Abstract
Pancreatic cancer is a leading cause of cancer death, and it has the poorest prognosis of any major tumour type. Familial pancreatic cancer registries are important for investigating the genetic aetiology of this devastating disease. Using data from our familial pancreatic cancer registry and other registries, this Review discusses the usefulness of family registries in the study of pancreatic and other cancers, and also how such registries provide a unique opportunity for laboratory, population and clinical research.
Pancreatic cancer is the fourth leading cause of cancer death in the United States. An estimated 99,901 and 37,685 new cases will be diagnosed in Europe and the United States, respectively, in 2012 (see the GLOBOCAN 2008 website; see Further information). Although considerable progress has been made in improving cancer survival rates over the past few decades, the 5-year survival rate for pancreatic cancer has remained mostly unchanged, rising only from 3.0% in 1975 to 5.4% in 2005 (according to data from surveillance, epidemiology and end results (SEER)1). The poor prognosis of pancreatic cancer is primarily due to its advanced stage at diagnosis, with more than 80% of patients presenting with locally advanced or metastatic disease1. Therefore, death rates from pancreatic cancer might be improved if this disease could be detected and diagnosed at an earlier stage2. One group of individuals for whom this might be possible is familial pancreatic cancer kindreds, as they are at a high risk of developing pancreatic cancer3. The National Familial Pancreas Tumour Registry (NFPTR) was first established in 1994 at Johns Hopkins Hospital in the United States to help to demonstrate that pancreatic cancer clusters in some families. Not only has this registry quantified the risk of pancreatic cancer in individuals with a family history3,4 but it has also led to the discovery of several pancreatic cancer susceptibility genes5–7, increasing our understanding of the aetiology of this disease. The genes involved in familial pancreatic cancer also have an important role in the non-familial forms of the disease and could provide important targets for personalized therapy.
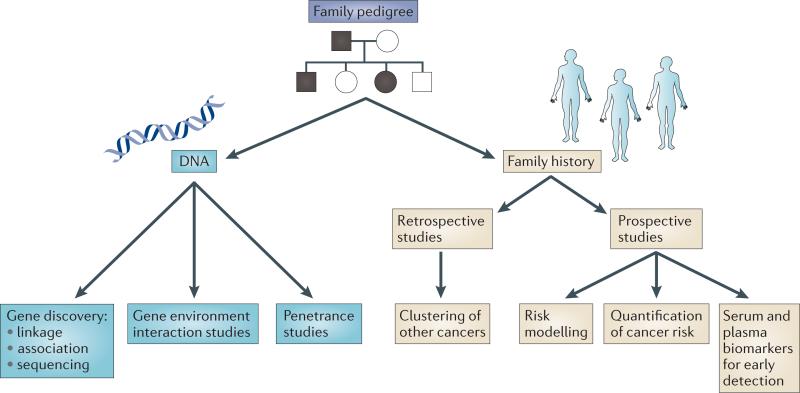
The goal of this Review is to provide an understanding of the usefulness of family registries in the study of pancreatic and other cancers, and to show how such registries provide a unique opportunity for laboratory, population and clinical research, from traditional approaches to genome sequencing and personalized medicine (fig. 1).
Figure 1. Overview of the research conducted with pancreatic cancer family registry data.
Family registries represent a collection of data that typically includes a detailed family history, as well as blood or saliva for DNA studies. This information can be used in a variety of research studies, including gene discovery and cancer risk. The family data can be expanded to capture pathology samples and prospective data collection that enable other studies, such as prospective studies and biomarker studies, to be carried out.
Familial registries in cancer
The study of cancer in families began more than 100 years ago. In 1895, Warthin began to identify families with a clustering of cancer through the careful examination of pathology records from the University of Michigan Hospital, USA8. Warthin clearly showed the aggregation of cancers in families, in particular colon and breast cancers, at a time when many in the scientific community believed that cancer did not have a hereditary component. Although each family in Warthin's research contributed valuable information, a single family included in the study, Family G, has now been followed for more than a century, thus contributing to a wealth of knowledge on familial colon cancer9. This work set the precedent for the importance of studying cancer families and how long-term follow-up of these families can greatly affect our understanding of the aetiology and the natural history of these cancers, as well as paving the way for targeted risk assessment and counselling. Despite substantial changes in the technologies used to study cancer over the 100 years since Warthin began his studies, the importance of studying hereditary cancer remains.
Today, family registries are research studies in which the unit of observation is not a single case, but the entire family unit. Data collection typically includes a detailed family history, as well as blood or saliva for DNA studies, and this can be expanded to capture pathology samples and prospective data collection. The advantages and disadvantages of family studies are detailed in Box 1.
I discuss below how familial cancer registries provide an important resource to aid the identification of cancer susceptibility genes, to characterize how mutations in these genes affect cancer risk and to provide a population for early detection studies using examples from familial pancreatic cancer.
Family aggregation
The first studies to document a familial clustering of pancreatic cancer were case reports in the early 1970s. These ranged from the report of a single pancreatic cancer kindred to summaries of a small series of families with pancreatic cancer10–17. These anecdotal reports were then followed by case–control studies and cohort studies that further demonstrated that pancreatic cancer risk is increased in individuals with a family history of pancreatic cancer and they also quantified the risk of pancreatic cancer that is due to a family history18–26. These studies estimated that individuals with a family history of pancreatic cancer had a 1.5–13-fold increased risk of developing pancreatic cancer. Although these population-based studies provide good evidence that family history has a role in pancreatic cancer risk, because familial pancreatic cancer is rare, and so few individuals in these studies carry high-penetrance pancreatic cancer genes, these risk estimates do not accurately reflect the risk in familial pancreatic cancer kindreds.
The early case reports of the clustering of pancreatic cancer in families led to the creation of family registries that aim to study the familial aggregation of pancreatic cancer. One of the largest is the NFPTR, partly because Johns Hopkins Hospital is the largest surgical referral centre in the world for pancreatic resection. Coupled with this large patient base, the NFPTR was also one of the first registries to recruit families using the internet27. Enrolment into the NFPTR is ongoing, and as of 13 September 2012 there were more than 4,482 families enrolled. Of these families, 1,693 have multiple cases of pancreatic cancer. The NFPTR has helped to develop the suggested guideline for defining a family with familial pancreatic cancer — a kindred with a pair of first-degree relatives (parent–child or sibling pair) with pancreatic cancer28,29. In the NFPTR, 1,425 families meet this definition. Although such families are more likely to harbour a pancreatic cancer susceptibility gene, it is important to note that a substantial proportion of familial pancreatic cancer kindreds will not carry a mutation in a moderate- to high-penetrance pancreatic cancer predisposition gene. Furthermore, not all individuals who carry a mutation in a pancreatic cancer predisposition gene will have a family history of pancreatic cancer. The use of a common definition across research studies facilitates the comparison of findings across studies, including gene discovery studies that aim to estimate the prevalence of pancreatic cancer gene mutations in familial pancreatic cancer kindreds and risk-quantification studies.
In addition to the observation of the familial clustering of pancreatic cancer, there is statistical evidence that an autosomal dominant gene or genes have a role in pancreatic cancer risk through a population-based segregation analysis of pancreatic cancer that was conducted in the NFPTR30. This study provided statistical evidence that there is a dominantly inherited major gene or genes responsible for the clustering of pancreatic cancer in some families. Overall, six of 1,000 people in the population were estimated to carry a moderate- to high-risk pancreatic cancer genotype, and the lifetime risk of developing pancreatic cancer (by the age of 85) was 32%. Thus, through the family registries we were able to establish the importance of hereditary factors in the development of pancreatic cancer.
In addition to the NFPTR, several other familial pancreatic cancer registries have been established in the United States and Europe, including the European Registry of Familial Pancreatic Cancer and Hereditary Pancreatitis (EUROPAC) and the German National Case Collection for Pancreatic Cancer (FaPaCa)31–33. The United States-based Pancreatic Cancer Genetic Epidemiology Consortium is a collaboration of familial pancreatic cancer registries in the United States and Canada, including those at the Mayo Clinic, Rochester, USA; the Dana-Farber Cancer Institute, Boston, USA; Mount Sinai Hospital, Toronto, Canada; the Karmanos Cancer Center, Detroit, USA; the MD Anderson Cancer Center, Houston, USA; and the NFPTR at Johns Hopkins Hospital34.
Risk quantification using family history data
In addition to establishing the aggregation of pancreatic cancer in families, family registries can help to quantify the risk of pancreatic cancer and identify higher risk populations. A family history of pancreatic cancer is an imperfect surrogate of genetic risk because, although individuals are either born with an inherited mutation that predisposes to pancreatic cancer or not, the effect of family history changes over an individual's lifetime depending on whether additional family members develop the disease. However, family history is still an important risk factor that should be assessed when examining an individual's risk of pancreatic cancer. Families in the NFPTR are prospectively followed and updated health information is collected annually. As of 13 September 2012, 107 new (incident) cases of pancreatic cancer had developed in family members who were disease-free at enrolment. In 2010, we estimated the prospective risk of pancreatic cancer in NFPTR participants by computing the standardized incidence ratio (SIR) to compare the observed and the expected rates of pancreatic cancer using this prospective data. Overall, the risk of pancreatic cancer was 6.79-fold higher (95% confidence interval (CI) = 4.54–9.75) in the relatives of patients with familial pancreatic cancer; 2.41-fold higher (95% CI = 1.04–4.74) in relatives of patients with sporadic pancreatic cancer (individuals from kindreds without a pair of first-degree relatives with pancreatic cancer); and a non-significant 2.14-fold higher (95% CI = 0.58–5.49) in spouses of patients with pancreatic cancer4. These findings support a strong role for genetic factors, as shown by the high risk in relatives of patients with familial cases followed by a moderate risk in relatives of patients with sporadic cases. However, there is also the suggestion of a weaker effect for shared non-genetic factors among household members, as shown by the non-significant but twofold increased risk in spouses of patients with pancreatic cancer.
In the same study, we examined whether the risk of pancreatic cancer varies by age of onset of pancreatic cancer in the family. Although previous studies had examined this question, the results were inconsistent, with some studies suggesting that risk may be higher in relatives of young-onset cases35 but others showing no association31. Using prospective data from the NFPTR we found that having a young-onset patient (<50 years old) in the family increased pancreatic cancer risk for members of familial kindreds but did not alter the risk for sporadic pancreatic cancer kindred members4.
Prospective data from a family registry were crucial for conducting these studies. Retrospective studies of high-risk families cannot provide an unbiased estimate of the risk of pancreatic cancer. Selection bias for the quantification of risk is introduced as families with multiple cases of pancreatic cancer are targeted for enrolment in these studies, and it is likely that as the number of pancreatic cancers in the family increases the probability of joining a family study also increases. Studies which use prospective data are not subject to these specific selection biases.
In addition to quantifying the risk of pancreatic cancer in high-risk families, family registries allow for the study of the aggregation of other diseases, particularly other cancers. For example, the aggregation of other cancers is evident in high-risk colon cancer kindreds with Lynch syndrome9. Because most familial cancer registries often select families on the basis of the occurrence of cancer at a single site, the incidence of other cancers in these families can be examined using either retrospective or prospective data. These studies can provide important insights into the aetiology of these diseases by pointing to shared predisposition genes or to common biological pathways between the diseases. In addition, the co-aggregation of cancers at multiple sites in the same family has important implications for risk assessment and counselling.
Using both retrospective and prospective data from the NFPTR we found an increased risk of colon cancer among the relatives of young-onset (<50 years old) pancreatic cancer probands, with a weighted standardized mortality ratio (wSMR) of 2.31 and 95% CI = 1.30–3.81, supporting an association between hereditary nonpol posis colorectal cancer (HNPCC) and pancreatic cancer36. In addition, we found an increased risk of dying from cancers of the breast (wSMR = 1.98; 95% CI = 1.01–3.52) and prostate (wSMR = 2.31; 95% CI = 1.14–4.20) in relatives of young-onset cases, and a significantly increased risk of dying from breast cancer (wSMR = 1.66; 95% CI = 1.15–2.34) or ovarian cancer (wSMR = 2.05; 95% CI = 1.10–3.49) in relatives of familial pancreatic cancer probands36, which may be partly due to the shared genetic aetiology between these diseases. Both the relatives of patients with familial pancreatic cancer and the relatives of patients with sporadic pancreatic cancer had an increased risk of dying from bile duct cancers (wSMR = 2.89; 95% CI = 1.04–6.39 and wSMR = 3.01; 95% CI = 1.09–6.67, respectively)36. These findings support a shared susceptibility between pancreatic cancer and a number of other cancers. These findings stress the importance of at-risk relatives following, at a minimum, the current screening recommendations for breast and colon cancer, as well as considering the usefulness of screening for prostate and ovarian cancers. The occurrence of other cancers in familial pancreatic cancer kindreds was also examined in the FaPaCa in which they reported 28 families with breast cancer, 20 with colon cancer and 11 with lung cancer of the 56 familial pancreatic cancer kindreds studied32. However, the frequency of these cancers in the families was not compared with the expected number based on incidence rates in the general population.
Genetic risk factors
Although most of the genetic reasons for the clustering of pancreatic cancer in families remain unclear, several genes have been identified that increase the risk of pancreatic cancer (table 1). Family registries were a key component of many of these discoveries. Germline BRCA2 mutations were first described in patients with pancreatic cancer by Goggins et al.5, who reported that 7% (four of 41) of patients with pancreatic cancer carried BRCA2 mutations. The mutation prevalence has been shown to be higher in familial pancreatic cancer kindreds — 16% in probands from families with three or more pancreatic cancers37 and 10–12% in families with two or more first-degree relatives with pancreatic cancer38,39. When evaluating pancreatic cancer kindreds, BRCA2 mutations cannot be ruled out by the lack of a family history of breast and/or ovarian cancer, as a substantial proportion of mutation-positive pancreatic cancer families report neither cancer5,37.
Table 1.
Pancreatic cancer predisposition genes
| Genes | Risk of pancreatic cancer |
|---|---|
| BRCA2 | OR = 3.5 (95% CI = 1.87–6.58) |
| STK11 | SIR = 132 (95% CI = 44–261) |
| PALB2 | Increased |
| PRSS1 and SPINK1 | SIR = 67 (95% CI = 8–80) |
| ATM | Increased |
| CDKN2A | SIR = 13–38 |
| Unknown* | SIR=6–32 |
| Mismatch repair genes (MLH1, MSH2, MHS6 and PMS2) | No effect up to SIR = 8.6 (95% CI = 4.7–15.7) |
| BRCA1 | No effect up to OR= 2.26 (95% CI = 1.26–4.06) |
CI, confidence interval; OR, odds ratio; SIR, standardized incidence ratio.
Kindred with familial pancreatic cancer but without mutations in an established pancreatic cancer gene.
Although the increased prevalence of BRCA2 mutations in patients with pancreatic cancer is clear, the evidence supporting a role for BRCA1 in pancreatic cancer has been mixed. Early studies suggested a 2.26-fold (95% CI = 1.26–4.06) increased risk of pancreatic cancer in BRCA1 mutation carriers40, but other studies have reported no increase in the prevalence of BRCA1 mutations in patients with pancreatic cancer41,42.
Several other less commonly mutated genes also increase pancreatic cancer risk. Germline mutations in the CDKN2A locus that encodes the tumour suppressors ARF and INK4A are associated with familial melanoma. People with these mutations also have a 38-fold increased risk of developing pancreatic cancer compared with the general population, so that their lifetime risk (by the age of 75) of developing pancreatic cancer is 17%43,44.
Lynch syndrome mutation carriers have been estimated to have a 3.68% (95% CI = 1.45–5.88) lifetime (by the age of 70) risk of pancreatic cancer45, and individuals with Peutz–Jeghers syndrome, which is caused by mutations in the STK11 (also known as LKB1) gene, have a 132-fold (95% CI = 44–261) increased risk of pancreatic cancer compared with the general population, and the lifetime risk of pancreatic cancer in these individuals has been estimated to be 11–32%46. In addition to these cancer syndromes, individuals with hereditary pancreatitis, a rare inherited form of chronic pancreatitis that is characterized by repeated attacks of acute pancreatitis that begin early in childhood, are also at an increased risk of developing pancreatic cancer47. Germline mutations in PRSS1 and SPINK1 explain a proportion of hereditary pancreatitis families48–50. The lifetime risk (by the age of 70) of developing pancreatic cancer in patients with hereditary pancreatitis has been estimated to be 30–44%47,51,75. This risk is even higher among smokers with hereditary pancreatitis who develop this disease an average of 20 years before non-smokers51.
Environmental risk factors
In addition to a family history of pancreatic cancer, several environmental risk factors have been associated with an increased risk of this disease. Active cigarette smoking has been shown to be associated with a 1.74-fold increased risk of pancreatic cancer (95% CI = 1.61–1.87)52. Type 2 diabetes has also been associated with an increased risk of pancreatic cancer (overall odds ratio (OR) = 1.8; 95% CI = 1.5–2.1) compared with non-diabetics. However, the relationship between diabetes and pancreatic cancer is complex because patients with a long history of diabetes have an increased risk of pancreatic cancer, whereas a new diagnosis of diabetes in older individuals could actually be an early symptom of pancreatic cancer53. The risk of pancreatic cancer also increases with increasing body mass index (BMI). Individuals with a BMI of >35 have an increased risk (OR = 1.55; 95% CI = 1.16–2.07) of pancreatic cancer compared with individuals with a BMI of 18.9 to 24.9 (ref. 54). Heavy alcohol consumption (≥ six alcoholic drinks per day) has also been associated with increased pancreatic cancer risk (OR = 1.46; 95% CI = 1.16–1.83) compared with individuals who drink fewer than one alcoholic drink per day55.
Although these risk factors have been established through the case–control and cohort studies, they have not been extensively studied in the familial setting. As the genetic basis of pancreatic cancer is elucidated, studies of the importance of these risk factors among individuals who harbour pancreatic cancer susceptibility genes will need to be conducted. Because the clustering of pancreatic cancer can be due to genetic factors, environmental factors or both, it is difficult to disentangle the relative effects of these factors in the absence of genetic data. Traditional genetic epidemiological methods, such as sibling relative risk and segregation analysis, can provide some suggestion as to the relative importance of genetic and environmental factors. However, large sample sizes and detailed information on the risk factors for all family members are needed.
Gene discovery
Family registries provide an invaluable resource for gene discovery. Many registries were partly designed to collect families for traditional linkage analysis. Linkage analysis is based on the search for co-segregation of genetic markers as measures of genomic position along with disease in families. Linkage analysis produces robust results in terms of allelic heterogeneity but the power of these studies is limited when there is a moderate to large amount of locus heterogeneity56. Many cancer susceptibility genes were found using linkage approaches; however, the lack of moderate to strong linkage genes for pancreatic cancer could be partly due to the fact that a mutation in one of several genes can lead to the same hereditary cancer phenotype. In addition, the unavailability of DNA from affected family members because of the rapidly fatal course of pancreatic cancer might also explain a lack of strong linkage genes. These problems have been partly overcome by using exome and whole-genome sequencing approaches for gene discovery, to which the data and samples from high-risk families in cancer registries are well suited.
The first successful use of whole-exome sequencing to identify the cause of a hereditary disease was the identification of PALB2 as a hereditary pancreatic cancer susceptibility gene. This finding came from the use of traditional Sanger sequencing of more than 20,000 protein-coding genes6. The germline and tumour DNA from a NFPTR patient with familial pancreatic cancer was sequenced, and more than 15,460 genetic variants that were not present in the human reference sequence were identified. The list of potential disease-causing mutations was narrowed using several criteria: the variant must cause premature inactivation or truncation of the protein; the gene must function as a tumour suppressor gene with a heterozygous germline variant coupled with a somatic mutation in the tumour; and the variant must not be present in control databases. After applying these criteria three candidate genes were identified — SERPINB12, RAGE and PALB2 — the strongest of which was PALB2. PALB2 is the binding partner of BRCA2, an established pancreatic cancer susceptibility gene, and, like BRCA2, mutations in PALB2 had previously been associated with familial breast cancer57. To validate this finding PALB2 was sequenced in an additional 96 familial pancreatic cancer probands from the NFPTR, and three were found to carry protein-truncating mutations. Although subsequent studies have validated the role of PALB2 mutation in familial pancreatic cancer, PALB2 mutations explain only a small proportion (1–3%) of familial pancreatic cancer kindreds58,59.
Although this first success relied on Sanger sequencing, the improvement in quality and decreased cost of next-generation sequencing technologies has allowed for large-scale studies. Whole-genome sequencing has been conducted in 16 patients with familial pancreatic cancer from six families, and whole-exome sequencing has been carried out on samples from 22 patients with familial pancreatic cancer from ten families. On average, more than six million variants and 35,000 variants per individual were detected using whole-genome sequencing and exome sequencing, respectively. Candidate disease-associated changes were identified by excluding variants that already existed in single nucleotide polymorphism databases and by concentrating on variants that were rare in the population, were inactivating and heterozygous in the patient, and were shared among all affected pedigree members. After filtering, 156 variants remained, including mutations in the ataxia telangiectasia mutated (ATM) gene7. In two of the families, variants in ATM co-segregated with pancreatic cancer, and these variants had previously been reported in patients with ataxia telangiectasia. ATM was sequenced in an additional 166 NFPTR probands, and four additional mutations were found7. Again, these variants had also previously been associated with ataxia telangiectasia.
Genome sequencing is a powerful gene discovery tool, but identification of the single genetic change that causes a hereditary disease can be elusive. The discovery of PALB2 and ATM as pancreatic cancer susceptibly genes was partly due to the successful application of filters to narrow the list of candidate variants. However, these filters are imperfect and other causal variants could have been eliminated through the filtering approach. Although the publically available genomic databases (such as the Single Nucleotide Polymorphism database (dbSNP) and the 1000 Genomes database) that are used to filter gene variations provide valuable data on the occurrence and frequency of variants in the population, phenotypic data are not available from these databases. This makes it possible that disease-causing variants are included in these data sets. Furthermore, many causative mutations, including those that lead to pancreatic cancer, are likely to be of only moderate penetrance. Therefore, it is possible that these mutations may be observed in databases that are used for control data. More than 11 genes have already been found to have a role in the aggregation of pancreatic cancer in families, and given the lack of strong linkage signals it is likely that rather than there being only a few genes that explain the remainder of the aggregation of pancreatic cancer in families, there are instead many genes, each of which only explains a small proportion of familial pancreatic cancer.
The use of family data in genome-sequencing studies allows for the use of filters that are based on the sharing of mutations among affected relatives and/or non-sharing among affected–unaffected relative pairs. Although these filters can be very powerful, the presence of phenocopies — sporadic cancers that occur in kindreds with familial cancer — can lead to the exclusion of disease-causing variants, as not all cancer cases in a kindred will necessarily carry the disease-causing mutation.
Despite these challenges, it is anticipated that genome sequencing will lead to the discovery of additional pancreatic cancer predisposition genes in the coming years. Once such genes are identified and their contribution to the risk of developing pancreatic cancer, along with other non-genetic factors, is quantified, risk models can be developed to facilitate the transition of these research findings to the clinic.
Risk model development and validation
The primary goal of risk modelling is to develop a tool for identifying individuals at a high risk of disease. Well-developed models can provide accurate risk assessment for individuals and can inform both genetic testing and screening decisions. Risk models can be developed using a variety of statistical frameworks ranging from traditional regression and Bayasian modelling to machine learning approaches. Some of the first risk models that were developed for cancer syndromes included the Gail and Claus models60,61. The Gail model for breast cancer was developed using a regression framework with several predictors: current age, age at menarche, age at first live birth, number of first-degree relatives with breast cancer, a previous breast biopsy, race or ethnicity, and a history of ductal carcinoma in situ or lobular carcinoma in situ. The Gail model was used to identify women at a high risk of breast cancer for the National Surgical Adjuvant Breast and Bowel Project Breast Cancer Prevention Trail, which evaluated the efficacy of tamoxifen for breast cancer prevention in high-risk women. However, one limitation of the Gail model and other models built using population-based data is that the low prevalence of familial cases provides limited data on the risk of cancer in members of these high-risk families.
Alternative approaches that are well suited to the study of cancer families are models that use information on the patterns of inheritance, prevalence and penetrance of genetic factors. This information can be obtained from the analysis of familial cancer registry data. The Bayes Mendel software package was developed as a generalized tool for the development of risk models using family data. In brief, the model uses four primary steps. First, a model of inheritance for the genes underlying the aggregation of cancer in families is specified; and second, estimates of the prevalence of disease in mutation carriers and non-carries in conjunction with mutation frequency is estimated. Using Bayes’ rule, penetrance and prevalence estimates are converted to the probability of a phenotype given a particular genotype, and cancer risk is obtained through a weighted average of the penetrance in carriers and non-carriers, with weights derived from the probability that a given individual is a carrier or a non-carrier of the disease-causing mutation based on their family history data62. For our pancreatic cancer model, PancPRO, estimates of penetrance and prevalence were obtained from our prior segregation studies of pancreatic cancer30. To use the model, detailed family history data from each high-risk individual, including personal and family history of cancer, current age, age of onset of cancer, as well as any other model covariates, is provided as model input. Model output can include the probability of carrying a susceptibility gene or genes based on model assumptions and the risk, by age, of developing the cancer63.
Although risk model development is an important step, models should be validated before being applied to high-risk populations. Various statistics have been developed to examine model fit, most of which address the discriminatory ability of the model and model calibration. Discrimination is often assessed by estimating model sensitivity and specificity, and examining area under the receiver operator characteristic (ROC) curve (AUC). Model calibration assesses the agreement between predicted outcome and observed outcome. However, the choice of study population for validation is also important, as the population should reflect the clinical population that is to be served by the model. Data from family registries or genetic clinics can provide valuable validation populations, as these are sources of high-risk families that are likely to be seeking clinical evaluation. Although clinic-based samples often represent the target population and are likely to have genetic testing data available, often these data are cross-sectional in nature. Family registries can provide prospective data, if the registry conducts uniform follow-up of all families enrolled. The PancPRO model was validated in an independent set of NFPTR families on whom prospective follow-up was available. We compared model predictions made using family history data collected at baseline with the observed incidence of pancreatic cancer during follow-up. The overall AUC for the PancPRO model was 0.75 (95% CI = 0.68–0.81), which was higher than the AUC for risk predictions made using counts of the number of affected relatives alone, which was 0.61 (95% CI = 0.51–0.71).
Early detection tools and trials
Although recent studies have suggested that it takes at least 15 years from the initiating mutation to the development of metastatic pancreatic cancer, indicating that many years are available for the detection of early stage disease, the vast majority of patients with pancreatic cancer present with locally advanced or metastatic disease. Such advanced disease is not responsive to chemotherapy or radiation therapy64. The approximately 5% of patients with pancreatic cancer who are alive 5 years after diagnosis are typically patients who were diagnosed with small, resectable early stage cancers. Thus, improved detection of early stage cancers, or ideally of high-grade pre-neoplastic precursors, such as intraductal papillary mucinous neoplasm (IPMN) and pancreatic intraepithelial neoplasia (PanIN), provides the best opportunity to reduce mortality from pancreatic cancer.
Numerous studies are currently underway to identify novel early detection tools for pancreatic cancer. These tools typically fall into one of two approaches: imaging-based technologies or biomarkers. Given the rarity of pancreatic cancer and the consequences of false-positive findings, these studies need high specificity coupled with high sensitivity.
Although several promising blood-based and tumour-based biomarkers have been reported in recent years, the individual sensitivity and specificity of these markers falls short of what is needed for a practical, population-based screening test. For example, the PAM4 antibody (clivatuzumab) has been reported to detect 64% of stage I pancreatic cancers with a specificity of 85% in patients with benign pancreatic disease65. As novel biomarkers are developed, tests combining several markers either jointly or sequentially may provide the best opportunity for a successful blood-based screening tool.
Imaging technology can be a powerful early detection tool. Endoscopic ultrasound (EUS), magnetic resonance cholangiopancreatography (MRCP) and computed tomography (CT) can all be used for the clinical evaluation of the pancreas. A recent comparison of these modalities as part of the Cancer of the Pancreas Screening (CAPS) study found that 93% of cystic lesions of the pancreas could by identified by EUS, 81% by MRCP and 27% by CT scan66. EUS not only has good sensitivity but this technique also allows for the collection of pancreatic fluid, which can be used for biomarker studies. Targeted imaging agents, such as agents that detect plectin, a cell-surface protein that is expressed in PanIN3 lesions67, have the potential to improve imaging for non-cystic pancreatic lesions.
Clinical trials examining the use of imaging techniques, in particular EUS, to screen for pre-cancerous changes in the pancreas among individuals with a strong family history of pancreatic cancer began in the late 1990s. Families who were participating in ongoing familial pancreatic cancer registries were invited to take part in these studies. As part of the initial phase of the CAPS study 38 individuals — 37 from kindreds with three or more cases of pancreatic cancer and one patient with Puetz–Jeghers syndrome — were screened using EUS. Six were found to have pancreatic masses, including one IPMN and one invasive ductal adenocarcinoma68. In a similar study, Brentnall et al.69 reported that seven of 14 patients from familial pancreatic cancer kindreds screened using EUS followed by surgery had widespread pancreatic dysplasia. However, it is important to note that six of the seven individuals were from a single family, and that these individuals reported pancreatic insufficiency at a young age in addition to their diagnosis of pancreatic cancer, a feature that is not typical of most familial pancreatic cancer kindreds. Recently, the results of the third phase of the CAPS study have been published. This phase examined individuals from familial pancreatic cancer kindreds or individuals with a documented mutation in a pancreatic cancer predisposition gene. This was a multicentre study that included Johns Hopkins Hospital, the Mayo Clinic, the Dana-Farber Cancer Institute and the MD Anderson Cancer Center. The majority of the 225 asymptomatic individuals analysed in this phase of the study were participants in familial registries at one of the hospitals. Of the individuals screened, 92 (42%) had at least one pancreatic mass, and of these 92 individuals 85 were reported to have either proven or suspected neoplasms (82 IPMN and three neuroendocrine tumours). Five of the 225 individuals underwent surgical resection of the pancreas of which three were found to have high-grade pancreatic dysplasia66. These findings demonstrate that early detection screening in high-risk families can detect pre-cancerous changes in the pancreas. However, many questions remain, including the identification of the most appropriate screening populations and, most importantly, which criteria for the selection of patients for surgery maximizes benefit and minimizes risk. As such, early detection screening should be limited to well-developed research protocols and/or clinical trials.
Personalized therapy
Understanding the inherited genetic changes that can lead to the development of pancreatic cancer can have important therapeutic implications not only for familial cases but also for sporadic cases, as the same genes and pathways are often involved in both cases. For example, sporadic and familial pancreatic cancers can be deficient in BRCA2 or PALB2, and tumours with these mutations are especially sensitive to DNA crosslinking agents and poly(ATP-ribose) polymerase (PARP) inhibitors70–74. Clinical trials using both DNA crosslinking agents and PARP inhibitors in patients with pancreatic cancer are currently underway. As our knowledge of the pathways involved in pancreatic cancer expands, more targeted agents for this disease should be revealed. These will hopefully provide improved disease outcomes.
Conclusions
Family registries are a powerful resource in cancer research. In particular, through the study of pancreatic cancer kindreds, we and others have demonstrated the familial aggregation of pancreatic cancer, quantified the risk of pancreatic cancer, as well as other cancers, in these high-risk families, identified pancreatic cancer susceptibility genes and begun the first screening trials for pancreatic cancer. Collaboration across family registries is ongoing, with several of the North American family registries participating in the Pancreatic Cancer Genetic Epidemiology Consortium. The collection of detailed family history data, as well as data on the established pancreatic cancer risk factors, facilitates the harmonization of data across different sites. In addition, special consideration of the consent under which the family data was collected is needed so that any collaboration is consistent with the original informed consent. The recent technological advances that have allowed us to begin to conduct whole-genome and whole-exome sequencing and collaborative ongoing sequencing efforts will enable the identification of several new pancreatic cancer susceptibility genes in the coming years. Furthermore, personalized treatments that target these gene defects and improved screening of high-risk individuals provide an opportunity to reduce the mortality from this disease.
Prospective.
A term used in epidemiological studies, when a population is defined and then followed in time so that the occurrence of particular events or outcomes can be observed.
At a glance.
Familial cancer registries have had a key role in our understanding of the aetiology of many cancer types, such as breast and colon cancer.
As pancreatic cancer is a leading cause of cancer death and because it has the poorest prognosis of any major tumour type, familial pancreatic cancer registries are an important tool for investigating the genetic aetiology of this devastating disease.
By studying the families that are enrolled in our familial pancreatic cancer registry we have been able to identify several familial pancreatic cancer susceptibility genes, conduct some of the first early detection screening trials for pancreatic cancer and have begun to understand the potential of personalized treatment for this disease.
Registries also allow for the prospective follow-up of a population at a high risk of pancreatic cancer, a disease that is traditionally difficult to study owing to its rarity.
The adaptive nature of family registries has allowed for the rapid adoption of new technologies, such as genome sequencing for gene discovery.
Case–control studies.
Retrospective observational epidemiological studies of individuals with a given condition (case) who are identified along with a comparison group of individuals without the disease (controls). The frequency or, more specifically, the odds of exposure are then compared between these two groups.
Penetrance.
The probability of developing a given phenotype that is conditional on a given genotype. For example, the probability of developing breast cancer given that one carries a mutation in BRCA1.
Retrospective studies.
Identification of a group of individuals from historical data and reconstruction of their exposure history and occurrence of disease until the present time. Disease outcome between exposed and unexposed individuals is compared.
Lynch syndrome.
Also known as hereditary non-polyposis colorectal cancer. An autosomally dominant inherited cancer syndrome that is most notably known for a high risk of colon cancer in both genders and of endometrial cancer in women.
Box 1 Advantages and disadvantages of family-based studies.
Advantages
Detailed pedigree data are available.
A population that is enriched for those who develop disease owing to an inherited genetic basis increases the power of gene discovery studies.
An ability to search for the co-aggregation of other diseases and cancers in the family.
Relatives of patients represent a high-risk group that can enable early detection studies.
At-risk relatives can be followed as a high-risk cohort.
Highly motivated population.
Disadvantages
Ascertainment (selection) bias.
Studies often include only highly selected families so that results may not be able to be generalized to an unselected population.
More difficult to recruit families versus single cases.
Reconciling reported family history across relatives can be challenging.
Ascertainment (selection) bias.
When families are selected based on a particular phenotype (that is, multiple cases of cancer), any analyses using these families that do not properly control for this selection can lead to biased results.
Peutz–Jeghers syndrome.
An autosomally dominant inherited syndrome that is characterized by hamartomatous polyps of the gastrointestinal tract and pigmented macules on the buccal mucosa.
Allelic heterogeneity.
Mutations located in different alleles in the same gene that can cause a given genetic disease of phenotype.
Locus heterogeneity.
When the same genetic disease or phenotype occurs but can be due to a single genetic mutation in more than one gene.
Ataxia telangiectasia.
An autosomal recessive condition caused mutations in the ATM gene. This syndrome is characterized by immunodeficiency, decreased DNA damage repair and neurological abnormalities.
Phenocopies.
Non-genetic or sporadical occurrences of a phenotype when the same phenotype has been shown to occur owing to a genetic mutation in other individuals.
Area under the receiver operator characteristic (ROC) curve.
(AUC). A graphical plot created by plotting the fraction of true positives out of the positives versus the fraction of false positives out of the negatives, at various threshold settings. The true positive rate is also known as sensitivity, and the false-positive rate is one minus the specificity or true negative rate. The area under the curve represents the probability that an individual with the disease has a higher score than an individual without the disease.
Cross-sectional.
A type of epidemiological study in which outcomes and exposures are assessed at a single point in time.
Acknowledgements
A special thank you to all of the patients and families who participate in the National Familial Pancreas Tumor Registry and other pancreatic cancer family studies without whom this research would not be possible. The author would also like to thank R. Hruban, K. Brune, E. Palmisano and D. Echavarria. A.P.K. receives support from the NCI SPORE in Gastrointestinal Cancer CA62924, RO1 CA154823 and the Sol Goldman Pancreatic Cancer Research Center.
Footnotes
Competing interests statement
The author declares competing financial interests. See Web version for details.
FURTHER INFORMATION
Alison P. Klein's homepage: www.nfptr.org
GLOBOCAN 2008: http://globocan.iarc.fr/
References
- 1.Howlader N, et al. SEER Cancer Statistics Review, 1975–2005. National Cancer Institute. 2010 [online], http://seer.cancer.gov/csr/1975_2005/
- 2.Cleary SP, et al. Prognostic factors in resected pancreatic adenocarcinoma: analysis of actual 5-year survivors. J. Am. Coll. Surg. 2004;198:722–731. doi: 10.1016/j.jamcollsurg.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Klein AP, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. This study quantified the risk of pancreatic cancer among individuals with a strong family history of the disease paving the way for gene discovery and screening studies. [DOI] [PubMed] [Google Scholar]
- 4.Brune KA, et al. Importance of age of onset in pancreatic cancer kindreds. J. Natl Cancer Inst. 2010;102:119–126. doi: 10.1093/jnci/djp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goggins M, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 6.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. This was the first identification of a cause of a hereditary disease using exome sequencing and also demonstrated mutations in PALB2 at increased frequency in patients with familial pancreatic cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts NJ, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. This study used exome and whole-genome sequencing to show that mutations in the ATM gene were at an increased frequency in patients with familial pancreatic cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warthin AS. Heredity with reference to carcinoma. Arch. Intern. Med. 1913;12:546–555. This is a historical study of the power of studying cancer. [Google Scholar]
- 9.Douglas JA, et al. History and molecular genetics of Lynch syndrome in family G: a century later. JAMA. 2005;294:2195–2202. doi: 10.1001/jama.294.17.2195. [DOI] [PubMed] [Google Scholar]
- 10.MacDermott RP, Kramer P. Adenocarcinoma of the pancreas in four siblings. Gastroenterology. 1973;65:137–139. [PubMed] [Google Scholar]
- 11.Friedman JM, Fialkow PJ. Carcinoma of the pancreas in four brothers. Birth Defects Orig. Artic. Ser. 1976;12:145–150. [PubMed] [Google Scholar]
- 12.Reimer RR, Fraumeni JF, Jr, Ozols RF, Bender R. Pancreatic cancer in father and son. Lancet. 1977;1:911. doi: 10.1016/s0140-6736(77)91244-2. [DOI] [PubMed] [Google Scholar]
- 13.Danes BS, Lynch HT. A familial aggregation of pancreatic cancer. An in vitro study. JAMA. 1982;247:2798–2802. [PubMed] [Google Scholar]
- 14.Dat NM, Sontag SJ. Pancreatic carcinoma in brothers. Ann.Intern.Med. 1982;97:282. doi: 10.7326/0003-4819-97-2-282_1. [DOI] [PubMed] [Google Scholar]
- 15.Katkhouda N, Mouiel J. Pancreatic cancer in mother and daughter. Lancet. 1986;2:747. doi: 10.1016/s0140-6736(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 16.Ghadirian P, Simard A, Baillargeon J. Cancer of the pancreas in two brothers and one sister. Int. J. Pancreatol. 1987;2:383–391. doi: 10.1007/BF02788437. [DOI] [PubMed] [Google Scholar]
- 17.Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer J. 2001;7:266–273. [PubMed] [Google Scholar]
- 18.Falk RT, Pickle LW, Fontham ET, Correa P, Fraumeni JF. Life-style risk factors for pancreatic cancer in Louisiana: a case-control study. Am. J. Epidemiol. 1988;128:324–336. doi: 10.1093/oxfordjournals.aje.a114972. [DOI] [PubMed] [Google Scholar]
- 19.Friedman GD, Van Den Eeden SK. Risk factors for pancreatic cancer: an exploratory study. Int. J. Epidemiol. 1993;22:30–37. doi: 10.1093/ije/22.1.30. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez E, La Vecchia C, d'Avanzo B, Negri E, Franceschi S. Family history and the risk of liver, gallbladder, and pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 1994;3:209–212. [PubMed] [Google Scholar]
- 21.Price TF, Payne RL, Oberleitner MG. Familial pancreatic cancer in south Louisiana. Cancer Nurs. 1996;19:275–282. doi: 10.1097/00002820-199608000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Ghadirian P, et al. Reported family aggregation of pancreatic cancer within a population- based case-control study in the Francophone community in Montreal, Canada. Int. J. Pancreatol. 1991;10:183–196. doi: 10.1007/BF02924156. [DOI] [PubMed] [Google Scholar]
- 23.Coughlin SS, Calle EE, Patel AV, Thun MJ. Predictors of pancreatic cancer mortality among a large cohort of United States adults. Cancer Causes Control. 2000;11:915–923. doi: 10.1023/a:1026580131793. [DOI] [PubMed] [Google Scholar]
- 24.Schenk M, et al. Familial risk of pancreatic cancer. J. Natl Cancer Inst. 2001;93:640–644. doi: 10.1093/jnci/93.8.640. [DOI] [PubMed] [Google Scholar]
- 25.Silverman DT. Risk factors for pancreatic cancer: a case-control study based on direct interviews.Teratog. Carcinog. Mutagen. 2001;21:7–25. doi: 10.1002/1520-6866(2001)21:1<7::aid-tcm3>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs EJ, et al. Family history of cancer and risk of pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan). Int. J. Cancer. 2010;127:1421–1428. doi: 10.1002/ijc.25148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goggins M, et al. Use and benefits of a Web site for pancreatic cancer. JAMA. 1998;280:1309–1310. doi: 10.1001/jama.280.15.1309-a. [DOI] [PubMed] [Google Scholar]
- 28.Petersen GM, Hruban RH. Familial pancreatic cancer: where are we in 2003? J. Natl Cancer Inst. 2003;95:180–181. doi: 10.1093/jnci/95.3.180. [DOI] [PubMed] [Google Scholar]
- 29.Hruban RH, Petersen GM, Ha PK, Kern SE. Genetics of pancreatic cancer. From genes to families. Surg. Oncol. Clin. N. Am. 1998;7:1–23. [PubMed] [Google Scholar]
- 30.Klein AP, et al. Evidence for a major gene influencing risk of pancreatic cancer. Genet. Epidemiol. 2002;23:133–149. doi: 10.1002/gepi.1102. [DOI] [PubMed] [Google Scholar]
- 31.Bartsch DK, et al. Prevalence of familial pancreatic cancer in Germany. Int. J. Cancer. 2004;110:902–906. doi: 10.1002/ijc.20210. [DOI] [PubMed] [Google Scholar]
- 32.Schneider R, et al. German national case collection for familial pancreatic cancer (FaPaCa): ten years experience. Fam. Cancer. 2011;10:323–330. doi: 10.1007/s10689-010-9414-x. [DOI] [PubMed] [Google Scholar]
- 33.Greenhalf W, Malats N, Nilsson M, Bartsch D, Neoptolemos J. International registries of families at high risk of pancreatic cancer. Pancreatology. 2008;8:558–565. doi: 10.1159/000159214. [DOI] [PubMed] [Google Scholar]
- 34.Petersen G, et al. Pancreatic cancer genetic epidemiology consortium. Cancer Epidemiol. Biomarkers Prev. 2006;15:704–710. doi: 10.1158/1055-9965.EPI-05-0734. [DOI] [PubMed] [Google Scholar]
- 35.James TA, et al. Risk factors associated with earlier age of onset in familial pancreatic carcinoma. Cancer. 2004;101:2722–2726. doi: 10.1002/cncr.20700. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, et al. Elevated cancer mortality in the relatives of patients with pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 2009;18:2829–2834. doi: 10.1158/1055-9965.EPI-09-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy KM, et al. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17%. Cancer Res. 2002;62:3789–3793. [PubMed] [Google Scholar]
- 38.Hahn SA, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J. Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 39.Couch FJ, et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:342–346. doi: 10.1158/1055-9965.EPI-06-0783. [DOI] [PubMed] [Google Scholar]
- 40.Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J. Natl Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 41.Ferrone CR, et al. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J. Clin. Oncol. 2009;27:433–438. doi: 10.1200/JCO.2008.18.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Axilbund JE, et al. Absence of germline BRCA1 mutations in familial pancreatic cancer patients. Cancer Biol. Ther. 2009;8:1–5. doi: 10.4161/cbt.8.2.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutter JL, et al. Heterogeneity of risk for melanoma and pancreatic and digestive malignancies: a melanoma case-control study. Cancer. 2004;101:2809–2816. doi: 10.1002/cncr.20669. [DOI] [PubMed] [Google Scholar]
- 44.Vasen HF, et al. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int. J. Cancer. 2000;87:809–811. [PubMed] [Google Scholar]
- 45.Kastrinos F, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302:1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Lier MG, et al. High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am. J. Gastroenterol. 2010;105:1258–1264. doi: 10.1038/ajg.2009.725. [DOI] [PubMed] [Google Scholar]
- 47.Lowenfels AB, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J. Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 48.Whitcomb DC, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nature Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 49.Whitcomb DC, et al. A gene for hereditary pancreatitis maps to chromosome 7q35. Gastroenterology. 1996;110:1975–1980. doi: 10.1053/gast.1996.v110.pm8964426. [DOI] [PubMed] [Google Scholar]
- 50.Witt H, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nature Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 51.Lowenfels AB, Maisonneuve P, Whitcomb DC, Lerch MM, DiMagno EP. Cigarette smoking as a risk factor for pancreatic cancer in patients with hereditary pancreatitis. JAMA. 2001;286:169–170. doi: 10.1001/jama.286.2.169. [DOI] [PubMed] [Google Scholar]
- 52.Iodice S, Gandini S, Maisonneuve P, Lowenfels AB. Tobacco and the risk of pancreatic cancer: a review and meta-analysis. LangenbecksArch. Surg. 2008;393:535–545. doi: 10.1007/s00423-007-0266-2. [DOI] [PubMed] [Google Scholar]
- 53.Chari ST, et al. Probability of pancreatic cancer following diabetes: a population-based study. Gastroenterology. 2005;129:504–511. doi: 10.1053/j.gastro.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arslan AA, et al. Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan). Arch. Intern. Med. 2010;170:791–802. doi: 10.1001/archinternmed.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lucenteforte E, et al. Alcohol consumption and pancreatic cancer: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4). Ann. Oncol. 2012;23:374–382. doi: 10.1093/annonc/mdr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suarez BK, Hampe GL, Van Eerdewegh P. In: Genetic Approaches to Mental Disorders. Gershon ES, Cloning CR, editors. Amereican Psyscological Association; 1994. pp. 23–46. [Google Scholar]
- 57.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nature Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tischkowitz MD, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slater EP, et al. PALB2 mutations in European familial pancreatic cancer families. Clin. Genet. 2010;78:490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 60.Claus EB, Risch NJ, Thompson WD. Age at onset as an indicator of familial risk of breast cancer. Am. J. Epidemiol. 1990;131:961–972. doi: 10.1093/oxfordjournals.aje.a115616. [DOI] [PubMed] [Google Scholar]
- 61.Gail MH, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J. Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 62.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. BayesMendel: and R Environment for Mendelian Risk Prediction. Stat. Appl. Genet. Mol. Biol. 2004;3:21. doi: 10.2202/1544-6115.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W, et al. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J. Clin. Oncol. 2007;25:1417–1422. doi: 10.1200/JCO.2006.09.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gold DV, et al. PAM4 enzyme immunoassay alone and in combination with CA 19–19 for the detection of pancreatic adenocarcinoma. Cancer. 2012 Aug 16; doi: 10.1002/cncr.27762. (doi:10.1002/cncr.27762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canto MI, et al. Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology. 2012;142:796–804. doi: 10.1053/j.gastro.2012.01.005. This is currently the largest published study of early detection screening in individuals at a high risk of pancreatic cancer owing to the family history. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kelly KA, et al. Targeted nanoparticles for imaging incipient pancreatic ductal adenocarcinoma. PLoS Med. 2008;5:e85. doi: 10.1371/journal.pmed.0050085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Canto MI, et al. Screening for pancreatic neoplasia in high-risk individuals: an EUS-based approach. Clin. Gastroenterol. Hepatol. 2004;2:606–621. doi: 10.1016/s1542-3565(04)00244-7. [DOI] [PubMed] [Google Scholar]
- 69.Brentnall TA, Bronner MP, Byrd DR, Haggitt RC, Kimmey MB. Early diagnosis and treatment of pancreatic dysplasia in patients with a family history of pancreatic cancer. Ann.Intern.Med. 1999;131:247–255. doi: 10.7326/0003-4819-131-4-199908170-00003. [DOI] [PubMed] [Google Scholar]
- 70.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 71.McCabe N, et al. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer Biol. Ther. 2005;4:934–936. doi: 10.4161/cbt.4.9.2141. [DOI] [PubMed] [Google Scholar]
- 72.van der Heijden MS, et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin. Cancer Res. 2005;11:7508–7515. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 73.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 74.Villarroel MC, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol. Cancer Ther. 2010;10:3–8. doi: 10.1158/1535-7163.MCT-10-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howes N, et al. Clinical and genetic characteristics of hereditary pancreatitis in Europe. Clin. Gastroenterol. Hepatol. 2004;2:252–261. doi: 10.1016/s1542-3565(04)00013-8. [DOI] [PubMed] [Google Scholar]