Abstract
The enterococci evolved over eons as highly adapted members of gastrointestinal consortia of a wide variety of hosts, but for reasons that are not entirely clear, emerged in the 1970s as leading causes of multidrug resistant hospital infection. Hospital-adapted pathogenic isolates are characterized by the presence of multiple mobile elements conferring antibiotic resistance, as well as pathogenicity islands, capsule loci and other variable traits. Enterococci may have been primed to emerge among the vanguard of antibiotic resistant strains because of their occurrence in the GI tracts of insects and simple organisms living and feeding on organic matter that is colonized by antibiotic resistant, antibiotic producing micro-organisms. In response to the opportunity to inhabit a new niche – the antibiotic treated hospital patient – the enterococcal genome is evolving in a pattern characteristic of other bacteria that have emerged as pathogens because of opportunities stemming from anthropogenic change.
Introduction
Enterococci began to emerge as leading causes of multidrug resistant hospital acquired infection in the 1970’s and 1980’s (1,2). They now rank among leading causes of hospital acquired infection of the bloodstream, urinary tract, surgical wounds and other sites (3). Prior to that, enterococci isolated from infections were generally regarded as contaminants (1). Two species of Enterococcus are mainly responsible for hospital infections – E. faecalis and E. faecium (Figure 1). The most recent data available on enterococcal infection from all infection sites and all classes of hospitals in the US, which covers the period 01/01/10 – 06/30/12, identified 9,309 bloodstream isolates, 54,709 urinary tract isolates, and 20,032 wound isolates (84,050 isolates total) (TSN® Database, Eurofins, Inc., personal communication Daniel F. Sahm, Ph.D.). Of the total, 17,360 are vancomycin resistant (20.6%), and 64,015 (76%) are E. faecalis. Although 24% of isolates are E. faecium, they represent 14,998 of the 20,038 (75%) of the vancomycin resistant isolates. Similar trends have been reported for the European Union (4, 5). E. faecium infection appears to be highly dependent on resistance to last line drugs, whereas E. faecalis has a greater innate capacity to cause infection irrespective of resistance (2,6). Interestingly, even though E. faecium are more dependent on vancomycin resistance to cause infection, and are more likely to be vancomycin resistant, E. faecalis is the primary species that has transmitted vancomycin resistance to Staphylococcus aureus (7).
Figure 1.
Dendrogram of the genus Enterococcus. A dendrogram of all available 16S rDNA sequences for members of the Enterococcus genus was compiled using the Geneious software (Biomatters Ltd) using the neighbour-joining algorithm and the 16S sequence of Tetragenococcus solitarius as an outgroup. Boostrap values were generated over 1000 iterations. The species E. faecalis and E. faecium are highlighted in blue.
Despite emergence as leading causes of antibiotic resistant infection, enterococci evolved over eons to be members of a broad range of GI tract consortia. In the 1960’s and 1970’s Mundt et al. (8,9) grew enterococci from the GI tracts of mammals (71.3%), reptiles (85.7%), birds (31.8%) and insects (53%) (culture-positive rates typically underestimate carriage several fold (10), meaning that the actual rate of colonization may be closer to 100%). This indicates that since at least the early Devonian period ~ 412 MYA (11) (time of last common ancestor of mammals, reptiles, birds and insects), enterococci have been ubiquitous members of gut microbiomes. Enterococci may in fact have been among the earliest members GI tract consortia. The phylogeny of the genus Enterococcus is shown in Figure 1.
Reflecting their highly evolved role as members of a consortium in an extremely competitive environment, enterococci have reduced genomes. Enterococcus faecalis (cited as Streptococcus zymogenes (12)) has long been known to require a number of amino acids (including Val, Leu, Ile, Ser, Met, Glu, Arg, His and Trp) and vitamins (including biotin, nicotinic acid, pantothenate, pyridoxine, riboflavin, and sometimes folic acid) for maximal growth, with other species being similar in their fastidiousness. The implication is that it is more efficient (i.e., there is a selective advantage) for enterococci to acquire these nutrients from their habitats (e.g., diet of the host or cross-feeding relationships with other microbes in the gut consortium), as opposed to carrying the additional genetic material necessary for their biosynthesis from simpler precursors. In other words, the intense competition in the complex milieu of the gastrointestinal tract has led to a well-adapted, streamlined enterococcal genome.
Commensalism for microbes is not simply the absence of virulence traits, but the active production of factors that lead to stable relationships and limit entry into potentially pathogenic pathways. The transition from commensal to pathogen appears to be associated with changes in ecologies that open new habitats or routes of transmission. Mechanisms that contribute to destabilization of the commensal/host relationship include the acquisition of toxins, or blocks of genes related to pathogenesis, such as pathogenicity islands (13). Alternatively, virulence can result from the loss of key commensal functionality, such as appears to have occurred Bordetella pertussis. B. pertussis is 99.8% identical at the 16S rRNA level to the much less virulent species B. bronchoseptica, but through deletion of 20% of the genome, and conversion of 10% more into pseudogenes by proliferation of the insertion element IS481, it has lost the ability to colonize the host in a non-pathogenic manner (14,15). Similarly, Yersinia pestis evolved from less virulent Y. pseudotuberculosis, mainly by the inactivation of 15% of its genome by deletion and IS element proliferation and acquisition of a virulence plasmid. It is likely in both B. pertussis and Y. pestis, that rapid proliferation of IS elements led to the destabilization of the chromosome, contributing to numerous inversion and deletion events (14,15,16).
Evidence for the recent devolution of the commensal enterococcal genome, leading to a hospital adapted pathogen
We (17) and others (18,19,20) observed that most multidrug resistant hospital infections caused by enterococci were caused by hospital endemic, clonal lineages. For example, in a retrospective study of an outbreak of 206 enterococcal bacteremias over a 36 month period (1984 – 1987), we found that 190 were caused by E. faecalis, most of which were resistant to high levels of aminoglycosides and macrolides (17). Moreover, nearly half of the 190 infections were caused by a single E. faecalis strain that was multidrug resistant as well as hemolytic, whereas the remainder were caused by largely non-hemolytic idiosyncratic strains with few identities to each other or the hospital endemic clone, based on pulsed field electrophoresis pattern (17). The prototype strain, termed MMH594, had become highly hospital adapted and unusually pathogenic, capable of repeated bed-to-bed transmission. In parallel studies, colleagues at Barnes-Jewish Hospital at Washington University isolated the first vancomycin resistant Enterococcus in the US from the bloodstream, urine and feces of a chronically infected HIV/AIDS patient who had received vancomycin therapy (21). E. faecalis V583 manifests a novel vancomycin resistance phenotype, termed VanB, but otherwise possessed numerous similarities to MMH594 (21).
Identification of novel virulence traits and a pathogenicity island (PAI) in hospital adapted enterococci
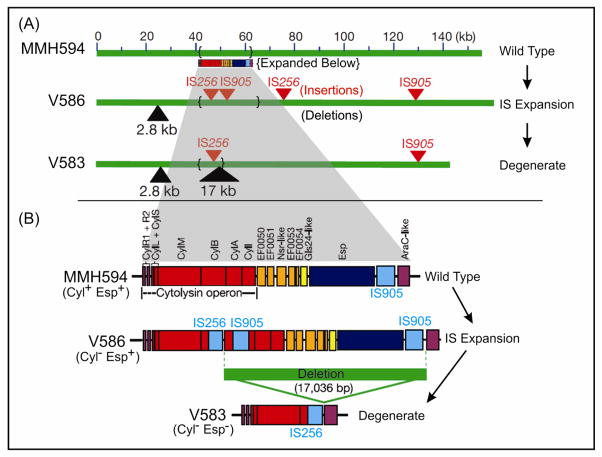
In examining the chromosomes of these multidrug resistant hospital outbreak strains, we identified several traits that had not been known to occur in enterococci, including a capsule (22,23), and a novel adhesin termed Enterococcal Surface Protein (Esp) (24,25). Additionally, we observed that MMH594 expressed the enterococcal cytolysin (17), a toxin that renders E. faecalis infection lethal for humans (17) and animals (26–31), which is regulated by a novel quorum sensing mechanism (32) enabling enterococci to detect target cells at a distance (33). In contrast, V583 possessed a defective cytolysin operon. Exploring the chromosomal organization of virulence traits further in these strains, we found that Esp, the cytolysin, and another factor, termed aggregation substance (an enterococcal surface factor that causes clumping (34) and has been shown to be lethally synergistic with the cytolysin in endocarditis (29)), were encoded by a pathogenicity island (PAI [35]) unlike anything previously seen in gram positives (Figure 2). The E. faecalis PAI discovered in MMH594 is over 150 kb in size, includes a phage like integrase and excisionase, is inserted at a lysyl-tRNA locus, and possesses terminal 10 bp direct repeats. Close comparison of the PAI in V583, and sister isolates from the same patient (V586), and ward (V587), with Wisconisin outbreak strain MMH594, revealed a pattern of genome evolution in E. faecalis similar to that which had been observed for B. pertussis evolving from B. brochoseptica, and Y. pestis evolving from Y. pseudotuberculosis. Specifically, the E. faecalis genome appeared to be undergoing an expansion of IS elements (IS256 and IS905), which volatilized regions of the pathogenicity island (and possibly other regions of the chromosome), leading to an extremely high rate of pseudogene (and intervening functional gene) loss (35). A similar evolutionary trajectory has been identified in E. faecium, in which a large pathogenicity island carrying the esp gene and several IS elements are characteristic for clinical isolates (36,37).
Figure 2.
Microevolution of the E. faecalis pathogenicity island showing the basis for variation between PAI copies in CC2 strains MMH594, V583 and V586 (35). MMH594 is a highly virulent clone that caused approximately 85 E. faecalis bacteremias over a 17 month period (17). V583 and V586 were obtained from the bloodstream of a chronically infected patient (21), and show a process of steady attenuation common in strains associated with chronic infection. In evolving from the prototype PAI of MMH594, IS expansion occurred resulting in new insertions of IS256 and IS905 in strain V586, followed by high frequency excision of a 17 kb fragment encoding half of the cytolysin operon and the esp gene (35). Adapted from (35).
Genome sequence of V583 reveals a profound influx of mobile elements
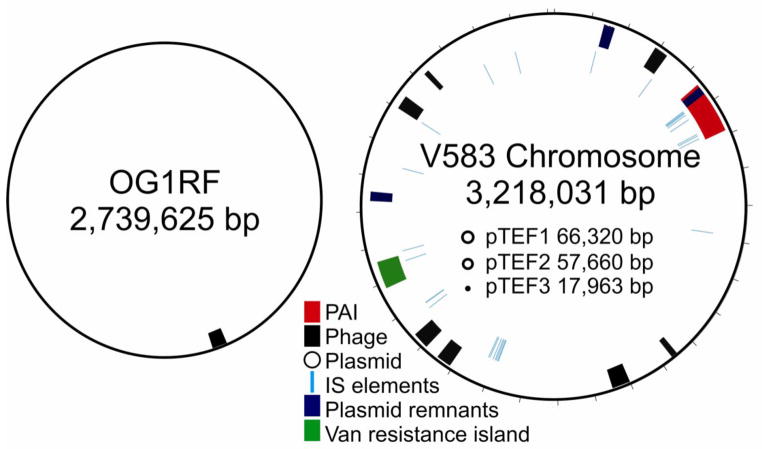
Strain V583 was provided to The Institute for Genomic Research (TIGR) in 1997 for collaborative genome sequence determination. Sequence analysis revealed that 26% of the 3.36 Mb genome consisted of mobile elements, including 7 putative phages, 38 insertion elements, the above mentioned pathogenicity island, remnants of 3 integrated plasmids, as well as 3 independently replicating plasmids (Figure 3) (38). Most of the inferred mobile elements are atypical in nucleotide composition and codon usage compared to that in the rest of the genome. In addition to the plethora of mobile elements, the genome is notable for encoding 35 PTS-type sugar transporters, as well as pathways for the utilization of 15 different sugars (38). The importance of V583 as representative of the leading hospital adapted lineage of E. faecalis was subsequently verified by others (39,40,41).
Figure 3.
Comparative mobile element content of the genomes of the commensal-like strain OG1RF, and the hospital-adapted strain V583. (V583 diagram adapted from (38)).
Sequence of OG1RF provides a basis for comparison
Following publication of the closed sequence of V583 in 2003, there was little basis for comparison until publication of the closed genome sequence of strain OG1RF in 2008 (42). OG1RF is derived from strain 2SaR (which occurred in the oral consortium of a pediatric dental caries lesion (43)). For laboratory use, spontaneous resistances to rifampicin and fusidic acid were selected (44). It was determined to be plasmid-free, and not to possess native resistance to any antibiotic (44). Of oral cavities examined (43), 69% were found to carry E. faecalis strains independent of caries status indicating that enterococci are commensals of the oral cavity as well. Since OG1RF had no known or suspected association with antibiotic resistance or hospital adaptation, as a likely commensal it provided a good comparator for V583. Moreover, its extensive use in the laboratory made its nucleotide sequence of special interest. In contrast to the 3.36 Mb genome of V583, that of OG1RF was found to be 2.74 Mb (42).
One interesting difference between OG1RF and V583 is that OG1RF harbors a clustered, regularly interspaced short palindromic repeat (CRISPR) locus (42). CRISPRs provide bacteria and archaea with sequence specific, acquired defense against plasmids and phage (45). As noted above, hospital endemic strains of E. faecalis, exemplified by V583, are replete with mobile elements, constituting over 25% of the genome. Since V583 lacked a CRISPR locus, it was of interest to determine whether presence or absence of CRISPR correlated with mobile element accumulation in E. faecalis. We therefore generated draft genome sequences for 16 additional E. faecalis strains (46), selecting those that represented the deepest phylogenetic nodes spanning the diversity of the species (47). These draft genome sequences were used to identify polymorphisms in the location and content of CRISPR loci among these strains (48). From among the CRISPR loci identified, we found CRISPR spacer identity to pheromone-responsive plasmids as well as phage, indicating that in E. faecalis, CRISPR influences the movement of these elements between strains. We found the location of CRISPR loci to be highly conserved, and therefore screened additional E. faecalis strains, including isolates that predated the use of antibiotics, for CRISPR content by PCR. A highly significant correlation was found between the absence of a CRISPR-cas locus and the presence of acquired antibiotic resistance in E. faecalis. Similar results were found for a more limited set of E. faecium genomes from US isolates (48). The near complete absence of CRISPR-cas in multidrug resistant strains suggests that antibiotic use inadvertently selects for enterococcal strains with compromised genome defense (48).
E. faecium genomics
The first release of an E. faecium genome sequence was noted in a press release in 2000, which reported generation of draft genome sequence for E. faecium strain DO in a day (Press release May 9, 2000, Joint Genome Institute, David Gilbert; http://www.jgi.doe.gov/News/news_5_9_00.htm ). However, that data was not assembled in a practically useful form and analyzed until 2012 (49). The first genome sequence-based analysis for E. faecium appeared in 2010 (50). van Schaik and coworkers reported the generation, assembly and analysis of high quality draft genomes for 7 strains, including 4 infection derived strains, 1 hospital-associated fecal strain, and 2 commensal strains of E. faecium without known connection to a health care setting. Compared to E. faecalis, E. faecium strains appeared to possess more metabolic pathways for the utilization of carbohydrates of plant origin, but lacked the pathway for ethanolamine utilization, and also lacked genes for the detoxification of reactive oxygen species. Instead, E. faecium appears to have a glutathione peroxidase to protect the cell from oxidative damage (50). It was noted that one commensal strain, E980 was substantially diverged from the other six strains (50). This study also revealed that the pathogenicity island carrying the esp gene is highly variable in clinical isolates and can be transferred by conjugation (50).
A Broad Institute collaboration in 2010 also announced the initial generation and public release of draft genome sequence data for the above mentioned diverse 16 E. faecalis strains, and additionally 8 E. faecium strains, 3 E. casseliflavus strains and 1 E. gallinarum strain (46). A complete analysis of these genomes was reported in 2012 (51). Based on comparison of the 2113 core genes common to each of the 8 E. faecium strains, it was noted that the strains fell into two distinct clades – Clade A, which consisted entirely of infection derived isolates, and Clade B, which consisted mainly of commensal strains and one infection derived isolate. Closer examination of the core genes shared by E. faecium strains showed that average nucleotide identity between Clade A and B (93.9% – 95.6%) approximated the degree of divergence that typically occurs between species. Moreover, strain E980, the most divergent strain from the previous study, was included in this commensal clade. Further, the most divergent strains of Clade A were shown to be mosaics of Clades A and B. Similar results were found comparing a much smaller subset of 100 core genes occurring in these genomes, the genomes sequenced by the van Schaik group, and several other genome sequences in the public domain (52). Of potential but yet untested importance, the hospital adapted vancomycin-resistant E. faecium strains possessed an elaborate cell wall carbohydrate locus that included putative sialic acid biosynthesis (neuABCD) genes in a region found to vary among E. faecium isolates, along with a divergently transcribed, which is annotated as a β-lactamase gene, but is likely to be involved in cell wall transpeptidation (51). The observation that hospital-adapted strains have the genetic potential for altered cell surfaces, possibly camouflaged by a coating of sialic acid, a molecule common in host tissues, raises the possibility of enhanced immune evasion.
Recently, the first complete E. faecium genome was reported (53). Analysis showed that the MLST type 17 Clade A strain contained a proliferation of 77 insertion elements in the chromosome, including a Tn1549 vanB element, along with 3 novel phages. Inverted copies of a prophage appear to have mediated an inversion of 683 kb around the replication terminus, resulting in an imbalance in the replichore. Even though this genome is representative of the leading hospital-adapted clade of E. faecium, its genome is approximately 10% smaller than that of E. faecalis V583, with much of that difference attributable to the presence of fewer phages that are lysogenic in the genome.
Conclusion
In conclusion, the enterococci are ancient, highly evolved members of GI tract consortia of all animals studied, suggesting that they were part of an early GI tract consortium in the last common ancestor. Their ability to survive the grinder of the nematode Caenorhabditis elegans and bind to its intestinal wall (54) may provide a mechanism for the genesis of the original GI tract consortium. Enterococci may have remained resident in the GI tract of a simple organism, such as C. elegans, where it was bathed in the lysates of other bacteria from which it derived nutrition, perhaps allowing for genome reduction and further specialization. Speculatively, as other types of microbes survived the grinder and adhered to either the intestine or to enterococci, the complexity of the consortium may have evolved. Since enterococci colonize the guts of insects (and potentially non-insect fauna of organic soils such as nematodes), enterococci in these gut reservoirs may well be continuously exposed to antibiotics and antibiotic resistance genes occurring in antibiotic producing organisms that are co-resident in the organic matter on which they feed. This may provide an initial opportunity for entry of resistance genes into enterococci, and subsequent selection for resistant strains. Insect enterococci then may comingle with the enterococci of reptile, bird and mammalian predators, up the food chain, creating a tenuous connection between antibiotic production and resistance in the soil, and antibiotic resistance in mammalian-adapted strains of enterococci. The large scale application of antibiotics in humans and agriculture during the last sixty years may then have selected for the outgrowth of rare antibiotic resistant lineages, and the occurrence of strains lacking CRISPR protection with enhanced ability to exchange resistances. This natural pipeline may have set the stage for the ongoing emergence of enterococci among the vanguard of multidrug resistant hospital pathogens.
As for other microorganisms that rapidly adapted new niches stemming from anthropological changes (e.g., B. pertussis (14,15) and Y. pestis (16)), hospital adapted lineages of the enterococci appear to be undergoing a similar changes to the genome. IS elements are proliferating in hospital-adapted enterococcal genomes (38,53) leading to the generation of many pseudogenes as the result of inactivation of genes that were important for competitive growth in the GI tract, but which are no longer required for non-competitive growth in the antibiotic-cleared ecology of the hospitalized patient ([35], unpublished, van Schaik et al. In preparation). It is likely that this IS element expansion will be followed by active genome reduction and loss of these pseudogenes (55), especially as IS elements proliferate to an extent where smaller and smaller spacing between copies increases the chance of penalty-free deletion by recombination across identical copies of an IS element. The ultimate outcome may be a highly hospital adapted pathogenic Enterococcus with altered biology including refined mechanisms for transmission in the hospital environment, analogous to the evolution of human pathogen B. pertussis from the generalist B. bronchoseptica.
Highlights.
Enterococci are ancient commensals in the GI tracts of hosts ranging from insects to man
Enterococci are now leading causes of multidrug resistant infection and are spreading resistances
Genomes of hospital adapted strains of enterococci consist of over 25% mobile elements
Loss of CRISPR protection of the genome correlates with the accumulation of resistance and virulence traits
Pathogens evolve from generalists by adapting to new ecologies created by anthropogenic change
Acknowledgments
Portions of this work were supported by NIH grant AI072360, and the Harvard-wide Program on Antibiotic Resistance, AI083214. Willem van Schaik is supported by the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) “Evolution and Transfer of Antibiotic Resistance” (EvoTAR) under grant agreement number 282004.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1*.Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–78. doi: 10.1128/cmr.7.4.462. Outstanding historical review of enterococcal virulence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huycke MM, Sahm DF, Gilmore MS. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–49. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK National Healthcare Safety Network Team and Participating National Healthcare Safety Network Facilities. NHSN Annual Update-Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Annual Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 4.de Kraker ME, Jarlier V, Monen JC, Heuer OE, van de Sande N, Grundmann H. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect. 2012 doi: 10.1111/1469–0691. [DOI] [PubMed] [Google Scholar]
- 5.Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top J, Vuopio-Varkila J, Willems RJ, Witte W, Woodford N. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008;13(47):pii, 19046. [PubMed] [Google Scholar]
- 6.Mundy LM, Sahm DF, Gilmore MS. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13:513–22. doi: 10.1128/cmr.13.4.513-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, Godfrey P, Palmer KL, Bodi K, Mongodin EF, Wortman J, Feldgarden M, Lawley T, Gill SR, Haas BJ, Birren B, Gilmore MS. Comparative Genomics of Vancomycin-Resistant Staphylococcus aureus Strains and Their Positions within the Clade Most Commonly Associated with Methicillin-Resistant S. aureus Hospital-Acquired Infection in the United States. MBio. 2012;3(3):pii, e00112–12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8**.Mundt JO. Occurrence of enterococci in animals in a wild environment. Appl Microbiol. 1963;11:136–40. doi: 10.1128/am.11.2.136-140.1963. Landmark paper showing the broad distribution of enterococci in animals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin JD, Mundt JO. Enterococci in insects. Appl Microbiol. 1972;24(4):575–80. doi: 10.1128/am.24.4.575-580.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 11.Seldon PA. Terrestrialization of animals 1.3.6. In: Briggs DEG, Crowther PR, editors. Paleobiology II. Blackwell Science, Ltd; 2003. pp. 71–74. [Google Scholar]
- 12**.Niven CF, Jr, Sherman JM. Nutrition of the Enterococci. J Bacteriol. 1944;47:335–342. doi: 10.1128/jb.47.4.335-342.1944. Seminal work defining the auxotrophies of E. faecalis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–79. doi: 10.1146/annurev.micro.54.1.641. Excellent source for the broad view of pathogenicity islands and their evolution. [DOI] [PubMed] [Google Scholar]
- 14**.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. Landmark paper describing the evolution of a pathogen from a generalist in genomic terms, and relating this to anthropogenic changes. [DOI] [PubMed] [Google Scholar]
- 15*.Bentley Stephen D, Parkhill Julian. Comparative genomics structure of prokaryotes. Annual Rev Genet. 2004;38:771–791. doi: 10.1146/annurev.genet.38.072902.094318. Insightful review of microbial genome structure and evolution. [DOI] [PubMed] [Google Scholar]
- 16.Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 2004;101:13826–31. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huycke MM, Spiegel CA, Gilmore MS. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow JW, Kuritza A, Shlaes DM, Green M, Sahm DF, Zervos MJ. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J Clin Microbiol. 1993;31:1609–11. doi: 10.1128/jcm.31.6.1609-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray BE, Singh KV, Markowitz SM, Lopardo HA, Patterson JE, Zervos MJ, Rubeglio E, Eliopoulos GM, Rice LB, Goldstein FW, et al. Evidence for clonal spread of a single strain of beta-lactamase-producing Enterococcus (Streptococcus) faecalis to six hospitals in five states. J Infect Dis. 1991;163:780–5. doi: 10.1093/infdis/163.4.780. [DOI] [PubMed] [Google Scholar]
- 20.Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11:821–8. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. Description of the first vancomycin resistant Enterococcus in the US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock LE, Gilmore MS. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc Natl Acad Sci U S A. 2002;99:1574–9. doi: 10.1073/pnas.032448299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock LE, Shepard BD, Gilmore MS. Molecular analysis of the Enterococcus faecalis serotype 2 polysaccharide determinant. J Bacteriol. 2003;185:4393–401. doi: 10.1128/JB.185.15.4393-4401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun. 2001;69:4366–4372. doi: 10.1128/IAI.69.7.4366-4372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ike Y, Hashimoto H, Clewell DB. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immun. 1984;45:528–30. doi: 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ike Y, Hashimoto H, Clewell DB. High incidence of hemolysin production by Enterococcus (Streptococcus) faecalis strains associated with human parenteral infections. J Clin Microbiol. 1987;25:1524–8. doi: 10.1128/jcm.25.8.1524-1528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jett BD, Jensen HG, Nordquist RE, Gilmore MS. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect Immun. 1992;60:2445–52. doi: 10.1128/iai.60.6.2445-2452.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, Zervos MJ. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37(11):2474–7. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh KV, Qin X, Weinstock GM, Murray BE. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. Infect Dis. 1998;178:1416–20. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 31.Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, Ausubel FM. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001;98:10892–7. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature. 2002;415:84–7. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- 33*.Coburn PS, Pillar CM, Jett BD, Haas W, Gilmore MS. Enterococcus faecalis senses target cells and in response expresses cytolysin. Science. 2004;306:2270–2. doi: 10.1126/science.1103996. First description of a telesensing mechanism that permits a microbe to sound the environment for targets. [DOI] [PubMed] [Google Scholar]
- 34.Yagi Y, Kessler RE, Shaw JH, Lopatin DE, An F, Clewell DB. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J Gen Microbiol. 1983;129:1207–15. doi: 10.1099/00221287-129-4-1207. [DOI] [PubMed] [Google Scholar]
- 35*.Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417:746–50. doi: 10.1038/nature00802. First description of a canonical PAI in a gram positive microbe. [DOI] [PubMed] [Google Scholar]
- 36.Leavis H, Top J, Shankar N, Borgen K, Bonten M, van Embden J, Willems RJ. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J Bacteriol. 2004;186:672–82. doi: 10.1128/JB.186.3.672-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leavis HL, Willems RJ, van Wamel WJ, Schuren FH, Caspers MP, Bonten MJ. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 2007;3(1):e7. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–4. doi: 10.1126/science.1080613. Seminal study on the genomics of enterococci providing a rigorous and insightful analysis of the genome of hospital adapted strain V583. [DOI] [PubMed] [Google Scholar]
- 39.Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J Bacteriol. 2005;187:5709–18. doi: 10.1128/JB.187.16.5709-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner G, Klare I, Spencker FB, Witte W. Intra-hospital dissemination of quinupristin/dalfopristin- and vancomycin-resistant Enterococcus faecium in a paediatric ward of a German hospital. J Antimicrob Chemother. 2003;52:113–5. doi: 10.1093/jac/dkg305. [DOI] [PubMed] [Google Scholar]
- 41.Leavis HL, Bonten MJ, Willems RJ. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006;9:454–60. doi: 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9(7):R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gold OG, Jordan HV, van Houte J. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch Oral Biol. 1975;20:473–477. doi: 10.1016/0003-9969(75)90236-8. [DOI] [PubMed] [Google Scholar]
- 44.Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinberger AD, Gilmore MS. CRISPR-Cas: to take up DNA or not - that is the question. Cell Host Microbe. 2012;12:125–6. doi: 10.1016/j.chom.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer KL, Carniol K, Manson JM, Heiman D, Shea T, Young S, Zeng Q, Gevers D, Feldgarden M, Birren B, Gilmore MS. High-quality draft genome sequences of 28 Enterococcus sp isolates. J Bacteriol. 2010;192:2469–2470. doi: 10.1128/JB.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Jr, Gilmore MS. Genetic diversity among Enterococcus faecalis. PLoS One. 2007;2(7):e582. doi: 10.1371/journal.pone.0000582. Comprehensive phylogenetic analysis of the species E. faecalis and the distribution of resistance and virulence traits throughout the species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48**.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1(4):pii, e00227–10. doi: 10.1128/mBio.00227-10. First association of CRISPR absence with the emergence of antibiotic resistant lineages of any species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin X, Galloway-Peña JR, Sillanpaa J, Roh JH, Nallapareddy SR, Chowdhury S, Bourgogne A, Choudhury T, Muzny DM, Buhay CJ, Ding Y, Dugan-Rocha S, Liu W, Kovar C, Sodergren E, Highlander S, Petrosino JF, Worley KC, Gibbs RA, Weinstock GM, Murray BE. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012;12:135. doi: 10.1186/1471-2180-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H, Willems RJ. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics. 2010;11:239. doi: 10.1186/1471-2164-11-239. First analysis of the genome content of the species E. faecium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. Comparative Genomics of Enterococci: Variation in Enterococcus faecalis, Clade Structure in E. faecium, and Defining Characteristics of E. gallinarum and E. casseliflavus. mBio. 2012;3(1):e00318–11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. Genomic and SNP Analyses Demonstrate a Distant Separation of the Hospital and Community-Associated Clades of Enterococcus faecium. PLoS ONE. 2012;7(1):e30187. doi: 10.1371/journal.pone.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam MM, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, Howden BP, Stinear TP. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol. 2012;194:2334–41. doi: 10.1128/JB.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, Ausubel FM. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001;98(19):10892–7. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuo C-H, Ochman H. The Extinction Dynamics of Bacterial Pseudogenes. PLoS Genet. 2010;6(8):e1001050. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]