Abstract
Tumors recruit mesenchymal stem cells (MSCs) to facilitate healing, which induces their conversion into cancer-associated fibroblasts that facilitate metastasis. However, this process is poorly understood on the molecular level. Here we show that the CXCR6 ligand CXCL16 facilitates MSC or Very Small Embryonic-Like (VSEL) cells recruitment into prostate tumors. CXCR6 signaling stimulates the conversion of MSCs into cancer-associated fibroblasts, which secrete stromal-derived factor-1, also known as CXCL12. CXCL12 expressed by cancer-associated fibroblasts then binds to CXCR4 on tumor cells and induces an epithelial to mesenchymal transition, which ultimately promotes metastasis to secondary tumor sites. Our results provide the molecular basis for MSC recruitment into tumors and how this process leads to tumor metastasis.
Introduction
Tumors have long been considered as wounds that do not heal 1. Wound healing normally requires the participation of many different cell types as well as the activation of a vast number of cellular processes including matrix degradation, proliferation, and recruitment of inflammatory cells. In addition, cells such as fibroblasts, epithelial and endothelial cells are also recruited and they too must coordinate their activities with inflammatory cells to pattern regeneration of normal tissues. As in normal wound healing, tumors also activate the recruitment of host cells into tumor beds to regulate survival and proliferation 2. In this context recent attention has focused on the roles of dendritic, tumor associated macrophages and other early hematopoietic lineage populations that establish niches within tumors that foster and protect cancer stem cells from cytotoxic and metabolic stresses 3. Moreover, many of these same cell populations are thought to promote and establish premetastatic niches at distant sites which ultimately facilitate the ability of disseminated tumor cells to establish metastatic foci 4,5.
MSCs are multipotent cells that contribute to tissue homeostasis and regeneration. Normally, MSCs are rapidly recruited into sites of injury and inflammation where they differentiate into a variety of connective tissue cell types 6,7. Recently, marrow-derived MSCs were shown to participate in tumor progression by establishing a favorable tumor microenvironment, differentiating into cancer-associated fibroblasts (CAFs) which establish cytokine networks that promote progression and migration 8-14. Precisely how MSCs are recruited into primary tumor sites, how they contribute to the development of tumor niches for cancer stem cells, what regulates the conversion of MSCs into CAFs, and how CAFs promote metastasis is not entirely understood.
Skeletal metastases are one of the most serious complications of prostate cancer15. Growing evidence suggests that the CXC chemokine ligand 16 (CXCL16) and its receptor CXCR6 play important roles in tumor progression and bone metastasis 16-19. CXCL16 is one of a small number of chemokines expressed as both soluble and cell surface molecules and it functions as a chemoattractant for many cell types20. CXCL16 is secreted by cells in response to IFN-γ, TNF-α and IL-1β 21-28. CXCL16 is the sole ligand for CXCR6, a member of the seven transmembrane G protein-coupled receptor family which signals through the AKT/mTOR pathways 17. Our group has shown that in primary and metastatic prostate cancer, CXCL16 is highly expressed compared to normal prostate epithelial cells 17,29. In addition, CXCL16/CXCR6 is involved in prostate cancer migration and invasion17,20,25,29.
In the present study we demonstrate that tumor growth is dependent on the recruitment of MSCs into human and mouse prostate cancer in response to CXCL16. Once in the tumor, CXCL16 binding to CXCR6 expressed by MSCs, stimulates their conversion into CAFs, which subsequently secrete enhanced levels of CXCL12. CXCL12 expression by CAFs promotes an epithelial to mesenchymal transition (EMT) of the cancer cells, which supports metastasis to secondary sites. Together, these studies provide the molecular basis for MSC recruitment into primary tumors, and the conversion of MSCs into CAFs that ultimately lay the foundations for the EMT required establishing distant metastasis.
Results
CXCL16 secreted by prostate cancer recruits MSCs
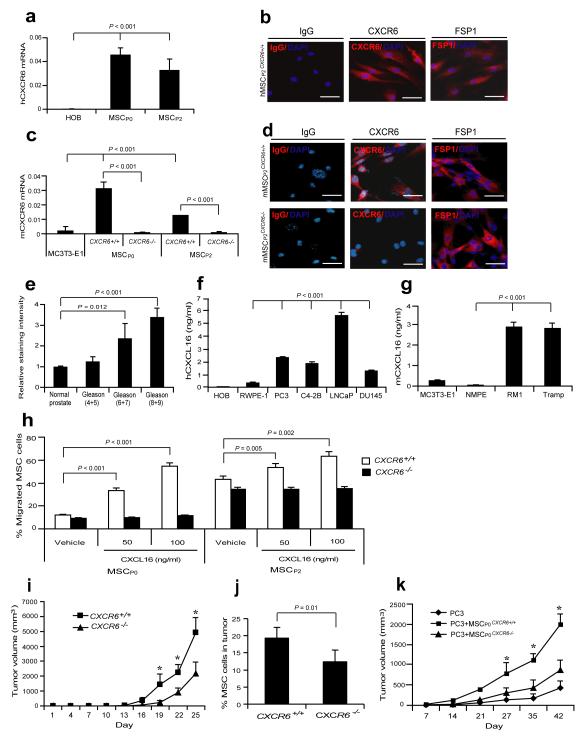
We reasoned that cells with stem cell-like properties must rapidly migrate into wounds to initiate tissue regeneration. We hypothesized that CXCR6-expressing MSCs from the bone marrow are likely rapidly recruited into tumors in response to CXCL16. Therefore, human and mouse bone marrow MSCs (Supplementary Fig. S1a) were evaluated for CXCR6 expression. Human (Fig. 1a,b) and freshly isolated non-passaged (P0) murine MSCs (Lin−Sca-1+CD45− or very small embryonic-like (VSEL) stem cells)7,30,31 and second passage MSCs (P2) expressed CXCR6, while MSCs isolated from CXCR6−/− (MSCCXCR6−/−) mice did not (Fig. 1c,d). Tissue microarrays (TMAs) from prostate cancer patients demonstrated that CXCL16 expression correlated with tumor aggressiveness (Fig. 1e; Supplementary Fig. S1b). Prostate cancer and breast cancer cell lines expressed significant levels of CXCL16 (Fig. 1f,g; Supplementary Fig. S1c-g). In vitro, P0 or P2 MSCs isolated from CXCR6 wild-type mice (MSCCXCR6+/+) migrated toward CXCL16, while MSCCXCR6−/− did not (Fig. 1h). To determine what role CXCL16 has in recruiting MSCs into tumors, prostate cancer was implanted s.c. into CXCR6+/+ or CXCR6−/− mice (Supplementary Fig. S1h). Significantly greater tumor volume was observed when the tumors were grown in CXCR6+/+ vs. CXCR6−/− mice suggesting that CXCR6 expressing host cells modulate tumor growth (Fig. 1i). Surprisingly, more MSCs were found in the tumors grown in the CXCR6+/+ mice than in the tumors grown in CXCR6−/− mice (Fig. 1j; Supplementary Table S1) though there were no differences in MSC numbers in the marrow of the CXCR6+/+ vs. CXCR6−/− mice (Supplementary Fig. S1i), suggesting a specific recruitment of MSCs into tumors facilitates growth. To validate that these results were representative of other tumors and not specific to subcutaneous tumor growth, the studies were repeated with human prostate cancer and breast cancer cell lines in an orthotopic setting. As seen previously, robust MSC recruitment into the tumors occurred when prostate cancer or breast cancer cell lines were implanted in an orthotopic setting (Supplementary Fig. S1j-r; Supplementary Table S1). To confirm that MSCs signaling through CXCR6 plays a critical role in tumor growth, prostate cancer cells were mixed with MSCP0CXCR6+/+ or MSCP0CXCR6−/− and tumor growth was monitored. As predicted, significantly larger tumor growth occurred when the tumor cells were mixed with MSCs expressing CXCR6 (MSCP0CXCR6+/+) compared with tumors established with MSCs not in which CXCR6 expression is knocked out (MSCP0CXCR6−/−) (Fig. 1k). Together these findings suggest a key role for CXCL16/CXCR6 in recruiting MSCs into tumors, and supporting tumor growth.
Figure 1. Expression of CXCL16 by prostate cancer recruits MSCs into tumors to support tumor growth.
(a) CXCR6 mRNA expression by human MSCs (P1 and P2). (b) Expression of CXCR6 protein by human MSCs. Controls included isotype matched controls and fibroblast-specific protein 1 (FSP1) for MSCs. Scale bars, 100μm. (c) CXCR6 mRNA by mMSCs. CXCR6 expression was determined in freshly isolated, non-cultured (P0) or P2 murine MSCs from CXCR6+/+ or CXCR6−/− mice. Human and murine osteoblasts (HOB and MC3T3-E1) were used as a negative control. (d) Expression of CXCR6 by murine P2 CXCR6+/+ or CXCR6−/− MSCs by IHC staining. Scale bar 100μm. Data in (a-c) are representative of mean with standard deviation for triplicates in each of three independent experiments (Student’s t-test). (e) CXCL16 expression in human prostate cancer tissue microarray in Supplementary Fig. S1b. Differences noted between normal prostate (n = 30), Gleason 4+5 (n = 9), Gleason 6+7 (n = 18), and Gleason 8+9 (n = 15) (mean±s.d. Student’s t-test). Secretionof CXCL16 by human prostate cancer cell lines (f) and murine prostate cancer cell lines (g) as determined by ELISA (mean±s.d., n = 3 independent experiments, Student’s t-test). (h) Migration of freshly isolated, non-cultured (P0) or P2 murine MSCs from CXCR6+/+ or CXCR6−/− mice in response to CXCL16. The % migrated MSC was determined by hemocytometer counting (mean±s.d., n = 3 independent experiments, Student’s t-test). (i) CXCR6+/+ or CXCR6−/− mice were implanted s.c. with RM1 cells and caliper measurements of tumor growth performed over 25 days. *Significant differences between tumors grown CXCR6+/+ and CXCR6−/− mice (mean±s.d, for 7 animals/group, n = 3 independent experiments, P < 0.05; Student’s t-test). (j) % MSCs (P0) present in RM1 tumors grown in CXCR6+/+ or CXCR6−/− mice at day 25 (mean±s.d. for 7 animals/group, n = 3 independent experiments, Student’s t-test). (k) SCID mice were implanted s.c. with PC3 cells mixed with MSCP0CXCR6+/+ or MSCP0CXCR6−/− cells and tumor growth was evaluated by caliper measurements over 42 days. *Significant differences between tumors grown with PC3 cells mixed with MSCP0CXCR6+/+ and MSCP0CXCR6−/− cells (mean±s.d. for n = 5 animals/group, n = 1 independent experiment, P < 0.05, Student’s t-test).
CXCL16/CXCR6 signalling induces CAF formation and CXCL12
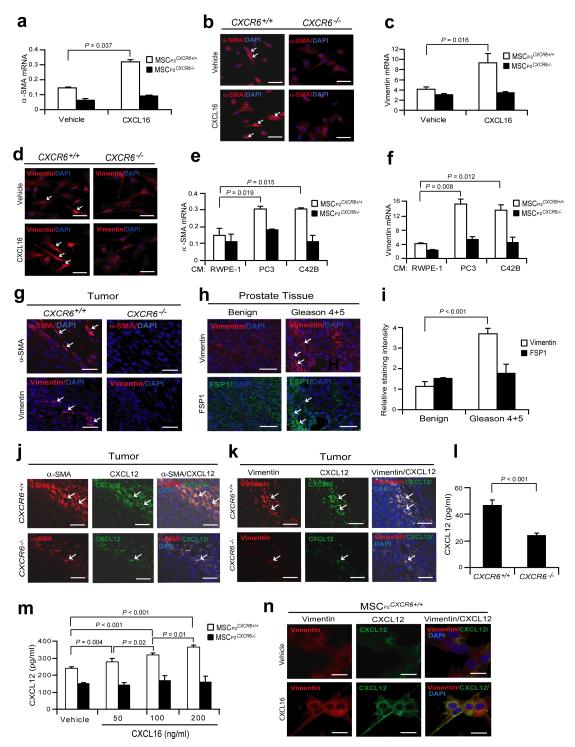
Local and recruited MSCs are known to convert into tumor associated fibroblasts (TAF) or CAFs in close proximity to tumor cells 32,33. To test whether prostate cancer-derived CXCL16 facilitates the conversion of MSCs into CAFs, MSCs were treated with CXCL16 and examined for expression of α-SMA and vimentin. MSCsCXCR6+/+ converted to α-SMA+ and vimentin+ expressing cells after CXCL16 stimulation while MSCsCXCR6−/− did not (Fig. 2a-d). To further investigate the role that CXCL16/CXCR6 signaling plays in tumor growth, MSCs isolated from wild-type or CXCR6−/− mice were treated with conditioned media derived from human and murine prostate cancer cell lines and examined for expression of α-SMA and vimentin. MSCCXCR6+/+ cells expressed significant levels of α-SMA and vimentin after treatment with conditioned media derived from prostate cancer cell lines, while MSCCXCR6−/− cells did not (Fig. 2e,f; Supplementary Fig. S2a,b). To validate these observations, prostate tumors grown in CXCR6+/+ or CXCR6−/− mice were probed for the CAF phenotype (Supplementary Fig. S2c). Paralleling the in vitro findings, fewer α-SMA+ and vimentin+ cells were identified in tumors grown in the CXCR6−/− mice compared with CXCR6+/+ mice (Fig. 2g). Previously we demonstrated that CXCL16 expression in human tumors corresponds with increasing Gleason grade 29. Therefore to validate the murine observations in a human setting, tumor tissue microarrays derived from human prostate cancer samples were stained for vimentin. The data demonstrate that more CAFs expressing vimentin were detected in the Gleason 4+5 prostate cancer than in the benign prostate cancer tissues (Fig. 3h,i; Supplementary Fig. S2d). A second critical feature of the CAF phenotype is the expression of stromal derived factor-1 (SDF-1 or CXCL12), which facilitates metastases34,35. Colocalization studies identified that more α-SMA+/CXCL12+ and vimentin+/CXCL12+ expressing cells were observed in tumors isolated from CXCR6+/+ vs. CXCR6−/− mice (Fig. 2j,k) and greater levels of CXCL12 were identified in the extracellular milieu of tumors grown in CXCR6+/+ vs. CXCR6−/− mice (Fig. 3l) or when mMSC are treated with CXCL16 (Supplementary Fig. S2e). Next, CXCL12 secretion by MSCs was examined in response to CXCL16. MSCCXCR6+/+ but not MSCsCXCR6−/− secreted CXCL12 response to CXCL16 (Fig. 2m,n), which was regulated through Erk and NF-κB signaling (Supplementary Fig. S2f-h).
Figure 2. Expression of CXCL12 by CAFs is dependent on CXCL16/CXCR6.
MSCs isolated from CXCR6+/+ or CXCR6−/− mice (P2) were exposed to vehicle or CXCL16 (100 ng/ml) for 7 days. The expression of α-SMA (a) mRNA by qRT-PCR or (b) protein by IHC (red, α-SMA, white arrows; blue, DAPI nuclear stain), or for vimentin (c) mRNA or (d) protein (red, vimentin, white arrows; blue, DAPI nuclear stain) were evaluated. Scale bars, 100μm. MSCs isolated from CXCR6+/+ or CXCR6−/− mice were exposed to human prostate cancer cell conditioned media for 7 days. The expression of α-SMA (e), and vimentin (f) mRNA were evaluated by qRT-PCR. (g) IHC of localization of α-SMA and vimentin postitive cells within tumors grown in CXCR6+/+ and CXCR6+/+ mice (red, α-SMA or vimentin, white arrow; blue, DAPI nuclear stain). Scale bars, 100μm. Data in (a,c,e,f) are representative of mean with standard deviation for triplicates in each of three independent experiments (Student’s t-test). (h) IHC of vimentin or FSP1 positive cells within benign or Gleason 4+5 prostate cancers in human tissue microarrays (TMAs) (red, vimentin, white arrows; green, FSP1,white arrows; blue, DAPI nuclear stain). Staining for FSP1 served as a positive control of MSCs. Scale bars, 100μm. (i) Quantification of Fig. 3h. Mean expression scores were multiplied by percent positive cells in the field. Significant differences were noted between benign (n = 30) or Gleason 4+5 prostate (n = 6) (mean±s.d., Student’s t-test). Colocalization of CXCL12 expression with α-SMA (j) and vimentin (k) positive cells (white arrows) within tumors grown in CXCR6+/+ or CXCR6−/− mice. Scale bars, 100μm. (l) CXCL12 protein expression in the extracellular milieu within tumors grown in CXCR6+/+ or CXCR6−/− mice (mean±s.d., for triplicates in each of three independent experiments, Student’s t-test). (m) Secretion of CXCL12 from MSCP2CXCR6+/+ cells or MSCP2CXCR6−/− cells were observed following exogenous CXCL16 treatment by ELISA (mean±s.d. for triplicates in each of three independent experiments, Student’s t-test, ANOVA). (n) Colocalization of CXCL12 with vimentin following exposure of MSCP2CXCR6+/+ cells or MSCP2CXCR6−/− cells to CXCL16 in vitro. Scale bars, 100μm. Mean±s.d. Student’s t-test
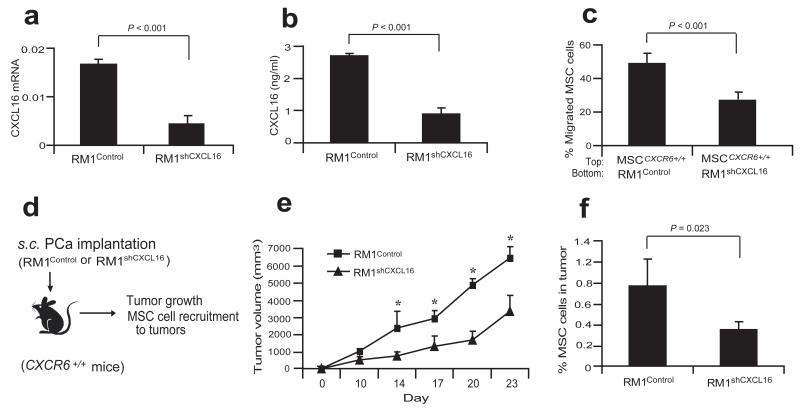
Figure 3. Knockdown of in prostate cancer cells reduces the tumor growth and MSC cell recruitment.
(a) Expression of CXCL16 mRNA in the RM1Control or RM1shCXCL16 cells by qRT-PCR. (b) Secretion of CXCL16 by RM1Control or RM1shCXCL16 cells was determined by ELISA. (c) Migration of MSCCXCR6+/+ cells was determined toward RM1Control cells or RM1shCXCL16 cells. Data in (a-c) are representative ofmean with standard deviation for triplicates in each of three independent experiments (Student’s t-test). Significance was determined using a Student’s t-test. (d) Experimental scheme of RM1Control or RM1shCXCL16 cell implantation to CXCR6+/+ mice for examining tumor growth and MSC cell recruitment to tumors. (e) The tumor growth of RM1Control or RM1shCXCL16 cells on CXCR6+/+ mice was evaluated by caliper measurements over 23 days. *Significant differences between tumors grown with RM1Control and RM1shCXCL16 cells (mean±s.d., for n = 5 animals/group, n = 2 independent experiments, P < 0.05;ANOVA). (f) % MSCs present in RM1Control or RM1shCXCL16 tumors grown in CXCR6+/+ mice (mean±s.d., for n = 5 animals/group, n = 2 independent experiments, Student’s t-test).
Knockdown of CXCL16 reduces MSC recruitment and CAF formation
To further explore the role of CXCL16 secreted by tumor cells and MSC cell recruitment, lentiviral vectors were used to silence CXCL16 expression in RM1 cells (RM1shCXCL16). After clonal selection, individual clones were pooled and assayed by qRT-PCR and ELISA for the reduction of CXCL16 expression (Fig. 3a,b). We then tested whether RM1shCXCL16 cells have the same capabilities to stimulate migration of MSCs compared to control (RM1Control) cells. As expected, the knocked-down of CXCL16 expression in RM1 cells inhibited the migration of MSC cells in vitro (Fig. 3c). In conjunction with these studies, tumor growth over time was evaluated in CXCR6+/+ mice (Fig. 3d). As shown in Fig. 3e, tumors generated from the RM1Control cells rapidly developed, while tumor growth of the RM1shCXCL16 cells was dramatically suppressed. Importantly, more MSCs were identified in the tumors grown with RM1Control cells than in tumors grown with RM1shCXCL16 cells (Fig. 3f; Supplementary Table S1). Taken together, the data suggest that CXCL16 expression by prostate tumors is critical for tumor growth and MSC cell recruitment.
Further studies examined the generation of the CAF phenotype in response to prostate cancer expressing CXCL16. In in vitro studies, MSCCXCR6+/+ cells expressed high levels of α-SMA and vimentin after treatment with conditioned media from RM1Control cells but not after treatment with conditioned media isolated from RM1shCXCL16 cells (Supplementary Fig. S3a,b). In in vivo studies, the tumors were generated from RM1Control or RM1shCXCL16 cells in CXCR6+/+ mice (Supplementary Fig. S3c). Fewer α-SMA+ and vimentin+ cells were identified in tumors generated from RM1shCXCL16 cells compared with tumors generated from RM1Control cells (Supplementary Fig. S3d). Colocalization studies identified that more α-SMA+/CXCL12+ and vimentin+/CXCL12+ cells were observed in tumors from RM1Control vs. RM1shCXCL16 cells (Supplementary Fig. S3e,f).
CAF CXCL12 promotes prostate cancer cell EMT
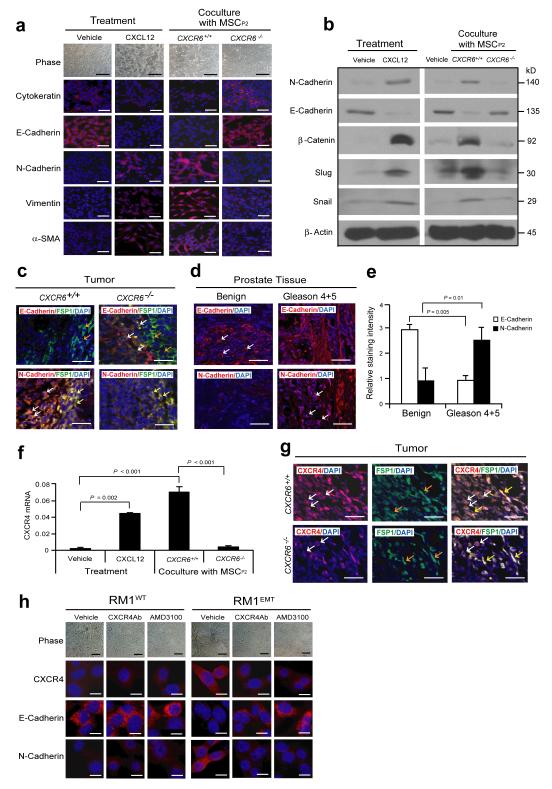
To explore the extent to which CXCL16 drives metastasis, we determined whether CAF-derived CXCL12 activates an EMT in prostate cancer cells (Supplementary Fig. S4a). Loss of cell-cell contacts and the emergence of a spindle-shaped morphology was observed following CXCL12 treatments of prostate cancer cells or when they were cocultured with MSCsCXCR6+/+, but not when cocultered with MSCCXCR6−/− (Fig. 4a). In fact, when prostate cancer cells were treated with CXCL12 or cocultured with MSCsCXCR6+/+, but not MSCsCXCR6−/− cells, a near complete loss of the epithelial transcriptome occurred including E-cadherin, reduced cytokeratin, enhanced expression of N-cadherin, vimentin, α-SMA, β-catenin, snail, and slug were observed (Fig. 4a-c). When tumor microarrays were stained for E-cadherin or N-cadherin, more E-cadherin expressing prostate cancer cells were detected in the benign prostate tissues, whereas more N-cadherin expressing prostate cancer cells were detected in the Gleason 4+5 prostate cancers (Fig. 4d,e; Supplementary Fig. S4b)36-38. Enhanced expression of the CXCL12 receptor CXCR4 is known to facilitate migration and metastasis in vivo39,40. We observed that CXCR4 expression by prostate cancer was enhanced following induction of an EMT phenotype in vitro and was associated with enhanced tumor growth in vivo (Fig. 4f,g). Studies with anti-CXCR4 antibody and the CXCR4 inhibitor AMD3100 showed that CXCL12 induces prostate cancer towards an EMT phenotype (Fig. 4h; Supplementary Fig. S4c-e). In fact, prostate cancer cells which had undergone an EMT were significantly more responsive than their parental counterparts to CXCL12 or serum (Fig. 5a). Such that CXCR4 blockade prevented prostate cancer migration in vitro (Fig. 5b).
Figure 4. CAF-mediated CXCL12 promotes EMT in primary tumor.
(a) Vehicle or CXCL12 treated RM1 cells, or RM1 cells co-cultured with MSCs from CXCR6+/+ or CXCR6−/− mice were examined by phase contrast microscopy and IHC staining for cytokeratin, E-cadherin, N-cadherin, vimentin, and α-SMA. Scale bars, 100μm. Representative images from 2 independent studies. (b) Western blots analysis for epithelial (E-cadherin) and mesenchymal (N-cadherin, β-catenin, snail, slug) markers. Representative images from 2 independent studies. (c) EMT markers in the primary tumor were examined by IHC. Colocalization of E-cadherin or N-cadherin with FSP1 was observed. More E-cadherin by prostate cancer cells (red; white arrows) was detected in close proximity to FSP1 expressing MSC cells (green; orange arrows) in tumors grown in CXCR6−/− mice compared to tumors grown in CXCR6+/+ mice. In contrast, more N-cadherin expressing prostate cancer cells (red; white arrows) were detected in close proximity to N-cadherin and FSP1 co-expressing CAF cells (yellow; yellow arrows) when the tumors were grown in CXCR6+/+ mice compared to tumors grown in CXCR6−/− mice. Blue, DAPI nuclear stain. Scale bars, 100μm. Representative images derived from n=10 mice/group). (d) IHC of E-cadherin or N-cadherin positive cells within benign or Gleason 4+5 prostate cancers in human prostate tissue microarrays (TMAs) (red, E-cadherin or N-cadherin, white arrows; blue, DAPI nuclear stain). Scale bars, 100μm. (e) Quantification of Fig. 4d. Mean expression scores were multiplied by percent positive cells in the field. Significant differences were noted between benign (n = 30) or Gleason 4+5 prostate (n = 6) (mean±s.d ANOVA). (f) CXCR4 mRNA was determined for EMT-induced RM1 cells following CXCL12 treatment or co-culture with MSCs derived from CXCR6+/+ or CXCR6−/− mice (mean±s.d., n = 3 independent experiments). (g) More CXCR4 expressing RM1 cells (red; white arrows) were detected in close proximity to CXCR4 and FSP1 (green; orange arrows) co-expressing CAF cells (yellow; yellow arrows) when the tumors were grown in CXCR6+/+ mice compared to tumors grown in CXCR6−/− mice. Scale bars, 100μRepresentative images from an experiment with n=10 animals/group). (h) AMD3100 or anti-CXCR4 antibody prevents the development of EMT by RM1 cells following CXCL12 exposure. Scale bars, 100μm. Representative images from an experiment with n=10 animals/group.
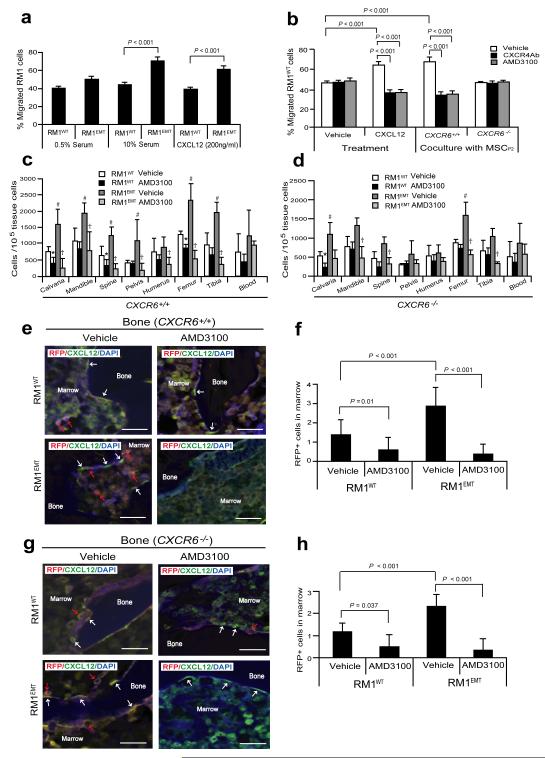
Figure 5. EMT-mediated CXCR4 is highly involved in prostate cancer metastasis.
(a) Migration assays were performed in Transwell® plates using 10% serum or CXCL12 as chemoattractants. Migration toward 0.5% serum was used as a negative control. (b) Blockade of CXCR4 by AMD3100 or anti-CXCR4 antibody prevents prostate cancer migration towards CXCL12 or MSCs isolated from CXCR6+/+, but not CXCR6−/− animals. Data in (a,b) are representativedata from two independent studies (mean±s.d., ANOVA). Significance was determined using a Student’s t-test. RFP-labeled RM1WT or RM1EMT cells (Supplementary Fig. S5a) were incubated with vehicle or AMD3100 in vitro, and then inoculated by intra-cardiac (i.c.) injection into CXCR6+/+ or CXCR6−/− (n = 7). Metastasis was assessed by qPCR for RFP in a number of tissues. (c,d) Number of metastatic RM1 cells following i.c. injection. *Significance between RM1WT treated with vehicle and RM1WT treated with AMD3100 (P < 0.05). #Significance between RM1WT treated with vehicle and RM1EMT cells treated with vehicle (P < 0.05). †Significance between RM1EMT treated with vehicle and RM1EMT treated with AMD3100 (P < 0.05). Error bars represents mean±s.d., n = 2 independent experiments, P < 0.05; Student’s t-test. (e-h) RM1 cells expressing RFP were identified in the femur of CXCR6+/+ or CXCR6−/− mice following i.c. injection. Red arrows identify RM1 cells. White arrows identify osteoblast on the bone surface staining positive for CXCL12 expression. Scale bars, 100μm. (f,h) Quantification of Fig. 5e and Fig. 5g, respectively. The numbers of RM1 cells were quantified on the endosteal region of the 7 long bones. Endosteal regions were defined as 12 cell diameters from bone surfaces. ((Mean±s.d. (n = 3)., ANOVA).
EMT-induced CXCR4 expression promotes metastasis
In an animal model of bone metastasis, RFP expressing wild-type (RM1WT) or EMT induced RM1 cells (RM1EMT) (Supplementary Fig. S5a) were incubated with AMD3100 or vehicle in vitro, and then injected by an intra-cardiac (i.c.) route into CXCR6+/+ or CXCR6−/− mice to establish prostate cancer bone metastases (Supplementary Fig. S5b). First, we examined CXCL12 levels in a number of osseous sites and in blood (Supplementary Fig. S5c). All the animals from CXCR6+/+ or CXCR6−/− mice had significant numbers of disseminated tumor cells (DTCs) in their bones 10 days following injection of RM1WT or RM1EMT cells (Fig. 5c-h). In contrast, RM1WT cells pretreated with AMD3100 had a reduced number of disseminated tumor cells (DTCs) in their calvaria, spine, and femur from CXCR6+/+ mice (Fig. 5c,e,f). Strikingly, animals receiving RM1EMT cells showed a significant increase of in the total DTC load in most osseous tissues compared to animals injected with the RM1WT cells alone. Critically, the number of DTCs were significantly reduced following CXCR4 blockade (Fig. 5c,e,f). However, fewer DTCs were identified in the bones of the CXCR6−/− vs. CXCR6+/+ mice (Fig. 5d,g,h). Together these data suggest CXCL16 initiated induction of an EMT-and CXCR4 expression via MSC activation plays an important role and critical role in prostate cancer cell dissemination and metastasis.
Discussion
Tumors arise from cells that have sustained and multiple genetic mutations resulting in deregulation of normal growth-control mechanisms 41. Recent evidence also suggests that the microenvironment itself regulates crucial neoplastic progression steps in hematologic tumors 42. Cancer cells not only interact with each other, their extracellular matrix and inflammatory cells, but also with recruited and resident cells of mesenchymal origin. The characteristic transformation of stromal cells that accompanies, or precedes the malignant conversion of epithelial cells has been linked to CAFs 43-45. Several cancer types demonstrate the concept that these fibroblasts can determine the fate of the epithelial cells, promote malignant progression either through soluble factors, and cell-cell interactions and/or alterations of the extracellular matrix 45. The complexity of these interactions has been amplified by studies showing alterations in resident cells may be drivers in cancer progression 46.
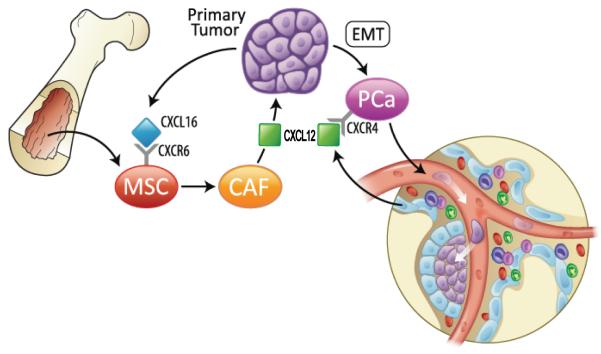
MSCs are multipotent cells that contribute to tissue homeostasis and regeneration. Tumors recruit MSCs to facilitate tumor progression and metastasis. Here, we provide evidence that the recruitment of MSCs into prostate cancer is dependent on the expression of the CXCR6 ligand, CXCL16 by tumor cells (Fig. 6). CXCR6 signaling supports the recruitment, conversion and activation of MSC into CXCL12 secreting CAFs. Moreover, enhanced CXCL12 secretion supports an EMT conversion of the prostate cancer cells and an increase in the expression of the CXCL12 receptor, CXCR4. These events result in enhanced tumor progression and ultimately extravasation and metastasis. Targeting MSCs and the CXCL16/CXCR6 axis may prevent tumor progression and metastasis of prostate cancer and provide a more effective therapeutic strategy for prostate cancer.
Figure 6. Bone marrow-derived MSCs promote prostate cancer growth and metastasis.
Model showing putative potential mechanisms underlying primary prostate cancer progression by the recruitment of mescenchymal cells (MSCs) and bone metastasis. Secretion of CXCL16 by cancer cells recruits MSCs into tumor sites. Tumor-derived CXCL16 interacts with its receptor, CXCR6 on MSCs and activates signal transduction, leading MSCs to convert into cancer-associated fibroblasts (CAFs), which secrete the high levels of CXCL12. CXCL12 promotes the malignant transformation of proliferating cancer cells to an epithelial-mesenchymal transition (EMT). EMT enhances CXCR4 expression in prostate cancer cells. CXCR4 expression facilitates metastasis.
Several lines of evidence demonstrate that cells with stem cell-like properties must be able to migrate into wound sites rapidly in order for regeneration to occur 47-50. In the context of neoplasia, bone marrow derived MSCs have been shown to increase the metastatic potential of weakly metastatic human breast cancer cells, in part through the conversion to a CAF phenotype 51. We demonstrate that primary small Lin−Sca-1+CD45− cells (VSEL stem cells) isolated from marrow and MSCs passaged in vitro express high mRNA levels of CXCR6 and migrate towards CXCL16. CXCL16 exists both in soluble and transmembrane forms 52. CXCL16 is expressed on monocytes, macrophages, B cells, and dendritic cells 53,54 and both forms of CXCL16 are expressed by human tumor cells 16,17,55,56. The precise role of soluble vs. transmembrane CXCL16 in tumor progression remains unclear. Transmembrane CXCL16 may function to suppress tumor proliferation, while soluble CXCL16 induces proliferation and migration of cancer cells 20. Which form of CXCL16 regulates prostate cancer growth within tumors is still unclear.
Previous work by our group demonstrated that CXCR6 expression in tumors correlated with Gleason score. Using lentiviral vectors to overexpress CXCR6 in PC3, LNCaP C4-2B, and LNCaP cells, or by reducing CXCR6 expression by siRNA, we also found that modifying CXCR6 expression altered the ability of prostate cancer cell lines to invade and grow both in vitro and in vivo 17. In addition, CXCR6 regulates the expression of several proangiogenic factors including IL-8 and vascular endothelial growth factor (VEGF), both of which are likely to participate in the regulation of tumor angiogenesis 17. In part, binding of CXCL16 to CXCR6 induces activation of Akt, p70S6K, and eukaryotic initiation factor 4E binding protein 1 in prostate cancer cells in addition to mammalian target of rapamycin (mTOR) pathways 17. Moreover, rapamycin not only drastically inhibited CXCL16-induced prostate cancer cell invasion and growth but also reduced secretion of IL-8 or VEGF levels and inhibited expression of other CXCR6 targets including CD44 and matrix metalloproteinase 17.
The present study further adds to the complexity of CXCL16/CXCR6 signaling and the role that paracrine/autocrine loops play in tumor progression. These findings are similar to previous observations showing that CXCL12/CXCR4 signaling 57,58 supports cross-talk between CAFs and prostate cancer 59. Our results illustrate the molecular basis for MSC recruitment into tumors and how this process leads to tumor metastasis by coupling activities of the CXCL12/CXCR4 and CXCL16/CXCR6 axes. By demonstrating that MSCs play an active role in establishing an EMT in cancer which ultimately facilitates metastasis, MSCs are critical components of the host-response network in tumors and represent viable entities for the design of targeting therapies to prevent the establishment of distant metastasis.
Methods
Animals
Five to seven week-old male CXCR6+/+ or CXCR6−/− mice (Jackson Laboratory, Bar Harbor, ME) and SCID mice (CB.17. SCID; Taconic, Germantown, NY) were used as transplant recipients. All animal procedures were performed in compliance with the institutional ethical requirements and approved by the University of Michigan Committee for the Use and Care of Animals (UCUCA).
Cell lines
The human prostate cancer cell lines PC3, LNCaP, and DU145, and murine prostate cancer cell lines RM1 and Tramp were used (American Type Culture Collection (ATCC), Rockville, MD). C4-2B were originally isolated from a lymph node of a patient with disseminated bony and lymph node involvement. Prostate cancer cell lines were cultured in RPMI 1640 (Invitrogen, Grand Island, NY) with 10% fetal bovine serum (FBS, Invitrogen), 1% penicillin-streptomycin (P/S, Invitrogen). The human prostate epithelial cell line RWPE-1 (ATCC) was cultured in Keratinocyte-SFM with supplements (Invitrogen). The mouse prostate epithelial cell line NMPE (CHI Scientific, Maynard, MA) was cultured in Mouse Prostate PrimaCell™ medium (CHI Scientific). The human breast epithelial cell line MCF-10A and breast cancer cell line MCF-7 were kindly provided by Dr. Max Wicha (University of Michigan). MCF-10A cell line was cultured in DMEM/F12 with supplements (Invitrogen) and MCF-7 cell line was cultured in DMEM with supplements (Invitrogen). Conditioned media were collected and frozen after filtration through a 0.22 μm filter. For induction of EMT, RM1 cells were cultured to confluence, and serum-starved for 24 hours. The cells were cultured in RPMI with 0.1% FBS supplemented with 200 ng/ml CXCL12 (cat. 350-NS, R&D Systems, Minneapolis, MN) for 48 hours, and termed RM1EMT vs. RM1WT. RM1 cells used in metastasis assays were labeled with red fluorescent protein (RFP) by lentiviral transfection (Supplementary Fig. S5a) and selected by FACS.
MSC cells
Human MSCs (hMSCs, Lonza, Walkersville, MD), and freshly isolated mouse MSC cells (non-passaged (P0)) (Lin−Sca-1+CD45− cells or very small embryonic-like (VSEL) stem cells) and primary bone marrow derived-MSC cells (passaged once (P1) or twice (P2)) from CXCR6+/+ or CXCR6−/− mice were used for this study. The hMSC cells were cultured in DMEM supplemented with 10% FBS, and 1% P/S. For mouse MSCs, after sacrifice, marrow was flushed from femurs and tibias of both CXCR6+/+ and CXCR6−/− mice into α-MEM (Invitrogen) with 2% FBS, to generate primary MSC cells and cultured in α-MEM containing 10% FBS and 1% P/S. Once confluent, the cells were passaged 2-3 times to minimize macrophage contamination. Subsequently, repopulated bone marrow derived-MSCs were obtained, and cells were cultured in DMEM containing 10% FBS and 1% P/S. For MSC differentiation assays, mouse primary bone marrow derived-MSC cells were cultured in adipogenic, osteogenic, or chondrogenic conditions for 2 weeks, and cells were stained with Alizarin Red S, Oil Red O, and Alcian Blue, respectively.
Isolation of MSCP0 (VSEL) cells
Small Lin−Sca-1+CD45− (or very small (less than 8 microns) embryonic-like (VSEL) stem cells) referred to as MSC cells (P0) herein were isolated from mononuclear bone marrow cells (1 × 108 cells/ml) from both CXCR6+/+ and CXCR6−/− mice were resuspended in PBS containing 2% heat-inactivated FBS and 1% P/S. The cells were incubated with the following antibodies: biotin-conjugated rat anti-mouse Ly-6A/E (Sca-1) (cat. 553334, 1:50 dilution, BD Pharmingen, San Diego, CA), streptavidin-PE-Cy5 (cat. 554062, 1:50 dilution, BD Pharmingen), anti-CD45-APC (cat. 557659, 1:30 dilution, BD Pharmingen), anti-CD45R/B220-PE (cat. 553089, 1:200 dilution, BD Pharmingen), anti-Gr-1-PE (cat. 553128, 1:200 dilution, BD Pharmingen), anti-TCRαβ PE (cat. 553172, 1:200 dilution, BD Pharmingen), anti-TCRγζ PE (cat. 553178, 1:200 dilution, BD Pharmingen), anti-CD11b PE (cat. 557397, 1:200 dilution, BD Pharmingen), and anti-Ter-119 PE (cat. 553673, 1:200 dilution, BD Pharmingen). MSCP0 cells were freshly isolated by cell sorting (FACSAria II, Becton Dickinson, Mountainview, CA).
Western blot analyses
Cells were lysed in RIPA buffer with protease inhibitors and lysates separated on 10% SDS-polyacrylamide gel and transferred to PVDF membranes. The membranes were incubated with 5% milk for 1 hour and probed overnight at 4°C with antibodies targeting N-cadherin (cat. 610921, 1:2500 dilution, BD Transduction laboratory, Lexington, KY), E-cadherin (cat. sc-7870, 1:1000 dilution, Santa Cruz, Santa Cruz, CA), β-catenin (cat. 9582, 1:1000 dilution, Cell Signaling Technology, Danvers, MA), snail (cat. 3879, 1:1000 dilution, Cell Signaling), slug (cat. 9585, 1:1000 dilution, Cell Signaling), and β-actin (cat. 4970, 1:1000 dilution, Cell Signaling). After washing, blots were incubated with peroxidase-conjugated anti-rabbit IgG HRP secondary antibodies (cat. W401B, 1:7500 dilution, Promega, Madison, WI) for 1 hour. Protein expression was detected with SuperSignal West Pico Chemiluminescent Substrate (cat. 34080, Thermo Scientific, Rockford, IL).
Immunohistochemistry
Cells were fixed and tissue sections were de-waxed and re-hydrated, then blocked with Image-iT FX signal enhancer for 30 min, and incubated 2 hours at room temperature in dark with 10 μg/ml primary antibodies combined with reagents of Zenon Alexa Fluor 488 (green) or 555 (red) labeling kit. CXCR6 (cat. NLS-1102, 1:100 dilution, Novus Biologicals, Littleton, CO), fibroblast-specific protein 1 (FSP1, cat. 07-2274, 1:100 dilution, Millipore, Temecula, CA), CXCL12 (cat. ab25117, 1:100 dilution, Abcam, Cambridge, MA), cytokeratin (cat. ab9377, 1:200 dilution, Abcam), E-cadherin (cat. sc-7870, 1:25 dilution, Santa Cruz), N-cadherin (cat. 610921, 1:25 dilution, BD Transduction laboratory,), vimentin (cat. ab 8978, 1:100 dilution, Abcam), anti-mouse α-SMA (cat. ab5694, 1:20 dilution, Abcam), CXCR4 (cat. ab2074, 1:100 dilution, Abcam), and RFP (cat. ab62341, 1:50 dilution, Abcam) antibodies were diluted in PBST (PBS plus 0.2% Triton X-100). The cells and tissue sections were post-fixed with 10% formalin for 10 min followed by processing with ProLong Gold antifade reagent with DAPI medium. Images were acquired with an Olympus FV500 confocal microscope.
Human prostate tissue microarrays were purchased from US Biomax, Inc. (Rockville, MD). Anti-human CXCL16 (ab17537, 1:20 dilution,, Abcam), FSP1 (cat. 07-2274, 1:100 dilution, Millipore), vimentin (cat. ab8978, 1:100 dilution, Abcam), E-cadherin (cat. sc-7870, 1:25 dilution, Santa Cruz), and N-cadherin (cat. 610921, 1:25 dilution, BD Transduction laboratory) antibodies were used. Staining intensity was ranked from 1 to 4 (1, negative; 2, weak; 3, moderate; and 4, strong intensity staining).
Subcutaneous tumor growth
Tumors were established by injecting RM1 (1 × 104) cells in growth factor-reduced Matrigel (cat. 354236, BD Bioscience, Bedford, MA) s.c. into 5-7 week-old male CXCR6+/+ or CXCR6−/− mice (Supplementary Fig. S1h; Supplementary Table S1). Tumors were also established by injecting RM1Control or RM1shCXCL16 (1 × 104) cells in growth factor-reduced Matrigel s.c. into 5-7 week-old male CXCR6+/+ mice (Fig. 3d; Supplementary Table S1). In some cases, tumors were also established by injecting human PC3 cells (2×105) mixed with MSCP0CXCR6+/+ or MSCP0CXCR6−/− cells (2×103) in growth factor-reduced Matrigel s.c. into 5-7 week-old male SCID mice. The animals were monitored daily and tumor volumes were evaluated every 3-7 days. Tumor volumes were calculated using the formula V= (the shortest diameter) × (the longest diameter) × height. After 23-42 days the animals were sacrificed. The tumors were measured and prepared for Lin−Sca-1+CD45− cell analyses and histology.
Orthotopic tumor growth
After anesthesia with 2-4% isoflurane inhalation, a low midline incision was made in the lower abdomen. The human and murine prostate cancer cells (5×104-5×105) in 20 μl of PBS were injected into the right or left dorsolateral lobe of the prostate of 5-7 week-old male SCID or CXCR6+/+ mice, and the wound was closed with surgical clips. The human breast cell lines (1×106) in growth factor-reduced Matrigel were injected abdominal fat pad of 5-7 week-old female SCID mice (Supplementary Fig. S1j; Supplementary Table S1).
In vivo metastasis assays
Cells (2×105 RFP labeled RM1 cells/mouse) were incubated with 10 μM AMD3100 (cat. A5602, Sigma) or vehicle for 30 min at 4°C, and then introduced into 5-7 week-old male CXCR6+/+ or CXCR6−/− mice by intra-cardiac (i.c.) injection. qPCR and immunohistochemistry were used to identify the location of the cells. Metastasis was first assessed in osseous tissues and blood samples at day 10 by qPCR using a probe for the red fluorescent protein gene [AICSVE0-F AGAGCATCTACATGGCCAAGAAG (forward), AICSVE0-R TCGTTGTGGCTGGTGATGTC (reverse), and FAM CTTGCTGTCCACGTAGTAGT (TaqMan probe; Applied Biosystems)]. The data were normalized to mouse tissue β-actin (Mm00607939_s1). Immunohistochemistry for prostate cancer cells in the marrow was also used for metastasis assays. The numbers of RM1 cells were quantified on the endosteal region of the 7 long bones defined as 10 cell diameters from the bone surfaces.
Statistical analyses
All in vitro experiments were performed at least three times with similar results and representative assays are shown. Statistical analysis was performed by ANOVA or Student’s t-test using GraphPad Instat (GraphPad, San Diego, CA) with significance at P<0.05.
Supplementary Material
Acknowledgements
This work is directly supported by the National Institutes of Health (CA163124, CA093900, CA166307, CA069568, DK082481, MH095589, Y.S., E.T.K. P.H.K, K.J.P. and R.S.T.), the Fund for Cancer Research (R.T.), the Department of Defense (E.T.K., Y.S., and R.S.T.), the Prostate Cancer Foundation (Y.S, K.J.P. and R.S.T), National Natural Funding of China (81071747) (J.W), National Key Program (973) for Basic Research of China (2011CB510106) (J.W.), and Shanghai Pujiang Program (10PJ1406400) (J.W). Logistical support was also received from the University of Michigan FACS and Imaging Cores, and the Dental School’s Molecular Biology Core.
Footnotes
Author contributions
Y.J., J.K.K., Y.S., and R.S.T. designed experiments. Y.J., J.K.K., Y.S., J.W., A.M., J.J., J.E.B., S.M., E.L., H.S., T.J., H.Z., and J.D. performed experiments and analyzed the data. J.W., P.H.K., E.T.K., and K.J. P. discussed the results and gave valuable critique on the paper. Y.J. and R.S.T. wrote the manuscript.
Competing financial interest
The authors declare no competing interests.
Reference List
- 1.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 2.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes Dev. 2008;22:559–574. doi: 10.1101/gad.1636908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang ZJ, Wechsler-Reya RJ. Hit ’em where they live: Targeting the cancer stem cell niche. Cancer Cell. 2007;11:3–5. doi: 10.1016/j.ccr.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafii S, Lyden D. S100 chemokines mediate bookmarking of premetastatic niches. Nat. Cell Biol. 2006;8:1321–1323. doi: 10.1038/ncb1206-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quante M, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taichman RS, et al. Prospective identification and skeletal localization of cells capable of multilineage differentiation in vivo. Stem Cells Dev. 2010;19:1557–1570. doi: 10.1089/scd.2009.0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergfeld SA, DeClerck YA. Bone marrow-derived mesenchymal stem cells and the tumor microenvironment. Cancer Metastasis Rev. 2010;29:249–261. doi: 10.1007/s10555-010-9222-7. [DOI] [PubMed] [Google Scholar]
- 10.Mi Z, et al. Osteopontin promotes CCL5-mesenchymal stromal cell-mediated breast cancer metastasis. Carcinogenesis. 2011;32:477–487. doi: 10.1093/carcin/bgr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, et al. Effects of inflammatory factors on mesenchymal stem cells and their role in the promotion of tumor angiogenesis in colon cancer. J. Biol. Chem. 2011;286:25007–25015. doi: 10.1074/jbc.M110.213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abarrategi A, Marinas-Pardo L, Mirones I, Rincon E, Garcia-Castro J. Mesenchymal niches of bone marrow in cancer. Clin. Transl. Oncol. 2011;13:611–616. doi: 10.1007/s12094-011-0706-x. [DOI] [PubMed] [Google Scholar]
- 13.Mishra PJ, Mishra PJ, Glod JW, Banerjee D. Mesenchymal stem cells: flip side of the coin. Cancer Res. 2009;69:1255–1258. doi: 10.1158/0008-5472.CAN-08-3562. [DOI] [PubMed] [Google Scholar]
- 14.Mi Z, et al. Osteopontin promotes CCL5-mesenchymal stromal cell-mediated breast cancer metastasis. Carcinogenesis. 2011;32:477–487. doi: 10.1093/carcin/bgr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, et al. CXCL16 functions as a novel chemotactic factor for prostate cancer cells in vitro. Mol. Cancer Res. 2008;6:546–554. doi: 10.1158/1541-7786.MCR-07-0277. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, et al. CXCR6 induces prostate cancer progression by the AKT/mammalian target of rapamycin signaling pathway. Cancer Research. 2008;68(24):10367–76. doi: 10.1158/0008-5472.CAN-08-2780. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Chandrasekar B, Bysani S, Mummidi S. CXCL16 signals via Gi, phosphatidylinositol 3-kinase, Akt, I kappa B kinase, and nuclear factor-kappa B and induces cell-cell adhesion and aortic smooth muscle cell proliferation. J. Biol. Chem. 2004;279:3188–3196. doi: 10.1074/jbc.M311660200. [DOI] [PubMed] [Google Scholar]
- 19.Ha HK, et al. Clinical significance of CXCL16/CXCR6 expression in patients with prostate cancer. Mol. Med. Report. 2011;4:419–424. doi: 10.3892/mmr.2011.446. [DOI] [PubMed] [Google Scholar]
- 20.Deng L, Chen N, Li Y, Zheng H, Lei Q. CXCR6/CXCL16 functions as a regulator in metastasis and progression of cancer. Biochim. Biophys. Acta. 2010;1806:42–49. doi: 10.1016/j.bbcan.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 21.van, d. V, et al. An alternatively spliced CXCL16 isoform expressed by dendritic cells is a secreted chemoattractant for CXCR6+ cells. J. Leukoc. Biol. 2010;87:1029–1039. doi: 10.1189/jlb.0709482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama T, et al. Cutting edge: profile of chemokine receptor expression on human plasma cells accounts for their efficient recruitment to target tissues. J. Immunol. 2003;170:1136–1140. doi: 10.4049/jimmunol.170.3.1136. [DOI] [PubMed] [Google Scholar]
- 23.Meijer J, et al. The chemokine receptor CXCR6 and its ligand CXCL16 are expressed in carcinomas and inhibit proliferation. Cancer Res. 2008;68:4701–4708. doi: 10.1158/0008-5472.CAN-08-0482. [DOI] [PubMed] [Google Scholar]
- 24.Matsushita K, et al. Soluble CXCL16 in Preoperative Serum is a Novel Prognostic Marker and Predicts Recurrence of Liver Metastases in Colorectal Cancer Patients. Ann. Surg. Oncol. 2011;19(Suppl. 3):S518–527. doi: 10.1245/s10434-011-1993-8. [DOI] [PubMed] [Google Scholar]
- 25.Hu W, et al. CXCR6 is expressed in human prostate cancer in vivo and is involved in the in vitro invasion of PC3 and LNCap cells. Cancer Sci. 2008;99:1362–1369. doi: 10.1111/j.1349-7006.2008.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Held-Feindt J, et al. Overexpression of CXCL16 and its receptor CXCR6/Bonzo promotes growth of human schwannomas. Glia. 2008;56:764–774. doi: 10.1002/glia.20651. [DOI] [PubMed] [Google Scholar]
- 27.Ha HK, et al. Clinical significance of CXCL16/CXCR6 expression in patients with prostate cancer. Mol. Med. Report. 2011;4:419–424. doi: 10.3892/mmr.2011.446. [DOI] [PubMed] [Google Scholar]
- 28.Darash-Yahana M, et al. The chemokine CXCL16 and its receptor, CXCR6, as markers and promoters of inflammation-associated cancers. PLoS One. 2009;4:e6695. doi: 10.1371/journal.pone.0006695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, et al. CXCL16 Functions as a Novel Chemotactic Factor for Prostate Cancer Cells In vitro. Mol. Cancer Res. 2008;6:546–554. doi: 10.1158/1541-7786.MCR-07-0277. [DOI] [PubMed] [Google Scholar]
- 30.Ratajczak MZ, Zuba-Surma EK, Machalinski B, Ratajczak J, Kucia M. Very small embryonic-like (VSEL) stem cells: purification from adult organs, characterization, and biological significance. Stem Cell Rev. 2008;4:89–99. doi: 10.1007/s12015-008-9018-0. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Song J, Taichman RS, Krebsbach PH. Ablation of proliferating marrow with 5-fluorouracil allows partial purification of mesenchymal stem cells. Stem Cells. 2006;24:1573–1582. doi: 10.1634/stemcells.2005-0399. [DOI] [PubMed] [Google Scholar]
- 32.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 33.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 34.Begley L, Monteleon C, Shah RB, Macdonald JW, Macoska JA. CXCL12 overexpression and secretion by aging fibroblasts enhance human prostate epithelial proliferation in vitro. Aging Cell. 2005;4:6–291. doi: 10.1111/j.1474-9726.2005.00173.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, et al. Characterization of phosphoglycerate kinase-1 expression of stromal cells derived from tumor microenvironment in prostate cancer progression. Cancer Res. 2010;70:471–480. doi: 10.1158/0008-5472.CAN-09-2863. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Tran NL, Nagle RB, Cress AE, Heimark RL. N-Cadherin expression in human prostate carcinoma cell lines. An epithelial-mesenchymal transformation mediating adhesion withStromal cells. Am. J. Pathol. 1999;155:787–798. doi: 10.1016/S0002-9440(10)65177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Algaba F, Arce Y, Fernandez S, Oliver A, Alcaraz A. Adhesion molecules expression as a potential marker of prostate cancer aggressivity. A TMA study of radical prostatectomy specimens. Arch. Ital. Urol. Androl. 2006;78:130–134. [PubMed] [Google Scholar]
- 38.Rubin MA, et al. E-cadherin expression in prostate cancer: a broad survey using high-density tissue microarray technology. Hum. Pathol. 2001;32:690–697. doi: 10.1053/hupa.2001.25902. [DOI] [PubMed] [Google Scholar]
- 39.Taichman RS, et al. Use of the Stromal Cell-derived Factor-1/CXCR4 Pathway in Prostate Cancer Metastasis to Bone. Cancer Res. 2002;62:1832–1837. [PubMed] [Google Scholar]
- 40.Muller CA, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 41.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U. S. A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raaijmakers MH, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Experimental Cell Research. 2001;264(1):169–84. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa H, et al. Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene. 2004;23(44):7366–77. doi: 10.1038/sj.onc.1208013. [DOI] [PubMed] [Google Scholar]
- 45.Micke P, Ostman A. Tumour-stroma interaction: cancer-associated fibroblasts as novel targets in anti-cancer therapy? Lung Cancer. 2004;45(Suppl 2):S163–75. doi: 10.1016/j.lungcan.2004.07.977. [DOI] [PubMed] [Google Scholar]
- 46.Hill R, Song Y, Cardiff RD, Van Dyke T. Selective evolution of stromal mesenchyme with p53 loss in response to epithelial tumorigenesis. Cell. 2005;123(6):1001–11. doi: 10.1016/j.cell.2005.09.030. see comment. [DOI] [PubMed] [Google Scholar]
- 47.Kollet O, et al. Rapid and efficient homing of human CD34(+)CD38(−/low)CXCR4(+) stem and progenitor cells to the bone marrow and spleen of NOD/SCID and NOD/SCID/B2m(null) mice. Blood. 2001;97:3283–3291. doi: 10.1182/blood.v97.10.3283. [DOI] [PubMed] [Google Scholar]
- 48.Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 49.Peled A, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 50.Sackstein R, et al. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nature Medicine. 2008;14(2):181–7. doi: 10.1038/nm1703. [DOI] [PubMed] [Google Scholar]
- 51.Coffelt SB, et al. The pro-inflammatory peptide LL-37 promotes ovarian tumor progression through recruitment of multipotent mesenchymal stromal cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3806–3811. doi: 10.1073/pnas.0900244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilbanks A, et al. Expression cloning of the STRL33/BONZO/TYMSTRligand reveals elements of CC, CXC, and CX3C chemokines. J. Immunol. 2001;166:5145–5154. doi: 10.4049/jimmunol.166.8.5145. [DOI] [PubMed] [Google Scholar]
- 53.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat. Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 54.Shimaoka T, et al. Critical role for CXC chemokine ligand 16 (SR-PSOX) in Th1 response mediated by NKT cells. J. Immunol. 2007;179:8172–8179. doi: 10.4049/jimmunol.179.12.8172. [DOI] [PubMed] [Google Scholar]
- 55.Wente MN, et al. Expression and potential function of the CXC chemokine CXCL16 in pancreatic ductal adenocarcinoma. Int. J. Oncol. 2008;33:297–308. [PubMed] [Google Scholar]
- 56.Meijer J, et al. The chemokine receptor CXCR6 and its ligand CXCL16 are expressed in carcinomas and inhibit proliferation. Cancer Res. 2008;68:4701–4708. doi: 10.1158/0008-5472.CAN-08-0482. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Loberg R, Taichman RS. The pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis. Cancer Metastasis Rev. 2006;25:573–587. doi: 10.1007/s10555-006-9019-x. [DOI] [PubMed] [Google Scholar]
- 58.Wang J, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, et al. Characterization of phosphoglycerate kinase-1 expression of stromal cells derived from tumor microenvironment in prostate cancer progression. Cancer Res. 2010;70:471–480. doi: 10.1158/0008-5472.CAN-09-2863. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.