) the relative ranking of individual expression for a given gene could be maintained across tissues. Because of the direct regulatory effect of proximal variants, it may be easier to detect this type of correlation in cis-regulated genes.
For genetic analysis, we were especially interested in genes whose relative ranks of individual variation in lymphocyte expression levels most reliably predicted rank order in other tissues. We treated the 3 tissue types as repeated within-individual measures of expression and computed the reliability of each transcript as its ICC (28, 29). In contrast to Pearson’s r—the ICC in Fisher’s terminology—the ICC assumes that repeated measures are drawn from the same distribution; when this assumption is justified, the ICC should be a more accurate measure of association than r because it is based on fewer df (28). Also unlike r, the ICC is not limited to pairwise comparisons. For all 3 tissues, ∼10% of transcripts had an ICC = 0.6; this is a suggested reliability threshold for the ICC and corresponded in our sample to a transcript-wise P = 0.048 (29). Not surprisingly for biological reasons, reliability was greater in pairwise comparisons: lymphocyte-fat, 29.2% with an ICC = 0.6; lymphocyte-muscle, 23.1% with ICC ≥ to 0.06 for both comparisons. Again, given our sample size, we expect that these are minimum estimates of reliability.
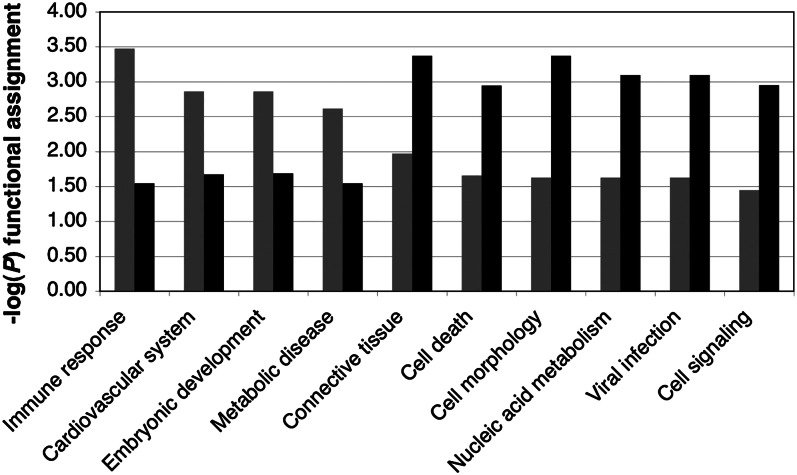
Bioinformatic analysis showed that the reliable transcripts represented a broad range of functions not limited to housekeeping genes (36). The pool of highly reliable transcripts (ICC = 0.6, 3 tissues) was enriched for genes related to immunity, development, and metabolic disease, whereas the least reliable transcripts were enriched for genes with tissue-specific functions (e.g., connective tissue development) or related to cell turnover/cell death (Fig. 4). The latter relationship may reflect the higher turnover rate of lymphocytes relative to the other tissue types.
Figure 4.
The intraclass correlation coefficient (ICC) is positively correlated with the proportion of total variance due to cis variants: r = 0.07, P = 1.1 × 10−7.
Comparison of gene expression across tissues is an area of intense research for a wide variety of purposes. Expression of specific obesity-related genes (leptin, adiponectin, PPARγ, FAT/CD36, and HSD) has been measured recently in samples of abdominal subcutaneous, omental, and mesenteric adipose tissue acquired during gastric bypass surgery in 18 obese patients (37). The comparison of interest in this case was expression in each depot in diabetic versus nondiabetic patients; except for adiponectin, the mean expression levels in mesenteric fat were significantly up-regulated in diabetes. Neither individual differences in expression nor concordance among fat depots were reported. Numerous investigators have sought to develop networks of coexpressed genes to define functional pathways (38, 39). Using banked tissue samples, expression profiles of RNA from 24 human tissues have been analyzed to identify clusters of coexpression related to gene function; this study includes considerable investigation of analytical methods (40). The banked tissues are presumably from different individuals, and samples were pooled; consequently, any evidence of individual variation in expression is absent from these data (and was, of course, not a focus of the study).
In addition to the international benefits of the GEMM Family Study to both Mexico and the United States in terms of biomedical research and health care, the study is structured to enhance the scientific capacity of the centers in Mexico by providing new equipment, technical training, and research opportunities for Mexican investigators in collaborative projects using GEMM data. Establishment of the diagnostic facilities will involve providing equipment and training to the centers. On completion of the data collection for GEMM, these resources will be made available to the sponsoring institutions for future medical research and diagnostic work.
The long-term aim of this project is to study gene expression before and after a well-defined meal to characterize normal variation in postprandial metabolism. This expression profiling is expected to find genes contributing to the metabolic flexibility of individuals in the Mexican population, by using the latest advances in genomic science focused on studies based on an integrated systems approach to human biology. Such a focus on the genetic response following the consumption of a nutritionally defined meal at the level of the specific tissues involved (i.e., fat and muscle), will produce new insights into the genetic architecture of individual variation in metabolism of carbohydrates, fats and proteins. It will also shed light on how this variation in response relates to the risk of a variety of chronic diseases including obesity, diabetes, and heart disease. Although significant challenges remain in the interpretation of large-sample expression data, such data will be crucial for the formal integration of positional and transcriptomic information (41).
This integrative global gene expression profiling approach is proving extremely useful for identifying genes and pathways that contribute to complex clinical traits. Clearly, the coincidence of clinical trait QTL and expression QTL can help in the prioritization of positional candidate genes. More important, mathematical modeling of correlations between levels of transcripts and clinical traits can allow the identification of master key regulatory genes and provide the data needed to develop models of biological networks that better explain disease pathogenesis of complex traits such as CVD, T2DM, and obesity.
Acknowledgments
The authors thank Raul Rangel-Rangel, MD, Department of Surgery, and Carolina Benitez-Mendoza, RN, Department of Nursing, of the Hospital Metropolitano “Dr. Bernardo Sepulveda”, Monterrey, NL, Mexico for their excellent care of the participants during the biopsy procedures. They also thank J. Eduardo Cervantes, Coca Cola Co.-Mexico, for his leadership in obtaining funding for the GEMM Family Study. A special thanks to HEB Grocery Co., San Antonio, TX, and the International Life Sciences Institute, Mexico, which generously made private donations. All authors have read and approved the final manuscript.
Footnotes
Published in a supplement to Advances in Nutrition. Presented at the conference “2nd Forum on Child Obesity Interventions” held in Mexico City, Mexico, August 22–24, 2011. The conference was organized and cosponsored by Fundación Mexicana para la Salud A.C. (FUNSALUD). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of FUNSALUD. The supplement coordinator for this supplement was Frania Pfeffer, FUNSALUD. Supplement Coordinator disclosures: Frania Pfeffer is employed by FUNSALUD, which received a research donation from Coca Cola, PEPSICO, and Peña Fiel, 3 major beverage companies in Mexico, to support the program of childhood obesity research and communication. The supplement is the responsibility of the Guest Editor to whom the Editor of Advances in Nutrition has delegated supervision of both technical conformity to the published regulations of Advances in Nutrition and general oversight of the scientific merit of each article. The Guest Editor for this supplement was Nanette Stroebele, University of Colorado, Denver. Guest Editor disclosure: Nanette
Financial support for this project was provided by private donations from the HEB Grocery Co. of San Antonio, TX, and The International Life Sciences Institute in Mexico. Analyses at Texas Biomedical Research Institute were conducted in facilities constructed with support from the U.S. NIH (Research Facilities Improvement Program grant C06-RR017515) and from the AT&T Foundation. There was no involvement or competing interest in the study design, analysis, or interpretation of the data by these funding sources.
Author disclosures: R. A. Bastarrachea, E. C. Gallegos-Cabriales, E. J. Nava-González, K. Haack, V. Saroja Voruganti, J. Charlesworth, H. A. Laviada-Molina, R. A. Veloz-Garza, V. M. Cardenas-Villarreal, S. B. Valdovinos-Chavez, P. Gomez-Aguilar, G. Meléndez, J. C. López-Alvarenga, H. H. H. Göring, S. A. Cole, J. Blangero, A. G. Comuzzie, J. W. Kent, Jr., no conflicts of interest.
Stroebele had no conflicts to disclose. Publication costs for this supplement were defrayed in part by the payment of page charges. This publication must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact. The opinions expressed in this publication are those of the authors and are not attributable to the sponsors or the publisher, Editor, or Editorial Board of Advances in Nutrition.
Abbreviations used: CVD, cardiovascular disease; GEMM, Genética de las Enfermedades Metabólicas en México (Genetics of Metabolic Diseases in Mexico); ICC, interclass correlation; NCEP-III, Third Report of The National Cholesterol Education Program Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults; QTL, quantitative trait loci; SAFHS, San Antonio Family Heart Study; T2DM, type 2 diabetes mellitus.
Literature Cited
- 1.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–91 [DOI] [PubMed] [Google Scholar]
- 2.Curran JE, Johnson MP, Göring HHH, Dyer TD, Rainwater DL, Cole SA, Mahaney MC, Jowett JBM, MacCluer JW, Collier GR, et al. Genetic analysis of transcriptional profiles for the identification of genes influencing common complex diseases. HUGO’s 11th Genome Meeting, Helsinki, Finland, p33 (A72). 2006.
- 3.Curran JE, Johnson MP, Göring HHH, Dyer TD, Stern MP, Cole SA, Comuzzie AG, Jowett JBM, MacCluer JW, Collier GR, et al. Genetic analysis of transcriptional profiles for the identification of genes influencing risk of diabetes. 66th Scientific Sessions of the American Diabetes Association. Washington DC. June 9–13, Late Breaking Abstracts, p. 7 (A25-LB). 2006.
- 4.Curran JE, Johnson MP, Charlesworth JC, Goring HHH, Dyer TD, Comuzzie AG, Cole SA, Mahaney MC, Jowett JBM, MacCluer JW, et al. Large scale transcriptional profiling for the identification of genes influencing obesity. Int J Obes. 2007;31: Suppl 1:S20 [Google Scholar]
- 5.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–70 [DOI] [PubMed] [Google Scholar]
- 6.Göring HHH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–16 [DOI] [PubMed] [Google Scholar]
- 7.Deutsch S, Lyle R, Dermitzakis ET, Attar H, Subrahmanyan L, Gehrig C, Parand L, Gagnebin M, Rougemont J, Jongeneel CV, et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum Mol Genet. 2005;14:3741–9 [DOI] [PubMed] [Google Scholar]
- 8.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–4 [DOI] [PubMed] [Google Scholar]
- 9.Executive summary of the third report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). JAMA. 2001;285:2486–97 [DOI] [PubMed] [Google Scholar]
- 10.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the Third National Health and Nutrition Examination Survey. JAMA. 2002;287:356–9 [DOI] [PubMed] [Google Scholar]
- 11.Aguilar-Salinas CA, Rojas R, Gomez-Perez FJ, Mehta R, Franco A, Olaiz G, Rull JA. The metabolic syndrome: a concept hard to define. Arch Med Res. 2005;36:223–31 [DOI] [PubMed] [Google Scholar]
- 12.Ramirez R, De la Cruz GP. The Hispanic Population in the United States. Washington (DC): US Census Bureau. Current Population Reports 2002;P20–545.
- 13.Rivera JA, Barquera S, Campirano F, Campos I, Safdie M, Tovar V. Epidemiological and nutritional transition in Mexico: rapid increase of non-communicable chronic diseases and obesity. Public Health Nutr. 2002;5:113–22 [DOI] [PubMed] [Google Scholar]
- 14.Sánchez-Castillo CP, Velasquez-Monroy O, Lara-Esqueda A, Berber A, Sepulveda J, Tapia-Conyer R, James WP. Diabetes and hypertension increases in a society with abdominal obesity: results of the Mexican National Health Survey 2000. Public Health Nutr. 2005;8:53–60 [DOI] [PubMed] [Google Scholar]
- 15.Hunt KJ, Lehman DM, Arya R, Fowler S, Leach RJ, Göring HH, Almasy L, Blangero J, Dyer TD, Duggirala R, et al. Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: the San Antonio Family Diabetes/Gallbladder Study. Diabetes. 2005;54:2655–62 [DOI] [PubMed] [Google Scholar]
- 16.Butte NF, Cai G, Cole SA, Comuzzie AG. Viva la Familia Study: genetic and environmental contributions to childhood obesity and its comorbidities in the Hispanic population. Am J Clin Nutr. 2006;84:646–54 [DOI] [PubMed] [Google Scholar]
- 17.Bastarrachea RA, Kent JW, Jr, Rozada G, Cole SA, López-Alvarenga JC, Aradillas C, Brito-Zurita O, Cerda-Flores RM, Ibarra-Costilla E, Gallegos E, et al. Heritability and genetic correlations of metabolic disease-related phenotypes in Mexico: Preliminary report from the GEMM Family Study. Hum Biol. 2007;79:121–9 [DOI] [PubMed] [Google Scholar]
- 18.Bastarrachea RA, Kent JW, Jr, Comuzzie AG. Study of the genetic component of cardiovascular disease risk phenotypes in a Mexican population. Med Clin (Barc). 2007;129:11–3 [DOI] [PubMed] [Google Scholar]
- 19.Almasy L, Blangero J. Variance component methods for analysis of complex phenotypes. Cold Spring Harb Protoc. 2010; 2010(5):pdb.top77. [DOI] [PMC free article] [PubMed]
- 20.Blangero J. Statistical genetic approaches to human adaptability.1993. Hum Biol. 2009;81:523–46 [DOI] [PubMed] [Google Scholar]
- 21.Voruganti VS, Lopez-Alvarenga JC, Nath SD, Rainwater DL, Bauer R, Cole SA, Maccluer JW, Blangero J, Comuzzie AG. Genetics of variation in HOMA-IR and cardiovascular risk factors in Mexican-Americans. J Mol Med (Berl). 2008;86:303–11 [DOI] [PubMed] [Google Scholar]
- 22.Arya R, Duggirala R, Jenkinson CP, Almasy L, Blangero J, O’Connell P, Stern MP. Evidence of a novel quantitative-trait locus for obesity on chromosome 4p in Mexican Americans. Am J Hum Genet. 2004;74:272–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duggirala R, Blangero J, Almasy L, Arya R, Dyer TD, Williams KL, Leach RJ, O’Connell P, Stern MP. A major locus for fasting insulin concentrations and insulin resistance on chromosome 6q with strong pleiotropic effects on obesity-related phenotypes in nondiabetic Mexican Americans. Am J Hum Genet. 2001;68:1149–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197 [DOI] [PubMed] [Google Scholar]
- 25.Kent JW, Jr, Bastarrachea RA, Haack K, Charlesworth J, Higgins P, López-Alvarenga JC, Laviada-Molina HA, Gallegos-Cabrales EC, Nava-González EJ, Voruganti VS, et al. Synchronous in-vivo large scale transcriptional profiling in human peripheral blood mononuclear cells myocytes and adipocytes. 28th Annual Scientific Meeting of the Obesity Society, San Diego, CA, Oct 8–12, 2010. Abstract Suppl., Vol. 18, Suppl. 2, p. S71 (103P)
- 26.Bastarrachea RA, López-Alvarenga JC, Kent JW, Jr, Laviada-Molina HA, Cerda-Flores RM, Calderón-Garcidueñas AL, Torres-Salazar A, Nava-González EJ, Solis-Pérez E, Gallegos-Cabrales EC, et al. Transcriptome among Mexicans: a large scale methodology to analyze the genetics expression profile of simultaneous samples in muscle, adipose tissue and lymphocytes obtained from the same individual. Gac Med Mex. 2008;144:473–9 [PubMed] [Google Scholar]
- 27.The R Project for Statistical Computing. Available from: http://www.r-project.org. Accessed February 8, 2008.
- 28.Fisher RA. Statistical Methods for Research Workers. Available from: http://psychclassics.yorku.ca/Fisher/Methods/. Accessed February 8, 2008
- 29.Boyer KK, Verma R. Multiple raters in survey-based operations management research: a review and tutorial. Prod Oper Manag. 2000;9:128–40 [Google Scholar]
- 30.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–42 [DOI] [PubMed] [Google Scholar]
- 32.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–53 [DOI] [PubMed] [Google Scholar]
- 33.Lone Dog L. Whose genes are they? The Human Genome Diversity Project. J Health Soc Policy. 1999;10:51–66 [DOI] [PubMed] [Google Scholar]
- 34.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter D. Cellular transcriptomics–the next phase of endocrine expression profiling. Trends Endocrinol Metab. 2006;17:192–8 [DOI] [PubMed] [Google Scholar]
- 36.Ingenuity Pathways Analysis 5.5. Available from: www.ingenuity.com. Accessed February 1, 2008
- 37.Yang Y-K, Chen M, Clements RH, Abrams GA, Aprahamian CJ, Harmon CM. Human mesenteric adipose tissue plays unique role versus subcutaneous and omental fat in obesity related diabetes. Cell Physiol Biochem. 2008;22:531–8 [DOI] [PubMed] [Google Scholar]
- 38.Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konstantopoulos N, Foletta VC, Segal DH, Shields KA, Sanigorski A, Windmill K, Swinton C, Connor T, Wanyonyi S, Dyer TD, et al. A gene expression signature for insulin resistance. Physiol Genomics. 2011;43:110–20 [DOI] [PubMed] [Google Scholar]
- 40.Prieto C, Risueño A, Fontanillo C, De Las Rivas J. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS ONE. 2008;3:e3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlesworth JC, Peralta JM, Drigalenko E, Göring HH, Almasy L, Dyer TD, Blangero J. Toward the identification of causal genes in complex diseases: a gene-centric joint test of significance combining genomic and transcriptomic data. BMC Proc. 2009;3 Suppl 7:S92. [DOI] [PMC free article] [PubMed]